Abstract
Haemophilus parasuis is the etiological agent of Glässer's disease in swine, which associates with severe economic losses in the swine industry worldwide. A real‐time recombinase polymerase amplification assay (real‐time RPA) was developed for direct and rapid detection of H. parasuis basing on the translation‐initiation factor IF2 (infB) gene. The assay was performed successfully at 39°C for 20 min in Genie III, which is portable and chargeable by battery. The developed assay was highly specific for H. parasuis, and the limit of detection of the assay was 6.0 × 103 fg of H. parasuis genomic DNA, which was the same as that of a real‐time PCR developed previously. The assay was further evaluated on 68 pig tissue samples, and 18 (26.5%), 20 (29.4%), and 8 (11.8%) samples were positive for H. parasuis by the real‐time RPA, real‐time PCR and bacterial isolation, respectively. With the bacteria isolation as the reference method, the real‐time RPA showed a diagnostic specificity of 83.33% and a diagnostic sensitivity of 100%. The above data demonstrated the well‐potentiality and usefulness of the developed real‐time RPA assay in reliable diagnosis of swine Glässer's disease, especially in resource limited settings.
Keywords: diagnosis, Glässer's disease, Haemophilus parasuis, infB gene, real‐time RPA
Haemophilus parasuis is the etiological agent of Glässer’s disease in swine, which associates with severe economic losses in the swine industry worldwide. A real‐time recombinase polymerase amplification assay (real‐time RPA) was developed for direct and rapid detection of H. parasuis basing on the infB gene.
1. INTRODUCTION
Haemophilus parasuis, recently renamed Glaesserella parasuis after detailed phylogenetic analysis, is a non‐motile, pleomorphic, gram‐negative bacillus of the Pasturellacea family, and is the nicotinamide adenine dinucleotide (NAD) dependent bacterium (Dickerman, Bandara, & Inzana, 2020; Zhang, Tang, Liao, & Yue, 2014). Haemophilus parasuis is the etiological agent of Glässer's disease, which is characterized by fibrinous polyserositis, peritonitis, polyarthritis and meningitis (Oliveira & Pijoan, 2004). Presently, 15 serovars of H. parasuis have been identified worldwide (McCaig, Loving, Hughes, & Brockmeier, 2016), and the most common serovars in China are serovars 4 and 5 (Cai et al., 2005; Ma et al., 2016; Zhang, Xu, et al., 2012). Haemophilus parasuis could cause a infection rate of 50%–70% and a mortality rate above 10%, thus inducing considerable production losses due to the mortality and unthrifty of pigs (McCaig et al., 2016; Zhang et al., 2014). Glässer's disease has caused severe economic losses in the swine industry worldwide.
The H. parasuis infection can be controlled by vaccination and antibiotic treatment. However, one of the key elements for controlling the H. parasuis infection is the rapid and accurate detection of the bacterium (Aarestrup, Seyfarth, & Angen, 2004; Oliveira & Pijoan, 2004). Isolation and microbiological culture of H. parasuis is the gold standard for diagnosis of Glässer's disease, but it could be ineffective due to the fastidious growth of the bacteria, easily being overgrown by other bacterial contaminants and the previous antibiotic treatment of affected animals (Angen, Oliveira, Ahrens, Svensmark, & Leser, 2007, Oliveira et al., 2001, Turni, Pyke, & Blackall, 2010). A series of molecular diagnostic methods have been described for sensitive and specific detection of H. parasuis, such as polymerase chain reaction (PCR), real‐time PCR, loop‐mediated isothermal amplification (LAMP) and cross‐priming amplification (CPA) (Angen et al., 2007, Chen, Chu, Liu, Zhang, & Lu, 2010, Frandoloso, Martinez‐Martinez, Rodriguez‐Ferri, & Gutierrez‐Martin, 2012, Gou et al., 2018, Oliveira et al., 2001, Turni et al., 2010, Yang et al., 2010). The requirements of high‐precision thermocycler, a centralized laboratory facility and experienced technicians limit the widespread use of PCR assays in under‐equipped laboratories, in clinic settings, and on farm diagnosis. Although the developed LAMP assay shows the advantages in respects to convenience and minimal equipment requirement, it still requires four primers, 60 min at 61°C or 45 min at 65°C for reaction and 10 min at 80°C to terminate the reaction (Chen et al., 2010; Yang et al., 2010). The CPA assay combines with a vertical flow visualization nucleic acid detection strip showing good specificity and sensitivity, thus reducing the requirement of specialized instrument, however, the reaction time is 1h (Gou et al., 2018). A rapid test may assist the microbiological diagnosis for peritonitis in pigs, so a more convenient and rapid detection method is still required for detection of H. parasuis on farm and in low resource settings.
Recombinase polymerase amplification (RPA), which is an isothermal DNA amplification technique, has experienced rapid development in the field of molecular diagnostic since it was firstly reported in 2006 (Daher, Stewart, Boissinot, & Bergeron, 2016; Lei et al., 2020; Li, Macdonald, & von Stetten, 2018; Liu et al., 2018; Miao et al., 2019; Piepenburg, Williams, Stemple, & Armes, 2006; Wang, Yuan, Han, Wang, & Liu, 2017). Some studies showed that RPA may be the most applicable approach for the on farm diagnosis due to its convenience and rapidness (Amer et al., 2013; Li et al., 2018). In this study, the real‐time RPA assay targeting the translation‐initiation factor IF2 (infB) gene was developed for rapid and reliable detection of H. parasuis, and the performance of the assay was further evaluated and compared to the bacteria isolation and a real‐time PCR by detecting the swine tissue samples.
2. MATERIALS AND METHODS
2.1. Bacteria strains and clinical samples
To determine the specificity of the real‐time RPA, a total of 20 H. parasuis strains and 17 other bacterial strains were tested. For the H. parasuis strains, the genomic DNA of 15 serovar reference strains were kindly provided by Lanzhou Veterinary Research Institute (Lanzhou, China), two reference strains were obtained from CIVDC (China Institute of Veterinary Drug Control, Beijing, China), and three field strains were identified by species‐specific PCR described previously (Oliveira et al., 2001, Turni et al., 2010). Details of all the bacterial strains used in this study were provided in Table 1.
Table 1.
Bacterial strains used in this study
| Organism | Serovar | Reference/field strain | Detection results | |
|---|---|---|---|---|
| Real‐time RPA | Real‐time PCR | |||
| Haemophilus parasuis | 1 | NO.4 | + | + |
| 2 | SW140 | + | + | |
| 3 | SW114 | + | + | |
| 4 | CVCC3894 | + | + | |
| SW124 | + | + | ||
| 5 | CVCC3895 | + | + | |
| Nagasaki | + | + | ||
| 6 | 131 | + | + | |
| 7 | 174 | + | + | |
| 8 | C5 | + | + | |
| 9 | D74 | + | + | |
| 10 | H367 | + | + | |
| 11 | H465 | + | + | |
| 12 | H425 | + | + | |
| 13 | 17,975 | + | + | |
| 14 | 22,113 | + | + | |
| 15 | 15,995 | + | + | |
| ND | Field strain | + | + | |
| ND | Field strain | + | + | |
| ND | Field strain | + | + | |
| Actinobacillus pleuropneumoniae | 1 | CVCC259 | − | − |
| 3 | CVCC261 | − | − | |
| 5 | CVCC263 | − | − | |
| 7 | CVCC265 | − | − | |
| 8 | CVCC266 | − | − | |
| Pseudomonas aeruginosa | CICC21636 | − | − | |
| Field strain | − | − | ||
| Streptococcus hemolytic‐β | CMCC10373 | − | − | |
| Bacillus cereus | ATCC 14579 | − | − | |
| Field strain | − | − | ||
| Pasteurella multocida | Field strain | − | − | |
| Field strain | − | − | ||
| Bordetella bronchiseptica | Field strain | − | − | |
| Mycoplasma hyopneumoniae | Strain 168 | − | − | |
| Klebsiella pneumoniae | ATCC 4352 | ‐ | ‐ | |
| Field strain | ‐ | ‐ | ||
| Mannheimia haemolytwa | Field strain | ‐ | ‐ | |
| Listeria monocytogenes | ATCC 15313 | − | − | |
| Field strain | − | − | ||
| Staphylococcus aureus | ATCC 6538 | − | − | |
| Field strain | − | − | ||
Abbreviations: −, negative; +, positive; ATCC, America Type Culture Collection; CICC, China Center of Industrial Culture Collection; CMCC, National Center for Medical Culture Collection; CVCC, China Veterinary Culture Collection Center; ND, not determined.
Thirty swine tonsils were collected at slaughterhouse, and 38 swine fresh lungs were collected from the markets for agricultural products in Shijiazhuang city, Hebei province from November to December 2018.
2.2. DNA extraction
All the bacterial genomic DNA were extracted using the TIANamp Bacteria DNA kit (Tiangen), which were performed according to the manufacturer's instructions. The tonsil and lung samples were homogenized with phosphate‐buffered saline (PBS, pH 7.4) as a 10% (w/v) suspension and centrifuged for 10 min at 10,000 g at 4°C. After centrifugation, the supernatant was discarded and the remaining pellet was suspended in 200 μl of PBS for DNA extraction using the TIANamp Bacteria DNA kit (Tiangen). Total DNA extracted from tissue samples was finally eluted in 50 µl of nuclease‐free water. All DNA were quantified using a ND‐2000c spectrophotometer (NanoDrop) and stored at −80°C until use.
2.3. Real‐time RPA primers and probes
The infB gene is highly conserved in all serovars of H. parasuis and is determined as the detection target of real‐time RPA. Nucleotide sequence data for all 15 serovars of H. parasuis strains available in GenBank were aligned to identify the conserved regions in the infB gene according to the reference sequences of H. parasuis (accession number: HM243087, DQ781806, DQ781808, DQ781810–DQ781817, CP001321.1, CP005384.1), the primers and exo probe were designed following the RPA manufacturer guidelines (TwistDx), and the amplicon size is 182 bp. Primers and probe are listed in Table 2 and synthesized by Sangon Biotech Co., Shanghai, China.
Table 2.
Sequences of the primers and probes for Haemophilus parasuis real‐time RPA and PCR assays
| Assay | Primers and probes | Sequence 5´−3´ | Amplicon size (bp) | References |
|---|---|---|---|---|
| Real‐time RPA | infB‐exo‐F | ACCAGAAGCAAACCTAGAGCGTGTAGAGCAA | 182 | This study |
| infB‐exo‐R | CCTCTTTCACTGCGCTTAATTCTAATACTTCC | |||
| infB‐exo‐P | CACGAAGTGATTTCTGAGAAATTCGGTGG (FAM‐dT)G(THF)(BHQ1‐dT)GTTCAATTTGTTCC ‐C3spacer | |||
| Real‐time PCR | CTinfF1 | CGACTTACTTGAAGCCATTCTTCTT | 75 | Turni et al. (2010) |
| CTinfR1 | CCGCTTGCCATACCCTCTT | |||
| CTinfP | FAM‐ ATCGGAAGTATTAGAATTAAGTGC ‐TAMRA | |||
| PCR | HPS‐forward | GTGATGAGGAAGGGTGGTGT | 821 | Oliveira et al. (2001) |
| HPS‐reverse | GGCTTCGTCACCCTCTGT |
2.4. Real‐time RPA assay
The H. parasuis RPA assay was performed using a ZC BioScienceTM exo kit (ZC BioScience, Hangzhou, China). Firstly, the freeze‐dried enzyme pellet in the reaction tube was rehydrated by 40.5 μl of rehydration buffer. Then, other reaction components including 2.1 μl of forward primer (infB‐exo‐F, 10 μM), 2.1 μl of reverse primer (infB‐exo‐R, 10 μM), 0.8 μl of exo probe (ecfX‐exo‐P, 10 μM), 1 μl of genomic DNA and 1 μl of ddH2O were added into the reaction tube and vortexed thoroughly. Finally, 2.5 μl of magnesium acetate (280 mM) was added into the reaction tube and the total reaction volume was 50 μl. The reaction tube was vortexed briefly and spun down, and the assay was performed immediately at 39°C for 20 min in a Genie III scanner device (OptiGene Limited). Samples produced an exponential amplification curve above the threshold of the negative control were considered positive.
2.5. Analytical specificity and sensitivity of the real‐time RPA assay
The analytical specificity of H. parasuis real‐time RPA assay was determined by testing the genomic DNA extracted from a panel of bacteria listed in Table 1. Three independent reactions were performed.
The analytical sensitivity of H. parasuis real‐time RPA assay was determined and compared with that of real‐time PCR using a 10‐fold serial dilution of genomic DNA of H. parasuis ranging from 6.0 × 107 to 6.0 × 100 fg/μl. One microliter of each dilution was amplified by RPA to determine the limit of detection (LOD) of the assay, and eight independent reactions were performed.
2.6. Real‐time PCR
A real‐time PCR assay for H. parasuis was performed on a ABI 7500 instrument described previously (Turni et al., 2010). Sequences for the primers (CTinfF1 and CTinfR1) and TaqMan probe (CTinfP) are provided in Table 2. The Premix Ex Taq (Takara) was applied in real‐time PCR assay and the reaction was performed as follows: 95°C for 30 s; then 40 cycles of 95°C for 10 s and 60°C for 40 s.
2.7. Validation with tissue samples
The H. parasuis real‐time RPA assay were assessed on 38 swine fresh lungs and 30 swine tonsils, and all samples tested by RPA assay were also tested by a real‐time RT‐PCR (Turni et al., 2010), which was run in parallel. Bacteria isolation of the H. parasuis was also performed for the above tissue samples, which was performed in detail as the following protocol. The surfaces of the tonsils and lungs were seared with a hot iron and cut to obtain scrapings of sample tissue for culture. Scraping of tissue was spread on TSA plates supplemented with 5% foetal bovine serum and 0.01% NAD (Sigma). The plates were incubated at 37°C for 48 hr. All suspect colonies of H. parasuis were passaged twice before identification by PCR (Oliveira et al., 2001).
3. RESULTS
3.1. Analytical specificity of the real‐time RPA assay
Specific amplification was only observed with H. parasuis, including the reference strains and the field isolates, and there was no cross‐detections of other bacteria tested (Table 1). Three independent reactions were repeated and similar results were observed, which demonstrated the high specificity and good repeatability of the assay.
3.2. Analytical sensitivity of the real‐time RPA assay
Using a dilution range of 6.0 × 107 to 6.0 × 100 fg of H. parasuis genomic DNA as template, the data showed that the LOD of the real‐time RPA assay was 6.0 × 103 fg, which was the same as that of the real‐time PCR (Figure 1). The real‐time RPA assay was performed eight times on the molecular standard, in which 6.0 × 107–6.0 × 103 fg DNA molecules were detected in 8/8 runs, and 6.0 × 102–6.0 × 100, 0/8 (Figure 2). Two‐fold serial dilutions of the genomic DNA were made from 6.0 × 103 to 7.5 × 102 fg/μl. The above twofold dilutions of the genomic DNA were further used in the RPA, and the LOD of the assay was still 6.0 × 103 fg per reaction.
Figure 1.
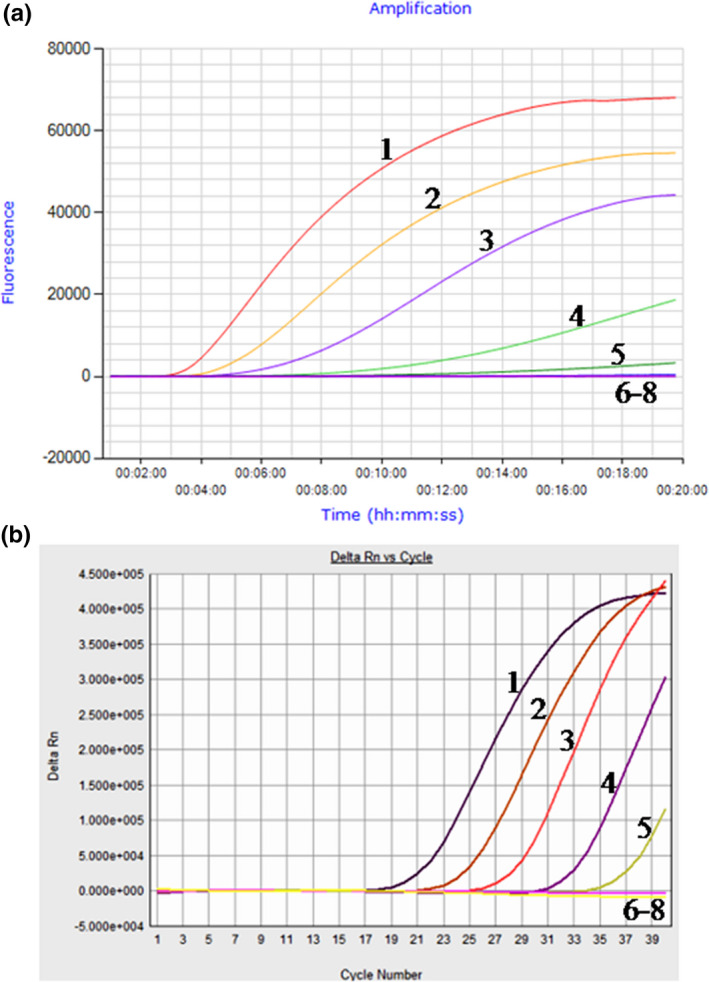
Comparative analytical sensitivity of real‐time RPA and real‐time PCR assays for Haemophilus parasuis. The assays were performed using the 10‐fold dilution of H. parasuis genomic DNA from 6.0 × 107fg to 6.0 × 100 fg per tube, and the assays showed the same analytical sensitivity, 6.0 × 103 fg. A: Limit of detection of real‐time RPA assay with a Genie III. B: Limit of detection of real‐time PCR assay with an ABI 7500 system. Lane 1. 6.0 × 107 fg; lane 2. 6.0 × 106 fg; lane 3. 6.0 × 105 fg; lane 4. 6.0 × 104 fg; lane 5. 6.0 × 103 fg; lane 6. 6.0 × 102 fg; lane 7. 6.0 × 101 fg; lane 8. 6.0 × 100 fg
Figure 2.
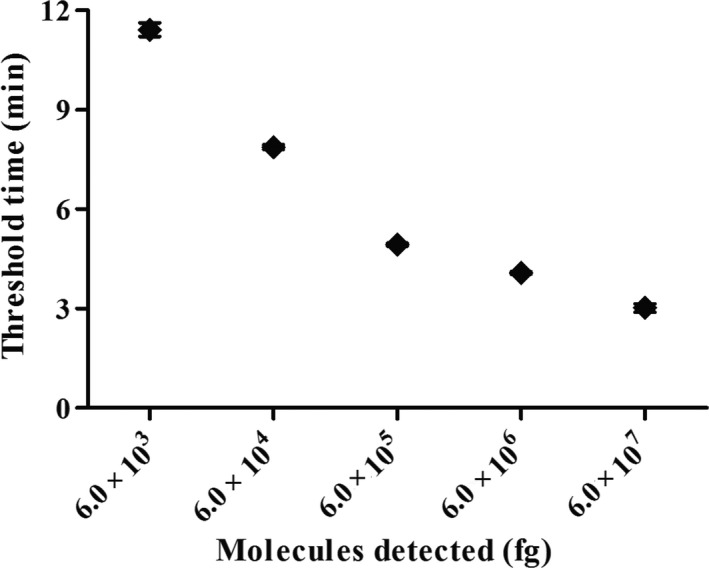
Reproducibility of Haemophilus parasuis real‐time RPA assay. Semi‐logarithmic regression of the data collected from eight real‐time RPA runs tested on the H. parasuis genomic DNA standards using Prism Software. The run time of the assay was between 3 and 12 min for 6.0 × 107 fg‐6.0 × 103 fg genomic DNA
3.3. Validation of the real‐time RPA assays on tissue samples
Of the 68 swine tissue samples, 18 (26.5%), 20 (29.4%) and 8 (11.8%) samples were positive for H. parasuis by the real‐time RPA, real‐time PCR and bacterial isolation, respectively (Table 3). All the positive samples by bacterial isolation were also positive by both real‐time RPA and real‐time PCR. The Ct values of two samples tested positive in real‐time PCR while negative in real‐time RPA were over 35.0, which contained low amounts of H. parasuis DNA. Compared to the bacteria isolation, the real‐time RPA assay showed a diagnostic sensitivity (DSe) of 100%, a diagnostic specificity (DSp) of 83.33%, a positive predictive value (PPV) of 44.44% and a negative predictive value (NPV) of 100% (Table 4). It took approximately 5–15 min in the real‐time RPA assay to obtain the positive results, while it took approximately 30–50 min in the real‐time PCR with the Ct values ranging from 22.73 to 36.65. The above data indicated that the performance of the real‐time RPA assay was comparable to the real‐time PCR, but the real‐time RPA assay is faster.
Table 3.
Comparison of Haemophilus parasuis bacteriology, real‐time RPA and real‐time PCR assays for detection of tissue samples
| Samples | Number of samples | Real‐time RPA | Real‐time PCR | Bacteriology | |||
|---|---|---|---|---|---|---|---|
| P | N | P | N | P | N | ||
| Tonsil | 30 | 5 | 25 | 7 | 23 | 2 | 28 |
| Lung | 38 | 13 | 25 | 13 | 25 | 6 | 32 |
| T | 68 | 18 | 50 | 20 | 48 | 8 | 60 |
Abbreviations: P, positive; N, negative; T, total.
Table 4.
Diagnostic sensitivity, diagnostic specificity and predictive values of real‐time RPA assay and bacteria isolation method for detection of Haemophilus parasuis
| Bacteria isolation | |||
|---|---|---|---|
| P | N | T | |
| Real‐time RPA | |||
| P | 8 | 10 | 18 |
| N | 0 | 50 | 50 |
| T | 8 | 60 | 68 |
| DSe: 100% | DSp: 83.33% | ||
| PPV: 44.44% | NPV: 100% | ||
Abbreviations: DSe, diagnostic sensitivity; DSp: diagnostic specificity; N, negative; NPV, negative predictive value.; P, positive; PPV, positive predictive value; T, total.
4. DISCUSSION
Although isolation of H. parasuis is the gold standard for diagnosis of Glässer's disease, but it is usually difficult as the bacteria is very sensitive to pH changes and heat (Morozumi & Hiramune, 1982) and it is also a slow growing, fastidious organism with specific nutritional requirements (Angen et al., 2007, Oliveira et al., 2001, Turni et al., 2010). Furthermore, H. parasuis is easily overgrown by other faster growing bacteria. This makes recovery of the H. parasuis very difficult after sample collection and transport to the laboratory. Therefore, the method of identification by culture is not always optimal, and nucleic acid amplification‐based methods are the attractive alternatives. In this study, we developed a real‐time RPA assay for rapid detection of H. parasuis, and the positive results could be obtained within 5–15 min. The assay for H. parasuis was performed on the Genie III, which is portable, lightweight and could work a whole day charging by battery. The assay was further demonstrated to be specific, sensitive and easy to perform in the detection of the swine lung and tonsil samples. RPA reagents are provided in the form of lyophilized powder and cold chain independent, and RPA is tolerant to most of the PCR inhibitors during the amplification process (Daher et al., 2016; Lillis et al., 2016; Moore & Jaykus, 2017). The above facts make the real‐time RPA assay a good potentiality of rapid detection of H. parasuis in the farm conditions, which could clarify the microbiology in a difficult diagnostic situation of serositis in pigs.
Several studies had demonstrated the efficacy of the PCR and LAMP to detect 16S rRNA, infB gene and other conserved regions of genomic DNA of H. parasuis in different clinical specimens (Angen et al., 2007, Chen et al., 2010, Gou et al., 2018, McDowall, Slavic, MacInnes, & Cai, 2014, Oliveira et al., 2001, Turni et al., 2010, Yang et al., 2010). The PCR targeting on 16S rRNA gene has problems in specificity giving a weak positive with Actinobacillus indolicus (Turni et al., 2010), and a real‐time PCR using the 16S rRNA as the target could not differentiate Pasteurella mairii from H. parasuis (Turni et al., 2010). The infB gene codes for the two forms of the translation initiation factor IF2 – IF2 alpha and IF2 beta (Hedegaard et al., 2000). The developed real‐time PCR and LAMP based on the infB gene could detect all the 15 serovars of H. parasuis and demonstrated good performance (Chen et al., 2010; Turni et al., 2010; Zhang, Shen, et al., 2012), which shows that the infB gene is a good target for molecular detection methods for H. parasuis and could separate H. parasuis from all other closely related species. In this study, the real‐time RPA primers and probe were also designed based on the infB gene in this study, just as the PCR and LAMP assays developed previously (Chen et al., 2010; Turni et al., 2010; Zhang, Shen, et al., 2012), and the real‐time RPA assay demonstrated very good specificity in the detection of H. parasuis.
Of the 68 tissue samples, 18 (26.5%) and 20 (29.4%) samples were positive for H. parasuis by the real‐time RPA and real‐time PCR, respectively, which were much higher than the bacterial isolation (8, 11.8%). Compared to the bacterial isolation, the PPV and NPV of real‐time RPA was 44.44% and 100%. All the negative samples in real‐time RPA were also negative in bacteria isolation, while the H. parasuis could be only isolated in less than half of the positive samples in real‐time PRA. One possible reason is that the bacteria isolation of H. parasuis is difficult, especially in the tissue samples collected in the slaughterhouse and market. It is also possible that only the dead H. parasuis or genomic DNA but not the live bacteria were present in the samples. The low positive predictive value of RPA was a deficiency of this study, and the assay should be assessed on more clinical positive samples in the following study. The diagnostic performance of the real‐time RPA assay for H. parasuis was comparable to the real‐time PCR assay, while the RPA shows distinct advantages of rapidness and convenience. Therefore, we believe that real‐time RPA will enhance the diagnosis of H. parasuis infection, especially for those laboratories without access to real‐time PCR instrumentation, those are not experienced in the culture of H. parasuis or in situation where culture is not possible.
In summary, the developed real‐time RPA assay is rapid and reliable for detection of H. parasuis, and demonstrates great promise in the diagnosis of Glässer's disease in laboratory and in the field, which is of great importance to eliminate the infected pigs from the herds.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTION
Qiaoyi Han: Data curation; Methodology. Jinfeng Wang: Methodology. Ruiwen Li: Data curation; Validation. Qingan Han: Funding acquisition; Resources. Wanzhe Yuan: Conceptualization; Funding acquisition; Writing‐review & editing. Wang Jianchang: Conceptualization; Funding acquisition; Supervision; Writing‐original draft.
ETHICAL STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The US National Research Council's guidelines for the Care and Use of Laboratory Animals were followed.
ACKNOWLEDGEMENTS
This work was supported by the Project for Key Common Technologies for High Quality Agricultural Development of Hebei Province (19226612D), Earmarked Fund for Hebei Pig Innovation Team of Modern Agro‐industry Technology Research System (HBCT2018110207), and partially supported by the Fund for One‐hundred Outstanding Innovative Talents from Hebei Institution of Higher Learning (SLRC2017039). The authors also thank Dr. Yuefeng Chu from Lanzhou Veterinary Research Institute for kindly providing the genomic DNA of 15 serovar H. parasuis reference strains.
Han Q, Wang J, Li R, Han Q, Yuan W, Wang J. Development of a recombinase polymerase amplification assay for rapid detection of Haemophilus parasuis in tissue samples. Vet Med Sci. 2020;6:894–900. 10.1002/vms3.287
Qiaoyi Han and Jinfeng Wang contributed equally to this work.
The peer review history for this article is available at https://publons.com/publon/10.1002/VMS3.287
[Correction added on 20 June 2020, after first online publication: URL for peer review history has been corrected.]
Contributor Information
Wanzhe Yuan, Email: yuanwanzhe2015@126.com.
Jianchang Wang, Email: jianchangwang1225@126.com.
DATA AVAILABILITY STATEMENT
The dataset analysed during this study is available from the corresponding author on reasonable request.
REFERENCES
- Aarestrup, F. M. , Seyfarth, A. M. , & Angen, O. (2004). Antimicrobial susceptibility of Haemophilus parasuis and Histophilus somni from pigs and cattle in Denmark. Veterinary Microbiology, 101, 143–146. 10.1016/j.vetmic.2004.02.012 [DOI] [PubMed] [Google Scholar]
- Amer, H. M. , Abd El Wahed, A. , Shalaby, M. A. , Almajhdi, F. N. , Hufert, F. T. , & Weidmann, M. (2013). A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. Journal of Virological Methods, 193, 337–340. 10.1016/j.jviromet.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angen, O. , Oliveira, S. , Ahrens, P. , Svensmark, B. , & Leser, T. D. (2007). Development of an improved species specific PCR test for detection of Haemophilus parasuis . Veterinary Microbiology, 119, 266–276. 10.1016/j.vetmic.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Cai, X. , Chen, H. , Blackall, P. J. , Yin, Z. , Wang, L. , Liu, Z. , & Jin, M. (2005). Serological characterization of Haemophilus parasuis isolates from China. Veterinary Microbiology, 111, 231–236. [DOI] [PubMed] [Google Scholar]
- Chen, H. T. , Chu, Y. F. , Liu, Y. S. , Zhang, J. , & Lu, Z. X. (2010). Loop‐mediated isothermal amplification for the rapid detection of Haemophilus parasuis . FEMS Immunology and Medical Microbiology, 60, 283–285. [DOI] [PubMed] [Google Scholar]
- Daher, R. K. , Stewart, G. , Boissinot, M. , & Bergeron, M. G. (2016). Recombinase polymerase amplification for diagnostic applications. Clinical Chemistry, 62, 947–958. 10.1373/clinchem.2015.245829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerman, A. , Bandara, A. B. , & Inzana, T. J. (2020). Phylogenomic analysis of Haemophilus parasuis and proposed reclassification to Glaesserella parasuis, gen. nov., comb. nov. International Journal of Systematic and Evolutionary Microbiology, 70, 180–186. 10.1099/ijsem.0.003730 [DOI] [PubMed] [Google Scholar]
- Frandoloso, R. , Martinez‐Martinez, S. , Rodriguez‐Ferri, E. F. , & Gutierrez‐Martin, C. B. (2012). Comparison of real‐time PCR and culture isolation in colostrum‐deprived pigs immunized and challenged with Haemophilus parasuis . Letters in Applied Microbiology, 54, 149–152. 10.1111/j.1472-765X.2011.03187.x [DOI] [PubMed] [Google Scholar]
- Gou, H. , Li, J. , Cai, R. , Song, S. , Li, M. , Yang, D. , … Li, C. (2018). Rapid detection of Haemophilus parasuis using cross‐priming amplification and vertical flow visualization. Journal of Microbiological Methods, 144, 67–72. 10.1016/j.mimet.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Hedegaard, J. , Hauge, M. , Fage‐Larsen, J. , Mortensen, K. K. , Kilian, M. , Sperling‐Petersen, H. U. , & Poulsen, K. (2000). Investigation of the translation‐initiation factor IF2 gene, infB, as a tool to study the population structure of Streptococcus agalactiae . Microbiology, 146(Pt 7), 1661–1670. [DOI] [PubMed] [Google Scholar]
- Lei, R. , Wang, X. , Zhang, D. , Liu, Y. , Chen, Q. , & Jiang, N. (2020). Rapid isothermal duplex real‐time recombinase polymerase amplification (RPA) assay for the diagnosis of equine piroplasmosis. Scientific Reports, 10, 4096 10.1038/s41598-020-60997-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Macdonald, J. , & von Stetten, F. (2018). Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. The Analyst, 144, 31–67. 10.1039/C8AN01621F [DOI] [PubMed] [Google Scholar]
- Lillis, L. , Siverson, J. , Lee, A. , Cantera, J. , Parker, M. , Piepenburg, O. , … Boyle, D. S. (2016). Factors influencing Recombinase polymerase amplification (RPA) assay outcomes at point of care. Molecular and Cellular Probes, 30, 74–78. 10.1016/j.mcp.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Wang, J. , Zhang, R. , Lin, M. , Shi, R. , Han, Q. , … Yuan, W. (2018). Visual and equipment‐free reverse transcription recombinase polymerase amplification method for rapid detection of foot‐and‐mouth disease virus. BMC Veterinary Research, 14, 263 10.1186/s12917-018-1594-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Wang, L. , Chu, Y. , Li, X. , Cui, Y. , Chen, S. , … Liu, Y. (2016). Characterization of Chinese Haemophilus parasuis isolates by traditional serotyping and molecular serotyping methods. PLoS ONE, 11, e0168903 10.1371/journal.pone.0168903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig, W. D. , Loving, C. L. , Hughes, H. R. , & Brockmeier, S. L. (2016). Characterization and vaccine potential of outer membrane vesicles produced by Haemophilus parasuis . PLoS ONE, 11, e0149132 10.1371/journal.pone.0149132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall, R. , Slavic, D. , MacInnes, J. I. , & Cai, H. Y. (2014). Evaluation of a real‐time polymerase chain reaction assay of the outer membrane protein P2 gene for the detection of Haemophilus parasuis in clinical samples. Canadian Journal of Veterinary Research, 78, 150–152. [PMC free article] [PubMed] [Google Scholar]
- Miao, F. , Zhang, J. , Li, N. , Chen, T. , Wang, L. , Zhang, F. , … Hu, R. (2019). Rapid and sensitive recombinase polymerase amplification combined with lateral flow strip for detecting African Swine fever virus. Frontiers in Microbiology, 10, 1004 10.3389/fmicb.2019.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M. D. , & Jaykus, L. A. (2017). Development of a recombinase polymerase amplification assay for detection of epidemic human noroviruses. Scientific Reports, 7, 40244 10.1038/srep40244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozumi, T. , & Hiramune, T. (1982). Effect of temperature on the survival of Haemophilus parasuis in physiological saline. National Institute of Animal Health Quarterly, 22, 90–91. [PubMed] [Google Scholar]
- Oliveira, S. , Galina, L. , & Pijoan, C. (2001) Development of a PCR test to diagnose Haemophilus parasuis infections. Journal of Veterinary Diagnostic Investigation, 13, 495–501. [DOI] [PubMed] [Google Scholar]
- Oliveira, S. , & Pijoan, C. (2004). Haemophilus parasuis: New trends on diagnosis, epidemiology and control. Veterinary Microbiology, 99, 1–12. 10.1016/j.vetmic.2003.12.001 [DOI] [PubMed] [Google Scholar]
- Piepenburg, O. , Williams, C. H. , Stemple, D. L. , & Armes, N. A. (2006). DNA detection using recombination proteins. PLoS Biology, 4, e204 10.1371/journal.pbio.0040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turni, C. , Pyke, M. , & Blackall, P. J. (2010). Validation of a real‐time PCR for Haemophilus parasuis . Journal of Applied Microbiology, 108, 1323–1331. [DOI] [PubMed] [Google Scholar]
- Wang, J. C. , Yuan, W. Z. , Han, Q. A. , Wang, J. F. , & Liu, L. B. (2017). Reverse transcription recombinase polymerase amplification assay for the rapid detection of type 2 porcine reproductive and respiratory syndrome virus. Journal of Virological Methods, 243, 55–60. 10.1016/j.jviromet.2017.01.017 [DOI] [PubMed] [Google Scholar]
- Yang, W. , Ying, F. , Yingyu, L. , Pin, C. , Wentao, L. , Shuqing, L. , … Qigai, H. (2010). Development and evaluation of loop‐mediated isothermal amplification for rapid detection of Haemophilus parasuis. FEMS Microbiology Letters, 313, 54–60. 10.1111/j.1574-6968.2010.02126.x [DOI] [PubMed] [Google Scholar]
- Zhang, B. , Tang, C. , Liao, M. , & Yue, H. (2014). Update on the pathogenesis of Haemophilus parasuis infection and virulence factors. Veterinary Microbiology, 168, 1–7. 10.1016/j.vetmic.2013.07.027 [DOI] [PubMed] [Google Scholar]
- Zhang, J. M. , Shen, H. Y. , Liao, M. , Ren, T. , Guo, L. L. , Xu, C. G. , … Zhang, B. (2012). Detection of Haemophilus parasuis isolates from South China by loop‐mediated isothermal amplification and isolate characterisation. The Onderstepoort Journal of Veterinary Research, 79, E1–6. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Xu, C. , Guo, L. , Shen, H. , Deng, X. , Ke, C. , … Liao, M. (2012). Prevalence and characterization of genotypic diversity of Haemophilus parasuis isolates from Southern China. Canadian Journal of Veterinary Research, 76, 224–229. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset analysed during this study is available from the corresponding author on reasonable request.


