Abstract
North Borneo (NB) is home to more than 40 native populations. These natives are believed to have undergone local adaptation in response to environmental challenges such as the mosquito-abundant tropical rainforest. We attempted to trace the footprints of natural selection from the genomic data of NB native populations using a panel of ∼2.2 million genome-wide single nucleotide polymorphisms. As a result, an ∼13-kb haplotype in the Major Histocompatibility Complex Class II region encompassing candidate genes TSBP1–BTNL2–HLA-DRA was identified to be undergoing natural selection. This putative signature of positive selection is shared among the five NB populations and is estimated to have arisen ∼5.5 thousand years (∼220 generations) ago, which coincides with the period of Austronesian expansion. Owing to the long history of endemic malaria in NB, the putative signature of positive selection is postulated to be driven by Plasmodium parasite infection. The findings of this study imply that despite high levels of genetic differentiation, the NB populations might have experienced similar local genetic adaptation resulting from stresses of the shared environment.
Keywords: natural selection, North Borneo, malaria, BTNL2, TSBP1, HLA-DRA
Significance
We had identified a putative positive selection signal that spans a 13-kb haplotype in the MHC II region covering genes TSBP1–BTNL2–HLA-DRA in North Borneo native populations. The selection signal was estimated to appear 5,500 years ago and coincided with the period of agricultural expansion in Southeast Asia. Considering the long history of endemic malaria in North Borneo, we therefore hypothesized that the putative signature of positive selection is driven by Plasmodium parasite infection.
Introduction
The island of Borneo, geographically located in Southeast Asia, is the third-largest island in the world. Malaysia, Brunei, and Indonesia have sovereignty over the island, with the latter having the largest portion of the land area. The Malaysian section of Borneo comprises the states of Sabah and Sarawak. The Sabah state (known as North Borneo [NB]) is home to culturally diverse populations of more than 40 major ethnicities that converse in over 80 local dialects (Combrink et al. 2006). The people are broadly categorized into five major groups based on their linguistic and sociocultural practices, namely Dusunic, Paitanic, Murutic, Ida’anic, and Sama-Bajaw. Their vernacular is part of the Austronesian superfamily of languages. The northern region of Borneo was once linked to the southern Philippines as part of the larger Sundaland before the rise ofsea level ∼15 thousand years ago (kya) (Bellwood 2007).
Archaeological evidence suggests that this landmass may have been inhabited by the Australo-Melanesian group and was subsequently replaced by the Austronesian group (Bellwood 2007). Our recent study based on the genome-wide single nucleotide polymorphism (SNP) array suggested that the native populations from NB are closely related to the aborigines from Taiwan and the non-Austro–Melanesian Filipinos (Yew et al. 2018a). Further investigation using whole-genome sequencing technology suggested that the time of divergence of the NB natives predates the Austronesian expansion, implying a possible human habitation in this landmass during the pre-Neolithic period (Yew et al. 2018b).
NB houses part of the world’s oldest tropical rainforest. Owing to its unique geological situation and equatorial climate, the tropical rainforest in NB is enriched in biodiversity and natural resources, but this entails enormous environmental stresses to human habitation and survival, such as the hot and humid climate, limited food resources, and pathogen infections, especially Plasmodium parasites. This offers a unique and complex environment for selective pressure. Several investigations of positive natural selection of the indigenous populations inhabiting this region have been carried out. Notably, different signals of positive natural selection against malarial infection and other traits have been detected among the indigenous populations (Orang Asli) from Peninsular Malaysia (Deng et al. 2014; Liu et al. 2015), whereas a recent study reported a genetic adaptation signal for breath-holding diving capability among the Sea Bajau people from Borneo (Ilardo et al. 2018).
Historical demographic events such as population expansion or genetic drift will have an influence on the entire genome. In contrast, natural selection typically affects the diversity of local genomic regions and thus is distinguishable from the genome-wide pattern. Population genomics offers an approach that enables us to identify signatures of past events with footprints left in local genomic regions. Therefore, in this study, we investigated the forces of natural selection that could have had impacts on the unique genetic architecture of these populations. We utilized the genotyping array data set that was published earlier (Yew et al. 2018a) comprising five ethnic groups representing three major linguistic groupings in NB: Dusun, Rungus, Sonsogon (all three represent the Dusunic speaking family), Sungai-Lingkabau (representing the Paitanic speaking family), and Murut-Paluan (representing the Murutic speaking family). A strong putative signal of positive selection in the Major Histocompatibility Complex (MHC) Class II region was identified and was estimated to have occurred in the NB populations ∼5.5 kya (∼220 generations ago), which coincided with the period of Austronesian expansion. Considering the long history and high prevalence of malaria among the NB populations (Copeland, 1935; William et al. 2014), we postulate that the positive selection could have been driven by the endemic Plasmodium species.
Results
Genetic Relatedness of the Five NB Populations
Principal component analysis (PCA) revealed that the NB populations formed a closely related but distinct cluster from other Southeast Asian populations (fig. 1a, supplementary fig. S1, Supplementary Material online). The phylogenetic tree constructed based on the Fixation index (FST) of pairwise populations also showed that the NB populations formed a separate clade (fig. 1b). The NB populations and the metropolitan Singapore populations (e.g., Malays [MAS] and Han Chinese [CHS]) are in a closer relationship (FST = 0.020–0.042) than any of them with the Orang Asli from Peninsular Malaysia (e.g., Jakun, abbreviated as JKN, an Austronesian speaking indigenous population categorized under Proto-Malay, and Negrito, abbreviated as NGO, an anthropologically defined Austro-asiatic speaking hunter-gather population from Peninsular Malaysia, also locally known as Semang) (FST = 0.029–0.048 between the Singapore populations and the Orang Asli; FST = 0.054–0.089 between the NB populations and the Orang Asli; fig. 1b, supplementary table S1, Supplementary Material online). These findings are in agreement with those of Yew et al. (2018a). However, the genetic differentiation among the five NB populations, ranging from 0.014 (Dusun vs. Rungus) to 0.044 (Murut-Paluan vs. Sonsogon), was surprisingly higher when compared with that between the populations from Singapore (FST = 0.011 between MAS and CHS), indicating nontrivial genetic diversity among the NB populations (supplementary table S1, Supplementary Material online).
Fig. 1.
Genetic relatedness of the five native populations from NB. (a) PCA of Southeast Asian populations; (b) An unrooted population phylogenetic tree constructed under a neighbor-joining framework. The branch scores were obtained by 100 replication analyses. MAS, Metropolitan Malays from Singapore; CHS, Southern Han Chinese from Singapore; INS, Southern India from Singapore; NGO, Negrito from Peninsular Malaysia including Negrito Bateq, Negrito Mendriq, and Negrito Jahai; JKN, Proto-Malay Jakun from Peninsular Malaysia; CEU, Northern & Western European; YRI, Yoruba Nigeria.
Identification of Signatures of Positive Selection
We first estimated the site-specific FST (Weir and Hill 2002) to scan for putative signals of positive natural selection and subsequently corroborated the results using haplotype-based selection metrics, including the integrated haplotype score (iHS) (Voight et al. 2006) and the cross-population extended haplotype homozygosity (XP-EHH) (Sabeti et al. 2007) (see Materials and Methods for more details). Signals of positive natural selection for each of these metrics are tabulated in supplementary tables S2–S4, Supplementary Material online.
We identified a genomic region spanning ∼4.5 Mb of the MHC Class II region (chr6:29,545,208–34,083,564) that was highly differentiated between the NB populations and the surrounding neighbor populations, including MAS, CHS, NGO, and JKN. This putative signal consistently presented in the top 0.1% of the genome-wide FST in the comparisons between each NB population and each of the three reference populations (except for Sonsogon vs. MAS), but not between any two NB groups (supplementary fig. S2 and table S2, Supplementary Material online). Considering the geographical relatedness of the five NB populations as well as the shared signature of local adaptation, we reasoned that the NB populations might have collectively experienced similar forces of local adaptation in the tropical rainforest. Therefore, we pooled these populations as a single group (denoted in general as NB) in subsequent analyses to locate shared signatures for positive natural selection resulting from local adaptation. We repeated the pairwise FST analysis using the pooled data set and confirmed that this signal was consistent across pairwise population analyses between NB with MAS, CHS, NGO, and JKN, and was not affected by the Hardy–Weinberg equilibrium filtration of the data (fig. 2a–d; supplementary fig. S3a, Supplementary Material online). This putative selection signal harboring the MHC Class II region covered 193 genes. As expected, functional enrichment analysis performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang et al. 2009) showed that these genes were significantly enriched with associations to autoimmune diseases and viral infections (supplementary table S5, Supplementary Material online). We found ten SNPs in this region showing most significant and consistent signals of FST across population pairs (supplementary table S2, Supplementary Material online).
Fig. 2.
A putative signal of positive selection on Chromosome 6 in the North Borneo (NB) populations. Manhattan plot of FST showing profound differentiation on Chromosome 6 between NB and (a) Southern Chinese from Singapore (CHS); (b) Metropolitan Malays from Singapore (MAS); (c) Proto Malay Jakun from Peninsular Malaysia (JKN); (d) Negrito from Peninsular Malaysia (NGO). In each plot, the red-dashed line indicates the top 0.1% cutoff of the genome-wide FST in each population pair, and the red dots indicate signals in the MHC Class II region. (e) Haplotype decay around TSBP1-rs3129943 in NB and non-NB populations.
The putative signal at the MHC Class II region was further confirmed with iHS and XP-EHH analyses (supplementary tables S3–S4, Supplementary Material online). We found a 500-kb region (chr6:32,200,001–32,700,001) showing very high density of iHS signals (in the top 5% of the whole genome). Especially, the proportion of iHS signals in chr6:32,300,001–32,400,001 reaches the top 1% of the whole genome. Notable candidates in this region include rs9268605 with the top iHS value (|iHS| = 4.62), and rs3129943 and rs984778 amongst the ten candidate SNPs identified by the FST test. These variants could possibly mediate the gene expression changes according to the Genotype-Tissue Expression (GTEx) database (https://gtexportal.org/home/, last accessed October 17, 2020): rs984778 was reported as a splicing quantitative locus (sQTL) for HLA-DRA in multiple tissues; rs3129943 is an sQTL for TSBP1 predominantly in testis and is an eQTL for BTNL2 in nerve. The variant rs9268605 was only 255 bp upstream to rs984778, and 61.1 kb downstream to rs3129943. We observed an apparent extension of haplotypes with higher frequencies in this region in NB compared with the other populations (fig. 2e; supplementary figs. S3b, S4, and S5, Supplementary Material online). In addition, we found a region (chr6:26,300,001-26,500,001) upstream to the MHC Class II also showed outstanding iHS signal (in the top 1% of the whole genome; top |iHS| score = 4.97 at rs9467750). Since we aimed to locate the adaptive sites shared across the NB populations but highly differentiated between NB and other populations, we did not focus on this region in the subsequent analyses as it was not captured by cross-population analyses (FST or XP-EHH).
We observed an overall reduction in genome-wide heterozygosity in the five NB populations compared with other South and Southeast Asian populations, including the pooled NGO populations. Interestingly, the heterozygosity for the putative signal region of MHC Class II was slightly lower than at the genome-wide scale in the NB populations; this was unexpected assuming balancing selection could have occurred in the region (supplementary fig. S6 and table S6, Supplementary Material online). In contrast, pairwise FST exhibited an overall higher level of genetic differentiation in the MHC region than at the genome-wide scale in the NB populations (supplementary fig. S7, Supplementary Material online). These collective findings are consistent with the hallmark characteristics of positive natural selection (Meyer et al. 2018).
The three selected SNPs of interest (rs3129943-G, derived allele; rs9268605-G, ancestral allele; rs984778-G, derived allele) showed higher allele frequencies in the NB populations than in the non-NB populations (P = 6.85 × 10−5, 8.08 × 10−5, and 6.89 × 10−5, respectively; one-sided Wilcoxon rank-sum test; table 1; supplementary table S7, Supplementary Material online). The linkage between rs3129943 and rs984778 was moderately strong in NB (r2 = 0.491 in the pooled NB population; highest in Sungai-Lingkabau, r2 = 0.648) and JKN (r2 = 0.359) but was almost nil in others (MAS, CHS, INS, and NGO) (table 2). All three variants were homozygous in Sonsogon. Interestingly, rs9268605 and rs984778 exhibited strong linkage disequilibrium (LD) in the NB populations but not in NGO or JKN. We were not able to assess the LD between rs9268605 and rs984778 in MAS, CHS, and INS, as the rs9268605 genotype was not successfully captured in these data. Genes affected by these three SNPs were TSBP1, BTNL2, and HLA-DRA, in which TSBP1 and BTNL2 were in strong LD (supplementary fig. S8, Supplementary Material online).
Table 1.
Derived Allele Frequencies of the Four SNPs of Interest in the Five Native Populations from NB
| Population | rs9467750-C a | rs3129943-G a | rs9268605-G b | rs984778-Ca |
|---|---|---|---|---|
| NB | 0.964 | 0.934 | 0.913 | 0.893 |
| Dusun | 0.975 | 0.875 | 0.875 | 0.875 |
| Murut-Paluan | 0.975 | 0.875 | 0.875 | 0.750 |
| Rungus | 0.975 | 0.975 | 0.925 | 0.925 |
| Sonsogon | 1.000 | 1.000 | 1.000 | 1.000 |
| Sungai | 1.000 | 0.947 | 0.947 | 0.921 |
| Malay (MAS) - SGVP | NA | 0.534 | NA | 0.365 |
| Han Chinese (CHS) - SGVP | NA | 0.354 | NA | 0.240 |
| Southern Indian (INS) - SGVP | NA | 0.313 | NA | 0.301 |
| Negrito (NGO) | 0.982 | 0.264 | 0.482 | 0.218 |
| Jakun (JKN) | 0.967 | 0.300 | 0.333 | 0.133 |
| African Caribbean (ACB) | 0.370 | 0.313 | 0.729 | 0.594 |
| African American (ASW) | 0.410 | 0.328 | 0.664 | 0.484 |
| Bengali Bangladesh (BEB) | 0.791 | 0.390 | 0.616 | 0.413 |
| Chinese Dai (CDX) | 0.941 | 0.398 | 0.527 | 0.446 |
| Northern & Western European (CEU) | 0.874 | 0.258 | 0.596 | 0.349 |
| Northern Han Chinese Beijing (CHB) | 0.913 | 0.354 | 0.408 | 0.243 |
| Southern Han Chinese (CHS)—1kgp | 0.957 | 0.305 | 0.338 | 0.238 |
| Colombians Medellin (CLM) | 0.819 | 0.181 | 0.644 | 0.356 |
| Esan Nigeria (ESN) | 0.328 | 0.379 | 0.808 | 0.732 |
| Finnish Finland (FIN) | 0.869 | 0.273 | 0.717 | 0.354 |
| British England (GBR) | 0.885 | 0.225 | 0.632 | 0.357 |
| Gujarati Indian (GIH) | 0.786 | 0.354 | 0.524 | 0.384 |
| Gambian West Division of Gambia (GWD) | 0.336 | 0.155 | 0.553 | 0.319 |
| Iberian Spain (IBS) | 0.855 | 0.248 | 0.603 | 0.332 |
| Indian Telugu (ITU) | 0.878 | 0.451 | 0.583 | 0.343 |
| Japanese Tokyo (JPT) | 0.933 | 0.313 | 0.615 | 0.389 |
| Kinh Vietnam (KHV) | 0.904 | 0.485 | 0.525 | 0.222 |
| Luhya Kenya (LWK) | 0.288 | 0.424 | 0.727 | 0.551 |
| Mende Sierra Leone (MSL) | 0.318 | 0.288 | 0.435 | 0.359 |
| Mexican (MXL) | 0.703 | 0.211 | 0.594 | 0.359 |
| Peruvian Lima (PEL) | 0.800 | 0.112 | 0.365 | 0.182 |
| Punjabi Pakistan (PJL) | 0.828 | 0.260 | 0.474 | 0.365 |
| Puerto Ricans (PUR) | 0.760 | 0.188 | 0.567 | 0.389 |
| Sri Lankan Tamil (STU) | 0.809 | 0.368 | 0.456 | 0.304 |
| Toscani Italy (TSI) | 0.836 | 0.262 | 0.556 | 0.336 |
| Yoruba Nigeria (YRI) | 0.407 | 0.361 | 0.662 | 0.523 |
Note.—The derived allele frequencies for rs3129943 (G) and rs984778 (C) and the ancestral allele frequency for rs9268605 (G) are much higher in the NB populations than in other worldwide populations.
NA, not available in the data set.
Derived allele.
Ancestral allele.
Table 2.
LD Estimation between the SNPs of Interest
| Population | rs3129943 vs. rs9467750 | rs3129943 vs. rs9268605 | rs9268605 vs. rs984778 | rs984778 vs. rs3129943 | rs984778 vs. rs9467750 | rs9467750 vs. rs9268605 |
|---|---|---|---|---|---|---|
| NB | 0.025 | 0.625 | 0.791 | 0.491 | 0.042 | 0.059 |
| Dusun | 0.004 | 0.587 | 1 | 0.587 | 0.004 | 0.004 |
| Murut-Paluan | 0 | 0.673 | 0.636 | 0.429 | 0 | 0.036 |
| Rungus | 1 | 0.316 | 1 | 0.316 | 0.316 | 0.316 |
| Sonsogon | NA | NA | NA | NA | NA | NA |
| Sungai | 1 | 1 | 0.648 | 0.648 | 1 | NA |
| Malay (MAS) | NA | NA | NA | 0.068 | NA | NA |
| Han Chinese (CHS) | NA | NA | NA | 0.026 | NA | NA |
| Southern Indian (INS) | NA | NA | NA | 0.109 | NA | NA |
| Negrito (NGO) | 0 | 0.207 | 0.3 | 0.09 | 0.001 | 0.017 |
| Jakun (JKN) | 0.025 | 0.585 | 0.308 | 0.359 | 0.005 | 0.017 |
Note.—rs984778 and rs3129943 showed a moderately strong LD among the NB compared with the non-NB populations, except Jakun (JKN). The four SNPs are fixed in Sonsogon; Two SNPs, rs9467750 and rs9268605, are missing in MAS, CHS, and INS. rs9467750 was only captured by iHS and thus was not considered in subsequent analysis; it was used as a control.
NA, not available in the data set.
Dating Natural Selection
We next speculated as to the specific driving force(s) of positive natural selection in this region. To address this question, we first estimated the time since the natural selection occurred based on the extended haplotypes. Analysis on the haplotype diversity extended from the selection signal revealed a region ∼13 kb in length with exceedingly reduced haplotype diversity in the NB population. This region consisted of 71 SNPs, yet only seven haplotypes were observed, and they were further divided into three highly divergent haplotype groups: Haplotype group “A” (Hap 1–3) and “C” (Hap 5–7) consisting three haplotypes, respectively; Haplotype group “B” consisting one haplotype (Hap 4) (fig. 3; supplementary fig. S9, Supplementary Material online). The three haplotype groups could be differentiated using 59 of the 71 SNPs. The adaptive variants at rs9268605 (G) and rs984778 (C) were found in haplotype group “A.” Then we assigned samples from the 1000 Genomes Project Phase III dataset (1KGP; http://www.internationalgenome.org/, last accessed October 17, 2020) into the three haplotype groups (we allowed for variations up to three SNPs in the haplotype group,otherwise the haplotype would be was assigned to “others”), and this revealed that the three major groups explained most of the haplotypes of worldwide populations. It was noted that the frequency of the Haplotype group A reached 0.75–1 in the NB populations, largely attributed to Hap1 (0.4–0.61) (table 3; supplementary table S8, Supplementary Material online). However, the Haplotype group A frequency is much lower in other worldwide populations (0.18–0.73) (table 3). Assuming that the haplotype decay followed a Poisson process, our estimation revealed that the selection of Hap1 arose ∼5.5 kya (∼220 generations; 4.6–3.1 kya for each NB population) (table 4). The estimated onset of the positive natural selection matched the the emergence of agricultural society, which may have led to malarial expansion (Volkman 2001; Joy 2003).
Fig. 3.
Haplotypes of the ∼13 kb region in the NB populations. All 71 SNPs in this region are shown. The ancestral state information for each SNP is provided by dbSNP. For the three different haplotype groups, the dash “–” symbol represents a polymorphic nucleotide within all haplotypes. Sequences with one different nucleotide variation were defined as different haplotypes. The maximum-parsimony tree was constructed using the SPR algorithm implemented in MEGA7 (Kumar et al. 2016) with search level set to be 0, and the initial trees were obtained by the random addition of sequences (ten replicates). The branch length was calculated using the average pathway method (Nei and Kumar 2000) based on the number of nucleotide changes over the whole sequence.
Table 3.
Haplotype Frequencies in the NB and Global Populations from the 1000 Genomes Project
| Population | Region | Number of Haplotypes | Haplotype Group Frequency (%) |
|||
|---|---|---|---|---|---|---|
| A | B | C | Others | |||
| Dusun | NB | 40 | 87.5 | 0.0 | 12.5 | 0.0 |
| Murut-Paluan | NB | 40 | 75.0 | 7.5 | 17.5 | 0.0 |
| Rungus | NB | 40 | 92.5 | 0.0 | 7.5 | 0.0 |
| Sungai | NB | 38 | 92.1 | 2.6 | 5.3 | 0.0 |
| Sonsogon | NB | 38 | 100.0 | 0.0 | 0.0 | 0.0 |
| Sri Lankan Tamil (STU) | South Asia | 204 | 30.4 | 15.2 | 49.0 | 5.4 |
| Gujarati Indian (GIH) | South Asia | 206 | 38.4 | 14.1 | 47.1 | 0.5 |
| Indian Telugu (ITU) | South Asia | 204 | 34.3 | 24.0 | 40.2 | 1.5 |
| Punjabi Pakistan (PJL) | South Asia | 192 | 36.5 | 10.9 | 49.5 | 3.1 |
| Bengali Bangladesh (BEB) | South Asia | 172 | 41.3 | 20.4 | 38.4 | 0.0 |
| Southern Han Chinese (CHS) | East Asia | 210 | 23.8 | 10.0 | 54.8 | 11.4 |
| Japanese Tokya (JPT) | East Asia | 208 | 38.9 | 22.6 | 32.7 | 5.8 |
| Northern Han Chinese Beijing (CHB) | East Asia | 206 | 24.3 | 16.5 | 51.0 | 8.3 |
| Kinh Vietnam (KHV) | East Asia | 198 | 22.2 | 30.3 | 41.4 | 6.1 |
| Chinese Dai (CDX) | East Asia | 186 | 44.6 | 8.1 | 46.2 | 1.1 |
| Gambian West Division of Gambia (GWD) | Africa | 226 | 31.9 | 23.5 | 41.6 | 3.1 |
| Yoruba Nigeria (YRI) | Africa | 216 | 51.9 | 13.9 | 24.1 | 10.2 |
| Esan Nigeria (ESN) | Africa | 198 | 73.2 | 7.6 | 16.2 | 3.0 |
| Luhya Kenya (LWK) | Africa | 198 | 55.1 | 17.7 | 20.2 | 7.1 |
| Mende Sierra Leone (MSL) | Africa | 170 | 35.9 | 7.7 | 37.1 | 19.4 |
| African Caribbean (ACB) | America | 192 | 59.4 | 13.5 | 20.8 | 6.3 |
| African American (ASW) | America | 122 | 48.4 | 18.0 | 29.5 | 4.1 |
| Puerto Rican (PUR) | America | 208 | 38.5 | 18.3 | 42.3 | 1.0 |
| Colombian Medellin (CLM) | America | 188 | 35.6 | 28.7 | 34.6 | 1.1 |
| Peruvian from Lima (PEL) | America | 170 | 18.2 | 18.2 | 63.5 | 0.0 |
| Mexican (MXL) | America | 128 | 35.9 | 23.4 | 40.6 | 0.0 |
| Iberian in Spain (IBS) | Europe | 214 | 33.2 | 27.1 | 38.3 | 1.4 |
| Toscani Italy (TSI) | Europe | 214 | 33.6 | 22.0 | 43.9 | 0.5 |
| Northern & Western European (CEU) | Europe | 198 | 34.9 | 24.8 | 40.4 | 0.0 |
| Finnish Finland (FIN) | Europe | 198 | 35.4 | 36.4 | 28.3 | 0.0 |
| British England (GBR) | Europe | 182 | 35.7 | 27.5 | 36.8 | 0.0 |
Note.—A total of 59 SNPs were included in the haplotype group assignment. Haplotypes with more than three nucleotide differences with the three groups were assigned to “Others.” The NB populations were significantly enriched with haplotype group A.
Table 4.
Selection Time Estimation of the NB Populations
| Population | Time of selection in KYA (generations) |
||
|---|---|---|---|
| Haplotype 1 | Haplotype 2 | Haplotype 3 | |
| NB | 5.51 (220.4) | 5.93 (237.2) | 3.98 (159.2) |
| Dusun | 4.64 (185.6) | — | 3.87 (154.8) |
| Lingkabau | 3.46 (138.4) | — | — |
| Murut-Paluan | 3.29 (131.6) | — | — |
| Rungus | 3.06 (122.4) | — | 3.02 (120.8) |
| Sonsogon | 4.29 (171.6) | — | 2.30 (92) |
Note.—Selection time was estimated based on the extended haplotypes. NB, combination of the five NB populations. To minimize potential bias introduced by sample size, haplotypes with less than ten counts were excluded from the time estimation. Number of generations is shown in the brackets.
KYA, thousand years ago.
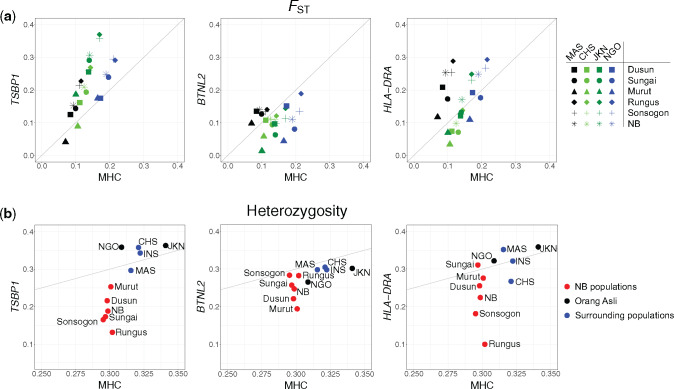
We then asked which of the three candidate genes (TSBP1, BTNL2, and HLA-DRA) could possibly be the key target of natural selection. We found that all these three genes, particularly TSBP1, in the NB populations showed greater genetic differentiation as measured by a higher pairwise FST yet with reduced heterozygosity when compared with the entire MHC region, or with genomic regions showing comparable length and SNP density with them (fig. 4; supplementary fig. S10 and table S9, Supplementary Material online). We suspect that TSBP1 was possibly the targeted candidate gene under positive natural selection in NB. Nonetheless, we do not rule out the possibility that BTNL2 and HLA-DRA may also have been under positive selection as epistatic effects might exist (Traherne 2008; Meyer et al. 2018), and they were functionally associated to TSBP1 according to the STRING network (https://string-db.org/network/9606.ENSP00000415517, last accessed October 17, 2020) (supplementary fig. S11, Supplementary Material online).
Fig. 4.
Genetic diversity of BTNL2, TSBP1, and HLA-DRA relative to the MHC Class II region. (a) FST for pairs of populations, averaging across 100 bootstrap replications. Each dot depicts the average FST of all sites in the MHC Class II region (x axis) and that in a candidate gene (y axis). The FST was calculated between the collective NB population and the non-NB populations (asterisk dots) and between each single NB population and non-NB populations (black dots): Metropolitan Malays from Singapore (MAS); red dots: Negrito (NGO); light green dots: Southern Han Chinese from Singapore (CHS); dark green dots: Proto-Malay Jakun (JKN). The dots above the grey line indicate candidate genes with higher average FST than that of the whole genome; (b) Heterozygosity of the candidate genes of interest relative to the MHC Class II heterozygosity. Heterozygosity of the candidate genes was lower than that of the MHC Class II region.
Discussion
The MHC region has been a classical landmark for balancing selection, likely under the pressure from pathogen diversity (Meyer et al. 2006; Traherne et al. 2006; Yasukochi and Satta 2013; Field et al. 2016). In this study, we have identified a region within the MHC Class II that was under positive natural selection among the NB populations. This putative signal was highly robust as it was consistently identified despite that different approaches were used.
We showed that although the genetic differentiation among the five NB populations was quite high, similar driving force(s) of natural selection could have affected their genetic structure, as supported by profoundly lower pairwise FST values in the MHC II region among the NB populations compared with that between NB and non-NB populations (supplementary table S6, Supplementary Material online). We reasoned that if selection favored similar sets of alleles or haplotypes within a broadly defined geographical region (in this case, the interior region of NB), reduced genetic differentiation (as measured by FST) would have been expected (Meyer et al. 2006).
The three candidate genes TSBP1, BTNL2, and HLA-DRA in the NB populations exhibited typical characteristics of positive selection, including the increased genetic differentiation, reduced heterozygosity relative to the genome-wide scale, and a strong LD block (Schierup et al. 2000). We reasoned that selection favors different alleles in distinct populations, thus driving locally adaptive MHC alleles to higher frequencies and resulting in increased population differentiation (Meyer et al. 2018). This pattern is particularly prominent in TSBP1. Essentially, there is no straightforward strategy for locating a single gene contribution to a trait of interest in which multiple linked interacting genes are at work. Although less likely, we also caution that the selection signal may be confounded by demographic history and genetic drift forces that would lead to sudden expansion of the haplotype frequency of interest. Further laboratory investigations may be required to rule out these possibilities.
We believe that this selection signal is likely a product of the local adaptation process for survival in the tropical rainforest—one of the toughest environments for human habitation characterized by unusually high protozoa and pathogen diversity (Fan et al. 2016) that may influence the reproductive success of the affected population. In this regard, we note that the parasitic protozoan Plasmodium species that causes malaria, resulting in significant mortality rates in tropical countries, likely exerts the strongest selection pressure (Kwiatkowski, 2005).
NB has been persistently recognized as a malaria-endemic region since the past several centuries (Copeland, 1935; William et al. 2013, 2014). Studies over the years have recorded a number of Plasmodium sp. in NB, including simian Plasmodium knowlesi (William et al. 2013, 2014). Notably, a high prevalence of malaria infection was recorded in Murut-Paluan and Dusun nearly a century ago (Copeland 1935). That study suggested a close attribution between malaria and the low juvenile populations and low birth rates in Murut-Paluan and Dusun in Ranau. Interestingly, the Ranau and Kudat districts, where the Rungus and Dusun samples were collected (Yew et al. 2018a), respectively, showed the highest density of knowlesi malaria (Barber et al. 2011; William et al. 2014).
Some evidence supported our postulation. First, gene expression of HLA-DRA was significantly upregulated in placental malaria (Muehlenbachs et al. 2007), whereas a selection signal in HLA-DRA was found in the low-altitude Ethiopians known to be affected by malaria and schistosomiasis (Alkorta-Aranburu et al. 2012). Second, the chimeric mice with BTNL2−/− had significantly decreased cerebral malaria survival rate (Subramaniam et al. 2015). In addition, the top candidate SNP rs3129943 located in TSBP1 showed substantially higher derived allele frequency (0.934) in the NB populations than in the other populations (0.11–0.53, see table 1). Although the ancestral allele A at this locus was reported to be associated with asthma (Hirota et al. 2011), we do not think that the selection signal was driven by asthma, owing to its lower prevalence in NB Malaysia (Lin and Kasim 1997). Analysis from the STRING database version 11.0 suggested plausible interactions of TSBP1, BTNL2, and HLA-DRA (the interaction score = 0.673–0.725; supplementary fig. S11, Supplementary Material online). Therefore, it is plausible to postulate that these neighboring genes may have demonstrated an epistatic effect, that is, these genes are tuned to work together as a set of alleles on a particular haplotype, hence the lack of detectable recombination due to the preference of favorable immunological function through selection (Traherne et al. 2006).
Owing to their natural habitat being similar to those of the NB populations, indigenous populations from Peninsular Malaysia (the Orang Asli) have been routinely exposed to malaria infection. Indeed, our earlier study had reported several putative signals of natural selection in the Orang Asli (Liu et al. 2015). We found that within a small geographical region in the tropical rainforest in Peninsular Malaysia, the Orang Asli exhibited differential evidence of positive selection against malaria. We found that none of the putative selection signals identified in the Orang Asli presented in the NB native populations, suggesting plausible attribution to their different population histories; presumably the adaptations occurred after the population diverged. In addition, recent reports suggested that the predisposition of Plasmodium parasites differed between NB and Peninsular Malaysia populations (Yap et al. 2018; Hussin et al. 2020). These factors may contribute to the different selection signals between the Orang Asli and native NB populations.
We acknowledge that different analyses of positive selection approaches differ in their power to detect a selection signal, depending on how long ago selection began, how close the selected allele is to fixation, and how different allele frequencies are in different populations. In this study, we selected the putative signals that showed a profound population differentiation. Although there were genomic regions that did not fit this criterion, the possibility of these regions being true selection signals should not be ruled out and warrants further investigation.
We note that all supporting evidence attributed to malaria so far has been indirect. However, we wish to reiterate that: 1) Malaria is believed to be the strongest selection pressure on human populations identified to date (Kwiatkowski 2005). There have been numerous records of malaria endemicity in NB populations since the last century. On a separate note, prevalence data for other parasitic infections among the NB populations are lacking. In the absence of other recorded evidence of parasitic or immune-related diseases among these native NB populations, malaria appears as the most likely driving force of the identified selection signal. 2) Functional studies in relation to malaria pathogenesis have been carried out on two of the candidate genes, namely HLA-DRA and BTNL2 (Muehlenbachs et al. 2007; Subramaniam et al. 2015). 3) It is generally accepted that the expansion of malaria infection in human populations is attributed to the expansion of agricultural technology. The estimated time of the selection signal identified in this study is in agreement with the period when agricultural expansion occurred in Southeast Asia, that is, between 4,000 and 6,000 years ago (Bellwood 2007).
In summary, an MHC Class II haplotype encompassing candidate genes TSBP1—BTNL2—HLA-DRA was identified as the putative signature of positive selection among the NB populations. This signal of selection is likely to have occurred during the period of agricultural expansion. With the supporting evidence from earlier studies, it is conceivable to hypothesize that the selection event was driven by Plasmodium parasite infection. Considering the prominent role of these candidate genes in the regulation of the autoimmune system, their plausibility in affecting the susceptibility to pathogen infection points to a fine balance between a strong and appropriate immune response to challenge by a pathogen and an excessive and inappropriate response leading to autoimmune disease (Hollox and Hoh 2014). However, further laboratory validation is required to explore this hypothesis and demonstrate the function(s) of the candidate genes in the response to Plasmodium infection. We also suggest that future studies expand the population range and involve full MHC sequences to assess the differential contributions of selection and recombination in shaping the contrasting evolutionary history of ancestral haplotypes.
Materials and Methods
Genotyping Data, Data Assemblage, and Quality Control (QC)
Genotyping data (comprising ∼2.2 million autosomal SNPs) of 98 unrelated samples representing Dusun, Rungus, Sonsogon, Sungai-Lingkabau, and Murut-Paluan from NB were included in this study as described in Yew et al. (2018a). Briefly, this study was approved by the Medical Research Ethics Committee of Universiti Malaysia Sabah (ref.no: JKEtika 4/10(3)), and the District Officers of Ranau, Pitas, Kota Marudu, Nabawan, and the respective village chiefs and chairpersons of the Committee for Village Development and Security, and complies with the Helsinki Declaration 1975 as revised in 2000. Genomic DNA was extracted from whole blood or buffy coat using the DNeasy Blood and Tissue kit (Qiagen, Germany). The DNA samples were genotyped with Illumina's Human Omni2.5 bead chip array following the manufacturer's protocol. Calling of SNP genotypes was performed in Genome Studio (Illumina) with the default GenCall score of 0.15.
Data assemblage and QC were carried out using PLINK version 1.07 (Purcell et al. 2007). Criteria for exclusion included: 1) individuals with missing rate >10%; 2) SNPs with missing rate >10%; 3) SNPs with minor allele frequency <0.01; 4) SNPs deviating from Hardy–Weinberg equilibrium (P < 0.0001). A total of 98 NB samples (84%) containing >1.2 million bi-allelic SNPs remained for subsequent analyses. Haplotype phasing for the final data sets was carried out using Shapeit2 without any reference population (Delaneau et al. 2011). SNPs were annotated using the human reference genome GRCh37. The coordinates of genes were provided by the UCSC hg19 RefSeq annotation.
Additional data sets analyzed in this study include metropolitan Malays (MAS) and Chinese from Singapore (CHS) provided by the Singapore Genome Variation Project (SGVP) (Teo et al. 2009), the Orang Asli from Peninsular Malaysia including Negrito (Bateq, Mendriq, and Jehai) and Proto-Malay (Jakun) (Aghakhanian et al. 2015; Liu et al. 2015), and the global populations from the 1000 Genomes Project Phase III dataset (1KGP; http://www.internationalgenome.org/, last accessed October 17, 2020). Data filtration was carried out independently for each population, using the same criteria as described above.
Analysis of Population Relatedness
PCA was performed using flashPCA version 2.0 (Abraham et al. 2017). Unbiased estimation of FST was computed according to Weir and Hill (2002) with 100 times bootstrap replications. A Neighbor-Joining tree was then generated based on FST using Phylip version 3.695 (http://evolution.genetics.washington.edu/phylip.html, last accessed October 17, 2020).
Estimation of Heterozygosity
Observed heterozygosity (Ho) of an SNP was calculated by the ratio of the number of heterozygous individuals to all genotyped individuals. Expected heterozygosity (He) was calculated following Nei (1973). The heterozygosity of one region was calculated by averaging across all the sites within this region.
Identifying Signatures of Positive Selection
We identified alleles or regions that were highly differentiated from other populations using pairwise FST. SNPs with the top 0.1% of the most extreme differentiation were considered as putative signals of positive selection. iHS and XP-EHH (Voight et al. 2006; Sabeti et al. 2007) were computed using the Selscan version 1.2.0 (Szpiech and Hernandez 2014). Default settings were used, and normalization was set at 100 bins with 100-kb nonoverlapping windows using the “norm” feature available in Selscan. Genomic regions within the top 1% in each calculation were considered as putative signals for positive selection.
Estimating the Time of Positive Natural Selection
An evolutionary history was inferred using the Maximum Parsimony method implemented in MEGA7 (Kumar et al. 2016). The tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm with search level set to be 0, and the initial trees were obtained by the random addition of sequences (10 replicates) (Nei and Kumar 2000).
The time since selection of Hap1 in NB populations was estimated based on the extended haplotype homozygosity (EHH). We assumed that the decay of haplotype homozygosity followed a Poisson process:
where Pr(Homozygosity) is the probability that two haplotypes are homozygous at a distance r to the selected haplotype, and g is the number of generations. Given a threshold of Pr(Homozygosity) to be 0.25 and a generation time of 25 years as previously reported (Voight et al. 2006; Tishkoff et al. 2007), g could be estimated.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Ethical Approval and Consent to Participate
This study was approved by the Research and Ethics Committee of Universiti Teknologi MARA [Ref no: 600-RMI (5/1/6)], the Department of Orang Asli Development (Jabatan Kemajuan Orang Asli Malaysia, JAKOA) [JHEOA.PP.30.052.Jld 5(17)], and the Universiti Malaysia Sabah Medical Research Ethics Committee [code: JKEtika 4/10(3)] as well as the district offices, village chief, and the chairperson of the Committee of Village Development and Security. It was also approved by the Biomedical Research Ethics Committee of Shanghai Institutes for Biological Sciences (ER-SIBS-261903). Informed written consent was obtained from the volunteers aged 18 years and above. Their family history, pedigree, and self-reported ethnicity were recorded via an interview using local dialect.
Data Availability
The genotyping data have been deposited in the National Omics Data Encyclopedia (NODE) (http://www.biosino.org, last accessed October 17, 2020) with accession number: OEP000154.
Supplementary Material
Acknowledgments
We thank all the participants, the Sabah state authorities for their full cooperation and assistance during the various field trips. We thank LetPub (www.letpub.com, last accessed October 17, 2020) for its linguistic assistance during the revision of this manuscript.
This study was supported by the National Natural Science Foundation of China (NSFC) grant (31525014, 32030020, 91731303, 31771388, 31900418, 31961130380, and 32041008), the Strategic Priority Research Program (XDB38000000), and Key Research Program of Frontier Sciences (QYZDJ-SSW-SYS009) of the Chinese Academy of Sciences (CAS), the UK Royal Society-Newton Advanced Fellowship [NAF\R1\191094]; the National Key Research and Development Program [2016YFC0906403]; the Shanghai Municipal Science and Technology Major Project [2017SHZDZX01]; and the Science and Technology Commission of Shanghai Municipality [19YF1455200]. This project was funded by the National Biotechnology Division of the Ministry of Science, Technology and Innovation of Malaysia [project code: 100-RMI/BIOTEK 16/6/2B(1/2011)], and by the Ministry of Higher Education of Malaysia [Fundamental Research Grant Scheme, project code: FRG0449-STG-1/2016; and FRGS/1/2015/ST03/UCSI/01/1]. H.B.P acknowledges the Chinese Academy of Sciences President’s International Fellowship Initiatives [2017VBA0008] awarded to him. Y.Y.T. acknowledge support by the National Research Foundation, Prime Minister’s Office, Singapore under its Research Fellowship [NRF-RF-2010-05] and administered by the National University of Singapore. S.X. is Max-Planck Independent Research Group Leader and member of CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges the support of the National Program for Top-notch Young Innovative Talents of the “Wanren Jihua” Project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors Contributions
B.P.H., V.K.S., and S.X. conceived the study; S.X. and B.P.H. designed and supervised the project; B.P.H., V.K.S., X.Z., and L.D. prepared the manuscript; B.P.H., X.Z., L.D., K.Y., C.W.Y., and W.Y.S. performed the data analysis; C.W.Y., M.Z.H., F.A., M.E.P., and V.K.S. involved in sample collection. All authors have read and approved the submission of the manuscript.
Literature Cited
- Abraham G, et al. 2017. FlashPCA2: principal component analysis of biobank-scale genotype datasets. Bioinformatics 33(17):2776–2778. [DOI] [PubMed] [Google Scholar]
- Aghakhanian F, et al. 2015. Unravelling the genetic history of Negritos and indigenous populations of Southeast Asia. Genome Biol Evol. 7(5):1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkorta-Aranburu G, et al. 2012. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 8(12):e1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BE, et al. 2011. Plasmodium knowlesi malaria in children. Emerg Infect Dis. 17(5):814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood P. 2007. Prehistory of the Indo-Malaysian Archipelago. Canberra, Australia: ANU Press. [Google Scholar]
- Combrink H, Soderberg C, Boutin ME, Boutin AY. 2006. Indigenous groups of sabah: an annotated bibliography of linguistic and anthropological sources. 2nd ed. Sabah: SIL International. [Google Scholar]
- Copeland AJ. 1935. The Muruts of North Borneo: malaria and racial extinction. Lancet. 225(5830):1233–1239. [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. 2011. A linear complexity phasing method for thousands of genomes. Nat Methods. 9(2):179–181. [DOI] [PubMed] [Google Scholar]
- Deng L, et al. 2014. The population genomic landscape of human genetic structure, admixture history and local adaptation in Peninsular Malaysia. Hum Genet. 133(9):1169–1185. [DOI] [PubMed] [Google Scholar]
- Fan S, Hansen ME, Lo Y, Tishkoff SA. 2016. Going global by adapting local: a review of recent human adaptation. Science 354(6308):54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, et al. 2016. Detection of human adaptation during the past 2000 years. Science 354(6313):760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, et al. 2011. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 43(9):893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollox EJ, Hoh BP. 2014. Human gene copy number variation and infectious disease. Hum Genet. 133(10):1217–1233. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Hussin N, et al. 2020. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar J. 19(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilardo MA, et al. 2018. Physiological and genetic adaptations to diving in sea nomads. Cell 173(3):569–580.e15. [DOI] [PubMed] [Google Scholar]
- Joy DA. 2003. Early origin and recent expansion of Plasmodium falciparum. Science 300(5617):318–321. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DP. 2005. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 77(2):171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HP, Kasim MS. 1997. Current trends in morbidity and mortality of children in Malaysia. Malaysian J Child Health. 9:104–132. [Google Scholar]
- Liu X, et al. 2015. Differential positive selection of malaria resistance genes in three indigenous populations of Peninsular Malaysia. Hum Genet. 134(4):375–392. [DOI] [PubMed] [Google Scholar]
- Meyer D, Aguiar VRC, Bitarello BD, Brandt DYC, Nunes K. 2018. A genomic perspective on HLA evolution. Immunogenetics 70(1):5–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G. 2006. Signatures of demographic history and natural selection in the human major histocompatibility complex Loci. Genetics 173(4):2121–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlenbachs A, Fried M, Lachowitzer J, Mutabingwa TK, Duffy PE. 2007. Genome-wide expression analysis of placental malaria reveals features of lymphoid neogenesis during chronic infection. J Immunol. 179(1):557–565. [DOI] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 70(12):3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press. [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, et al. ; The International HapMap Consortium. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup MH, Charlesworth D, Vekemans X. 2000. The effect of hitch-hiking on genes linked to a balanced polymorphism in a subdivided population. Genet Res. 76(1):63–73. [DOI] [PubMed] [Google Scholar]
- Subramaniam KS, et al. 2015. The T-cell inhibitory molecule butyrophilin-like 2 is up-regulated in mild Plasmodium falciparum infection and is protective during experimental cerebral Malaria. J Infect Dis. 212(8):1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD. 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 31(10):2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo YY, et al. 2009. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res. 19(11):2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 39(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traherne JA. 2008. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 35(3):179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traherne JA, et al. 2006. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2(1):e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4(3):e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman SK. 2001. Recent origin of Plasmodium falciparum from a single progenitor. Science 293(5529):482–484. [DOI] [PubMed] [Google Scholar]
- Weir BS, Hill WG. 2002. Estimating F-statistics. Annu Rev Genet. 36(1):721–750. [DOI] [PubMed] [Google Scholar]
- William T, et al. 2013. Increasing incidence of Plasmodium knowlesi malaria following control of P. falciparum and P. vivax Malaria in Sabah, Malaysia. PLoS Negl Trop Dis. 7(1):e2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William T, et al. 2014. Changing epidemiology of malaria in Sabah, Malaysia: increasing incidence of Plasmodium knowlesi. Malar J. 13(1):390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap NJ, et al. 2018. Genetic polymorphism and natural selection in the C-terminal 42 kDa region of merozoite surface protein-1 (MSP-1) among Plasmodium knowlesi samples from Malaysia. Parasit Vectors. 11(1):626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi Y, Satta Y. 2013. Current perspectives on the intensity of natural selection of MHC loci. Immunogenetics 65(6):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew CW, et al. 2018. a. Genetic relatedness of indigenous ethnic groups in northern Borneo to neighboring populations from Southeast Asia, as inferred from genome-wide SNP data. Ann Hum Genet. 82(4):216–226. [DOI] [PubMed] [Google Scholar]
- Yew CW, et al. 2018. b. Genomic structure of the native inhabitants of Peninsular Malaysia and North Borneo suggests complex human population history in Southeast Asia. Hum Genet. 137(2):161–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotyping data have been deposited in the National Omics Data Encyclopedia (NODE) (http://www.biosino.org, last accessed October 17, 2020) with accession number: OEP000154.