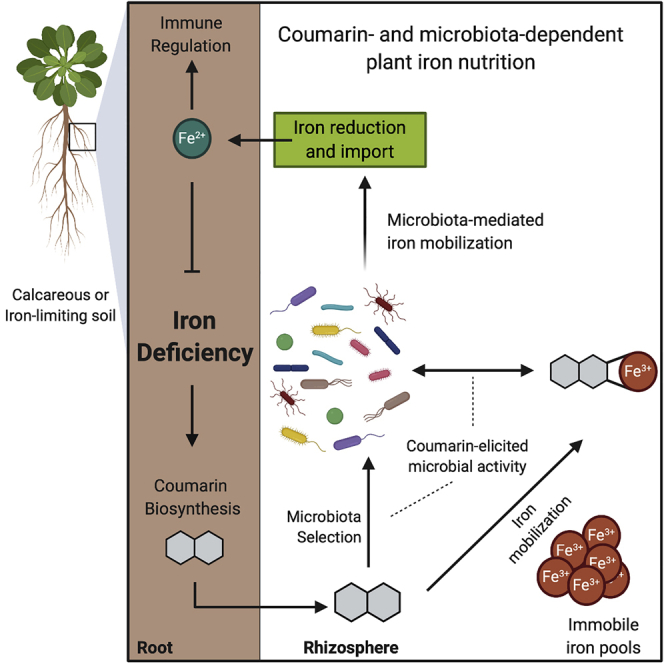
Summary
Plants benefit from associations with a diverse community of root-colonizing microbes. Deciphering the mechanisms underpinning these beneficial services are of interest for improving plant productivity. We report a plant-beneficial interaction between Arabidopsis thaliana and the root microbiota under iron deprivation that is dependent on the secretion of plant-derived coumarins. Disrupting this pathway alters the microbiota and impairs plant growth in iron-limiting soil. Furthermore, the microbiota improves iron-limiting plant performance via a mechanism dependent on plant iron import and secretion of the coumarin fraxetin. This beneficial trait is strain specific yet functionally redundant across phylogenetic lineages of the microbiota. Transcriptomic and elemental analyses revealed that this interaction between commensals and coumarins promotes growth by relieving iron starvation. These results show that coumarins improve plant performance by eliciting microbe-assisted iron nutrition. We propose that the bacterial root microbiota, stimulated by secreted coumarins, is an integral mediator of plant adaptation to iron-limiting soils.
Keywords: root microbiota, coumarins, iron nutrition, edaphic adaptation, secondary metabolites, microbiota, plant growth promotion, immune regulation
Graphical Abstract
Highlights
-
•
Coumarins alter the root microbiota and improve plant growth in iron-limiting soil
-
•
The microbiota improves plant iron nutrition via a coumarin-dependent mechanism
-
•
The iron-beneficial commensal trait is taxonomically widespread but strain specific
-
•
Coumarin-microbiota interaction resolves iron starvation and regulates immune response
Iron-limiting soils are widespread, causing significant losses in plant growth and productivity. Harbort et al. show that under iron limitation, plant-secreted coumarin compounds are mediators of a beneficial plant-microbiota interaction. These specialized metabolites alter root microbiota composition and are required for microbiota-mediated plant iron uptake and immune regulation.
Introduction
Plant roots are colonized by a diverse community of microbes, collectively termed the root microbiota, originating from the surrounding soil biome (Bai et al., 2015; Bulgarelli et al., 2012; Lundberg et al., 2012). The structure of these communities is shaped by soil edaphic factors and root-secreted photosynthates and secondary metabolites (Berendsen et al., 2012; Bulgarelli et al., 2013). The root microbiota provides indirect protection against soil-borne fungal pathogens (Carrión et al., 2019; Durán et al., 2018) and is thought to improve host nutrition by improving the bioavailability of nutrients (Hacquard et al., 2015). However, the extent to which plants can selectively alter their microbiota and harness these beneficial traits in response to nutritional stress is unknown.
Iron is an essential mineral nutrient of plants, acting as a catalyst in many biological processes including photosynthesis and respiration. Although it is an abundant element in most soils, its bioavailability is often limiting due to its extremely low solubility at neutral and alkaline pH, as in calcareous soils containing a high proportion of calcium carbonate. Iron deficiency results in stunted plant growth and leaf chlorosis and is responsible for decreased crop yields and nutrient content in ~30% of arable land (Morrissey and Guerinot, 2009). As such, there is great economic interest in improving plant iron nutrition. In response to iron-limiting conditions, non-graminaceous plants, such as A. thaliana, mount an iron starvation response that is coordinated by FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT) (Colangelo and Guerinot, 2004; Jakoby et al., 2004) and a suite of associated basic helix-loop-helix (bHLH)-type transcription factors (Ivanov et al., 2012). This response serves to improve the solubility of iron through rhizosphere acidification by H+-ATPase AHA2 (Santi and Schmidt, 2009) and reduction of iron(III) to more-soluble iron(II) by plasma membrane protein FERRIC REDUCTION OXIDASE 2 (FRO2) (Robinson et al., 1999). Iron(II) is imported into the root epidermis by IRON-REGULATED TRANSPORTER1 (IRT1) (Vert et al., 2002).
The secretion of coumarins, phenolic secondary metabolites deriving from the general phenylpropanoid pathway, is also induced by iron starvation and is thought to contribute to iron nutrition through direct mobilization of recalcitrant iron pools (Fourcroy et al., 2014, 2016; Rajniak et al., 2018; Rodríguez-Celma and Schmidt, 2013; Schmid et al., 2014; Schmidt et al., 2014; Sisó-Terraza et al., 2016; Siwinska et al., 2018; Tsai et al., 2018). Three main coumarin compounds are produced in A. thaliana via a linear biosynthetic pathway (Figure 1A) (Rajniak et al., 2018; Tsai et al., 2018). FERULOYL-COA 6-HYDROXYLASE1 (F6’H1) synthesizes scopoletin (Kai et al., 2008), which can be converted to fraxetin by SCOPOLETIN 8-HYDROXYLASE (S8H) (Rajniak et al., 2018; Tsai et al., 2018), and further converted to sideretin by CYTOCHROME P450, FAMILY 82C4 (CYP82C4) (Rajniak et al., 2018). Each of these coumarins can be exported by the ABC transporter PLEIOTROPIC DRUG RESISTANCE 9 (PDR9) (Fourcroy et al., 2014), though other efflux pumps may also contribute to the export of some coumarins (Ziegler et al., 2017). Coumarin secretion was recently shown to influence the structure of root microbial communities in artificially limed soil (Stringlis et al., 2018) and synthetic media (Voges et al., 2019). However, the impact of coumarin secretion on the root microbiota in soils with different mineral nutrient availabilities and the consequences for plant productivity remain undefined.
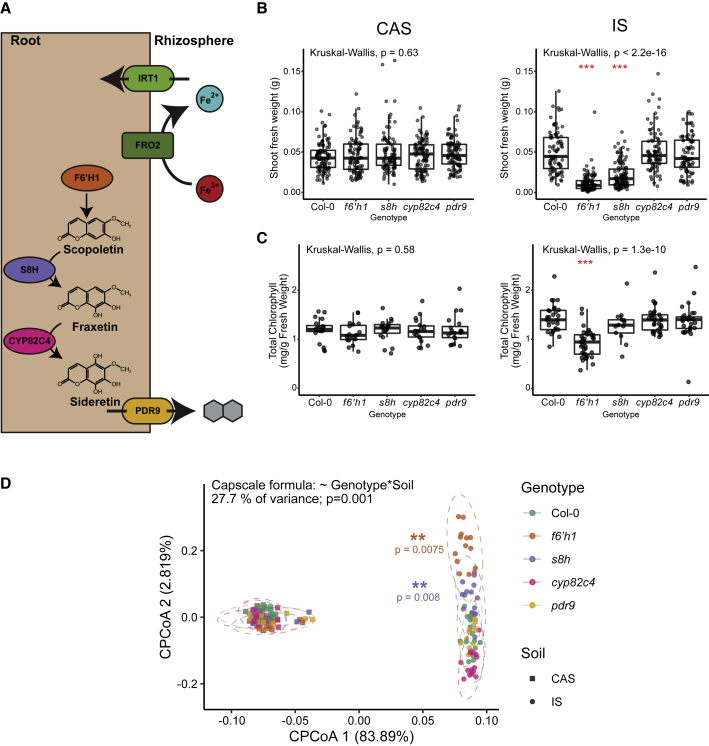
Figure 1.
Coumarin Biosynthesis Is Important for Plant Growth and Root Microbiota Composition in a Naturally Calcareous Soil
(A) Diagram of pathways for coumarin biosynthesis and export, and reductive uptake of iron in Arabidopsis.
(B and C) (B) SFW and (C) total chlorophyll content of coumarin pathway mutants grown in a non-calcareous (CAS) and a calcareous (IS) soil. Statistical significance was determined by Kruskal-Wallis; each mutant was compared with Col-0 by Wilcoxon Ranked Sum post-hoc. Significance is indicated by red asterisks (∗, ∗∗, ∗∗∗, indicate p < 0.05, 0.01, and 0.001, respectively). For shoot fresh weight measurements, Col-0 n = 171, 204; f6’h1 n = 168, 272; s8h n = 93, 113; cyp82c4 n = 164, 209; and pdr9 n = 172, 169 in CAS and IS, respectively. Chlorophyll content was measured from pooled leaf samples, (Col-0 n = 35, 29; f6’h1 n = 34, 36; s8h n = 19, 14; cyp82c4 n = 34, 30; and pdr9 n = 35, 30 in CAS and IS, respectively).
(D) Constrained ordination of root bacterial community composition of coumarin pathway mutants, constrained for the interaction between soil and genotype. Ellipses delineate multivariate normal distribution at 95% confidence. Data are from one representative experiment of three (Col-0 n = 17, 15; f6’h1 n = 18, 14; s8h n = 15, 14; cyp82c4 n = 18, 15; and pdr9 n = 17, 14 in CAS and IS, respectively). p values represent significance of separations between genotypes within each soil determined by pairwise PERMANOVA. Only f6’h1 (orange) and s8h (purple) were significantly separated from Col-0 in IS. See also: Figures S1 and S2.
Results
Coumarin Biosynthesis Is Important for Plant Growth and Root Microbiota Composition in Iron-Limiting Soil
To assess potential links between coumarin secretion, plant growth, and the root-associated microbiota, we grew A. thaliana Col-0 wild-type (WT) plants and mutants with defects in coumarin biosynthesis or export (Figure 1A) on two soils with contrasting iron availability. These mutants were previously shown to have abolished coumarin biosynthesis at their respective steps in the pathway (Rajniak et al., 2018; Schmid et al., 2014; Tsai et al., 2018; Ziegler et al., 2017). Cologne agricultural soil (CAS), obtained from a local site, is slightly acidic with pH 6.5, at which iron is sufficiently available. We also obtained soil from a vineyard in Italy, termed here Italian soil (IS), which is alkaline and calcareous (pH 7.5, 9.7% of active CaCO3). These conditions significantly reduce the availability of iron, despite this soil having a higher total iron content than CAS (Figure S1). We observed a decrease in shoot fresh weight (SFW) and leaf chlorophyll content in f6’h1 and s8h plants grown on IS, whereas the measured performance parameters of all genotypes were indistinguishable on CAS (Figures 1B and 1C). A similar growth deficit was observed in f6’h1 plants grown on other alkaline soils isolated from geographically diverse sites and could be improved by supplementation with solubilized iron (Figure S1). These results show that coumarin biosynthesis is important for growth in naturally iron-limiting soils. The performance of pdr9 plants, however, was indistinguishable from WT on both CAS and IS; thus, coumarin export via the ABC transporter PDR9 was not crucial for growth in iron-limiting soils (Figures 1 and S1). This is in contrast to reported germ-free growth on synthetic media (Fourcroy et al., 2014; Rodríguez-Celma et al., 2013) (and Figure 5 below). These data suggest that in soil, sufficient coumarin export may occur via additional members of the diversified and promiscuous ABC transporter family (Borghi et al., 2019; Ziegler et al., 2017).
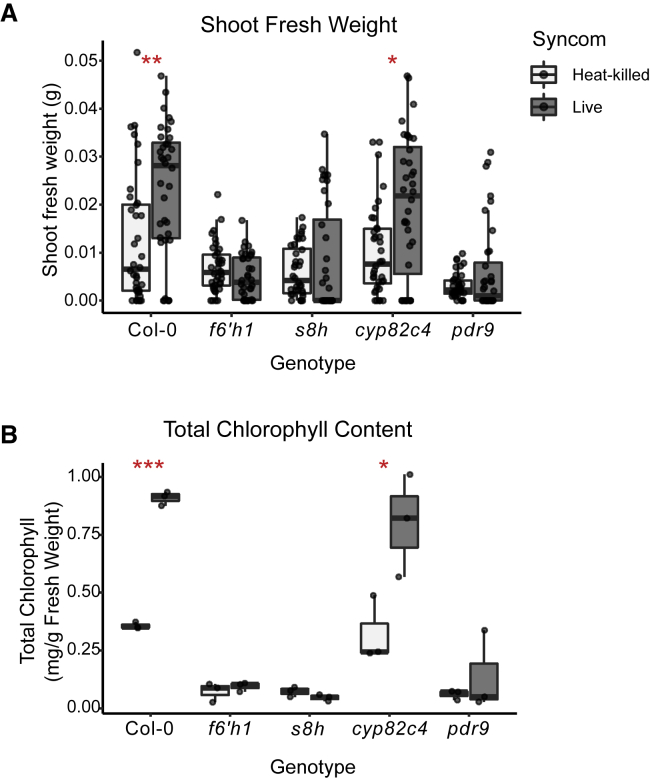
Figure 5.
Plant Biosynthesis and Secretion of Fraxetin Is Necessary for Microbiota-Mediated Growth Rescue
(A and B) (A) SFW and (B) leaf chlorophyll content of indicated coumarin biosynthesis and export mutants grown on unavFe media inoculated with heat-killed or live bacterial SynCom. SFW data are from two experiments (n = 36 plants). Chlorophyll content is from one experiment (n = 3 pooled leaf samples). Asterisks indicate significance between heat-killed- and live SynCom-inoculated groups by Wilcoxon ranked sum test for SFW and Student’s t test for chlorophyll content (∗, ∗∗, and ∗∗∗ indicate p ≤ 0.05, 0.01, and 0.001, respectively). See also Figure S5.
The root-associated bacterial microbiota of plants grown on CAS and IS were assessed by culture-independent 16S rRNA gene amplicon sequencing and analyzed at the amplicon sequence variant (ASV) level. Alpha diversity was greater in IS than CAS for both unplanted soil and root samples but did not vary by genotype (Figure S2). Unconstrained principal coordinate analysis (PCoA) of Bray-Curtis distances between samples showed that the soil type and batch were the largest drivers of divergence between samples (Figure S2). PCoA analysis of beta diversity constrained (CPCoA) for the interaction between genotype and soil type revealed a significant separation of f6’h1 and s8h plants from other genotypes when grown on IS, but not on CAS (Figure 1D). Analysis of bacterial community profiles from three batches of each soil type confirmed that f6’h1 plants separate significantly from other genotypes when grown in IS, but not in CAS (Figure S2). Together, these results indicate that coumarin biosynthesis, especially of scopoletin and fraxetin, is important for plant growth and determining root microbiota composition in a naturally iron-limiting calcareous soil but is dispensable in an iron-replete soil. Furthermore, this illustrates that the interaction between soil type and plant genotype can serve as a major determinant of root microbiota structure, explaining 27.7% of community variation (Figure 1D).
Coumarin Biosynthesis Restructures the Root Microbiota at the ASV Level
In order to explore the nature of the observed changes in community structure, we determined which ASVs were differentially enriched (deASVs) in each mutant genotype compared with WT in each soil. For this analysis, we pooled data from three batches of each soil (except s8h, which was included in only one batch). The greatest number of deASVs was observed in f6’h1 plants, with significantly more detected when grown in IS than CAS (260 deASVs in IS, 50 in CAS; Figure 2A). The impact of deASVs on the microbiota structure in terms of relative abundance was also greatest in f6’h1 plants grown in IS (Figure S3).
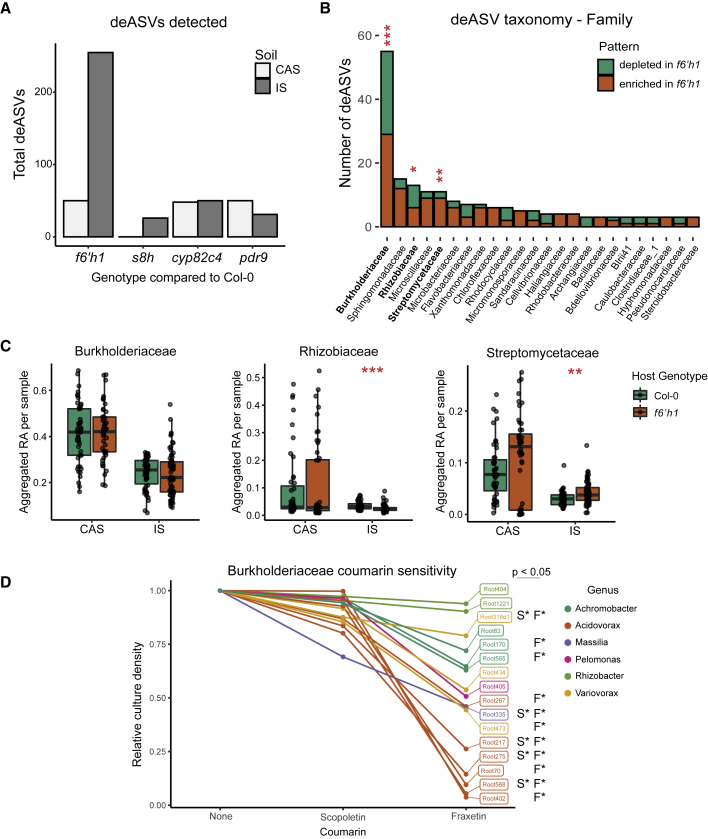
Figure 2.
Coumarin Biosynthesis Restructures the Root Microbiota at the ASV Level
(A) Number of deASVs detected in indicated mutants compared with Col-0 in each soil. Data are pooled from three experiments (except s8h, which was included in only one), and filtered for ASVs found in at least three samples with RA > 0.05%. Differential enrichment was calculated using a negative binomial generalized log-linear model at an FDR-adjusted p value of 0.05.
(B) Family-level taxonomic classification of deASVs in f6’h1 plants growing on IS. Colors indicate if deASVs were enriched or depleted in f6’h1 compared with Col-0. Hypergeometric enrichment test was performed to determine if each family was over- or under-represented in deASV list compared with all detected ASVs. Red asterisks indicate significance with FDR-adjusted p values.
(C) Sample-wise aggregated relative abundance of the top three families most significantly over-represented in deASVs: Burkholderiaceae, Rhizobiaceae, and Streptomycetaceae. Each data point represents the average RA aggregated at the family level in a single sample. Significance between genotypes in each soil was determined by Wilcoxon ranked sum test.
(D) Overnight growth of Burkholderiaceae bacterial strains in the presence of scopoletin or fraxetin. Optical density (OD) of cultures was normalized to the OD of each strain in the absence of coumarins. Significant differences (p ≤ 0.05 by Tukey’s HSD) in growth compared with the control are indicated for scopoletin (S∗) and fraxetin (F∗) to the right of each strain. Data are averages of 2–4 experiments, each with 2–3 technical replicates, per strain. ∗, ∗∗, and ∗∗∗ in (B) and (C) indicate p ≤ 0.05, 0.01, and 0.001, respectively. See also Figures S2 and S3.
Taxonomic analysis revealed that multiple bacterial families were significantly over- or under-represented within the deASV subset for f6’h1 plants in IS compared with the full list of detected ASVs (Figures 2B and S3). Burkholderiaceae was the most prevalent family detected within the deASVs (56 deASVs; 2.65-fold enriched in deASVs subset compared with all ASVs detected). However, a correlation between coumarin production and deASV relative abundance could not be generalized at family-level resolution; most families contained deASVs which were more abundant in f6’h1 plants than WT, as well as deASVs which were less abundant. Some patterns were observed at the genus level (Figure S3), but due to the overall low number of deASVs in each genus, their statistical significance remains unclear. This indicates that coumarin production restructures the root microbiota at the ASV level within multiple bacterial families. Despite being the family containing the most deASVs, the relative abundance of Burkholderiaceae was not significantly altered in coumarin-deficient plants (Figure 2C). The relative abundances of the next two most-impacted families, Rhizobiaceae and Streptomycetaceae, however, were slightly but significantly altered in f6’h1 plants on IS. This indicates that, at the ASV level, disruption of coumarin biosynthesis has a quantitative impact within multiple root-associated commensal families, but with relatively minor effects on the microbiota structure at higher taxonomic ranks. This suggests the existence of ASV-level compensatory mechanism(s) within bacterial families which, during root microbiota establishment, maintain higher taxonomic structure in coumarin-deficient plants on iron-limiting soil. An increase in the number of deASVs was also observed for s8h plants grown in IS compared with CAS (Figure 2A), though fewer deASVs overall were detected compared with other mutants as this genotype was only included in one experimental replicate. Consistently, the taxonomic profile of s8h deASVs was also enriched for Burkholderiaceae, and had considerable, but not complete, overlap with the deASVs detected on f6’h1 plants in the same experiment (Figure S3). This indicates that production of both scopoletin and fraxetin impact the root microbiota.
As various coumarin compounds have been shown to exert antimicrobial activity, we examined the coumarin sensitivity of a panel of root commensal Burkholderiaceae strains, the most prevalent family within the deASVs on f6’h1 and s8h plants. These strains were previously isolated from roots of Arabidopsis growing in CAS soil (Bai et al., 2015). Bacterial growth was quantified in the presence of 50 μM scopoletin or fraxetin, a concentration within the physiological range of coumarins observed within roots (Siwinska et al., 2014, 2018; Stringlis et al., 2018). We observed prevalent growth inhibition to a range of degrees in the presence of fraxetin, and to a lesser extent in response to scopoletin (Figure 2D). This indicates that fraxetin exerts variable antimicrobial activity on Burkholderiaceae strains. This strain-specific variation potentially explains part of the ASV-level shifts within Burkholderiaceae observed between WT and coumarin-deficient plants in iron-limiting soil.
Taxonomically Diverse Root Commensals Improve Iron-Limiting Plant Performance
To assess the impact of root commensals on plant performance under iron-limiting conditions, we employed an agar medium-based gnotobiotic system that allows control over both iron mobility and the presence of bacterial commensals. In this system, nutrient medium (1/2 MS) is strongly buffered at pH 7.4, similar to the pH of iron-limiting IS soil. Iron is provided at 100 μM in one of two forms: available iron (avFe) FeEDTA, a complex that retains solubility even at alkaline pH, or an unavailable form (unavFe) FeCl3, which forms Fe(OH)3 and is highly insoluble at alkaline pH. Providing unavFe mimics iron-limiting conditions in calcareous and alkaline soils such as IS; iron is present but recalcitrant due to extremely low solubility and must be actively mobilized for utilization. Using this gnotobiotic system, we reconstituted plants with a synthetic community (SynCom) of bacterial commensals to assess the impact of the microbiota on iron-limiting plant performance. To achieve this, we took advantage of a diverse culture collection of bacterial commensals isolated from A. thaliana roots grown in CAS (Bai et al., 2015). We designed a taxonomically diverse SynCom of 115 members, which reflects the root bacterial diversity observed by culture-independent methods at high taxonomic ranks (Figure 3A; Table S2). SFW and chlorophyll content were measured as readouts of plant performance and as a proxy measurement for iron nutritional status.
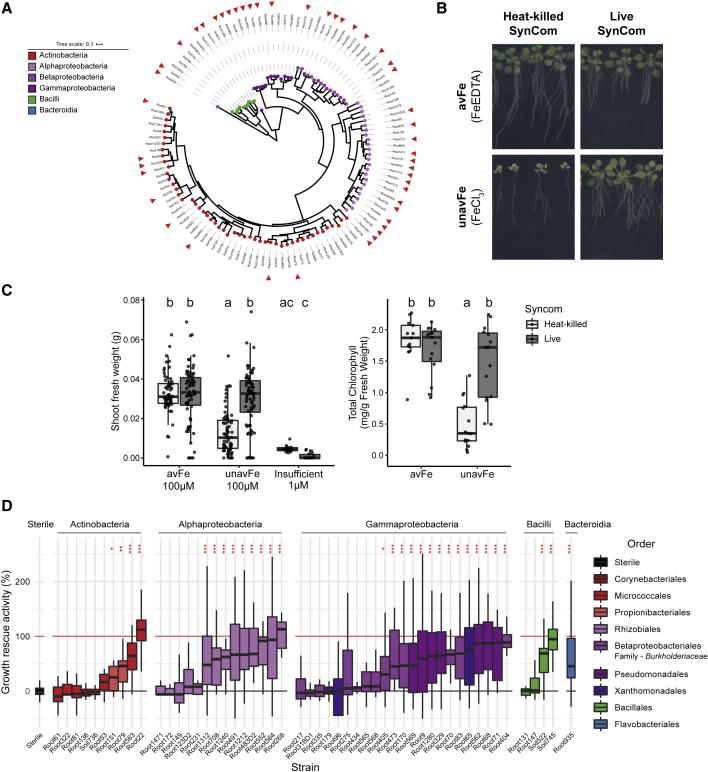
Figure 3.
Taxonomically Diverse Root Commensals Improve Iron-Limiting Plant Performance
(A) Phylogenetic tree of 115-strain SynCom derived from At-RSPHERE culture collection (Bai et al., 2015) used for microbiota reconstitution. Red arrows indicate strains used in (D).
(B) Representative images of plants grown for 2 weeks on media containing available (avFe) and unavailable (unavFe) forms of iron inoculated with live SynCom or heat-killed control.
(C) SFW and total chlorophyll quantification of Col-0 plants after 2 weeks of growth on indicated iron conditions. Data are pooled from three experiments with avFe and unavFe: n = 42–54 plants per condition, and chlorophyll measured in pooled samples, n = 13–15 per group. Insufficient iron data are from one experiment, n = 18 plants. Letters indicate significant pairwise differences between groups (p-adj ≤ 0.05 by Dunn’s pairwise comparison with Bonferroni correction for SFW, and Tukey’s HSD corrected for multiple comparisons for chlorophyll content).
(D) Iron-limiting growth rescue activity of SynCom strains in mono-association. SFW was measured and plotted as percent growth rescue of bacteria-inoculated plants on unavFe compared with the growth deficit between sterile plants on avFe versus unavFe. Black and red lines indicate 0% (axenic plants on unavFe) and 100% growth rescue (axenic plants on avFe), respectively. Data are pooled from 1–4 experiments per strain and normalized to respective sterile controls (n = 18 plants per experiment). Asterisks indicate significance from sterile plants by Wilcoxon ranked sum test with FDR adjustment (∗, ∗∗, and ∗∗∗ indicate p-adj ≤ 0.05, 0.01, and 0.001, respectively). See also Figure S4.
A growth deficit and leaf chlorosis (Figures 3B and 3C), symptoms of iron starvation, were observed in plants grown on unavFe. Furthermore, elemental and transcriptomic analyses revealed decreased leaf iron content and induction of iron import components FRO2 and IRT1 and repression of ferritins, iron storage proteins (Figures 7 and S7). Together, these data confirm a robust induction of iron deficiency in this experimental system. We found that reconstitution of the microbiota with a bacterial SynCom greatly improved both SFW and chlorophyll content of plants grown on unavFe. This beneficial interaction was unexpected, given that iron starvation-induced coumarins exert antimicrobial activity against some commensals (Stringlis et al., 2018; Voges et al., 2019; Wang et al., 2014; Yang et al., 2016) (Figure 2D). Iron starvation was also induced by reducing the supply of soluble iron to an insufficient amount (1 μM FeEDTA). However, commensal-mediated improvement of plant performance was not observed in plants grown on media containing only insufficient soluble iron (Figure 3C). Plant growth limitation on unavailable iron and growth rescue by a live SynCom were maintained when plant roots were shielded from light (Figure S4), supporting the robustness of this experimental system. These results suggest that bacterial commensals can improve plant performance by improving access to immobile sources of iron.
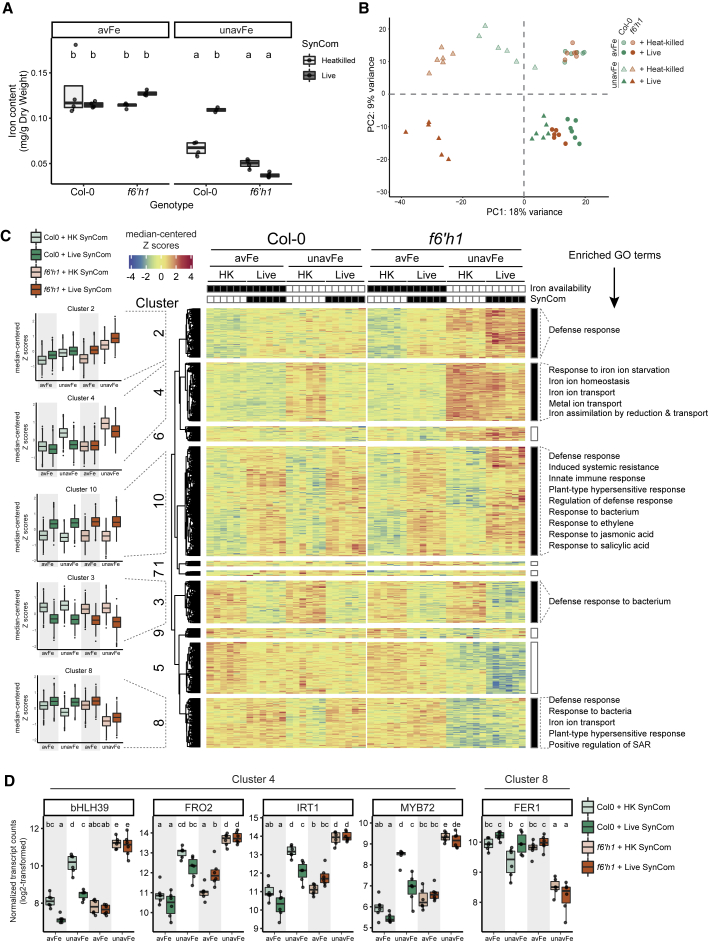
Figure 7.
A Bacterial SynCom Improves Plant Iron Nutrition, Relieves the Iron Deficiency Response, and Modulates a Subset of Defense Genes in a Coumarin-Dependent Manner
(A) Shoot iron content of Col-0 and f6’h1 plants grown on avFe and unavFe media with a live SynCom or heat-killed control measured by ICP-MS (n = 3–4 pooled plant samples per group).
(B) PCA ordination of sample distances between root transcriptional profiles of Col-0 and f6’h1 plants grown for 1 week on avFe or unavFe media inoculated with a live SynCom or heat-killed control. Data are from two pooled experiments (n = 6 samples pooled from 6 plant roots each).
(C) Heatmap of median-centered Z scores for 2,440 DEGs identified across samples, arranged by k-means clustering. Significantly enriched iron homeostasis-related and defense-related GO terms of pertinent clusters are indicated on right of heatmap. GO analysis was performed by comparing the indicated DEG cluster to the whole transcriptome (p-adj ≤ 0.05).
(D) Expression of select iron deficiency response marker genes. Data are log2-transformed, normalized counts. Letters in (A) and (D) indicate significant pairwise differences between groups (p-adj ≤ 0.05 by Tukey’s HSD corrected for multiple comparisons). See also Figures S6 and S7.
To survey the ability of various taxa to improve iron-limiting plant growth, we grew plants in mono-associations with bacterial strains on unavFe. Fifty-three taxonomically diverse SynCom strains were tested for their ability to rescue iron-limiting plant growth (Figure 3A, red arrows; Table S2). Within each broader taxonomic lineage, we observed growth-rescuing strains as well as strains lacking this ability (Figure 3D), demonstrating the ubiquity of this beneficial activity as well as the strain-specific variation within all core taxonomic lineages of the root microbiota. Thus, in a community context, the capacity of bacterial commensals to rescue plant growth under iron-starved conditions is functionally redundant. Furthermore, the ability of these strains to grow in the presence of scopoletin and fraxetin was found not to correlate with plant growth rescue capacity, indicating that this plant-beneficial trait does not require selection via coumarin antimicrobial activity (Figure S4).
Microbiota-Mediated Plant Growth Rescue Occurs via Reductive Import of Iron and Requires Fraxetin Secretion
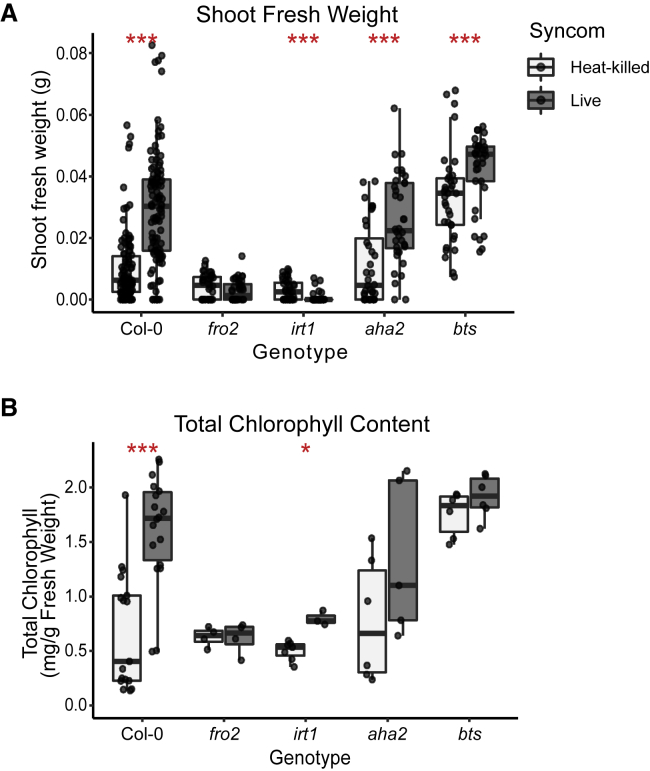
We utilized A. thaliana mutant lines deficient in genes involved in iron uptake and homeostasis to determine their importance for bacteria-mediated growth rescue under iron limitation. Mutants in components of the reduction-based iron uptake system (fro2 and irt1), rhizosphere acidification (aha2), and a negative regulator of the iron starvation response (bts) were grown on unavFe in the presence of a live SynCom, or a heat-killed SynCom as a negative control. Genotypes fro2 and irt1 displayed an exaggerated growth deficit and leaf chlorosis (Figure 4), consistent with their reported hypersensitivity to iron starvation (Robinson et al., 1999; Vert et al., 2002). In contrast to WT plants, addition of the bacterial SynCom was unable to improve the phenotype of these iron import mutants. Performance of irt1 plants was further reduced in the presence of the SynCom. This indicates that without the iron import channel, plants may be unable to compete with bacterial commensals for access to the already limited pool of available iron. Moreover, bts plants, which are tolerant to iron deficiency (Hindt et al., 2017; Selote et al., 2015), were larger than WT plants on unavFe, not chlorotic, and still displayed slightly improved performance when inoculated with the SynCom. No difference was observed between aha2 and WT controls, however, indicating that plant-mediated rhizosphere acidification is not rate limiting for commensal-mediated plant growth rescue in strongly buffered alkaline conditions. When grown on avFe, the SynCom did not improve plant performance in any of these genotypes (Figure S5). These results validate our gnotobiotic system for microbiota reconstitution under iron-limiting conditions, confirm that the growth limitation and chlorosis on unavFe is due to iron starvation, and suggest that improved plant performance in the presence of commensals depends on the plant’s endogenous system for iron reduction and import.
Figure 4.
Microbiota-Mediated Plant Growth Rescue Occurs via the Reductive Import of Iron
(A and B) (A) SFW and (B) leaf chlorophyll content of indicated mutants in the reductive import of iron pathway grown on unavFe media inoculated with heat-killed or live bacterial SynCom. Total chlorophyll content was measured in pooled leaf samples from six plants. Data are from two independent experiments per genotype (n = 36 plants, 6 chlorophyll samples). Each experiment included Col-0 control (n = 90 plants, 18 chlorophyll samples). Asterisks indicate significance between heat-killed- and live SynCom-inoculated groups by Wilcoxon ranked sum test for SFW and Student’s t test for chlorophyll content (∗, ∗∗, and ∗∗∗ indicate p ≤ 0.05, 0.01, and 0.001, respectively). See also Figure S5.
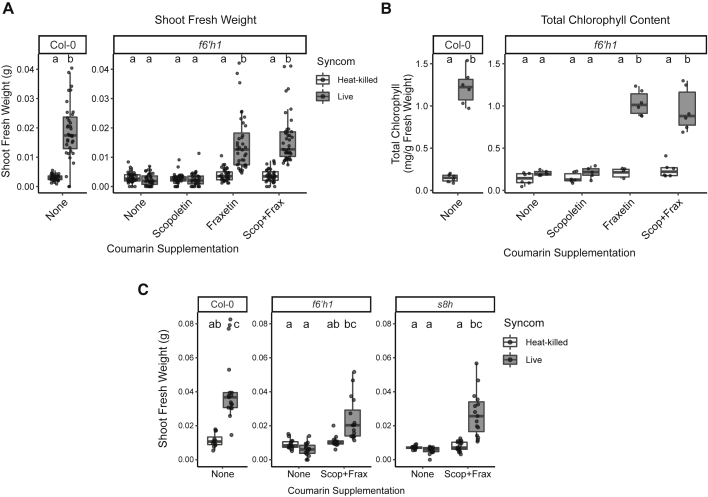
We next investigated the role of coumarins in commensal-mediated plant growth rescue under iron limitation. The addition of the SynCom provided no benefit to plant growth or chlorophyll content of f6’h1, s8h, or pdr9 plants grown on unavFe (Figure 5). In contrast, the SynCom improved performance of cyp82c4 plants similar to WT. These data suggest that plant biosynthesis of scopoletin and/or fraxetin (catalyzed by F6’H1 and S8H, respectively) and their secretion (through PDR9) are required for bacteria-mediated plant growth rescue under iron limitation, while sideretin (synthesized by CYP82C4) is dispensable. No growth promotion by the SynCom was observed in these genotypes grown on avFe (Figure S5).
To further assess the roles of scopoletin and fraxetin in commensal-mediated plant growth rescue, we chemically complemented f6’h1 plants by supplementing the growth medium with each compound (Figures 6A and 6B). Addition of scopoletin did not improve plant performance, while fraxetin fully restored the ability of the SynCom to improve both plant growth and leaf chlorophyll content in coumarin-deficient f6’h1 plants. That scopoletin was unable to complement f6’h1 plants suggests that external scopoletin is not sufficiently taken up by roots and converted to fraxetin after secretion. Furthermore, the ability of the SynCom to rescue growth of s8h plants was also fully restored by supplementation with scopoletin and fraxetin together (Figure 6C). This confirms that fraxetin is the necessary coumarin structure type for commensal-mediated growth rescue. Notably, supplementation with coumarins failed to rescue growth or chlorophyll content of germ-free f6’h1 or s8h plants at 50 μM, a concentration lower than what has been used to rescue iron starvation by directly mobilizing iron. This indicates that commensal-mediated improvement in iron-limiting plant growth is induced by fraxetin concentrations lower than those required for sufficient mobilization of iron in axenic conditions. Together, these results confirm that secreted fraxetin is both necessary and sufficient to elicit growth rescue activity from bacterial commensals under iron limitation. These findings argue for an indirect activity of fraxetin in recruiting commensal-mediated mobilization of recalcitrant iron pools in addition to its direct iron-mobilizing activity.
Figure 6.
Supplementation with Fraxetin Restores Microbiota-Mediated Growth Rescue of f6’h1 and s8h Plants
(A and B) (A) SFW and (B) leaf chlorophyll content of Col-0 plants, and f6’h1 plants grown on unavFe supplemented with 50 μM scopoletin and/or fraxetin and inoculated with heat-killed or live SynCom.
(C) SFW of Col-0, and f6’h1 and s8h plants grown on unavFe supplemented with 50 μM scopoletin and fraxetin and inoculated with heat-killed or live SynCom. Data in (A) and (B) are from two experiments (n = 30–42 plants, 6 pooled leaf chlorophyll samples). Data in (C) are from a single experiment (n = 18 plants). Letters indicate significant pairwise differences between groups (p-adj ≤ 0.05 by Dunn’s pairwise comparison with Bonferroni correction for SFW, and Tukey’s HSD corrected for multiple comparisons for chlorophyll content). See also Figure S5.
Coumarins and the Microbiota Interact to Alleviate Iron Starvation
To determine if the observed iron starvation symptoms and their improvement by root commensals correlated with plant iron status, we measured leaf elemental content by inductively coupled plasma mass spectrometry (ICP-MS) (Figures 7A and S6). Leaf iron concentration was significantly reduced on unavFe, confirming iron deficiency. Furthermore, addition of a live SynCom to WT, but not f6’h1 plants, restored plant iron content to replete avFe levels. However, addition of the SynCom had no impact on plant iron content on avFe. These results confirm that the plant growth rescue activity by commensals during iron starvation is due to improved iron nutrition. We next sought to determine if this nutritional benefit was a result of microbial stimulation of the plant iron deficiency response or increased iron availability in the presence of the SynCom. To identify plant pathways responding to the presence of coumarins and bacterial commensals under different iron regimes, we performed analysis of the whole-root transcriptome of WT and f6’h1 seedlings in our gnotobiotic system. An earlier time point (1 week) was chosen to observe potential stimulation of iron deficiency response genes by the SynCom (Verbon et al., 2019) that may lead to plant growth rescue. At this time point, SynCom-mediated plant growth rescue was observed in WT, but growth was still comparable between WT and f6’h1 plants (Figure S7).
Plotting transcriptome sample distances by PCA (Figure 7B) revealed that the supplied iron form was the largest determinant of dissimilarity (PC1, 18% of variance), followed by SynCom status (PC2, 9% of variance). When grown on avFe, both Col-0 and f6’h1 plants clustered together, but separation was observed based on SynCom status. This indicates that a live SynCom impacts host transcriptional responses when iron is available, independently of plant coumarin status. The transcriptomes of plants grown on unavFe, however, were distinct from those of plants grown on avFe, and further separated based on both genotype and SynCom status. When inoculated with a heat-killed SynCom, both WT and f6’h1 on unavFe separated from the avFe cluster (upper-left quadrant). A larger genotype-driven separation was observed between Col-0 and f6’h1 plants when inoculated with a live SynCom on unavFe. Remarkably, WT plants inoculated with a live SynCom on unavFe clustered closely with SynCom-inoculated WT plants on avFe (lower-right quadrant), while f6’h1 plants remained in the lower-left quadrant, clearly separated from the avFe cluster. This pattern indicates that the transcriptional responses to growth on unavFe are more pronounced in f6’h1 plants than WT, consistent with their hypersensitivity to iron starvation. Indeed, more iron-responsive differentially expressed genes (DEGs) were detected in f6’h1 than WT plants (Figure S7). Furthermore, the number of iron-responsive DEGs indicates that the iron starvation-induced response was mitigated by the addition of the SynCom in Col-0 but not f6’h1 plants.
We performed k-means clustering of all transcripts based on expression pattern and identified DEGs to investigate the interaction between iron availability, SynCom, and genotype (Figure 7C). The identified clusters showed iron- and SynCom-responsive gene sets (plots left of heatmap clusters). Cluster 4 revealed a set of genes activated in axenically grown Col-0 and f6’h1 plants in response to unavFe, which were more strongly induced in f6’h1 plants. Furthermore, their expression is reduced to homeostatic levels in Col-0 plants upon addition of the SynCom but remained elevated in f6’h1 plants. This gene set corresponds to iron-responsive genes that were also responsive to SynCom in a coumarin-dependent manner. Gene ontology analysis revealed that this cluster is enriched for genes belonging to the iron starvation response, iron homeostasis, and metal ion transport (Figure 7C, annotations right of heatmap). Genes in cluster 8 displayed the inverse pattern: downregulated on unavFe and restored in the presence of the SynCom in Col-0, but not f6’h1. We compared our DEGs to a list of 25 previously identified core iron starvation marker genes (Mai et al., 2016) (Figure S7). Of the 12 genes reported to be induced under iron starvation, 11 were found in cluster 4, while 7 out of 13 genes reported to be downregulated under iron starvation were present in cluster 8. The cluster assignment and expression patterns (Figures 7D and S7) of selected iron homeostasis regulators and coumarin biosynthesis genes revealed that these genes are iron- and SynCom responsive in a genotype-dependent manner. Importantly, the addition of live SynCom did not stimulate expression of iron deficiency genes. Iron starvation-upregulated genes (including bHLH39, FRO2, IRT1, and MYB72), as well as iron starvation-downregulated genes (including FER1) displayed expression patterns consistent with alleviation of the iron deficiency response by the SynCom in WT plants. This indicates that, rather than biostimulation of plant iron uptake, iron nutrition is improved by a commensal mechanism. In addition, the inability of the SynCom to alleviate the iron-starvation response in f6’h1 roots (cluster 4 and 8 genes; Figures 7C and 7D) supports a mechanism by which secreted coumarins are required to elicit microbiota-mediated iron nutrition, rather than two parallel mechanisms exerted by coumarins and the microbiota independently. Together, these data reveal a robust induction of iron starvation in plants grown on unavFe, which is alleviated by commensals in WT, but not in f6’h1 plants. This implies the existence of a coumarin- and microbiota-dependent mechanism that improves plant performance via iron nutrition.
Analysis of SynCom-responsive genes also revealed a core set of DEGs common to both genotypes independently of iron availability (clusters 3 and 10, Figures 7C and S7). These clusters were enriched for genes related to defense responses and response to bacteria. Interestingly, genes associated with these terms were also significantly enriched in clusters 2 and 8, the expression patterns of which are dependent on host genotype, iron status, and SynCom (Figure 7C). The presence of immune-related genes in clusters 2 and 8 suggests that, in addition to the core SynCom-responsive genes, a subset of defense genes is regulated by the presence of commensals in a coumarin-dependent manner. Collectively, these results show that both coumarin secretion and the root microbiota have profound impacts on plant transcriptional responses to iron deprivation. Furthermore, this emergent interaction between coumarins and the microbiota improves plant performance through bolstered iron nutrition, resolving the iron starvation response and regulating a subset of defense-related genes.
Discussion
Our results reveal unexpected impacts of root-secreted coumarins governing plant-bacteria interactions, including soil type-dependent alteration of root microbiota composition, elicitation of a commensal-mediated mechanism of plant iron nutrition, and regulation of a subset of defense genes. We show that f6’h1 and s8h plants, which are deficient in the biosynthesis of scopoletin and fraxetin, assemble an altered root bacterial community. Individual members of the Burkholderiaceae, core members of the plant root microbiota that often exert plant-beneficial activities (Eberl and Vandamme, 2016; Thiergart et al., 2020), are impacted by fraxetin in a strain-specific manner in both roots and growth in culture. This strain-specific sensitivity may in part underlie ASV-level changes in abundance on iron-limiting IS. The greater impact on the microbiota observed in f6’h1 compared with s8h plants, in terms of deASVs detected and their relative abundance (RA), suggests that both scopoletin and fraxetin impact bacterial microbiota assembly. Indeed, a metagenome analysis indicated an altered microbial multi-kingdom assemblage and provided evidence for selective scopoletin anti-fungal activity against soil-borne fungal pathogens in vitro (Ba et al., 2017; Carpinella et al., 2005; Kai et al., 2006; Stringlis et al., 2018). However, we cannot exclude that iron malnutrition in f6’h1 plants has additional consequences on the microbiota.
Plant performance data, coupled with elemental content and transcriptomic analysis, confirm that benefits conferred by commensals under iron limitation occur via improved iron nutrition. In contrast to this beneficial interaction, under low phosphate concentrations A. thaliana must compete with a bacterial SynCom for access to the macronutrient, requiring integration of phosphate starvation and defense responses (Castrillo et al., 2017). We similarly observe emergent effects of coumarins and the microbiota on a subset of defense-related genes, indicating potential trade-offs between growth and defense. Of note, the impact of commensal communities on plant performance when phosphate is present in unavailable forms, as is characteristic of many soils, has not been tested. Importantly, in our system, growth promotion by microbes is observed only when iron is present but immobile, conditions characterizing most iron-limiting soils. Thus, our results highlight the importance of studying plant nutritional phenotypes in systems closely mimicking natural conditions, including the presence of commensals and defined forms of soil minerals that are unavailable to the plant host. To the best of our knowledge, experimental evidence for clear plant nutritional benefits from commensals in a community context has not been reported before. This is a significant step forward in understanding how plant nutrition and productivity can be bolstered by harnessing endogenous soil microbes.
The presence of this beneficial activity across all core taxonomic lineages of the A. thaliana bacterial microbiota suggests that the underlying molecular mechanism(s) evolved independently rather than by common descent. As growth rescue depends on plant expression of the iron reductive import machinery, but does not involve microbial stimulation of its expression, the mechanism(s) must function upstream of reduction and import at the root surface. Multiple bacterial molecules can mobilize insoluble iron and are potentially utilized by plants, including siderophores and other metabolites, though the ability of plants to utilize microbially mobilized iron in the context of an intact microbiota has yet to be shown. The nutritional benefits provided by commensals requires plant-secreted fraxetin but is independent of its antimicrobial activity. This suggests that the impact of coumarins on root microbiota composition and on commensal-mediated iron nutrition may be independent mechanisms. This would indicate that coumarins have multiple roles in mediating host-microbe interactions. Importantly, our results were obtained using a bacterial culture collection derived from A. thaliana roots grown in CAS, in which coumarin status did not affect plant performance or microbiota structure. The observation that taxonomically diverse commensals isolated from an iron-replete soil are capable of rescuing iron-limiting plant growth further suggests the involvement of microbial functions that are ubiquitous across soil types and can be elicited by fraxetin. Future work with commensal culture collections derived from soils with contrasting iron availability are required to directly link plant phenotypes in natural calcareous soils and gnotobiotic systems.
Root-secreted coumarins are inducible under iron starvation and mediate an interaction between the host and commensals that improves host iron nutrition. This genotype-environment interaction strongly suggests that the root microbiota is an integral component of plant edaphic adaptation to growth in iron-limiting soil. Quantitative variation in coumarin production has been demonstrated among A. thaliana accessions (Siwinska et al., 2014; Tsai et al., 2018) and was shown to correlate with performance under iron limitation (Tsai et al., 2018). As coumarins are both ubiquitously present and chemically diverse among flowering plants (Bourgaud et al., 2014; Krieger et al., 2018; Rajniak et al., 2018), our findings provide an ecological framework for examining the consequences of their evolutionary diversification on microbiota-mediated mineral nutrition of plant hosts.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| AtSphere Arabidopsis thaliana-derived culture collection. Full list of strains in this study are in Table S2 | Bai et al., 2015 | http://www.at-sphere.com/ |
| Biological Samples | ||
| Cologne agricultural soil | Bai et al., 2015 | CAS |
| Italian soil | This study | IS |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DFS Taq Polymerase | Bioron | CAT#101005 |
| Agar Bacteriological | Difco | CAT#214530 |
| FeEDTA | Sigma-Aldrich | CAT#E6760 |
| FeCl3 hexahydrate | Merck | CAT#103943 |
| HEPES buffer | Roth | CAT#6763.1 |
| Scopoletin | Sigma-Aldrich | CAT#S2500 |
| Fraxetin | Sigma-Aldrich | CAT#18224 |
| Tryptic Soy Broth | Sigma-Aldrich | CAT#T8907 |
| Critical Commercial Assays | ||
| FastDNA SPIN kit for Soil | MP Biomedicals | CAT# SKU 116560200 |
| Quant_iT Pico Green dsDNA Assay | ThermoFischer | CAT#P7589 |
| AMPure XP DNA purification beads | Beckman Coulter | CAT#A63881 |
| RNEasy Plant Mini Kit | Qiagen | CAT#74904 |
| Deposited Data | ||
| 16S rRNA root profiling data | This paper https://www.ebi.ac.uk/ena | ENA: accession PRJEB38663 |
| Raw and processed data, scripts for analysis and figure generation | This paper | Mendeley Data DOI: 10.17632/tkdn6zbw7k.1 |
| Root RNA-Seq data | This paper https://www.ebi.ac.uk/ena | ENA: accession PRJEB38663 |
| Silva 132 rRNA database, Released Dec. 2017 | Quast et al., 2013 | RRID: SCR_006423 |
| Experimental Models: Organisms/Strains | ||
| A. thaliana: Col-0 wildtype | NASC | N60000 |
| A. thaliana: f6’h1-1 TDNA insertion mutant | NASC | SALK_132418C |
| A. thaliana: s8h-2 TDNA insertion mutant | NASC | SM_3_23443 |
| A. thaliana: cyp82c4-1 TDNA insertion mutant | NASC | SALK_001585 |
| A. thaliana: pdr9-2 TDNA insertion mutant | NASC | SALK_050885 |
| A. thaliana: irt1-1 TDNA insertion mutant | Vert et al., 2002 | irt1-1 |
| A. thaliana: fro2 (frd1-1) TDNA insertion mutant | NASC | N3777 |
| A. thaliana: aha2-4 TDNA insertion mutant | NASC | SALK_082786 |
| A. thaliana: bts-1 TDNA insertion mutant | NASC | SALK_016526 |
| Oligonucleotides | ||
| All primers are found in Table S1 | N/A | |
| Software and Algorithms | ||
| R statistical environment | https://www.r-project.org/ | V 4.0.1 |
| DADA2 | Callahan et al., 2016 | V1.12.1 |
| Interactive Tree of Life (iTOL) | Letunic and Bork, 2007 | https://itol.embl.de/ |
| Vegan, R package | Dixon, 2003 | V 2.5-6 |
| RVAideMemoire, R package | Hervé, 2020 | V 0.9-77 |
| edgeR, R package | Robinson et al., 2010 | V 3.24.3 |
| Fastp | Chen et al., 2018 | RRID: SCR_016962 |
| Kallisto | Bray et al., 2016 | RRID: SCR_016582 |
| Tximport, R package | Soneson et al., 2015 | V 1.16.1 |
| SVA, R package | Leek et al., 2012 | V 3.36.0 |
| DESeq2, R package | Love et al., 2014 | V 1.28.1 |
| ComplexHeatmap, R package | Gu et al. 2016 | V 2.4.2 |
| Goseq, R package | Young et al., 2010 | V 1.40.0 |
| Gene Ontology Consortium | Ashburner et al., 2000 | RRID: SCR_002811 |
| Ggpubr, R package | Kassambara, 2020 | V 0.3.0 |
| FSA, R package | Ogle et al., 2020 | V 0.8.30 |
| Rcompanion, R package | Mangiafico, 2020 | V 2.3.25 |
| Emmeans, R package | Lenth, 2020 | V 1.4.8 |
| BioRender (Graphical Abstract) | BioRender.com | RRID: SCR_018361 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be addressed to the Lead Contact, Paul Schulze-Lefert (schlef@mpipz.mpg.de)
Materials Availability
This study did not generate new unique reagents. All bacterial strains and Arabidopsis lines used in this study were previously described and are publically available.
Data and Code Availability
Raw sequencing data of 16S rRNA profiling experiments and RNA-Seq transcriptomic data have been deposited in the European Nucleotide Archive ENA: PRJEB38663. Datasets and scripts necessary to reproduce analyses and generate figures have been deposited to Mendeley Data: https://doi.org/10.17632/tkdn6zbw7k.1. Scripts for RNASeq analysis and heatmap generation are available at https://github.com/YulongNiu/MPIPZ_CJ_RNASeq.
Experimental Model and Subject Details
Soils
Cologne agricultural soil (CAS) was obtained from a local site (GPS code : 50.958 N, 6.856 E) that has not been exposed to agriculture for >15 years. Italian soil (IS) was obtained from an organic vineyard in Tebano, Italy (GPS code : 44.292 N, 11.784 E) which has been maintained since 2007 without irrigation or fertilization. Soil was homogenized, sieved, and stored at 4°C until used in experiments.
Plant Model
All A. thaliana genotypes used in this study were in the Columbia wild-type (Col-0, N60000) background. See Key Resources Table for full information on all genotypes. Mutants related to coumarin biosynthesis (f6’h1-1, SALK_132418C; s8h-2, SM_3_23443; cyp82c4-1, SALK_001585) and export (pdr9-2, SALK_050885), regulation of the iron starvation response (bts-1, SALK_016526), and iron reductive import (aha2-4, SALK_082786; fro2, also known as frd1-1 (Robinson et al., 1999);, and irt1-1 (Vert et al., 2002)) were employed in this study and are available from the Arabidopsis Biological Resource Center (ABRC). Each of these genes are expressed in roots.
Bacterial Strains
All bacterial strains used in this study were previously described (Bai et al., 2015) and are summarized in Table S2. Species phylogenetic trees were generated with iTOL version 5.5 (Letunic and Bork, 2007) from previous whole genome taxonomic classification (Bai et al., 2015). Each of these strains was originally isolated from A. thaliana roots grown in CAS soil. Strains were stored in 20% glycerol stocks and cultured on 50% tryptic soy agar (TSA) plates and 50% tryptic soy broth (TSB) at 25°C.
Plant Growth Conditions
Seeds were surface-sterilized with 70% ethanol for 15 min under agitation, followed by two washes with 70% ethanol, one with 100% ethanol, and three with sterile distilled water. Sterilized seeds were stratified at 4°C in the dark for 2–3 days either imbibed in water (for soil experiments) or on agar media plates (for agar-media experiments) before transfer to growth conditions.
Soil
Surface-sterilized, stratified seeds were germinated in 7x7 cm square pots filled with CAS or IS. Pots were watered from the top with non-sterile distilled water every 2 days. Plants were grown in the greenhouse under long-day conditions (16hrs light, 8 h dark). Pots were distributed at random within trays. and shuffled periodically to minimize edge and location effects.
Agar-media
Surface-sterilized seeds were sown on plates containing 1% agar (Agar, granulated, Difco) in 50% Murashige and Skoog (MS) medium with vitamins (2.2 g/L, Duchefa Biochemie) supplemented with 0.5% sucrose. After two days of stratification at 4°C, plates were positioned vertically in a climate chamber (Panasonic, MLR-352) and grown for six days (10 h light, 21°C; 14 h dark, 19°C). Uniform seedlings were then transferred to experimental condition plates prepared fresh on the day of seedling transfer.
Method Details
Soil Nutrient Content
Soil nutrient analysis was performed by Labor für Boden- und Umweltanalytik (Laboratory for Soil and Environmental Analyses, Switzerland). Plant-available iron was measured as iron mobilized by chelator diethylenetriaminepentaacetic acid (DTPA) extraction as in (Lindsay and Norvell, 1978). Briefly, 10g of air-dried soil was shaken for 2 h with 20 ml of extracting solution (0.005M DTPA, 0.1M triethanolamine, and 0.01M CaCl2, with a pH of 7.3). The leachate was centrifuged and filtered through a 0.22 micron membrane. Iron content in the leachate was measured by ICP-MS at 1:10 and 1:50 dilutions.
Leaf Chlorophyll Measurement
Chlorophyll extraction and quantification was adapted from (Hiscox and Israelstam, 1979). Samples were prepared from 20–40mg of leaf tissue pooled from ~6 plants per sample and weighed. Samples were either processed immediately or frozen at -80°C until processing. Chlorophyll was extracted by adding 1 ml DMSO per ~30mg tissue and incubating samples at 65°C with shaking for 45–60 min until plant tissue was transparent and chlorophyll completely extracted. Absorbance of tissue-free chlorophyll extract was measured at 652 nm on a spectrophotometer (NanoDrop One, Thermo Scientific). Alternatively, absorbance of 100 μl of samples was measured in 96-well microtiter plate in a microplate reader (Infinite M200 PRO, Tecan). Absorbance652nm cm-1 was converted to total chlorophyll per ml and normalized to input sample tissue mass with the following formula:
16S Profiling of the Root Microbiota
For 16S profiling, root samples were harvested and libraries were processed as in (Thiergart et al., 2020). Plant roots were harvested at the early flowering stage, average 37 days after sowing. The whole root system was carefully collected using tweezers, and large soil aggregates and debris were removed by gently washing with sterile water. Roots were transferred to a 15-ml conical with 5 ml PBS buffer (130 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, pH 7.0) and washed twice for 20 min on a tube rotator at 120 rpm. Washed roots were briefly dried on filter paper and stored in Lysing Matrix E tubes (MP Biomedicals) at -80°C until processing.
Total root DNA was extracted using the FastDNA SPIN Kit for Soil (MP Biomedicals). Samples were homogenized using the Precellys 24 homogenizer (Bertin Technologies) at 6,200 r.p.m. DNA was eluted in 80 μl of nuclease-free water and quantified using Quant-iT PicoGreen dsDNA Assay (ThermoFisher). Samples were diluted to 3.5 ng μl−1 and used in a two-step PCR amplification protocol. In the first step, the V5–V7 region of the bacterial 16S rRNA gene was amplified in triplicate reactions for each sample with primers 799F and 1192R (Table S1). Amplification was performed in a 25-μl reaction volume containing 2 U DFS-Taq DNA polymerase, 1× incomplete buffer (both Bioron), 2 mM MgCl2, 0.3% bovine serum albumin, 0.2 mM dNTPs (Life Technologies) and 0.3 μM forward and reverse primers. The same PCR parameters were used for each primer pair (94 °C for 2 min, 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s and 72 °C for 10 min for 25 cycles). Single-stranded DNA and proteins were digested by adding 1 μl of Antarctic phosphatase, 1 μl Exonuclease I and 2.44 μl Antarctic Phosphatase buffer (New England Biolabs) to 20 μl of the pooled replicate reactions. Digestion was performed at 37 °C for 30 min, followed by enzyme deactivation at 85 °C for 15 min. Samples were centrifuged for 10 min at 4,000 r.p.m., and 3 μl of supernatant were used for the second PCR step in triplicate reactions to add barcodes and Illumina adapters (primer sequences in Table S1). Reactions were prepared with barcoded primer pairs and performed as above with the number of cycles reduced to ten. PCR quality was controlled by loading 5 μl of each reaction on a 1% agarose gel. The remaining reaction volume was loaded on a 1.5% agarose gel and run at 80 V for 2 h; bands with the correct size of ~500 base pairs were cut out and purified using the QIAquick Gel Extraction Kit (Qiagen). Concentration of the purified DNA was determined, and 30 ng DNA from each of the barcoded amplicons were pooled into one library per experiment, then purified and re-concentrated two-fold with AMPure XP beads (Beckman Coulter). Paired-end Illumina sequencing was performed in-house using the MiSeq sequencer and custom sequencing primers.
Analysis of 16S Profiling Data
ASV Table Generation
Forward and reverse sequencing reads were demultiplexed separately according to the barcode sequence and output in individual fastq files per sample. A denoising pipeline, DADA2 (v1.12.1) (Callahan et al., 2016) was used to obtain the final ASV table. Raw sequencing reads were truncated to 260 bp for the forward reads and 240 bp for the reverse reads and filtered with the following parameters: maxN=0, maxEE=c(2,2), truncQ=2, rm.phix=TRUE. Subsequently, error rates were inferred from filtered reads until convergence or exceeding a maximum consistent number of 20. Sequence variants were then inferred from the trimmed, filtered, and error-corrected sequences and ASVs were obtained by merging the forward and reverse sequences together. Finally, chimeras were identified and removed when an ASV could be mapped to the left- and right-segments from two other, more abundant ASVs. Finally, the ASV table was generated by aggregating the number of reads mapped to each variant.
Alpha and Beta Diversity
Analyses and visualization were performed in the R statistical environment (Version 4.0.1). Analysis was performed on samples with a sequencing depth of at least 2,000 high-quality reads. Alpha and beta diversity were calculated on ASV count tables that were rarefied to 2,000 reads. Alpha diversity (Shannon index) was calculated with using the “diversity” function in vegan (Dixon, 2003) (R package version 2.5-6) and differences were tested with ANOVA.
Bray-Curtis dissimilarity index was calculated using the “vegdist” function in vegan and used for unconstrained ordination by Principal Coordinate Analysis (PCoA). Constrained PCoA (CPCoA) was performed with the “capscale” function in vegan, using the square-root distances of Bray-Curtis dissimilarity index. For ordination constrained on the interaction between genotype and soil type (Figure 1C), the formula used was “distance.matrix ~Host.Genotype∗Soil”. Statistical significance of genotype separations was determined using adonis function and pairwise PERMANOVA with 999 permutations using vegan and the RVAideMemoire package(Hervé, 2020).
Analysis of deASVs
Analyses and visualization were all performed in the R statistical environment (R Development Core Team, 2010). Analysis was performed on samples with a sequencing depth of at least 2,000 high-quality reads. Relative abundance (RA) was calculated using non-rarefied ASV count data. To calculate deASVs between coumarin pathway mutants and Col-0 WT, data were pooled from three experiments (except s8h, which was included in only one experiment), and filtered for ASVs found in at least three samples at a RA > 0.05%. Statistically significant differential enrichment was determined with the edgeR package (Robinson et al., 2010) (version 3.24.3) using pair-wise genotype comparisons in a negative binomial generalized log-linear model at an FDR-adjusted p value of 0.05.
Taxonomic classification of ASVs and culture collection strains was performed using the Silva 132 database (released Dec. 2017) (Quast et al., 2013). Hypergeometric enrichment test was performed using the stats package in R. Each family was tested for over- or under-representation in the deASVs set by comparing to the list of all detected ASVs. Red asterisks indicate significance at an FDR-adjusted p value of 0.05.
Coumarin Antimicrobial Activity
The antimicrobial activity of scopoletin and fraxetin (Sigma Aldrich) against single bacterial strains was assayed in liquid culture in 50% tryptic soy broth (TSB, 15g/L; Sigma Aldrich). Scopoletin and fraxetin stocks were prepared in sterile DMSO (Sigma Aldrich) and stored at -80°C. Bacterial colonies were picked from TSA plates into liquid TSB and grown for 2–3 days at 25°C with 180 rpm agitation. Liquid cultures were subcultured by diluting 1:100 into fresh TSB and incubated for 1–2 h. In a clear flat-bottom 96-well microtiter plate, 100 μl of subculture were added to 100 μl of fresh TSB media supplemented with scopoletin or fraxetin for a final 50 μM concentration, or equivalent DMSO negative control. The microtiter plate was sealed with a clear adhesive film to prevent evaporation. Growth was monitored kinetically in a microplate reader (Infinite M200 PRO, Tecan) with 30 seconds of shaking followed by measurement of optical density (OD) at 600 nm in four locations per well every 30–60 min for 18–20 h. The OD in each experiment was expressed as the average of triplicate wells per condition. Relative growth (Figure 2D) was calculated by dividing the average final OD measurement of each strain and indicated condition by the average OD in the coumarin-free control.
Gnotobiotic System for Iron Limitation
Iron limitation was achieved with a modified MS medium prepared from stock solutions. Stock solutions were prepared of ethylenediaminetetraacetic acid ferric sodium salt (Fe(III)EDTA, Sigma) in distilled water, and 100mM Fe(III)Cl3 (Merck) in 10 mM HCl (to prevent precipitation), sterile-filtered and stored at 4°C protected from light. A 2M stock solution of 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES) buffer (Roth) was prepared, and the pH was adjusted with KOH until a dilution to 10 mM in 50% MS resulted in a pH of 7.4 (approximately pH 8.2 for stock solution) and stored at 4°C.
As a base medium, modified 50% MS media without iron or pH buffer (750 μM MgSO4, 625 μM KH2PO4, 10.3 mM NH4NO3, 9.4 mM KNO3, 1.5 mM CaCl2, 55 nM CoCl2, 53 nM CuCl2, 50 μM H3BO3, 2.5 μM KI, 50 μM MnCl2, 520 nM Na2MoO4, 15 μM ZnCl2, and 9.4mM KCl) was prepared from individual stock solutions. Base media with 1% agar was autoclaved and cooled to 50°C before adding iron source (final 100 μM) and HEPES (final 10mM, pH 7.4) with constant stirring. Media were allowed to cool to ~45°C, and 45 ml were measured into a conical tube. Live or heat-killed bacteria or SynCom (preparation see below) were added to a final OD600nm = 0.0001, corresponding to approximately 105 cells/ml. For coumarin complementation experiments, scopoletin and fraxetin were added to a final concentration of 50 μM, or equivalent DMSO-only control. Media was mixed thoroughly by inverting, poured into 12∗12cm square petri dishes, dried with an open lid for 30 min, then allowed to solidify. Seedlings were transferred to experimental plates (six plants per plate, three replicate plates per experiment). For root light-shielding experiments, light-protected plates were placed in completely opaque black boxes with only shoots exposed to light as described in Silva-Navas et al., 2015. Plates were returned to the growth chamber and grown vertically with random shuffling and re-distribution every 2–3 days for uniformity. After two weeks, SFW was measured and chlorophyll and root samples were collected. Plant growth rescue in mono-association assays (Figure 3D) was expressed as percent growth rescue of the differential between axenic plant growth on avFe and unavFe using the formula:
Bacteria Preparation
SynCom
Cultures of 115 SynCom member strains were picked directly from glycerol stocks into 1 ml TSB in 96-well deep-well plates using a 96-well format microplate tip replicator and sealed with breathable plate sealer (AeraSeal, Sigma Aldrich). Cultures were grown at 25°C with 180 RPM agitation for five days and controlled to ensure that a majority of strains grew successfully. Fresh TSB (500 μl per well) was added and cultures were grown overnight, for 12–18 h to harvest metabolically active cells. Cultures were then centrifuged (4000 g, 20 min), washed once with MgCl2, and resuspended in 300μl/well. Cultures were combined, washed, resuspended in MgCl2, and adjusted to OD600 = 0.1. Heat-killed SynCom was prepared by incubating an aliquot of SynCom suspension at 99°C for 30 min. Heat-killed or Live suspensions were used 1000x to inoculate media (final OD600 = 0.0001).
Mono-associations
Individual colonies were picked from TSA plates and grown in liquid TSB at 25°C with 150RPM agitation for five days. Strains were subcultured 1:10 in fresh TSB for 2 h, washed twice with sterile 10 mM MgCl2 (Merck), resuspended, and adjusted to OD600 = 0.1. Strains forming clumps that could not be homogenously resuspended were diluted to a similar degree as other strains in the experiment.
Mineral Composition Analysis
The mineral composition of leaf tissue was determined by inductively coupled plasma mass spectrometry (ICP-MS) from 15-20 mg lyophilized shoot tissue by the CEPLAS Plant Metabolism and Metabolomics Laboratory, University of Cologne, using an Agilent 7700 ICP-MS (Agilent Technologies, Santa Clara, CA, USA)(Almario et al., 2017).
RNA Extraction and RNA-Seq Analysis
For transcriptomic analysis, 6-day-old A. thaliana seedlings were transferred to avFe or unavFe media with live or heat-killed SynCom as above and grown for eight days. Roots from six plants (one plate) were pooled for one replicate, with a total of three replicates per condition in each of two experiments (final n=6). Roots were homogenized with Lysing Matrix E and TissueLyser II (30 beats per second for 2x30 s; Qiagen) and RNA was extracted with the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA quality was determined using a 2100 Bioanalyzer (Agilent Technologies). Preparation of Illumina sequencing libraries was conducted by the Max Planck Genome Center. Sequences were generated using the Illumina HiSeq2500 platform. Approximately 20M paired-end reads with a length of 150 bp were obtained per sample in one experiment, and 8M per sample in the second.
Initial paired-end RNA-Seq reads were pre-processed using fastp (Chen et al., 2018). High quality reads were aligned to A. thaliana reference transcriptome (TAIR 10) using kallisto (Bray et al., 2016) with default settings. After removal of low abundant transcripts, 35,886 transcripts were imported using the tximport R package (Soneson et al., 2015). Batch effects were detected and removed using the SVA R package (Leek et al., 2012). Differential gene expression analysis was performed using DESeq2 (Love et al., 2014) by comparing the avFe and unavFe treatment with live or heat-killed SynCom in WT and f6’h1 plants, respectively. DEGs were selected with the threshold log2FoldChange > log2(1.5) and an adjusted p-value < 0.05.
Scaled counts normalized to library size were generated using DESeq2 (‘rlog’ function) and transformed as median-centered z-score (by transcripts, ‘scale’ function). Then, z-scores were used to conduct k-means clustering for all transcripts. The cluster number (k=10) was determined by sum of squared error and Akaike information criterion. Transcripts with similar expression patterns were grouped in the same cluster. Differentially expressed transcripts and cluster results were visualized using heatmaps generated with the ComplexHeatmap package in R (Gu et al., 2016). Gene expression in individual plots (Figures 7D and S7) was plotted using scaled counts data. Gene-set enrichment analyses were performed with the goseq package (Young et al., 2010) with gene ontology annotations from the Gene Ontology Consortium (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) (September 2019).
Quantification and Statistical Analysis
Details of data visualization, sample size and statistical analysis performed for each dataset can be found in the corresponding figure legend. All boxplots display individual data points, median values, interquartile range, and minimum and maximum values. Statistical analyses were performed in R using the packages and functions indicated above and in figure legends. For statistical analysis of SFW data, nonparametric tests were performed as data were not normally distributed as determined by Shapiro-Wilk test. Kruskal-Wallis one-way analysis of variance between groups was performed with the stats package, Wilcoxon signed-rank test to compare SynCom inoculation conditions was performed with ggpubr package (Kassambara, 2020), and Dunn’s pairwise comparison test was performed with packages FSA (Ogle et al., 2020) and rcompanion (Mangiafico, 2020). Leaf chlorophyll and elemental content were approximately normally distributed according to Shapiro-Wilk test, and were analysed by ANOVA and Tukey’s HSD with correction for multiple comparisons (emmeans package (Lenth, 2020)), or by Student’s T-test to compare SynCom inoculation conditions with small samples sizes. p values from analyses with multiple comparisons were adjusted using methods indicated in figure legends. Significance was indicated by asterisks (∗, ∗∗, and ∗∗∗, indicate p(adj) ≤ 0.05, 0.01, and 0.001, respectively) or by significance group (p ≤ 0.05) by indicated tests. Figures were assembled in Adobe Illustrator. The graphical abstract was created with BioRender.com.
Acknowledgments
This research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy—EXC-number 2048/1—project 390686111, a European Research Council advanced grant (ROOTMICROBIOTA), a RIKEN grant (SYMBIOLOGY), and a cooperative research project with Dong-A University funded by the Republic of Korea to P.S.-L., as well as funds to P.S.-L. from the Max Planck Society. M.H. was supported by JSPS KAKENHI grants 20K05955 and 15J04093. We thank the Max Planck-Genome-Centre Cologne for advising and performing the RNA-seq experiment in this study. We thank S. Ambrosius for technical support and the Biocenter MS Platform Cologne for the measurements of mineral composition. We also thank Sho Nishida for providing irt1 seeds, Erin Connolly for fro2, Terri Long for bts, and Sabrina Sabatini for aha2. Finally, thanks to Neysan Donnelly for editing the manuscript and Saurabh Pophaly for support with ENA data submission.
Author Contributions
P.S.-L., H.I., and M.H. conceived the project. C.J.H., M.H., H.I., and P.S.-L. designed the experiments. A.D.R. provided IS. M.H. and C.J.H. performed the root microbiota profiling experiments in natural soils. C.J.H., R.G., and R.G.-O. analyzed the microbiota profiling data. C.J.H. performed SynCom reconstitution experiments, bacterial growth assays, and RNA-seq experiments. RNA-seq data analysis was performed by Y.N., R.G.-O., and C.J.H.; R.G.-O., R.G., and Y.N. provided bioinformatic tools and support. S.K. advised and performed ICP-MS analyses. E.S.S. and M.J.E.E.E.V. provided scientific advice at an early stage of the project and feedback on the manuscript. P.S.-L. and R.G.-O. supervised the project. C.J.H. and P.S.-L. wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: October 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.09.006.
Contributor Information
Ruben Garrido-Oter, Email: garridoo@mpipz.mpg.de.
Paul Schulze-Lefert, Email: schlef@mpipz.mpg.de.
Supplemental Information
References
- Almario J., Jeena G., Wunder J., Langen G., Zuccaro A., Coupland G., Bucher M. Root-associated fungal microbiota of nonmycorrhizal Arabis alpina and its contribution to plant phosphorus nutrition. Proc. Natl. Acad. Sci. USA. 2017;114:E9403–E9412. doi: 10.1073/pnas.1710455114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba R., Alfa T., Gbaguidi F., Novidzro K.M., Dotse K., Koudouvo K., Houngue U., Hounsode M.T.D., Koumaglo K.H., Ameyapoh Y., Baba-Moussa L. Maize fungal growth control with scopoletin of cassava roots produced in Benin. Int. J. Microbiol. 2017;2017:5671942. doi: 10.1155/2017/5671942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Müller D.B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., Dombrowski N., Münch P.C., Spaepen S., Remus-Emsermann M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–369. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- Berendsen R.L., Pieterse C.M., Bakker P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Borghi L., Kang J., de Brito Francisco R. Filling the Gap: functional clustering of ABC proteins for the investigation of hormonal transport in planta. Front. Plant Sci. 2019;10:422. doi: 10.3389/fpls.2019.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Rott M., Schlaeppi K., Ver Loren van Themaat E.v., Ahmadinejad N., Assenza F., Rauf P., Huettel B., Reinhardt R., Schmelzer E. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Themaat E.v., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpinella M.C.C., Ferrayoli C.G., Palacios S.M. Antifungal synergistic effect of scopoletin, a hydroxycoumarin isolated from Melia azedarach L. fruits. J. Agric. Food Chem. 2005;53:2922–2927. doi: 10.1021/jf0482461. [DOI] [PubMed] [Google Scholar]
- Carrión V.J., Perez-Jaramillo J., Cordovez V., Tracanna V., de Hollander M., Ruiz-Buck D., Mendes L.W., van Ijcken W.F.J., Gomez-Exposito R., Elsayed S.S. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science. 2019;366:606–612. doi: 10.1126/science.aaw9285. [DOI] [PubMed] [Google Scholar]
- Castrillo G., Teixeira P.J., Paredes S.H., Law T.F., de Lorenzo L.d., Feltcher M.E., Finkel O.M., Breakfield N.W., Mieczkowski P., Jones C.D. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- Durán P., Thiergart T., Garrido-Oter R., Agler M., Kemen E., Schulze-Lefert P., Hacquard S. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell. 2018;175:973–983.e14. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl L., Vandamme P. Members of the genus Burkholderia: good and bad guys. F1000Res. 2016;5(F1000 Faculty Rev-1007) doi: 10.12688/f1000research.8221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourcroy P., Sisó-Terraza P., Sudre D., Savirón M., Reyt G., Gaymard F., Abadía A., Abadia J., Álvarez-Fernández A., Briat J.F. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 2014;201:155–167. doi: 10.1111/nph.12471. [DOI] [PubMed] [Google Scholar]
- Fourcroy P., Tissot N., Gaymard F., Briat J.F., Dubos C. Facilitated fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe(2+) transport system. Mol. Plant. 2016;9:485–488. doi: 10.1016/j.molp.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hacquard S., Garrido-Oter R., González A., Spaepen S., Ackermann G., Lebeis S., McHardy A.C., Dangl J.L., Knight R., Ley R. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Hervé M. 2020. RVAideMemoire: testing and plotting procedures for biostatistics, R package version 0; pp. 9–77.https://CRAN.R-project.org/package=RVAideMemoire [Google Scholar]
- Hindt M.N., Akmakjian G.Z., Pivarski K.L., Punshon T., Baxter I., Salt D.E., Guerinot M.L. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics. 2017;9:876–890. doi: 10.1039/c7mt00152e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J.D., Israelstam G.F. A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 1979;57:1332–1334. [Google Scholar]
- Ivanov R., Brumbarova T., Bauer P. Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant. 2012;5:27–42. doi: 10.1093/mp/ssr065. [DOI] [PubMed] [Google Scholar]
- Jakoby M., Wang H.Y., Reidt W., Weisshaar B., Bauer P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 2004;577:528–534. doi: 10.1016/j.febslet.2004.10.062. [DOI] [PubMed] [Google Scholar]
- Kai K., Mizutani M., Kawamura N., Yamamoto R., Tamai M., Yamaguchi H., Sakata K., Shimizu B.-i. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008;55:989–999. doi: 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- Kai K., Shimizu B.-i., Mizutani M., Watanabe K., Sakata K. Accumulation of coumarins in Arabidopsis thaliana. Phytochemistry. 2006;67:379–386. doi: 10.1016/j.phytochem.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Kassambara A. ggpubr: 'ggplot2' based publication ready plots. 2020. https://CRAN.R-project.org/package=ggpubr
- Krieger C., Roselli S., Kellner-Thielmann S., Galati G., Schneider B., Grosjean J., Olry A., Ritchie D., Matern U., Bourgaud F., Hehn A. The CYP71AZ P450 subfamily: a driving factor for the diversification of coumarin biosynthesis in apiaceous plants. Front. Plant Sci. 2018;9:820. doi: 10.3389/fpls.2018.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth R. emmeans: estimated marginal means, aka least-squares means. 2020. https://CRAN.R-project.org/package=emmeans
- Letunic I., Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Lindsay W.L., Norvell W.A. Development of a DTPA soil test for zinc, iron, manganese, and copper. Soil Science Society of America Journal. 1978;42:421–428. [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D.S., Lebeis S.L., Paredes S.H., Yourstone S., Gehring J., Malfatti S., Tremblay J., Engelbrektson A., Kunin V., del Rio T.G. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai H.J., Pateyron S., Bauer P. Iron homeostasis in Arabidopsis thaliana: transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biol. 2016;16:211. doi: 10.1186/s12870-016-0899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiafico S. rcompanion: functions to support extension education program evaluation, R package version 2.3.25. 2020. https://CRAN.R-project.org/package=rcompanion
- Morrissey J., Guerinot M.L. Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 2009;109:4553–4567. doi: 10.1021/cr900112r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle D.H., Wheeler P., Dinno A. FSA: fisheries stock analysis. 2020. https://github.com/droglenc/FSA
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2010. R: A language and environment for statistical computing.https://www.r-project.org/ [Google Scholar]
- Rajniak J., Giehl R.F.H., Chang E., Murgia I., von Wirén N.v., Sattely E.S. Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat. Chem. Biol. 2018;14:442–450. doi: 10.1038/s41589-018-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgaud F., Olry A., Hehn A. Recent advances in molecular genetics of furanocoumarin synthesis in higher plants. In: Jacob C., Kirsch G., Slusarenko A., Winyard P., Burkholz T., editors. Recent Advances in Redox Active Plant and Microbial Products. Springer; 2014. pp. 363–375. [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J., Lin W.D., Fu G.M., Abadía J., López-Millán A.F., Schmidt W. Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol. 2013;162:1473–1485. doi: 10.1104/pp.113.220426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J., Schmidt W. Reduction-based iron uptake revisited: on the role of secreted iron-binding compounds. Plant Signal. Behav. 2013;8:e26116. doi: 10.4161/psb.26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S., Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183:1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x. [DOI] [PubMed] [Google Scholar]
- Schmid N.B., Giehl R.F., Döll S., Mock H.P., Strehmel N., Scheel D., Kong X., Hider R.C., von Wirén N.v. Feruloyl-CoA 6’-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 2014;164:160–172. doi: 10.1104/pp.113.228544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Günther C., Weber M., Spörlein C., Loscher S., Böttcher C., Schobert R., Clemens S. Metabolome analysis of Arabidopsis thaliana roots identifies a key metabolic pathway for iron acquisition. PLoS One. 2014;9:e102444. doi: 10.1371/journal.pone.0102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D., Samira R., Matthiadis A., Gillikin J.W., Long T.A. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 2015;167:273–286. doi: 10.1104/pp.114.250837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Navas J., Moreno-Risueno M.A., Manzano C., Pallero-Baena M., Navarro-Neila S., Téllez-Robledo B., Garcia-Mina J.M., Baigorri R., Gallego F.J., del Pozo J.C. D-Root: a system for cultivating plants with the roots in darkness or under different light conditions. Plant J. 2015;84:244–255. doi: 10.1111/tpj.12998. [DOI] [PubMed] [Google Scholar]
- Sisó-Terraza P., Luis-Villarroya A., Fourcroy P., Briat J.-F.F., Abadía A., Gaymard F., Abadía J., Álvarez-Fernández A. Accumulation and secretion of Coumarinolignans and other coumarins in Arabidopsis thaliana Roots in response to iron deficiency at high pH. Front. Plant Sci. 2016;7:1711. doi: 10.3389/fpls.2016.01711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwinska J., Kadzinski L., Banasiuk R., Gwizdek-Wisniewska A., Olry A., Banecki B., Lojkowska E., Ihnatowicz A. Identification of QTLs affecting scopolin and scopoletin biosynthesis in Arabidopsis thaliana. BMC Plant Biol. 2014;14:280. doi: 10.1186/s12870-014-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwinska J., Siatkowska K., Olry A., Grosjean J., Hehn A., Bourgaud F., Meharg A.A., Carey M., Lojkowska E., Ihnatowicz A. Scopoletin 8-hydroxylase: a novel enzyme involved in coumarin biosynthesis and iron-deficiency responses in Arabidopsis. J. Exp. Bot. 2018;69:1735–1748. doi: 10.1093/jxb/ery005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis I.A., Yu K., Feussner K., de Jonge R., Van Bentum S., Van Verk M.C., Berendsen R.L., Bakker P.A.H.M., Feussner I., Pieterse C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA. 2018;115:E5213–E5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiergart T., Durán P., Ellis T., Vannier N., Garrido-Oter R., Kemen E., Roux F., Alonso-Blanco C., Ågren J., Schulze-Lefert P., Hacquard S. Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat. Ecol. Evol. 2020;4:122–131. doi: 10.1038/s41559-019-1063-3. [DOI] [PubMed] [Google Scholar]
- Tsai H.H., Rodríguez-Celma J., Lan P., Wu Y.C., Vélez-Bermúdez I.C., Schmidt W. Scopoletin 8-hydroxylase-mediated fraxetin production is crucial for iron mobilization. Plant Physiol. 2018;177:194–207. doi: 10.1104/pp.18.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbon E.H., Trapet P.L., Kruijs S., Temple-Boyer-Dury C., Rouwenhorst T.G., Pieterse C.M.J. Rhizobacteria-mediated activation of the fe deficiency response in Arabidopsis roots: impact on fe status and signaling. Front. Plant Sci. 2019;10:909. doi: 10.3389/fpls.2019.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges M.J.E.E.E., Bai Y., Schulze-Lefert P., Sattely E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. USA. 2019;116:12558–12565. doi: 10.1073/pnas.1820691116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zou D., Xie K., Xie M. Antibacterial mechanism of fraxetin against Staphylococcus aureus. Mol. Med. Rep. 2014;10:2341–2345. doi: 10.3892/mmr.2014.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ding W., Xu Y., Wu D., Li S., Chen J., Guo B. New insights into the antibacterial activity of hydroxycoumarins against Ralstonia solanacearum. Molecules. 2016;21:468. doi: 10.3390/molecules21040468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J., Schmidt S., Strehmel N., Scheel D., Abel S. Arabidopsis transporter ABCG37/PDR9 contributes primarily highly oxygenated Coumarins to root exudation. Sci. Rep. 2017;7:3704. doi: 10.1038/s41598-017-03250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data of 16S rRNA profiling experiments and RNA-Seq transcriptomic data have been deposited in the European Nucleotide Archive ENA: PRJEB38663. Datasets and scripts necessary to reproduce analyses and generate figures have been deposited to Mendeley Data: https://doi.org/10.17632/tkdn6zbw7k.1. Scripts for RNASeq analysis and heatmap generation are available at https://github.com/YulongNiu/MPIPZ_CJ_RNASeq.