Abstract
Background: Hepatocellular carcinoma (HCC) is one of the most leading causes of cancer-related mortality worldwide. Immune checkpoint inhibitors (ICIs) have been proved to be beneficial for advanced HCC. Tumor mutational burden (TMB) is an important predictor for efficacy of ICIs. However, the genetic landscape of Chinese HCC patients and the association between TMB and frequently mutated genes of HCC remain unclear.
Methods: Whole-exome sequencing data of 369 liver tumors from the Cancer Genome Altas (TCGA) and next generation sequencing (NGS) data of 657 liver tumors from Chinese clinical dataset were included.
Results: TP53 (61.8%) was the most frequently mutated gene in the Chinese cohort, followed by CTNNB1 (17.2%), RB1 (13.7%), and LRP1B (12.3%). The PI3K-Akt signaling (11.2%), the Rap1 signaling (8.1%), and Ras signaling (7.7%), were significantly mapped. LRP1B mutations were significantly associated with higher TMB in both TCGA cohort (P = 0.0003) and Chinese cohort (P = 0.0005). And TP53 mutations were also associated with higher TMB in the TCGA and Chinese cohort (P = 0.0005 and 0.0010, respectively). Prognosis analysis performed in TCGA cohort revealed LRP1B mutations were significantly associated with shorter overall survival (OS, median, 20.9 vs 61.7 months; HR, 2.22; P = 0.0012). TP53 mutation was an independent risk factor affecting both OS (HR 1.58, P = 0.0109) and PFS (HR 1.59, P = 0.0027).
Conclusions: The results suggest that LRP1B or TP53 mutations are associated with higher TMB and a poor prognostic factor in HCC.
Keywords: Hepatocellular carcinoma, LRP1B, TP53, tumor mutational burden, survival
Introduction
Hepatocellular carcinoma (HCC) is a heterogeneous disease with rising incidence and mortality rate 1. Surgery and liver transplantation are the potentially curative treatments; however, only less than 40% of the patients are diagnosed at the early stage 2. For patients with advanced stage, systemic therapies are the cornerstone. The multi-target tyrosine kinase inhibitor (TKI) Sorafenib was the unique systemic regimen which had been approved between 2007 and 2016 3. Recently, several promising drugs have showed clinical activity in patients with HCC, like levatinib in the frontline 4 and ramucirumab 5, regorafenib 6 and cabozantinib 7. However, the objective response rates (ORRs) were no more than 15%, which is still not enough to meet the clinical requirements. Immune checkpoint inhibitors (ICIs) have demonstrated clinical benefit in multiple solid tumors, which have been currently approved by FDA 8, 9. What's more, several researches have proven that patients with high tumor mutational burden (TMB) generally presented better response to ICIs than the rest 10-12.
TMB is a measurement of the number of mutations harbored by tumor cells, and is usually determined by using next-generation sequencing (NGS). Several studies have revealed that alternations in genomic integrity related genes might result in genomic instability and replication stress 13, 14. The transcription factor TP53, encoding p53 protein, activates in response to multiple stressors and regulate the expression of genes controlling proliferation, DNA repair, and cell death 15. As a major tumor suppressor, TP53 is highly prevalent on genic alternations in diverse cancer types 16. The low-density lipoprotein receptor-related protein 1B (LRP1B), encoding endocytic low density lipoprotein family receptor, commonly binds to extracellular ligands and is characterized as a candidate tumor suppressor 17.
Currently, the characteristic mutational landscape of HCC has been revealed by several researches 18-20. However, the association between the genomic feature and TMB or prognosis have not been deeply probed. Herein, we showed the genomic features in Chinese HCC patients based on NGS, analyzed the effect of gene mutations on TMB in a Chinese HCC dataset and the TCGA dataset. In addition, we further revealed the prognosis of patients harboring LRP1B or TP53 mutations in TCGA.
Materials and Methods
Sample collection and clinicopathologic data
From January 2017 to November 2018, tumor specimens and matched blood samples from 657 patients with primarily diagnosed as HCC were obtained in Fudan University Shanghai Cancer Center. Genomic profiling was tested in a CAP-certified/CLIA-accredited laboratory (3D Medicines Inc., Shanghai, China). Clinicopathologic information, including sex and age were collected. The present study was approved by the Ethics Committee of the hospital, and a waiver of informed consent form was signed by all the patients.
DNA extractions, targeted sequencing and data processing
The assay methodology of DNA extraction and sequencing was followed the methods published in previous paper with some modifications 21. Namely, tumor genomic DNA was extracted using QIAamp DNA FFPE Tissue kit (Qiagen GmbH, Hilden, Germany), and normal genomic DNA was extracted from peripheral blood mononuclear cells using QIAamp DNA Blood Mini kit (Qiagen GmbH, Hilden, Germany), respectively, following the manufacturer's protocols. Next-generation sequencing (NGS) targeted 381 cancer-related genes were performed on the NextSeq500 platform (Illumina, CA, USA), and samples with a mean coverage depth after de-duplicating reads of 500× was analyzed. Using BWA aligner (version 0.7.12), sequencing data were mapped to the human genome 19 (hg19). After that, the data experienced the process of PCR duplicate read removal and sequence metric collection with the aid of Picard (version 1.130) and Samtools (version 0.1.19). The clean data were analyzed to identify diverse genic alternations, like base substitution, indel, rearrangement, and copy number variant, by analysis pipelines developed by 3D Medicines Inc (Shanghai, China). Tumor mutational burden (TMB) was defined as total number of somatic non-synonymous mutations in coding region. Data from TCGA from cBioPortal (https://www.cbioportal.org/) was extracted in February 2019.
Statistical analysis
The demographic characteristics of patients were compared via the Chi-Square (χ2) test or T test. All P-values presented were two-sided, and associations were considered significant if the P-value was less than 0.05. Functional enrichment analysis, including Gene Oncology (GO) and signaling pathway, were performed by T test. Gene Oncology (GO) and pathway analysis on gene alternations were performed using DAVID (https://david.ncifcrf.gov/), and drawn in R by using the package “ggplot”. Overall survival (OS) and progression-free survival (PFS) were analyzed using the Kaplan-Meier method with a P value determined by the log-rank test, and drawn with GraphPad Prism version 6 (GraphPad Software Inc., LA, CA, USA). Hazard ratios (HR) were estimated using Cox proportional hazards regression. All statistical analyses were performed using the SPSS statistical package, version 20.0 (SPSS Inc®, Chicago, Illinois, USA).
Results
Patient characteristics
A total of 657 Chinese patients with HCC and 372 HCC patients from the TCGA database were included in this study. The basic characteristics were shown in Table 1, and not significant difference on sex and age existed between these two datasets. The median age was 53 years (range, 16-82) in the Chinese dataset and 61 years (range, 16-90) in the TCGA dataset. The percentage of male was respectively 87.2% and 68% in the Chinese dataset and TCGA dataset.
Table 1.
Characteristics of hepatocellular carcinoma patients from the clinical dataset and the TCGA dataset
| Characteristic | Clinical dataset | TCGA dataset |
|---|---|---|
| Cases | 657 | 369 |
| Median age, year (range) | 53 (16-82) | 61 (16-90) |
| Sex (male vs female) | 573 vs 84 | 251 vs 118 |
| LRP1B (mutation vs wild-type) | 81 vs 576 | 35 vs 334 |
| TP53 (mutation vs wild-type) | 406 vs 251 | 106 vs 263 |
Detection of gene alternations
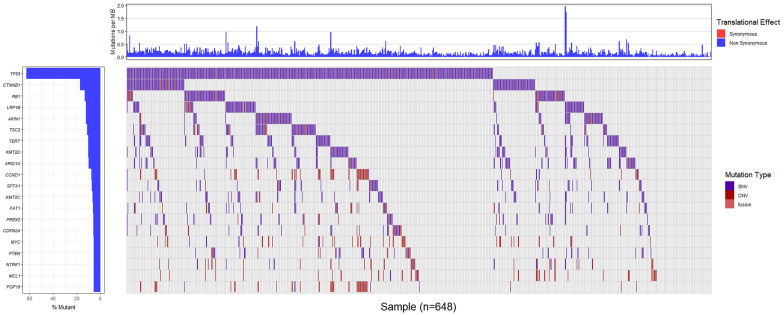
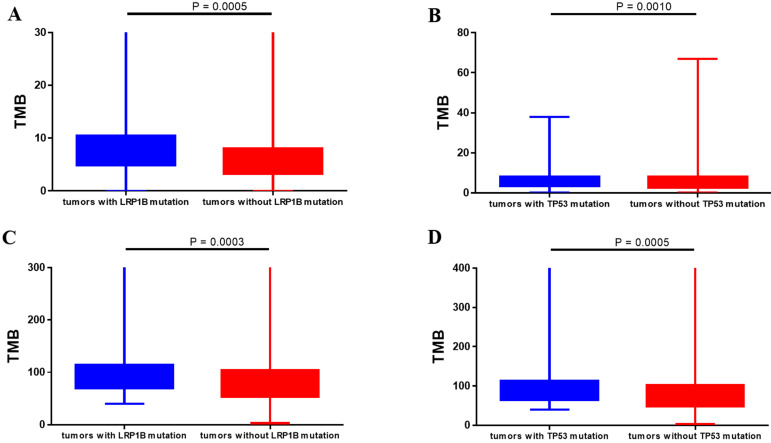
Totally, 648 patients (98.6%) in Chinese dataset detected somatic gene alternations. Significantly somatic genic alternations in tumor samples were identified and the genomic profile was drawn in Figure 1. TP53 (61.8%) was the most frequently mutated gene, followed by CTNNB1 (17.2%), RB1 (13.7%), and LRP1B (12.3%). Patients harboring LRP1B mutations presented significantly higher TMB than that with LRP1B wild-type (P=0.0005), and the same result was also observed in patients with TP53 mutations (P=0.0010, Figure 2A and 2B). What's more, the parallel results were also observed in the TCGA dataset. The frequency of TP53 mutations and LRP1B mutations in TCGA cohort was 28.7% (106/369) and 9.5% (35/369), respectively. The LRP1B and TP53 mutations were significantly associated with a higher TMB (P=0.0003 and 0.0005, respectively, Figure 2C and 2D).
Figure 1.
Genomic alternations of the Chinese patients with hepatocellular carcinoma.
Figure 2.
Correlation between gene alternations and tumor mutational burden. (A, B) Chinese clinical cohort. (C, D) TCGA dataset.
GO and pathway analysis
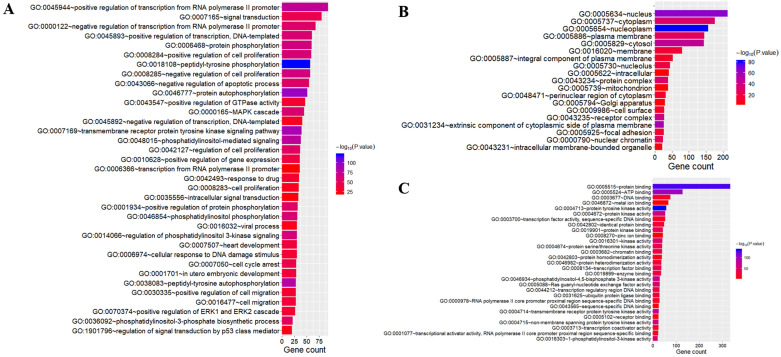
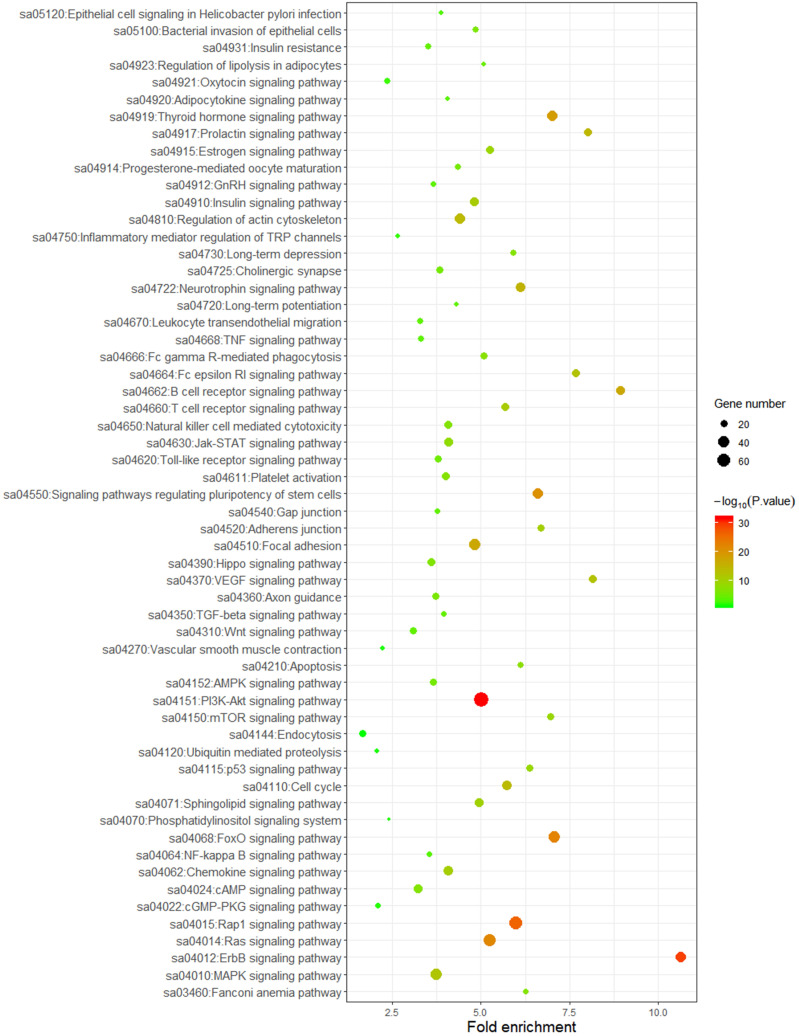
To better understand the biological function of these highly frequent somatic alternations in HCC, GO enrichment and signaling pathways analysis were performed. The significantly enriched GO terms of biological process associated with regulation of transcription, regulation of cell cycle, and the process of cellular signal transduction. Most of the genes located on nucleus and cytoplasm, and the top mapped molecular functions were protein binding, ATP binding, DNA binding, and metal ion binding (Figure 3). As shown in Figure 4, the pathways that may be implicated in HCC were commonly mapped. The PI3K-Akt signaling (11.2%) enriched the most altered genes, followed by the Rap1 signaling (8.1%), and Ras signaling (7.7%). In addition, several pathways related with cellular process, like cell cycle, apoptosis, immune microenvironment associated with pathways, and B cell/T cell receptor signaling pathways, were also significantly mapped. Although LRP1B and TP53 significantly impacted the TMB value, no obvious difference on signaling pathways existed between the patients with mutation and wild-type.
Figure 3.
Gene oncology analysis of highly altered genes. The terms of biological process (A); the terms of cellular component (B); the terms of molecular function (C).
Figure 4.
Mapping pathways by frequently mutated genes.
LRP1B or TP53 mutations predicted worse prognosis
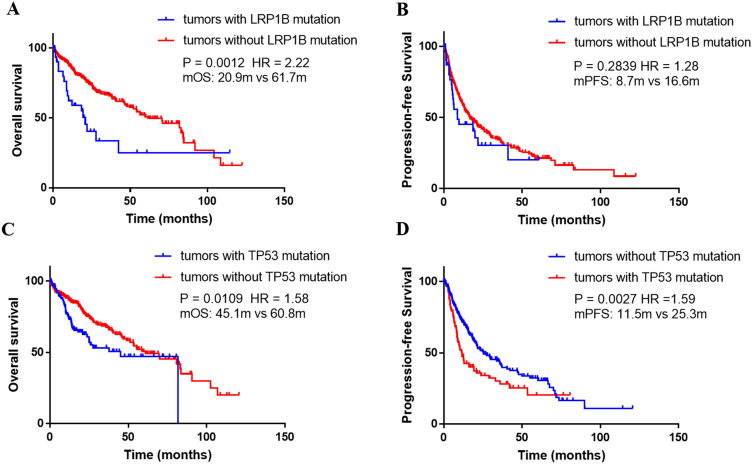
Using the Kaplan-Meier curve analysis, in the TCGA dataset, the overall survival (OS) was significantly shorter in the patients harboring LRP1B mutations than that with wild-type (median, 20.9m versus 61.7m, P=0.0012, HR=2.22, Figure 5A). In addition, median PFS was numerically shorter in the patients with LRP1B mutations than the rest of the patients (median, 8.7m versus 16.6m, P=0.2839, HR=1.28, Figure 5B). What's more, TP53 mutated patients presented significantly shorter OS and PFS compared with wild-type patients (median OS, TP53-mut versus TP53 wild-type: 45.1m vs 60.8m, P=0.0109, HR=1.58; median PFS, TP53-mut versus TP53 wild-type: 11.5 m vs 25.3 m, P=0.0027, HR=1.59, Figure 5C and 5D).
Figure 5.
Association between gene alternations and survival outcomes in the TCGA dataset. (A) Kaplan-Meier survival curves of overall survival comparing patients with wild-type LRP1B and patients with mutated LRP1B. (B) Kaplan-Meier survival curves of progression-free survival comparing patients with wild-type LRP1B and patients with mutated LRP1B. (C) Kaplan-Meier survival curves of overall survival comparing patients with wild-type TP53 and patients with mutated TP53. (D) Kaplan-Meier survival curves of progression-free survival comparing patients with wild-type TP53 and patients with mutated TP53.
Discussion
The present study revealed the genomic alternations of Chinese patients with HCC, the possibly biological function of altered genes, the association between the TMB value and LRP1B/TP53 mutations, and the impact of LRP1B/TP53 mutations on prognosis. These results indicate that LRP1B/TP53 mutations might be the prognostic predictors and associated with higher TMB, predicting better efficacy of immunotherapy in HCC patients.In our study, TP53 and CTNNB1 were the most frequently mutated genes, followed by RB1 and LRP1B. This result was similar with previous studies. A Whole-genome sequencing research indicated that TP53, CTNNB1, and RB1 were most common mutated among the protein-encoding genes, and frequently mutations were also observed in LRP1B which could be used as one of the mutational signatures to classify molecular subtypes in Japanese patients with liver cancer 18. Another study also showed the tumor suppressor genes, including TP53 and RB1, and the oncogenes, like CTNNB1, were the significantly mutated genes in HCC patients 19. Certainly, a relatively higher frequency of TP53/RB1 alternations were observed in our Chinese dataset, which is consistent with another research which included a large number of the Chinese HCC patients 22. For the mapped pathways of our work, besides the reported PI3K-AKT signaling and Ras signaling pathways 19, 23, we also reported the Rap1 signaling which plays an important role in tumor processes, such as cell migration, invasion, and metastasis 24.
We identified that patients harboring LRP1B and TP53 alternations presented higher TMB, respectively. This two genes were both frequently mutated in multiple types of human cancer. Chen and his colleagues have revealed that higher TMB was found in LRP1B mutated patients with melanoma and non-small cell lung cancer 25. An integrated analysis on the multiple-dimensional data types indicated that significantly increased non-synonymous mutations generally presented in the TP53-mutated group compared with the wild-type group 26. Although several researches have probed the relationship between LRP1B/TP53 and TMB, no solid conclusions existed on HCC. In addition, based on the public database, the LRP1B alternations correlating with poor survival was also revealed. In patients with glioblastoma, LRP1B is generally down expressed, and LRP1B deletion is associated with poor outcome 27. However, to our knowledge, no definite conclusion on the correlation between LRP1B and prognosis in HCC has been made, and certainly, our results are needed to be validated further.
One limitation of this paper is failure to collect the whole baseline clinical characteristic and therapeutic regimens. And another limitation was no prognosis data from the Chinese clinical dataset. Thus, the further research need to focus on the association between gene alterations and clinical outcomes in Chinese HCC cohort.
To sum up, our work revealed the genomic landscape of HCC with a large sample size, probed the association between gene alternations and TMB, and pointed out the impact of specific gene alternations on prognosis. These results might aid to screen potential therapeutic targets and better manage the whole disease process.
Acknowledgments
This work was jointly supported by Wu Jieping Medical Foundation (320.2710.1857), National Natural Science Foundation of China (81874182, 81874056), National Key Project of China (2017ZX10203204-007-004), Public Health Bureau Foundation of Shanghai (201840019), and Research Talent Foundation of Fudan University Shanghai Cancer Center (YJRC1604).
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The protocol was approved by the Ethics Committee of the hospital. The data that support the findings of this study are available on request from the corresponding author. A waiver of informed consent form was signed by all the patients.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF. et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 4.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F. et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM. et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:282–296. doi: 10.1016/S1470-2045(18)30937-9. [DOI] [PubMed] [Google Scholar]
- 6.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G. et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389(10064):56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 7.Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY. et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379(1):54–63. doi: 10.1056/NEJMoa1717002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu AX, Finn RS, Edeline J, Cattan S, Ogasawara S, Palmer D. et al. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;19(7):940–952. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 10.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C. et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V. et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stracquadanio G, Wang X, Wallace MD, Grawenda AM, Zhang P, Hewitt J. et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat Rev Cancer. 2016;16:251–65. doi: 10.1038/nrc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18:622–36. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2105; 16(7): 393e405. [DOI] [PubMed]
- 16.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 17.Liu CX, Li Y, Obermoeller-McCormick LM, Schwartz AL, Bu G. The putative tumor suppressor LRP1B, a novel member of the low density lipoprotein (LDL) receptor family, exhibits both overlapping and distinct properties with the LDL receptor-related protein. J Biol Chem. 2001;276(31):28889–96. doi: 10.1074/jbc.M102727200. [DOI] [PubMed] [Google Scholar]
- 18.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y. et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–9. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang N, Li Y, Liu Z, Qin H, Du D, Cao X. et al. The characteristics of ctDNA reveal the high complexity in matching the corresponding tumor tissues. BMC Cancer. 2018;18(1):319. doi: 10.1186/s12885-018-4199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding XF, He M, Chan AWH, Song QX, Sze SC, Chen H. et al. Genomic and Epigenomic Features of Primary and Recurrent Hepatocellular Carcinomas. Gastroenterology. 2019;157:1630–1645. doi: 10.1053/j.gastro.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YL, Wang RC, Cheng K, Ring BZ, Su L. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med. 2017;14(1):90–99. doi: 10.20892/j.issn.2095-3941.2016.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. 2019;10:1113. doi: 10.3389/fimmu.2019.01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY. et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 27.Tabouret E, Labussière M, Alentorn A, Schmitt Y, Marie Y, Sanson M. et al. LRP1B deletion is associated with poor outcome for glioblastoma patients. J Neurol Sci. 2015;358(1-2):440–3. doi: 10.1016/j.jns.2015.09.345. [DOI] [PubMed] [Google Scholar]