Abstract
Background
“Diffuse midline glioma (DMG), H3 K27M-mutant” is a new tumor entity established in the 2016 WHO classification of Tumors of the Central Nervous System that comprises a set of diffuse gliomas arising in midline structures and is molecularly defined by a K27M mutation in genes encoding the histone 3 variants H3.3 or H3.1. While this tumor entity is associated with poor prognosis in children, clinical experience in adults remains limited.
Methods
Patient demographics, radiologic and pathologic characteristics, treatment course, progression, and patient survival were collected for 60 adult patients with DMG, H3 K27M-mutant. A subset of tumors also underwent next-generation sequencing. Analysis of progression-free survival and overall survival was conducted using Kaplan–Meier modeling, and univariate and multivariate analysis.
Results
Median patient age was 32 years (range 18–71 years). Tumors were centered in the thalamus (n = 34), spinal cord (10), brainstem (5), cerebellum (4), or other midline sites (4), or were multifocal (3). Genomic profiling revealed p.K27M mutations exclusively in the H3F3A gene and an absence of mutations in HIST1H3B or HIST1H3C, which are present in approximately one-third of pediatric DMGs. Accompanying mutations in TP53, PPM1D, FGFR1, NF1, and ATRX were frequently found. The overall survival of this adult cohort was 27.6 months, longer than historical averages for both H3 K27M-mutant DMG in children and IDH-wildtype glioblastoma in adults.
Conclusions
Together, these findings indicate that H3 K27M-mutant DMG represents a heterogeneous disease with regard to outcomes, sites of origin, and molecular pathogenesis in adults versus children.
Keywords: adult, diffuse midline glioma, genetics, H3 K27M, survival
Key Points.
H3 K27M-mutant DMG has divergent genetic profiles in adults versus children.
Overall survival of the adult cohort reported here is longer than historical averages for H3 K27M-mutant DMG in children and IDH-wildtype glioblastoma in adults.
Importance of the Study.
We now recognize that glioblastoma (GBM) is not one disease, but rather a conglomerate of many different tumor entities with similar histologic features, each with their own distinct genetic drivers, anatomic site of origin, typical age of onset, and clinical outcomes. Here we provide evidence that DMG, H3 K27M-mutant has divergent clinical outcomes, genetic drivers, and anatomic site of origin in adults versus children. These findings have important ramifications for the clinical management of adults with this diagnosis. It is not simply the adult equivalent of the childhood disease diffuse intrinsic pontine glioma, nor directly analogous to IDH-wildtype GBM, but rather may be a distinct neoplasm in adults that is associated with more favorable survival and more frequent occurrence in the thalamus or spinal cord. In addition, this is the largest cohort to date detailing the genetic profile of these tumors, which differs from that of pediatric H3 K27M-mutant DMG and provides distinct therapeutic targets.
Recently, there is an increasing emphasis on specific genetic mutations in the pathological diagnosis of primary brain tumors. In the 2016 World Health Organization (WHO) classification system, “diffuse midline glioma (DMG), H3 K27M-mutant” replaced the prior entity of diffuse intrinsic pontine glioma (DIPG) and incorporated all diffuse gliomas of any midline structure in the brain and spinal cord harboring a lysine to methionine mutation at codon 27 in genes that encode histone 3 isoforms. H3F3A encodes for the H3.3 variant that is mutated in approximately 70% of H3 K27M-mutant DMGs in children, whereas HIST1H3B or HIST1H3C both encode for the H3.1 variant that is mutated in the remaining 30% of H3 K27M-mutant DMGs in children. This K27M amino acid substitution prevents the polycomb repressive methyltransferase complex 2 (PRC2) from modifying the lysine residue at codon 27 on the histone 3 tail, resulting in decreased trimethylation of H3 proteins, both mutated and wildtype.1 Decreased trimethylation at this critical residue on the histone H3 tail disrupts gene expression responsible for glial differentiation leading to a stem cell-like, oncogenic state.1
While H3 K27M mutations were initially discovered in pediatric DIPG, subsequent studies identified H3 K27M mutations in diffusely infiltrative astrocytomas arising in other midline structures of the central nervous system (CNS) including the diencephalon, cerebellum, and spinal cord, in both children and adults.2,3 The incidence of H3 K27M mutations in diffuse gliomas involving midline structures is estimated to be 80% in pediatric and 15–60% in adult patients.4–7 In children, these mutations often occur early in the oncogenic cascade during embryogenesis or the early postnatal period.8,9 Pediatric patients with H3 K27M-mutant DMGs have a poor prognosis, with a median overall survival (mOS) of 10–14 months,10–14 hence the rationale for the 2016 WHO Classification of Tumors of the CNS to designate these tumors as grade IV tumors, irrespective of histologic features such as necrosis or microvascular proliferation.15 Prognosis is less clear in adult DMG harboring the H3 K27M mutation16,17; this may in part depend on differences in location and spatially regulated gene expression.
This study elucidates a single institution’s experience with a series of 60 adults with pathologically confirmed DMG, H3 K27M-mutant. Herein, we describe the histopathology and concomitant genetic aberrations, radiologic features, and clinical outcomes of these patients.
Materials and Methods
Patient Population
The institutional review board at the University of California, San Francisco (UCSF) approved this study (UCSF IRB 10-03204). Patients included were 18 years or older at initial diagnosis and had pathological confirmation of DMG with H3 K27M mutation, during the period of 2014–2019 at UCSF. Cases were identified using immunohistochemistry (IHC) with a histone H3 K27M-mutant specific antibody and/or through next-generation sequencing (NGS) of the histone 3 genes (H3F3A, HIST1H3B, and HIST1H3C). Sixty-four adult patients were initially identified with gliomas harboring H3 K27M mutation. Four of these patients were excluded from analysis: 2 tumors were non-midline, located in the cerebral hemispheres, and 2 others were not diffuse glioma subtypes (ganglioglioma and pilocytic astrocytoma). Other histopathological markers for these cases were annotated if available, including IDH1 R132H-mutant protein status, ATRX protein expression, p53 protein expression, and BRAF V600E-mutant protein expression by IHC, EGFR copy number assessment by fluorescent in situ hybridization (FISH), and MGMT promoter methylation by bisulfite PCR sequencing.
UCSF500 Targeted NGS Panel
Targeted NGS for a subset of the tumors was performed using the institutional UCSF500 Cancer Panel as previously described and detailed here.18 Genomic DNA was extracted from formalin-fixed, paraffin-embedded blocks of tumor tissue using the QIAamp DNA FFPE Tissue Kit (Qiagen). Genomic DNA was also extracted from peripheral blood or buccal swab sample for a subset of the patients using the QIAamp DNA Blood Midi Kit (Qiagen). Capture-based next-generation DNA sequencing was performed using an assay that targets all coding exons of 479 cancer-related genes, select introns, and upstream regulatory regions of 47 genes to enable detection of structural variants including gene fusions, and DNA segments at regular intervals along each chromosome to enable genome-wide copy number and zygosity analysis, with a total sequencing footprint of 2.8 Mb (UCSF500 Cancer Panel). Multiplex library preparation was performed using the KAPA Hyper Prep Kit (Roche) according to the manufacturer’s specifications using 250 ng of sample DNA. Hybrid capture of pooled libraries was performed using a custom oligonucleotide library (Nimblegen SeqCap EZ Choice). Captured libraries were sequenced as paired-end 100 bp reads on an Illumina HiSeq 2500 instrument. Sequence reads were mapped to the reference human genome build GRCh37 (hg19) using the Burrows-Wheeler aligner. Recalibration and deduplication of reads were performed using the Genome Analysis Toolkit. Coverage and sequencing statistics were determined using Picard CalculateHsMetrics and Picard CollectInsertSizeMetrics. Single nucleotide variant and insertion/deletion mutation calling was performed with FreeBayes and PinDel. Structural variant calling was performed with Delly. Variant annotation was performed with ANNOVAR. Single nucleotide variants, insertions/deletions, and structural variants were visualized and verified using Integrative Genome Viewer. Genome-wide copy number analysis based on on-target and off-target reads was performed by CNVkit and visualized using Nexus Copy Number (Biodiscovery). Methods for external commercial NGS panels (Tempus, Foundation One, Caris) and the Stanford Solid Tumor Actionable Mutational Panel are published elsewhere.19–22
Magnetic Resonance Imaging
Neuro-imaging studies were analyzed by a neuroradiologist (J.E.V-M.) to characterize the anatomic location of tumor, tumor size, presence and patterns of contrast enhancement, and radiologic extent of resection. Tumor locations were classified based on either the largest site of disease or location of dominant enhancing focus in cases where there was an extension into or involvement of multiple sites. Tumor locations were designated as follows: “diencephalon” included thalamus, hypothalamus, midbrain, or the pineal region; “posterior fossa” included the pons, medulla, or cerebellum; “spinal cord” comprised the entire spinal cord.
Survival Analysis
Demographic and clinical course including the extent of surgical resection, radiation therapy type and dose, Karnofsky performance status (KPS) at time of diagnosis, time to tumor progression and number of tumor progressions, and treatments administered to each patient were collected as available and reviewed by a neuro-oncologist (R.A.B., S.L., and J.D.S.). Progression was defined on magnetic resonance imaging (MRI) using the modified Response Assessment in Neuro-oncology criteria,23 and/or clinical deterioration that was sufficient to trigger treatment modification (including discontinuing disease-modifying therapy and/or starting hospice).
Progression-free survival (PFS) was defined as the time from diagnosis until the date of first documentation of progression, or death due to any cause in the absence of documented progression,23 whichever occurred first. Patients known to be without progression and alive were censored at the date of last contact. Overall survival (OS) was defined as the time from diagnosis to the date of death. Patients known to be alive were censored at the date of last contact. The Kaplan–Meier method was used to estimate median PFS and OS with corresponding 2-sided 95% confidence intervals (CIs) and to draw survival curves. Median follow-up time was calculated with the reverse Kaplan–Meier estimator. Univariate and multivariate analysis was performed using Cox proportional hazards regression models. Association of common genetic features and presence or absence of enhancement were tested using Fisher’s exact test. All analyses were performed in R (version 3.6.1).
Results
Patient and Tumor Characteristics
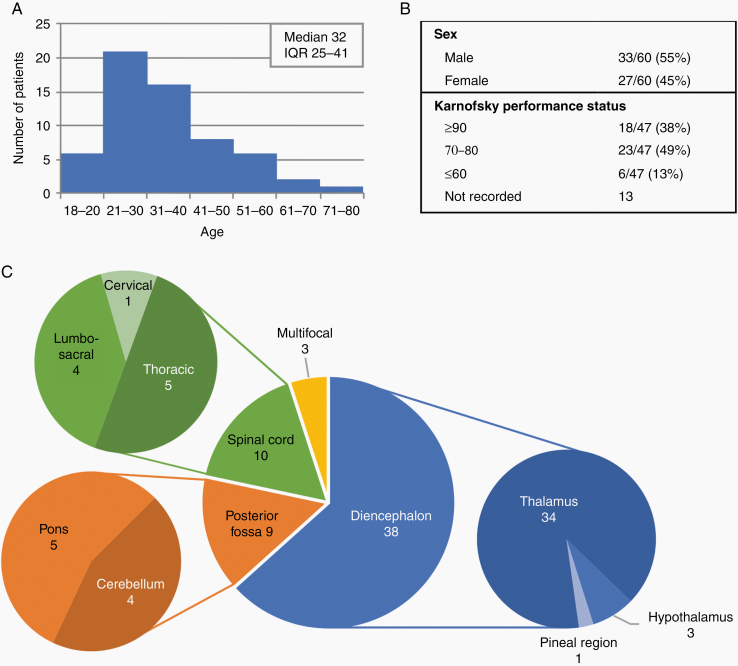
This study examined 60 adult patients with pathologically confirmed DMGs harboring H3 K27M mutation. The cohort was mostly in their third decade at the time of initial diagnosis with a median age of 32 years (interquartile range 25–41; Figure 1A). These patients were nearly all independently functioning at diagnosis (87% KPS ≥70) and there was a slight preponderance of males (55%; Figure 1B). Most tumors (63%) were supratentorial and located in the diencephalic region, with most originating in the thalamus (57% of all tumors). Of the 10 tumors located in the spinal cord, most were thoracic (50%) and 2 were in the conus medullaris (Figure 1C). Three cases were multifocal with both midline and non-midline sites of disease (2 patients) or diffuse craniospinal dissemination (1 patient).
Figure 1.
Patient demographics and tumor location for the 60 adults with DMG, H3 K27M-mutant: (A) Histogram shows the distribution of patient age at diagnosis, (B) sex and Karnofsky performance status at diagnosis, and (C) location of tumors.
Radiologic Features
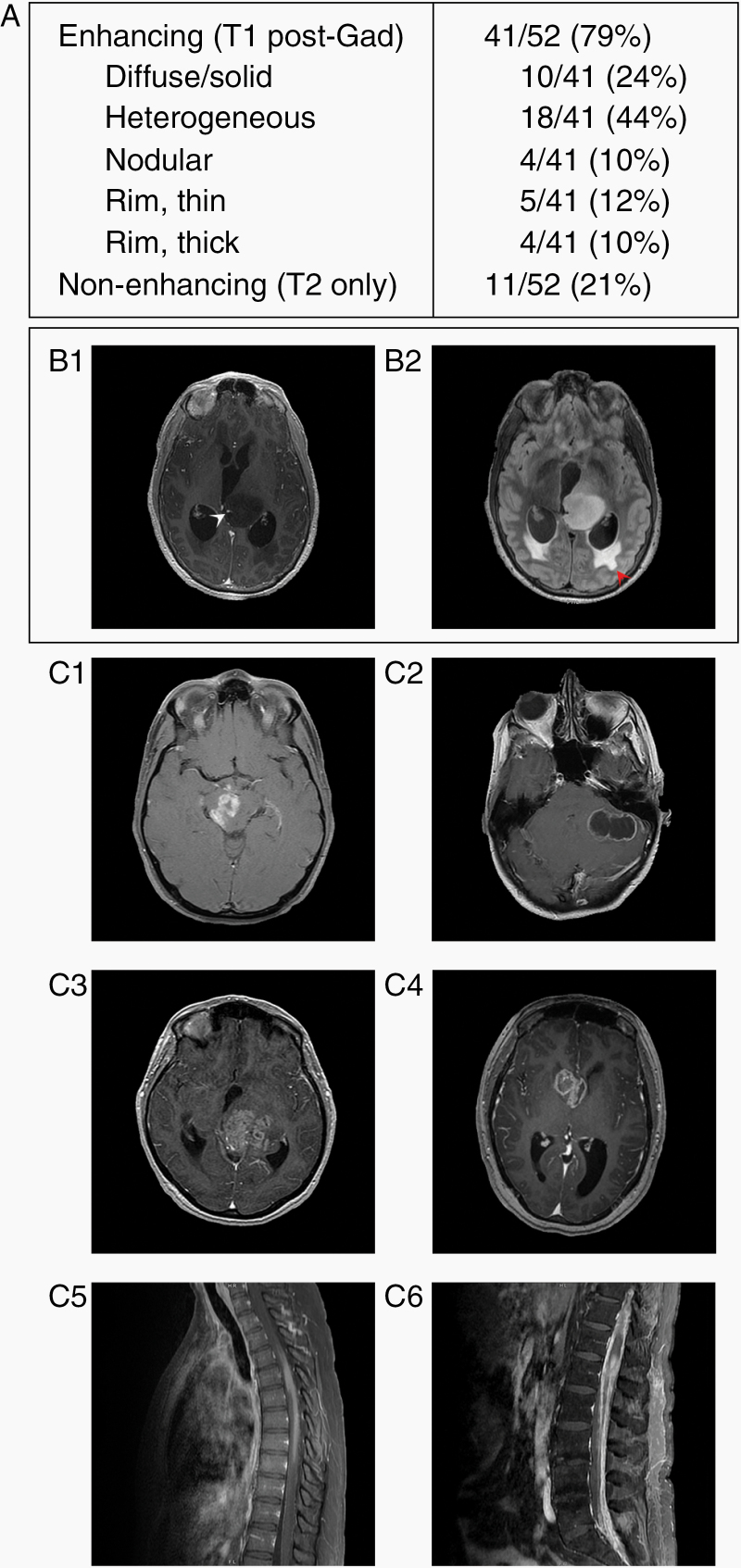
On MRI, tumors were principally contrast-enhancing (79%) on T1 post-gadolinium sequences, of which the most frequently observed enhancement pattern was heterogeneous, followed by diffuse/solidly enhancing (homogeneous) or rim-only enhancing patterns (Figure 2). The imaging characteristics of these H3 K27M-mutant DMGs were similar to those published elsewhere in both adult and pediatric patients.24,25 There was no association with the presence or absence of enhancement on MRI, and OS or PFS based on Kaplan–Meier and univariate Cox proportional hazards analysis (data not shown).
Figure 2.
MRI radiologic features of H3 K27M-mutant DMGs in adults at the time of initial diagnosis. (A) Description of gadolinium enhancement on MRI of the 60 patients. (B) Post-gadolinium (B1) and FLAIR (B2) images from a non-enhancing tumor. A blood vessel courses through the left thalamic tumor (white arrowhead), and periventricular FLAIR hyperintensity indicates transependymal flow secondary to acute obstructive hydrocephalus (red arrowhead). (C) Several examples of DMGs with H3 K27M mutation with different enhancement patterns: nodular (C1), thin rim (C2), heterogeneous (C3), thick rim (C4), diffuse/solid enhancement (C5), and disseminated disease with a conus lesion and leptomeningeal spread (C6).
Treatment
Of the 60 patients, most underwent biopsy only, but a subset received surgical resection (27%; Table 1). After surgery, most patients had external beam radiation (XRT, 98%) in combination with some other type of treatment, most commonly a regimen of temozolomide (TMZ) given concurrently with XRT, followed by TMZ alone in an adjuvant setting (74%). Median follow-up time was 59.2 months (95% CI: 23.8–NA). Of the patient cohort, 66% had known progression prior to death, and 91% had known tumor progression or known death (Table 1). At the time of progression, almost half of the patients were treated with bevacizumab, a monoclonal antibody directed at vascular endothelial growth factor (VEGF), and 26% underwent repeat XRT. These therapies were often given in combination with other treatments, including TMZ, lomustine/carmustine, and/or investigational drugs (Table 1). Of note, 16% of patients received no additional treatment beyond the initial treatment course.
Table 1.
Clinical Course, Treatment, and Outcomes for the 60 Adults with DMG, H3 K27M-Mutant
| Surgical intervention | |
| Any resection | 16/60 (27%) |
| Gross total | 4/60 (7%) |
| Subtotal | 12/60 (20%) |
| Biopsy only | 44/60 (73%) |
| Initial treatment at diagnosis | |
| Known treatment course at diagnosis | 50 |
| XRT only | 5/50 (10%) |
| XRT/TMZ | 7/50 (14%) |
| XRT/TMZ + adj TMZ | 29/50 (58%) |
| XRT/TMZ + adj TMZ + TTF | 4/50 (8%) |
| XRT/TMZ + adj TMZ+ investigational chemo | 3/50 (6%) |
| XRT + adj TMZ | 1/50 (2%) |
| No treatmenta | 1/50 (2%) |
| Tumor progression prior to death | |
| Known clinical course | 44 |
| No | 4/44 (9%) |
| Yes (median number of progression events per patient: 1) | 40/44 (91%) |
| Unknown | 16 |
| Second-line treatments b | |
| Known treatment course after initial therapy | 43 |
| BEV (monotherapy or with other chemo) | 20/43 (47%) |
| Re-XRT (including SRS) | 11/43 (26%) |
| Re-resection | 2/43 (5%) |
| TMZ (initial therapy) | 1/43 (2%) |
| TMZ (retrial) | 8/43 (19%) |
| CCNU/BCNU | 8/43 (19%) |
| Other chemo including investigational | 8/43 (19%) |
| Tumor treating fields | 1/43 (2%) |
| No treatment | 7/43 (16%) |
| Unknown | 17 |
| Survival status | |
| Alive | 14/60 (23%) |
| Deceased | 38/60 (63%) |
| Unknown | 8/60 (13%) |
| Median follow-up (95% CI), months | 59.2 (23.8–NA) |
| Median progression-free survival (95% CI), months c | 9.6 (7.4–14.3) |
| Median overall survival (95% CI), months d | 27.6 (19.1–36.7) |
The table describes any surgical intervention, initial treatment, treatment at progression, time to progression, and length of survival. If the denominator is less than 60, it indicates the number of patients for whom the information is available and documented. adj, adjuvant; Bev, Bevacizumab; CI, confidence interval; CCNU/BCNU; lomustine/carmustine; SRS, stereotactic radiosurgery; TMZ, temozolomide; TTF, tumor treating fields; XRT, radiation therapy.
aPatient died 3 weeks after surgery.
bPatients may have received more than one treatment, so categories are not mutually exclusive, and do not sum to 100%.
cProgression and death without known progression are recorded events.
dEstimated from Kaplan–Meier analysis.
Pathology and Genetic Features
With regard to the pathological features, 62% of tumors had necrosis and/or microvascular proliferation, while 38% lacked both of these features (Table 2). There was no difference in the survival of patients based on these histologic features (data not shown). In addition to the H3 K27M mutation, these tumors were universally negative for alterations in the IDH1 and IDH2 genes, either by IHC or by NGS (Table 2, Figure 3). Most tumors were MGMT promoter unmethylated (20 of 22 tested, or 91% patients; Table 2). No tumors demonstrated EGFR amplification by FISH or NGS (Table 2, Figure 3).
Table 2.
Histopathological Features of Adult H3 K27M-Mutant DMG
| Presence of necrosis and/or microvascular proliferation | |
| Absent | 22/58 (38%) |
| Present | 36/58 (62%) |
| Not reported | 2 |
| Immunohistochemical testing | |
| H3 K27M mutation | 59/59 (100%) |
| H3 K27 trimethylation complete loss | 15/16 (94%) |
| H3 K27 trimethylation mosaicism | 1/16 (6%) |
| IDH1 R132H mutation | 0/55 (0%) |
| p53 expression >10% | 32/47 (68%) |
| ATRX loss | 21/55 (38%) |
| BRAF V600E mutation | 0/20 (0%) |
| Molecular analysis | |
| MGMT promoter methylation | 2/22 (9%) |
| EGFR amplification (FISH) | 0/25 (0%) |
Summary table for histologic features, immunohistochemical stains, MGMT promoter analysis, and EGFR amplification status of tumors.
Denominator indicates the number of patients tested.
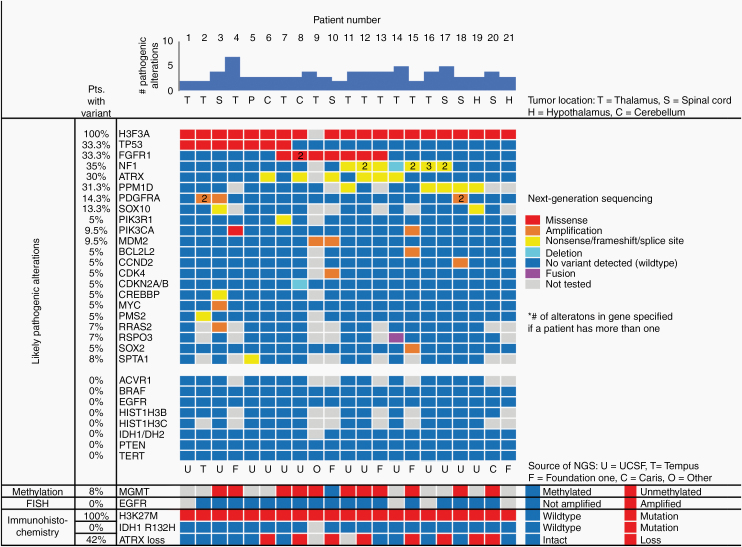
Figure 3.
Genetic profile of adult H3 K27M-mutant DMG. Summary plot of the next-generation sequencing results for the 21 evaluated patients, as well as immunohistochemical, FISH, and MGMT promoter methylation data for these tumors.
Targeted NGS was performed on tumors from 21 patients, utilizing either the described institutional panel at UCSF or a variety of commercial sources (Figure 3, Supplementary Tables 1–3). All 20 of the assessed tumors harbored K27M mutation in the H3F3A gene, which encodes the histone variant H3.3 (one tumor was interrogated by a targeted NGS panel that did not include the histone H3 genes). In contrast to brainstem DMGs in children, no tumors had mutations in the histone H3.1 genes HIST1H3B or HIST1H3C.
In addition to H3F3A K27M mutation, the most common additional alterations involved the TP53, PPM1D, FGFR1, NF1, and ATRX genes (Figure 3). Seven tumors (33%) harbored inactivating TP53 mutations. Of the 14 tumors lacking TP53 mutation, 5 harbored truncating mutations in exon 6 of PPM1D and 2 harbored focal amplification of MDM2; both genes encode p53 regulatory proteins. In total, 14 tumors (67%) demonstrated genetic alterations predicted to disrupt p53 function. Activating missense mutations in the kinase domain of FGFR1 (either p.N546 and p.K656) were identified in 7 tumors (33%). Inactivating mutations or focal deletions of the NF1 tumor-suppressor gene were also common, being present in 7 cases, 4 of which harbored multiple mutations in NF1. Mutations and/or focal high-level amplifications of PDGFRA were seen in 3 cases, including 2 that harbored amplification of a mutant allele (Figure 3). Six out of 20 tumors sequenced showed mutations in ATRX (30%), with 2 additional patients showing loss of ATRX by IHC without ATRX mutation identified, suggesting either cryptic alterations not detected by sequencing or alternative methods of protein expression regulation (Table 2, Figure 3).
Notably, there were no mutations, amplifications, or deletions involving the TERT promoter, EGFR, or PTEN, which are among the most frequently altered genes in IDH-wildtype glioblastomas (GBMs) in adults (Figure 3). In addition, there were no alterations detected in the BRAF oncogene, which commonly demonstrates fusions or mutations in low-grade gliomas such as ganglioglioma, pleomorphic xanthoastrocytoma, and pilocytic astrocytoma (Figure 3). ACVR1, which is commonly mutated in pediatric brainstem gliomas with H3.1 K27M mutation,12 was uniformly wildtype in the 14 evaluated cases. There were no clear associations of any of the frequently found genetic alterations and OS or PFS as assessed by Kaplan–Meier and Cox proportional hazards univariate analysis or the presence of enhancement as assessed by Fisher’s exact test (data not shown).
Two patients (#3 and #6, Figure 3) had repeat sequencing performed on resection specimens at the time of recurrence/progression, enabling assessment of clonal evolution following treatment. The recurrent tumor from patient 3 had acquired focal high-level amplification of PIK3CA not observed in the initial tumor resection, but no longer had the PDGFRA amplification found in the initial tumor (Supplementary Table 2). Next-generation sequencing for patient 6 demonstrated identical genetic alterations between the initial and recurrent tumors (Supplementary Tables 1–3). Additional information about the specifics of genetic and cytogenetic alterations for those 13 patients with NGS by the institutional UCSF500 panel is provided (Supplementary Tables 1–3).
Clinical Outcomes
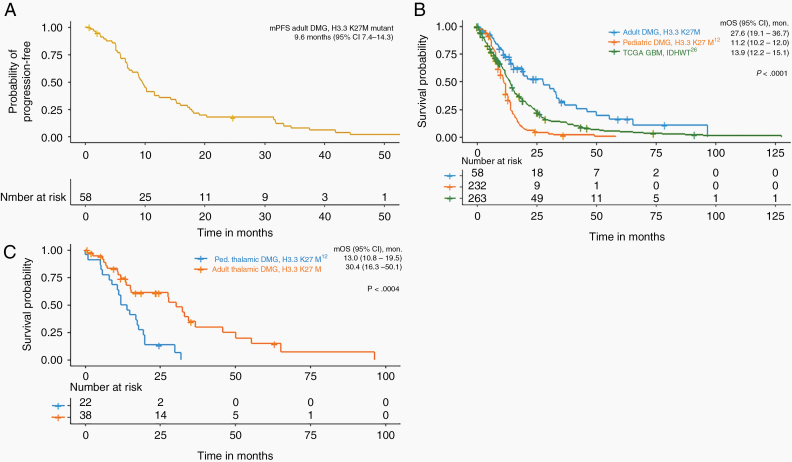
Median PFS in this series was 9.6 months (95% CI 7.4–14.3, Figure 4A) and mOS was 27.6 months (95% CI 19.1–36.7; Figure 4B, “Adult DMG, H3.3 K27M”). Sample size was too limited to determine if the location was predictive of OS (Supplementary Figure 1). For OS, univariate analysis showed factors causing a higher (ie, detrimental) hazard ratio (HR) were initial treatment of radiation only or no treatment at diagnosis compared to standard-of-care radiation and TMZ (HR 3.61 [CI 1.70–7.65, P = .001]) and no treatment at progression (HR 17.74 [CI 6.29–50.04, P < .001], Supplementary Table 4). Univariate analysis showed that OS was not affected by age (P = .222) or KPS (P = .074). No particular treatment at progression (i.e., bevacizumab, radiation, or lomustine/carmustine) was found to have a specific impact on survival. MGMT methylation was found to have a significant HR, but likely this was artifactual and related to the small sample size of the MGMT methylated group (n = 2, Supplementary Table 4). Multivariate analysis found that any treatment at progression significantly improved OS (P < .001), after accounting for age at diagnosis. For PFS, univariate analysis showed increased HR with age ≥50 (HR 2.51 [CI 1.18–5.34, P = .017]) and KPS ≤60 (HR 2.69 [CI 1.03–7.01, P = .043], Supplementary Table 4). Multivariate analysis showed that age at diagnosis had a significant impact on PFS when accounting for KPS ≤60 (P < .05).
Figure 4.
Progression-free and overall survival in adults with DMG, H3 K27M-mutant. (A) Kaplan–Meier plot progression-free survival for all adult patients in this study. (B) Kaplan–Meier analysis of overall survival for adult patients in this study (“Adult DMG, H3.3 K27M”), compared to a previously published cohort of pediatric patients with diffuse gliomas with H3.3 K27M mutation located in any midline structure12 (“Pediatric DMG, H3.3 K27M”) and adult patients with IDH-wildtype GBM included in the TCGA study26 (“TCGA GBM IDH-WT”). (C) Kaplan–Meier analysis of overall survival of patients with thalamic H3.3 K27M-mutant diffuse gliomas, comparing adults in this study cohort to a previously published cohort of pediatric patients.12
This cohort demonstrated prolonged survival compared to a historical series of adult patients with IDH-wildtype GBM from The Cancer Genome Atlas (TCGA) with unknown H3 K27M status,26 with mOS of 13.9 months (95% CI: 12.2–15.2, P < .001; Figure 4B, “TCGA GBM, IDH-WT”). A recently published meta-analysis of pediatric DMGs included 249 pediatric patients (aged <18 years) with H3F3A K27M mutation.12 In this meta-analysis, all patients tested for IDH were IDH-wildtype (150 of 150 patients tested; 89 patients not tested), and of those tested, nearly all had unmethylated MGMT promoter (74 of 79 patients tested; 170 patients not tested).12 For diffuse gliomas with H3.3 K27M mutation in any midline location, the cohort of adult patients reported here had better mOS of 27.6 months compared to this pediatric group with mOS 11.2 months (95% CI: 10.2–12.0, P < .001; Figure 4B, “Pediatric DMG, H3.3 K27M”).12 Focusing only on H3.3 K27M-mutant tumors located in the thalamus, there was still a significant difference in the median OS in this adult cohort compared to the pediatric group from the meta-analysis (30.4 months vs 13.0 months, P < .0004, Figure 4C).12
Discussion
Since the incorporation of DMG, H3 K27M-mutant in the 2016 WHO Classification of Tumors of the CNS,15 there is ever-increasing recognition and diagnosis of this entity. Though there is substantial molecular and phenotypic overlap with DIPG in children, much less was known about the natural history, molecular features, response to treatment, and clinical outcomes in adults.
Compared to pediatric cases, tumors in this adult cohort were more commonly located in the diencephalon (57% in the thalamus) and spinal cord and uncommonly located in the brainstem. A small number of tumors in our cohort were multifocal. While rare cases of non-midline H3 K27M-mutant gliomas have been reported, the diagnostic and prognostic ramifications of H3 K27M mutation are not well established outside midline locations.15,27 Median OS for our cohort was 27.6 months, substantially longer than previously published series of (1) adult patients with DMG, H3 K27M-mutant (8.4–19.6 months mOS),2,28,29 (2) adult patients with IDH-wildtype GBM (13–15 months mOS),26,30,31 and (3) children with DMG, H3.3 K27M-mutant (10–14 months mOS).10–14 Multivariate analysis shows that lack of treatment at progression had a significant impact on OS (P < .001), after accounting for age at diagnosis. Older age at diagnosis had a significant impact on PFS, even when accounting for poor KPS (KPS <60, P < .05).
Given that our cohort was pathologically confirmed, there may be an overrepresentation of tumors that were surgically accessible to biopsy and even accessible to some degree of resection (27% of patients). Furthermore, 72% of patients received upfront therapy that is standard of care for GBM (concurrent TMZ and radiation, followed by adjuvant TMZ). Of those that progressed, 83% of the patients received additional treatment, representing a potentially aggressive approach. It has been suggested that tumor location may account for better OS in adults, who have predominantly thalamic DMG, compared to children who have predominantly pontine DMG. However, this study shows that even when limiting analysis to H3 K27M-mutant DMG located in the thalamus, adult patients have significantly better survival than pediatric patients (30.4 months vs 13.0 months, P < .0004). This suggests that the difference in survival between adult and pediatric patients goes beyond location and may involve other factors including differences in concomitant genetic alterations. All these factors in concert may have potentially contributed to the longer OS demonstrated in this series.
Compared to published series of IDH-wildtype GBMs, this study suggests that H3 K27M-mutant DMG represents a tumor entity in adults with a more favorable prognosis. This corroborates findings from a recent publication comparing subgroups of midline high-grade gliomas, wherein survival of adults with H3 K27M-mutant DMG (n = 18, 17.6 months) was longer than H3-wildtype high-grade midline gliomas (n = 74, 7.7 months, P = .03).5 Improved survival in these tumors may be related to comparison of different biologic tumor entities or alternatively the concurrent genetic aberrations or poorly characterized epigenetic factors. Like other series of H3 K27M-mutant DMGs, our cohort was universally IDH-wildtype and lacked TERT promoter mutations, genetically distinguishing this tumor entity from diffuse lower-grade gliomas in adults as well as the majority of IDH-wildtype GBMs in adults.
In this series, there were frequent mutations in the p53 pathway, similar to that found in pediatric H3 K27M-mutant DMG.32 Our cohort also had frequent alterations in ATRX (38% ATRX loss by IHC and 30% mutation by NGS), which is more prevalent than the less than 1% incidence seen in IDH-wildtype GBMs in adults26 and 11% of pediatric H3 K27M-mutant DMGs.33 ATRX encodes a regulator of chromatin remodeling and transcription and directly interacts with the histone H3.3 variant at telomeres to facilitate alternative lengthening of telomeres.34 There were also frequent kinase domain hotspot mutations in FGFR1, which have been observed in pediatric H3 K27M-mutant DMGs located in the thalamus.35 In our cohort, these FGFR1 mutations were also primarily found in thalamic tumors (5 of the 11 thalamic cases studied by NGS). Three of 19 patients demonstrated mutations in the PI3-kinase pathway, including the PIK3CA catalytic subunit and the PIK3R1 negative regulatory subunit.
In this study, only 5% of tumors had CDKN2A/CDKN2B homozygous deletion, compared to approximately 60% of IDH-wildtype GBMs in adults.26 In contrast to IDH-wildtype GBMs in adults where approximately 40% of tumors harbor PTEN-inactivating mutations or homozygous deletion,26,36 this was absent in our cohort. EGFR alterations, also common in IDH-wildtype GBMs in adults, were absent in our cohort. Some recurrent molecular findings noted in pediatric gliomas were also absent from our cohort, including ACVR1 mutation (most often seen in pontine pediatric H3.1 K27M-mutant tumors12,34), MYCN amplification, MYB or MYBL1 fusion, and BRAF mutation or fusion.
This study emphasizes the importance of tissue biopsy both to confirm H3 status for its prognostic significance, and to obtain tissue for broad NGS, allowing further genetic characterization and identification of potential targets for treatment. In pediatric patients, there are a few clinical trials targeting DMG. GDC-0084, an inhibitor of PI3 kinase and mTOR, is under study in a phase I trial (Clinical Trials ID# NCT03696355). Histone deacetylase inhibitors panobinostat and fimepinostat (the latter also targets PI3 kinase) are also in phase I trials (NCT02717455 and NCT03893487). There is a phase I trial of a peptide vaccine targeting H3.3 K27M-mutant protein together with the PD-1 checkpoint inhibitor nivolumab (NCT02960230) and also a phase I trial in development using autologous chimeric antigen receptor T cells that express disialoganglioside GD2, which is overexpressed in H3 K27M-mutant DMGs.37 While this study illustrates genetic differences between adult and pediatric H3 K27M-mutant DMG, it is possible that therapies specifically targeting the H3 K27M mutation in children may be efficacious in adults. For both adult and pediatric patients, ONC201, an inhibitor of the dopamine receptor D2 that drives tumorigenic pathways including Akt and ERK, is currently undergoing phase I/II trial testing (NCT02525692). In addition to these targeted treatments, this study also identifies ATRX, NF1, and FGFR1 as potential therapeutic targets in adults with H3 K27M-mutant DMG harboring these mutations.
This retrospective analysis has limitations including a relatively small sample size and lack of a contemporaneous control cohort to compare clinical outcomes. There may be selection bias in defining the cohort by tumors that are accessible to surgical biopsy or resection and thus pathologic confirmation. Testing for H3 K27M was sometimes performed on archival tissue, generally for patients who were still alive, creating a longevity bias. In addition, some treatment and clinical follow-up information were missing for a subset of patients.
In summary, this series describes the clinical, radiologic, and genetic features of H3 K27M-mutant DMGs in adults. The patients described here lived longer compared to historical averages for IDH-wildtype GBMs in adults and H3.3 K27M-mutant DMGs in children. Thus, H3 K27M-mutant DMGs in adults may have a different prognosis and necessitate different strategies for optimal treatment compared to both their pediatric counterparts as well as other diffuse gliomas in adults. We suggest IHC and/or genetic testing for histone H3 K27M mutation in all midline gliomas in adults when feasible, as it may be important in determining patient prognosis and in guiding treatment decisions.
Supplementary Material
Acknowledgments
We thank the staff of the UCSF Clinical Cancer Genomics Laboratory for assistance with genetic profiling.
Funding
D.A.S. is supported by the NIH Director’s Early Independence Award from the Office of the Director, National Institutes of Health (DP5 OD021403). This work was supported in part by the National Cancer Institute, Specialized Programs of Research Excellence, UCSF Brain Tumor SPORE P50 CA097257 (to J.J.P.) and in part with philanthropic support from the UCSF Glioblastoma Precision Medicine Program (to A.M.M., Y.Z., S.C., D.A.S., and J.L.C.).
Conflict of interest statement. There are no conflicts of interest relevant to the enclosed work.
Authorship Statement. Data collection by J.D.S., R.A.B., S.L., and D.A.S; manuscript written by J.D.S. and R.A.B. and edited by all authors; imaging analysis by J.E.V.-M; pathological analysis by D.A.S., J.J.P., and A.P; compilation of next-generation sequencing data by D.A.S; statistical analysis by A.M.M. and Y.Z; figures created by J.D.S and Y.Z.
References
- 1. Harutyunyan AS, Krug B, Chen H, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun. 2019;10(1):1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aihara K, Mukasa A, Gotoh K, et al. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol. 2014;16(1):140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol. 2014;128(4):573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Broniscer A, Gajjar A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. Oncologist. 2004;9(2):197–206. [DOI] [PubMed] [Google Scholar]
- 5. Schreck KC, Ranjan S, Skorupan N, et al. Incidence and clinicopathologic features of H3 K27M mutations in adults with radiographically-determined midline gliomas. J Neurooncol. 2019;143(1):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solomon DA, Wood MD, Tihan T, et al. Diffuse midline gliomas with histone H3-K27M mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol. 2016;26(5):569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang L, Li Z, Zhang M, et al. H3 K27M-mutant diffuse midline gliomas in different anatomical locations. Hum Pathol. 2018;78:89–96. [DOI] [PubMed] [Google Scholar]
- 8. Larson JD, Kasper LH, Paugh BS, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019;35:140–155.e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pathania M, De Jay N, Maestro N, et al. H3.3K27M cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell. 2017;32:684–700.e689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gielen GH, Gessi M, Hammes J, Kramm CM, Waha A, Pietsch T. H3F3A K27M mutation in pediatric CNS tumors: a marker for diffuse high-grade astrocytomas. Am J Clin Pathol. 2013;139(3):345–349. [DOI] [PubMed] [Google Scholar]
- 11. Korshunov A, Ryzhova M, Hovestadt V, et al. Integrated analysis of pediatric glioblastoma reveals a subset of biologically favorable tumors with associated molecular prognostic markers. Acta Neuropathol. 2015;129(5):669–678. [DOI] [PubMed] [Google Scholar]
- 12. Mackay A, Burford A, Carvalho D, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32:520–537.e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janssens GO, Gandola L, Bolle S, et al. Survival benefit for patients with diffuse intrinsic pontine glioma (DIPG) undergoing re-irradiation at first progression: a matched-cohort analysis on behalf of the SIOP-E-HGG/DIPG working group. Eur J Cancer. 2017;73:38–47. [DOI] [PubMed] [Google Scholar]
- 14. Veldhuijzen van Zanten SEM, Baugh J, Chaney B, et al. ; Members of the SIOPE DIPG Network Development of the SIOPE DIPG network, registry and imaging repository: a collaborative effort to optimize research into a rare and lethal disease. J Neurooncol. 2017;132(2):255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 16. Feng J, Hao S, Pan C, et al. The H3.3 K27M mutation results in a poorer prognosis in brainstem gliomas than thalamic gliomas in adults. Hum Pathol. 2015;46(11):1626–1632. [DOI] [PubMed] [Google Scholar]
- 17. Mosaab A, El-Ayadi M, Khorshed EN, et al. Histone H3K27M mutation overrides histological grading in pediatric gliomas. Sci Rep. 2020;10(1):8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kline CN, Joseph NM, Grenert JP, et al. Targeted next-generation sequencing of pediatric neuro-oncology patients improves diagnosis, identifies pathogenic germline mutations, and directs targeted therapy. Neuro Oncol. 2017;19(5):699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beaubier N, Bontrager M, Huether R, et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat Biotechnol. 2019;37(11):1351–1360. [DOI] [PubMed] [Google Scholar]
- 20. Blumenthal DT, Dvir A, Lossos A, et al. Clinical utility and treatment outcome of comprehensive genomic profiling in high grade glioma patients. J Neurooncol. 2016;130(1):211–219. [DOI] [PubMed] [Google Scholar]
- 21. Hadd AG, Houghton J, Choudhary A, et al. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn. 2013;15(2):234–247. [DOI] [PubMed] [Google Scholar]
- 22. Xiu J, Piccioni D, Juarez T, et al. Multi-platform molecular profiling of a large cohort of glioblastomas reveals potential therapeutic strategies. Oncotarget. 2016;7(16):21556–21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics. 2017;14(2):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu T, Chanchotisatien A, Qin Z, et al. Imaging characteristics of adult H3 K27M-mutant gliomas. J Neurosurg. 2019;1–9. doi: 10.3171/2019.9.JNS191920. [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- 25. Aboian MS, Solomon DA, Felton E, et al. Imaging characteristics of pediatric diffuse midline gliomas with histone H3 K27M mutation. AJNR Am J Neuroradiol. 2017;38(4):795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. López G, Oberheim Bush NA, Berger MS, Perry A, Solomon DA. Diffuse non-midline glioma with H3F3A K27M mutation: a prognostic and treatment dilemma. Acta Neuropathol Commun. 2017;5(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kleinschmidt-DeMasters BK, Mulcahy Levy JM. H3 K27M-mutant gliomas in adults vs. children share similar histological features and adverse prognosis. Clin Neuropathol. 2018;37 (2018)(2):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyronet D, Esteban-Mader M, Bonnet C, et al. Characteristics of H3 K27M-mutant gliomas in adults. Neuro Oncol. 2017;19(8):1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yan H, Parsons DW, Jin G, et al. Mutations in gliomas. N Engl J Med. 2009;360:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 33. Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nandakumar P, Mansouri A, Das S. The role of ATRX in glioma biology. Front Oncol. 2017;7:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46(5):462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mount CW, Majzner RG, Sundaresh S, et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat Med. 2018;24(5):572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.