Abstract
Aims:
To investigate the feasibility of serum extra spindle pole bodies-like 1 (ESPL1) used as a biomarker for patients with hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC).
Methods:
131 chronic HBV-infection patients were recruited and divided into HBV S gene integration, non-HBV S gene integration, chronic hepatitis B (CHB), HBV-related liver cirrhosis (LC) and HBV-related HCC group, 24 non-HBV-related HCC patients were selected as HCC control group, 30 people without HBV-infection as healthy control group. Serum ESPL1 were detected and compared.
Results:
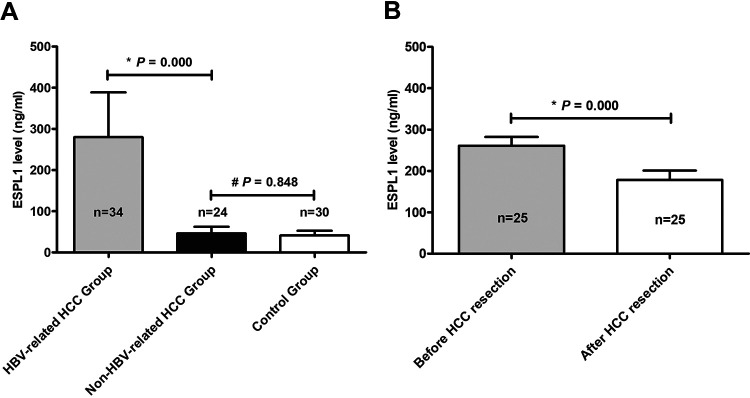
ESPL1 level of integration group was significantly higher than that of non-integration group (346.7 vs 199.6 ng/ml, P = 0.000) and healthy control group (346.7 vs 41.3 ng/ml, P = 0.000). ESPL1 level of non-integration group was significantly higher than that of healthy control group (199.6 vs 41.3 ng/ml, P = 0.000); ESPL1 levels in chronic HBV-infection related groups were increased in turn according to CHB group (95.8 ng/ml), HBV-related LC group (268.2 ng/ml), HBV-related HCC group (279.9 ng/ml) and integration group (346.7 ng/ml). Except that there was no significant difference in ESPL1 levels between HBV-related LC and HCC group (P = 0.662), pairwise comparisons between other groups showed significant differences (P < 0.05). ESPL1 level of HBV-related HCC group was significantly higher than that of non-HBV-related HCC group (279.9 vs 46.6 ng/ml, P = 0.000), there was no noticeable difference between non-HBV-related HCC and healthy control group (46.6 vs 41.3 ng/ml, P = 0.848). ESPL1 level of HBV-related HCC group after resection was significantly lower than that of before resection (178.4 vs 260.8 ng/ml, P = 0.000).
Conclusions:
Chronic HBV-infection patients with high ESPL1 level may indicate HBV S gene integration and is a high-risk population for HBV-related HCC. Serum ESPL1 can be used as a biomarker for screening HBV-related HCC high-risk population and monitoring recurrence.
Keywords: ESPL1, hepatitis B virus, integration, hepatocellular carcinoma, biomarker
Introduction
Liver cancer is the fifth most common cancer and the second most frequent cause of cancer-related death globally. Hepatocellular carcinoma (HCC) represents about 90% of primary liver cancers and constitutes a major global health problem.1,2 Approximately 90% of HCCs are associated with a known underlying etiology, most frequently chronic viral hepatitis (B and C), alcohol intake, aflatoxin exposure, liver cirrhosis, diabetes, and obesity.2-6 Previous studies have found that integrated hepatitis B virus (HBV) S gene and human gene were fused in the cancer tissues of HBV-related liver cancer to form HBV S-Human TERT, HBV S-Human UP3A and other fusion genes, which may be related to HCC occurrence.7 Recently, our team reported for the first time that Human ESPL1-HBV S fusion gene was detected in 75% of HCC tissues with HBV S gene integration. Therefore, we speculated that Human ESPL1-HBV S fusion gene may induce HCC in patients with HBV infection.8 However, detection of S gene integration needs liver biopsy, an invasive examination with certain known risk and contraindications, thus, it is not yet available for routine clinical application.9,10 As we known, ESPL1, which is expressed by human ESPL1 gene, mainly plays a role in the nucleus, is difficult to be secreted out of cells, and can’t be expressed in peripheral blood or only low expression to be detected.11-14 However, after ESPL1 gene fused with HBV S gene, how does the ESPL1 gene expression level change? Is it detectable in the peripheral blood? No relevant studies have been found so far. Based on the previous study background, on the basis of the long-term antiviral treatment cohort of chronic HBV infection, this study was designed to detect serum ESPL1 levels in patients with HBV infection-related groups, and explore the feasibility of serum ESPL1 level used as a biomarker for patients with HBV-related HCC.
Materials and Method
Patient Selection
In this study, all the chronic HBV-infection patients were selected from the long-term follow-up cohort, which established by our team since February 2002. They are all from the wards and outpatient departments of the Infectious Diseases Department, the First Affiliated Hospital of Guangxi Medical University. The inclusion criteria for each group were as follows:
HBV S gene integration group: serum hepatitis B surface antigen (HBsAg) positive over 6 months, diagnosis of CHB/LC before antiviral treatment, performed liver biopsy before treatment, and Alu-polymerase chain reaction (PCR) detection of liver tissue indicated HBV S gene integration in our previous study,15 and receiving antiviral treatment over 3 years;
Non-HBV S gene integration group: serum HBsAg positive over 6 months, diagnosis of CHB/LC before antiviral treatment, performed liver biopsy before treatment, and Alu-PCR detection of liver tissue indicated no HBV S gene integration in our previous study,15 and receiving antiviral treatment over 3 year;
Chronic hepatitis B (CHB) group: serum HBsAg positive over 6 months, CHB diagnosis meets the diagnostic criteria of the American Association for the Study of Liver Diseases (AASLD) 2018 Hepatitis B Guidance,16 receiving antiviral treatment over 7 years, and no LC and/or HCC occurred during follow-up;
HBV-related liver cirrhosis (HBV-related LC) group: serum HBsAg positive over 6 months, the treatment baseline LC diagnosis meets the diagnostic criteria of the AASLD 2018 Hepatitis B Guidance,16 receiving antiviral treatment over 1 years and no HCC occurred during follow-up;
HBV-related HCC group: serum HBsAg positive over 6 months, HCC occurred after more than 1 year of antiviral treatment, HCC diagnosis meets the diagnostic criteria of the 2018 Practice Guidance by the AASLD,17 continued regular follow-up over 6 months after HCC resection;
Healthy control group: healthy people who excluded hepatitis virus infection, other infectious diseases and malignant tumors;
Non-HBV-related HCC group: serum HBsAg detected negative, HCC diagnosis meets the diagnostic criteria of the 2018 Practice Guidance by the AASLD17;
Exclusion criteria: (1) Coinfection with other viruses such as HAV, HCV, HDV, HEV and HIV; (2) suffered from other malignant tumors during follow-up; (3) receiving nterferon therapy or immunosuppressive therapy during follow-up; (4) under 18 years old; (5) Loss of follow-up or death during follow-up.
Sample Collection and Storage
Five milliliter (5 ml) peripheral venous bloods were collected at the last follow-up period from Integration group and Non-integration group, CHB group, HBV-related LC group and non-HBV-related HCC group before HCC resection; HBV-related HCC group’s peripheral venous bloods were collected before and after HCC resection within 6 months, and the healthy control group’s sample were also obtained. After centrifuging at 3000 rpm for 15 minutes at room temperature, upper samples were kept frozen at -80˚C for ESPL1 detection. Five milligram (5 mg) HCC tissue specimens were collected by surgical resection in the HBV-related HCC and non-HBV-related HCC group.
Serum ESPL1 Levels Detection
Serum ESPL1 levels were detected by ESPL1 Enzyme-Linked Immunoassay (ELISA) Kit (Jianglai Biotechnology Co., Ltd., Shanghai, China) with a linear range of 0 -400 ng/ml following manufacturer’s protocol: 50ul of standard samples, unknown samples of each group and blank control samples were added into the 96-well plate. Except for the blank control wells, 100ul of enzyme labeling reagent was added to the remaining wells, mixed up and sealed, and then incubated at 37°C for 60 minutes. Washing the plate 5 times and patted dry. Adding 50µl of developer A and B respectively to each well, mixed up and sealed, and then incubate at 37°C in the dark for 15 minutes, finally adding 50µl of stop solution to each well to stop the reaction. The absorbance (OD value) of each sample was measured on a microplate reader (BioRad, Co., Ltd., USA) at a wavelength of 450 nm. All samples were detected with 3 replicate wells and the mean value was obtained. The concentration of serum ESPL1 in each group of unknown samples was calculated according to the standard curve. Experimental quality control required that the standard curve of every detection met R2 ≥ 0.99.
Histopathology and Immunohistochemistry
For histopathological examination, paraffin-embedded sections were stained with hematoxylin and eosin (HE) and were sealed with neutral gum. Immunohistochemical assay was performed using a Streptomyces antibiotic protein-peroxidase ligation method. In brief, 5µm tissue sections were cut from formalin-fixed, paraffin-embedded specimens. The expression of HBsAg protein was measured with UltraSensitive™ S-P Kit (Maixin, Fuzhou, Fujian, PR China). Before labeling with primary antibody, the sections were deparaffinized, antigens were restored, and the sections were incubated with hydrogen peroxide to block endogenous peroxidase. The specimen sections were incubated overnight at 4˚C with primary antibody to HBsAg (1:2000; Maixin) and then with a secondary antibody (1:2000; Maixin) conjugated to streptavidin-peroxidase. After color development with diaminobenzidine peroxidase substrate solution, the sections were counterstained with hematoxylin. For the negative control, phosphate buffered saline was used instead of primary antibody. The immunohistochemical results were examined independently by 2 pathologists. When all of the cytoplasm exhibited a brown reaction product, sections were considered positive. The sections were examined under a microscope at low power to identify evenly labeled areas. Images were acquired using a Leica DMRD optical microscope (Leica, Wetzlar, Germany) at 200× magnification.
Statistical Analysis
Statistical analysis was performed by SPSS, version 22.0. Continuous variables which normal distribution were expressed as mean ± standard deviation. Continuous variables which skewed distribution were expressed as median (P25∼P75). Categorical variables were summarized by counts and percentages. One-way Analysis of Variance (ANOVA) was used to compare the serum ESPL1 levels among multiple groups. Paired T test was applied to analyze the serum ESPL1 levels of HBV-related HCC group before resection and after resection. Statistical significance was defined as a P value of less than 0.05. All statistical tests were 2 sided.
Results
Patient Characteristics
All the patients entered in the study were Chinese. 131 cases of chronic HBV infection were recruited, 30 cases in the healthy control group, and 24 cases in the non-HBV-related HCC group. The characteristics of each group are shown in Table 1.
Table 1.
The Characteristics of Study Subjects.*
| Healthy control group | HBV S gene integration group | Non-HBV S gene integration group | CHB group | HBV-related LC group | HBV-related HCC group | Non-HBV related HCC group | |
|---|---|---|---|---|---|---|---|
| Number (n) | 30 | 20 | 25 | 27 | 25 | 34 | 24 |
| Age (years) | 47.2 ± 8.3 | 47.8 ± 7.5 | 50.8 ± 9.2 | 51.4 ± 13.1 | 49.3 ± 10.1 | 53.4 ± 9.2 | 56.9 ± 2.0 |
| Male (n, %) | 19 (63.3) | 18 (90.0) | 18 (72.0) | 24 (88.9) | 22 (88.0) | 30 (88.2) | 20 (83.3) |
| Average follow-up time (year) | / | 7.2 (6.4∼9.5) | 7.2 (5.9∼8.1) | 12.8 (11.7∼13.7) | 6.5 (4.9∼8.8) | 6.5 (3.7∼9.0) | / |
| First-line medication (n, %) | / | 18 (90.0) | 15 (60.0) | 26 (96.3) | 22 (88.0) | 30 (88.2) | / |
* Continuous variables which normal distribution were expressed as mean ± standard deviation. Continuous variables which skewed distribution were expressed as median (P25∼P75). Categorical variables were summarized by counts and percentages.
HBV, hepatitis B virus; CHB, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma; First-line medication, means treated with entecavir or tenofovir;
Imaging and Pathological Diagnosis of HBV-Related LC Group, HBV-Related HCC Group and Non-HBV-Related HCC Group
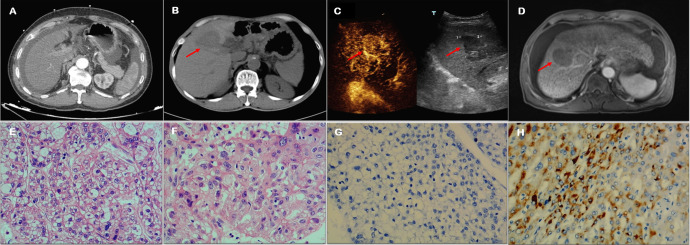
HBV-related LC group was diagnosed by multiphase computerized tomography (CT) examination (Figure 1A); HBV-related HCC and non-HBV-related HCC group were diagnosed by multiphase CT (Figure 1B), contrast-enhanced ultrasound (CEUS) (Figure 1C) or multiphase magnetic resonance imaging (MRI) (Figure 1D). The liver tissue samples from surgical resection in the HBV-related HCC group and non-HBV-related HCC group were confirmed to be HCC by HE staining (Figure 1E and F). Immunohistochemical staining of the non-HBV-related HCC group’s liver tissues showed HBsAg negative while the HBV-related HCC group positivity (Figure 1G and H).
Figure 1.
Imaging and pathological diagnosis of HBV-related LC group, HBV-related HCC group and non-HBV-related HCC group. (A) CT diagnosis of LC: A shrunken liver with rough edges, widened liver fissures, and a moderate amount of fluid in the peritoneal cavity; (B) CT diagnosis of HCC: A shrunken liver with rough outer edges, a lump with short T1 and long T2 signal lesions is seen in liver S8 (red arrow), the size is about 4.8cm × 4.2cm × 5.1cm, the internal signal is uneven; (C) CEUS diagnosis of HCC: Liver S8 has a hypoechoic mass with a size of 4.8×3.9cm (red arrow), with clear edges and regular morphology, with high enhancement in the diseased arterial phase, and low enhancement in the portal phase and delayed phase; (D) MRI diagnosis of HCC: An irregular low-density shadows are seen in liver S4 during plain scan (red arrow), the size is about 4.8cm×3.0cm×2.8cm, and the edges are not clear; (E) HE staining of non-HBVrelated HCC tissue (200×): hepatocellular carcinoma, 40% of them were solid type, 30% were coarse trabecular type and 30% were clear cell type; (F) HE staining of HBV-related HCC tissue (200×): moderately differentiated hepatocellular carcinoma, mostly clear cell type; (G) Immunohistochemical staining of non-HBV-related HCC tissue (200×): antibody to HBsAg is negative; (H) Immunohistochemical staining of HBVrelated HCC tissue (200×): antibody to HBsAg is positive.
Comparison of Serum ESPL1 level between Healthy Control Group, HBV S Gene Integration Group and Non-HBV S Gene Integration Group
The serum ESPL1 level of HBV S gene integration group was significantly higher than that of non-HBV S gene integration group (346.7 vs 199.6 ng/ml, P = 0.000) and healthy control group (346.7 vs 41.3 ng/ml, P = 0.000). The serum ESPL1 level of non-HBV S gene integration group was significantly higher than that of healthy control group (199.6 vs 41.3 ng/ml, P = 0.000) (Figure 2).
Figure 2.
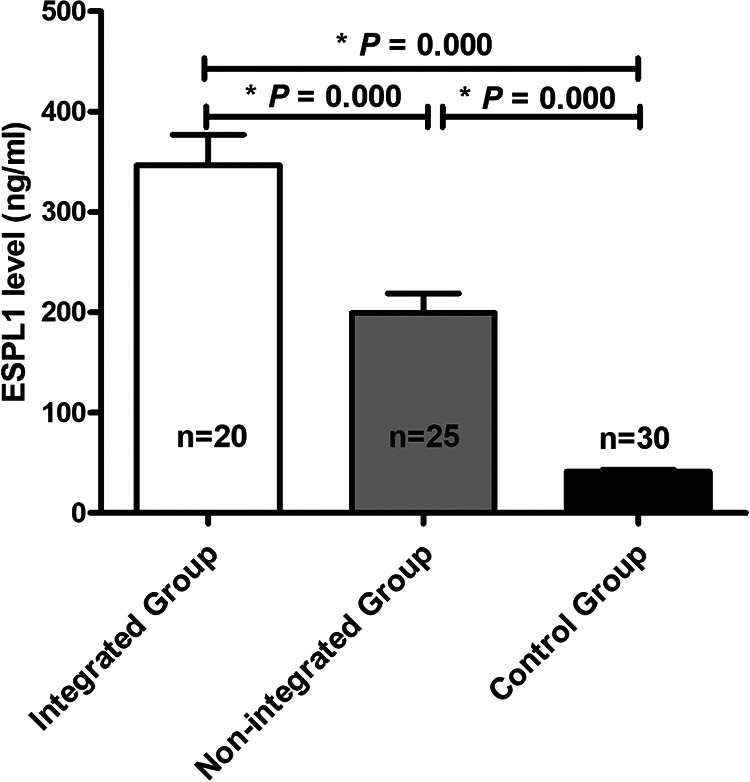
Comparison of serum ESPL1 level between healthy control group, HBV S gene integration group and non-HBV S gene integration group. One-way Analysis of Variance (ANOVA) was used to compare the serum ESPL1 levels among these groups. The serum ESPL1 level of HBV S gene integration group was significantly higher than that of non-HBV S gene integration group (346.7 vs 199.6 ng/ml, P = 0.000) and healthy control group (346.7 vs 41.3 ng/ml, P = 0.000). The serum ESPL1 level of non-HBV S gene integration group was significantly higher than that of healthy control group (199.6 vs 41.3 ng/ml, P =0.000).
Comparison of Serum ESPL1 Levels in Patients With Chronic HBV Infection
As we can see, the serum ESPL1 level of HBV S gene integration group was significantly higher than that of HBV-related HCC group (346.7 vs 279.9 ng/ml, P = 0.02), HBV-related LC group (346.7 vs 268.2 ng/ml, P = 0.011) and CHB group (346.7 vs 95.8 ng/ml, P = 0.000); The serum ESPL1 level of HBV-related HCC group was significantly higher than that of CHB group (279.9 vs 95.8 ng/ml, P = 0.000); The serum ESPL1 level of HBV-related LC group was significantly higher than that of CHB group (268.2 vs 95.8 ng/ml, P = 0.000); The HBV-related HCC group had a higher serum ESPL1 level than that of HBV-related LC group (279.9 vs 268.2 ng/ml, P = 0.662), but the difference was not statistically significant (Figure 3).
Figure 3.
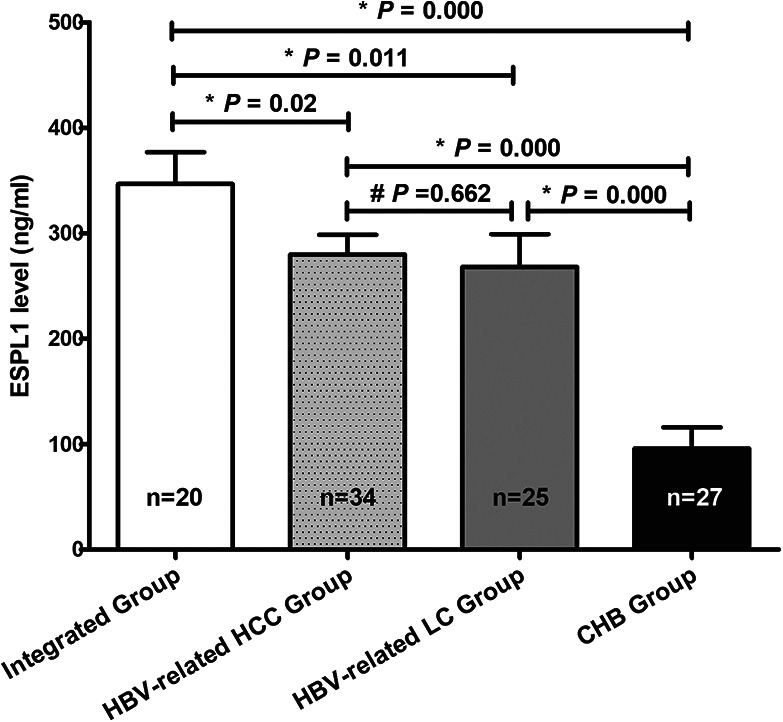
Comparison of serum ESPL1 levels in patients with chronic HBV infection. One-way Analysis of Variance (ANOVA) was used to compare the serum ESPL1 levels among these groups. The serum ESPL1 level of HBV S gene integration group was significantly higher than that of HBV-related HCC group 346.7 vs 279.9 ng/ml, P = 0.02), HBV-related LC group (346.7 vs 268.2 ng/ml, P = 0.011) and CHB group (346.7 vs 95.8 ng/ml, P = 0.000); The serum ESPL1 level of HBV-related HCC group was significantly higher than that of CHB group (279.9 vs 95.8 ng/ml, P = 0.000); The serum ESPL1 level of HBV-related LC group was significantly higher than that of CHB group (268.2 vs 95.8 ng/ml, P = 0.000); The HBV-related HCC group had a higher serum ESPL1 level than that of HBV-related LC group (279.9 vs 268.2 ng/ml, P = 0.662), but the difference was not statistically significant.
Comparison of Serum ESPL1 Levels in Healthy Control Group, Non-HBV-Related HCC Group and HBV-Related HCC Group Before and After Surgical Resection
The serum ESPL1 level of HBV-related HCC group was significantly higher than that of non-HBV-related HCC group (279.9 vs 46.6 ng/ml, P = 0.000), while there was no noticeable difference between non-HBV-related HCC group and healthy control group (46.6 vs 41.3 ng/ml, P = 0.848). Among the 34 patients in HBV-related HCC group, 25 patients had preserved serums before and after the HCC surgical resection. Compared the ESPL1 levels by paired t test, the serum ESPL1 level of HBV-related HCC group after resection was significantly lower than that of before resection (178.4 vs 260.8 ng / ml, P = 0.000) (Figure 4). It is worth noting that in the 3 (12%) HBV-related HCC patients, the serum ESPL1 level did not decrease within 6 months after surgical resection, and as a result, they all suffered HCC recurrence within 1 year after resection.
Figure 4.
Comparison of serum ESPL1 levels in healthy control group, non-HBV-related HCC group and HBVrelated HCC group before and after surgical resection. One-way Analysis of Variance (ANOVA) was used to compare the serum ESPL1 levels among these groups. The serum ESPL1 level of HBV-related HCC group was sgnificantly higher than that of non-HBV-related HCC group (279.9 vs 46.6 ng/ml, P = 0.000), while there was no noticeable difference between non-HBV-related HCC group and healthy control group (46.6 vs 41.3 ng/ml, P = 0.848). Compared the ESPL1 levels by paired t test, the serum ESPL1 level of HBV-related HCC group after resection was significantly lower than that of before resection (178.4 vs 260.8 ng / ml, = 0.000).
Discussion
The evidence linking HBV with HCC is unquestioned.18 Antiviral therapy can delay the progression of CHB and reduce the incidence of HBV-related HCC.19,20 However, even after long-term effective antiviral treatment, some patients still inevitably develop into HCC.21 The mechanism of HCC caused by chronic HBV infection is complex. Previous studies have found that HBV integration can be detected in 85-90% of HBV-related HCC tissues, which suggested that HBV integration may cause HCC.22-24 The occurrence of viral gene integrated into host gene is an early-to-mid event of viral infection, gene fusion occurs on the basis of gene integration. One of our previous studies has shown that the occurrence of HBV S gene integration was not related to factors such as gender, age, family history of HBV infection, ALT level, HBeAg status, genotype and antiviral therapy.15 In the integration group of this study, HBV S gene integration had occurred in the liver tissues before receiving antiviral treatment. These patients were followed up for an average of more than 7.2 years after antiviral treatment. It was found that 50% (10/20) of these patients still developed into HBV-related HCC. It can be seen that the existing antiviral drugs cannot act on the integrated gene, the different potency of antiviral therapies cannot affect the formation of integrated gene or even fusion gene, and the ESPL1 gene expression is still active even after long-term effective antiviral therapy.
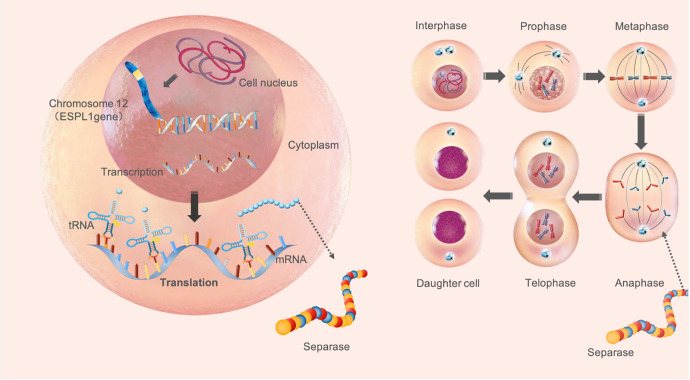
Human ESPL1 gene, also known as SEPA gene, exists at the human chromosome 12q13.13, with a total length of 6662 bp, is a protein coding gene for ESPL1, which also known as separase protein, whose main function is to catalyze fibronectin decomposition at mitosis anaphase in order to separate sister chromatids during cell division.11,12,25,26 In general, ESPL1 is mainly effective in the nucleus, which is difficult to be secreted out of the cell, the ESPL1 level in the peripheral blood is not high11-14 (Figure 5). However, Studies found ESPL1 levels is generally high in malignant tumor tissues such as liver cancer, breast cancer, osteosarcoma, prostate cancer, which may be due to the long-term effect of carcinogenic factors such as viral infection, chemical carcinogens, radiation and so on.27-30 Our previous study found a high detection rate of Human ESPL1-HBV S fusion gene in patients with HBV-related HCC,8 but ESPL1, which is highly expressed in the HCC tissue, whether can be detected in peripheral blood, it has not been reported yet. In this study, serum ESPL1 levels were measured in different groups. The results showed that serum ESPL1 levels in the healthy control group were significantly lower than those in HBV S gene integration group (41.3 vs 346.7 ng/ml, P = 0.000) and non-HBV S gene integration group (41.3 vs 199.6 ng/ml, P = 0.000), which further confirmed that ESPL1 is not expressed or low expressed in serum without HBV infection. Once HBV infection, especially after HBV S gene fused with human ESPL1 gene to form Human ESPL1-HBV S fusion gene, which may change the secretion characteristics of the ESPL1 gene expression product and makes it can be detected in peripheral blood.
Figure 5.
Mechanism of ESPL1 gene in normal cell cycle. Human ESPL1 gene, also known as SEPA gene, exists at the human chromosome 12q13.13, with a total length of 6662 bp, is a protein coding gene for ESPL1, which also known as separase protein, whose main function is to catalyze fibronectin ecomposition at mitosis anaphase in order to separate sister chromatids during cell division.
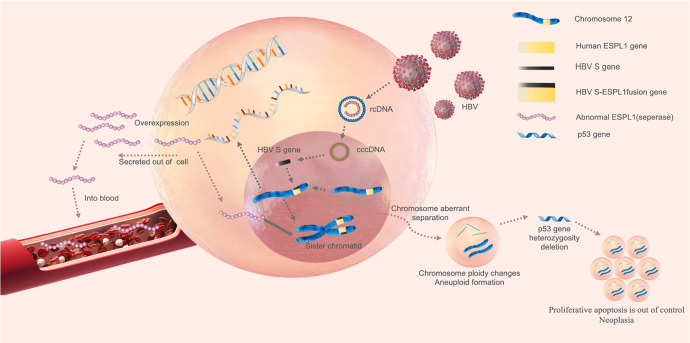
Chronic HBV infection usually follows the development of CHB, LC and HCC. This study found that serum ESPL1 levels in patients with chronic HBV infection showed a trend from quantitative to qualitative changes consistent with the development of disease progression. Serum ESPL1 levels increased in turn according to CHB group (95.8 ng/ml), HBV-related LC group (268.2 ng/ml), HBV-related HCC group (279.9 ng/ml) and HBV S gene integration group (346.7 ng/ml). It is suggested that the activity of ESPL1 gene may be consistent with the progression of HBV infection. The more active the expression of ESPL1 gene, the higher the risk of progression towards HCC. The mechanism of ESPL1 gene over-expression causing HCC may be due to genetic mutations (gene fusion, deletion, etc.) resulting in the inherent dysfunction of ESPL1. As a result, ESPL1 is unable to play a role in the anaphase of cell division, leading to abnormal sister chromatids separation and aneuploidy accumulation, which causes the heterogeneity of tumor suppression gene p53 missing and loss of heterozygosity, normal cells appear out of control of proliferative apoptosis, which may eventually lead to the formation of HCC31-34 (Figure 6). Human ESPL1-HBV S fusion gene may also have a synergistic effect with protooncogenes such as c-myc and further promote the occurrence and development of HCC.35 It is speculated that in patients with chronic HBV infection with high serum ESPL1 levels have a high probability of fusion of HBV S gene and ESPL1 gene, which are the high-risk population of HCC; chronic HBV infection patients with low serum ESPL1 level, there is a low probability of fusion of HBV S gene and ESPL1 gene, they are the low-risk population of HCC. In this study, serum ESPL1 level in CHB group was low, and no HCC occurred during an average of more than 12.8 years of follow-up, which further confirmed our hypothesis. It is worth noting that the serum ESPL1 level in HBV-related LC group also showed a high level, which was close to that of HBV-related HCC group (268.2 vs 279.9 ng / ml, P = 0.662). Of the 20 patients in the integration group in this study, 6 were diagnosed with HBV-related LC before antiviral therapy. We found the HBV S-Human ESPL1 fusion gene in the liver tissues of these patients, and also detected high expression of serum ESPL1. It indicates that ESPL1 gene is also actively expressed in HBV-related LC patients, which is a precancerous lesion of HCC and is consistent with the clinical outcome of CHB, that is, most HCC patients have a LC background. Whether the active expression of ESPL1 gene not only has a carcinogenic effect, but also initiates the related mechanism of liver fibrosis remains to be further studied.
Figure 6.
The mechanism of ESPL1 gene overexpression causing HCC. Hepatocyte is infected by hepatitis B virus (HBV), extracellular HBV DNA is a relaxed circular double-stranded DNA (relaxed circular DNA, rcDNA). During the replication of HBV, viral DNA enters the nucleus of the host cell, and under the action of DNA polymerase, the gaps of the 2 strands are filled to form a supercoiled covalently closed circular DNA (cccDNA). Under the influence of some carcinogenic factors, HBV S gene can fuse with human ESPL1 gene to form Human ESPL1-HBV S fusion gene, which resulting in the inherent dysfunction of ESPL1 gene, its expression product ESPL1 will be overexpressed and secreted into the blood to be detectable. On the other hand, Overexpressed ESPL1 is unable to play a role in the anaphase of cell division, leading to abnormal sister chromatids separation and aneuploidy ccumulation, which causes genetic instability and makes the heterogeneity of tumor suppression gene p53 missing and loss of heterozygosity, normal cells appear to be out of control of proliferative poptosis, which may eventually lead to the formation of hepatocellular carcinoma.
Previous studies on ESPL1 gene expression were mostly carried out on tumor tissues or animal models. Studies on the relationship between ESPL1 and HCC was also limited to the expression in HCC tissues.31,36 There were few reports on the relationship between serum ESPL1 level and HCC monitoring. The focus of this study is on HBV-related HCC. Of the 20 patients in the integration group in this study, the fusion gene had been detected in their liver tissue before antiviral treatment. After starting antiviral treatment, we followed up these patients for an average of more than 7.2 years, and found that 50% (10/20) of them eventually occurred HBV-related HCC during antiviral treatment. We collected their last follow-up serum of these patients for testing and found the serum ESPL1 was highly expressed. In order to confirm the abnormal expression of ESPL1 gene in HBV-related HCC patients is affected by HBV infection, we added a group of patients with non-HBV-related HCC for comparison. The result showed that serum ESPL1 level of non-HBV-related HCC group was not high, which was close to the healthy control group (46.6 vs 41.3 ng/ml, P = 0.848). Of the 24 patients in the non-HBV-related HCC group, 6 of them was infected by HCV, while their serum ESPL1 level was not high either. Above results suggested that it is not a common pathway for all HCC patients that ESPL1 gene expresses abnormal. Maybe because of HBV infection forms HBV S-Human ESPL1 fusion gene, which changes the secretion characteristics of ESPL1, the increase in serum ESPL1 level is HBV-related HCC specific. Moreover, serum ESPL1 levels were compared in 25 patients before and after resection in the HBV-related HCC group, the serum ESPL1 levels after tumor resection were significantly lower than those before resection (178.4 vs 260.8 ng/ml, P = 000), which may be associated with the active ESPL1 gene expression in HCC tissue before resection. After HCC resection, ESPL1 gene expression activity decreased or silenced, and serum ESPL1 levels also decreased. It is worth noting that 3 cases (12%) of patients after resection of serum ESPL1 levels did not decrease, still maintain a high level, results in this part of patients occurred HCC recurrence within 1 year after resection. It can be seen that serum ESPL1 can be used as a biomarker to monitor the recurrence of HBV-related HCC.
In conclusion, this study suggests that chronic HBV-infection patients with a high ESPL1 level may indicate HBV S gene integration and is a high-risk population for HBV-related HCC. Serum ESPL1 can be used as a biomarker for screening HBV-related HCC high-risk population and monitoring recurrence. The results of this study provide a new direction for the development of anti-tumor drugs targeting HBV integrated gene. In the next step, we will continue to expand clinical samples and longitudinally monitor the serum ESPL1 levels in patients with chronic HBV infection to further reveal the relationship between abnormal expression of ESPL1 and clinical outcomes.
Acknowledgments
We gratefully acknowledge our colleagues in Department of Hepatobiliary Surgery and Department of Pathology for their help during the study process.
Authors’ Note: Rongming Wang and Weiwei Zang contributed equally as joint first authors. All patients gave their written informed consent prior to their inclusion in the study. This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (approval number: 2019-KYE-193).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from National Natural Science Foundation of China (No.81960115), National Major Science and Technology Project of China (No.2017ZX10202201), the Key Laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (No. GKE2018-05, GKE2019-04) and Youth Science Foundation of Guangxi Medical University (No.GXMUYSF201910).
ORCID iD: Jianning Jiang  https://orcid.org/0000-0001-8961-417X
https://orcid.org/0000-0001-8961-417X
References
- 1. European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi:10.1016/j.jhep.2018.03.019 [DOI] [PubMed] [Google Scholar]
- 2. Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi:10.1001/jamaoncol.2017.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim MN, Kim SU, Kim BK, et al. Increased risk of hepatocellular carcinoma in chronic hepatitis B patients with transient elastography-defined subclinical cirrhosis. Hepatology. 2015;61(6):1851–1859. doi:10.1002/hep.27735 [DOI] [PubMed] [Google Scholar]
- 4. Sangiovanni A, Prati GM, Fasani P, et al. The natural history of compensated cirrhosis due to hepatitis C virus: a 17-year cohort study of 214 patients. Hepatology. 2006;43(6):1303–1310. doi:10.1002/hep.21176 [DOI] [PubMed] [Google Scholar]
- 5. Yu MW, Lin CL, Liu CJ, Yang SH, Tseng YL, Wu CF. Influence of metabolic risk factors on risk of hepatocellular carcinoma and liver-related death in men with chronic hepatitis B: a large cohort study. Gastroenterology. 2017;153(4):1006–1017.e5 doi:10.1053/j.gastro.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 6. Dyson J, Jaques B, Chattopadyhay D, et al. Hepatocellular cancer: the impact of obesity, type 2 diabetes and a multidisciplinary team. J Hepatol. 2014;60(1):110–117. doi:10.1016/j.jhep.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 7. Lau CC, Sun T, Ching AK, et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25(3):335–349. doi:10.1016/j.ccr.2014.01.030 [DOI] [PubMed] [Google Scholar]
- 8. Hu B, Huang W, Wang R, et al. High rate of detection of human espl1-hbv s fusion gene in patients with HBV-related liver cancer: a Chinese case-control study. Anticancer Res. 2020;40(1):245–252. doi:10.21873/anticanres.13946 [DOI] [PubMed] [Google Scholar]
- 9. Papatheodoridis GV, Manolakopoulos S. EASL clinical practice guidelines on the management of chronic hepatitis B: the need for liver biopsy. J Hepatol. 2009;51(1):226–227. doi:10.1016/j.jhep.2009.02.017 [DOI] [PubMed] [Google Scholar]
- 10. Cholongitas E, Senzolo M, Standish R, et al. A systematic review of the quality of liver biopsy specimens. Am J Clin Pathol. 2006;125(5):710–721. doi:10.1309/W3XC-NT4H-KFBN-2G0B [DOI] [PubMed] [Google Scholar]
- 11. Morgan DO. The Cell Cycle: Principles of Control. Primers in Biology: New Science; 2010. [Google Scholar]
- 12. Lin Z, Luo X, Yu H. Structural basis of Cohesin cleavage by Separase. Nature. 2016;532(7597):131–134. doi:10.1038/nature17402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hellmuth S, Böttger F, Pan C, Mann M, Stemmann O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 2014;33(10):1134–1147. doi:10.1002/embj.201488098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar R. Separase: function beyond cohesion cleavage and an emerging oncogene. J Cell Biochem. 2017;118(6):1283–1299. doi:10.1002/jcb.25835 [DOI] [PubMed] [Google Scholar]
- 15. Hu B, Wang R, Fu J, et al. Integration of hepatitis B virus S gene impacts on hepatitis B surface antigen levels in patients with antiviral therapy. J Gastroenterol Hepatol. 2018;33(7):1389–1396. doi:10.1111/jgh.14075 [DOI] [PubMed] [Google Scholar]
- 16. Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67(4):1560–1599. doi:10.1002/hep.29800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–750. doi:10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 18. Bréchot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127(5):S56–S61. doi:10.1053/j.gastro.2004. 09.016 [DOI] [PubMed] [Google Scholar]
- 19. Jiang JN, Li SH, Su MH, et al. The clinical outcomes of the CHB and LC patients treated with long term nucleos(t)ide analogs under whole-course: a real-life cohort study. J. Hepatol. 2014; 60:S428 doi:10.1016/S0168-8278(14)61215-0 [Google Scholar]
- 20. Dandri M, Petersen J. Latest developments in the treatment of hepatitis B. Minerva Gastroenterologica e Dietologica. 2015;62(1):88–102. PMID: 26448309. [PubMed] [Google Scholar]
- 21. Zang WW, Su M, Ling X, et al. Risk factors of liver cancer in patients with hepatitis B-related cirrhosis treated with long-term nucleos(t)ide analogues. Chinese J Hepatol. 2020, 28(08):679–685. doi:10.3760/ cma.j.cn501113-20200228-00074 [DOI] [PubMed] [Google Scholar]
- 22. Li X, Zhang J, Yang Z, et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. J Hepatol. 2014;60(5):975–984. doi:10.1016/j.jhep.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 23. Kawai-Kitahata F, Asahina Y, Tanaka S, et al. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J Gastroenterol. 2016;51(5):473–486. doi:10.1007/s00535-015-1126-4. [DOI] [PubMed] [Google Scholar]
- 24. Amaddeo G, Cao Q, Ladeiro Y, et al. Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut. 2015;64(5):820–829. doi:10.1136/gutjnl-2013-306228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wirth KG, Wutz G, Kudo NR, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172(6):847–860. doi:10.1083/jcb.200506119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haaß W, Kleiner H, Müller MC, Hofmann WK, Fabarius A, Seifarth W. Measurement of separase proteolytic activity in single living cells by a fluorogenic flow cytometry assay. PLoS One. 2015;10(8):e0133769 doi:10.1371/journal.pone.0133769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyer R, Fofanov V, Panigrahi A, Merchant F, Zhang N, Pati D. Overexpression and mislocalization of the chromosomal segregation protein separase in multiple human cancers. Clin Cancer Res. 2009;15(8):2703–2710. doi:10.1158/1078-0432.CCR-08-2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukherjee M, Byrd T, Brawley VS, et al. Overexpression and constitutive nuclear localization of cohesin protease separase protein correlates with high incidence of relapse and reduced overall survival in glioblastoma multiforme. J Neurooncol. 2014;119(1):27–35. doi:10.1007/s11060-014-1458-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Finetti P, Guille A, Adelaide J, Birnbaum D, Chaffanet M, Bertucci F. ESPL1 is a candidate oncogene of luminal B breast cancers. Breast Cancer Res Treat. 2014;147(1):51–59. doi:10.1007/s10549-014-3070-z [DOI] [PubMed] [Google Scholar]
- 30. Zhang N, Ge G, Meyer R, et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci U S A. 2008;105(35):13033–13038. doi:10.1073/pnas.0801610105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mukherjee M, Ge G, Zhang N, et al. Separase loss of function cooperates with the loss of p53 in the initiation and progression of T- and B-cell lymphoma, leukemia and aneuploidy in mice. PLoS One. 2011;6(7):e22167 doi:10.1371/journal.pone.0022167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baum P, Yip C, Goetsch L, Byers B. A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol. 1988;8(12):5386–5397. doi:10.1128/mcb.8.12.5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17(3):344–354. doi:10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uzawa S, Samejima I, Hirano T, Tanaka K, Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990;62(5):913–925. doi:10.1016/0092-8674(90)90266-h [DOI] [PubMed] [Google Scholar]
- 35. Prinzhorn W, Stehle M, Kleiner H, et al. c-MYB is a transcriptional regulator of ESPL1/separase in BCR-ABL-positive chronic myeloid leukemia. Biomark Res. 2016;4:5 doi:10.1186/s40364-016-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holland AJ, Taylor SS. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J Cell Sci. 2006;119(16):3325–3336. doi:10.1242/jcs.03083 [DOI] [PubMed] [Google Scholar]