Abstract
Objective
To identify serum microRNAs (miRNAs) as potential non-invasive biomarkers for patients with chronic kidney disease (CKD).
Methods
We collected serum samples from healthy controls, CKD stage 1 (CKD1), and stage 5 (CKD5) patients with primary glomerulonephritis (GN), screened differentially expressed miRNAs (DEMs) using next-generation sequencing (NGS), and confirmed the sequencing data using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
Results
We identified 20 and 42 DEMs in the CKD1 and CKD5 patients compared with the controls, respectively, and 70 DEMs in the CKD5 compared with the CKD1 patients. The qRT-PCR results showed that miR-483-5p was up-regulated in the CKD1 and CKD5 patients compared with controls (fold change = 2.56 and 18.77, respectively). miR-363-3p was down-regulated in the CKD5 patients compared with the controls and CKD1 patients (fold change = 0.27 and 0.48, respectively).
Conclusion
We identified a genome-wide serum miRNA expression profile in CKD patients, and serum miR-483-5p and miR-363-3p may act as potential diagnostic biomarkers for CKD.
Keywords: Chronic kidney disease, microRNA, biomarker, next-generation sequencing, noninvasive, primary glomerulonephritis
Introduction
Chronic kidney disease (CKD) is a significant public health issue because of its increasing prevalence, poor outcome, and economic burden on global health care resources. It is estimated that approximately 11% to 12% of the general population has CKD1 and its morbidity and mortality are increasing at an annual growth rate of 8%.2 Approximately 1.2 million people died of CKD worldwide in 2015.3 In China, the overall prevalence of CKD was 10.8%,4 and 155,390 people died of CKD in 2013.5
CKD is a progressive disease, and it is classified into five stages on the basis of the declining estimated glomerular filtration rate (eGFR): stage 1, ≥90 mL/minute/1.73 m2; stage 2, 60 to 89 mL/minute/1.73 m2; stage 3, 30 to 59 mL/minute/1.73 m2; stage 4, 15 to 29 mL/minute/1.73 m2; and stage 5 (end-stage renal disease, ESRD), < 15 mL/minute/1.73 m2. Early diagnosis of CKD is essential to slow the progression of kidney failure and reduce complications and mortality rates.1 Symptoms are usually not apparent in the early stages of CKD, and serum creatinine (sCr)-based eGFR has been shown to lack a reliable predictive value in early CKD stages.1 In addition, kidney biopsy, which is the gold standard for diagnosis of intrinsic renal disease currently, is an invasive procedure with the inherent risks of hemorrhage and infection. Thus, identification of novel non-traumatic biomarkers is critical for early CKD diagnosis and to determine the prognosis of CKD patients.
MicroRNAs (miRNAs) are a class of approximately 22 nucleotide non-coding RNAs that regulate post-transcriptional gene expression by binding to the 3ʹ untranslated region (UTR) of specific mRNA targets, and they are involved in a wide range of biological processes, such as cell proliferation, differentiation, apoptosis, and metabolism.6 An increasing amount of evidence indicates that miRNAs are present ubiquitously in serum, plasma, urine, and other human body fluids, which are known as circulating miRNAs.7 Owing to their noninvasive or minimally invasive nature, high stability, and resistance to storage handling (i.e. the miRNA does not degrade easily when stored and processed),7,8 serum- and urine-circulating miRNAs have been extensively investigated as novel and noninvasive biomarkers for diagnosis and prognosis of various human diseases.9 Currently, some studies have shown the role of urinary miRNAs in the diagnosis and prognosis of renal diseases.10–12 However, diagnosis data concerning serum miRNAs in CKD patients are still limited.
Accurate quantification of serum miRNAs presents several challenges because of their low abundance and small size. Most studies of serum miRNA quantification have been performed using quantitative reverse transcription polymerase chain reaction (qRT-PCR) or microarray. These methods have several strengths, but also contain some limitations, including the number of miRNAs that can be analyzed simultaneously, the sensitivity and dynamic range of detection, and reliance upon existing knowledge of targets.13 Fortunately, because of the advent and rapid advances of next-generation sequencing (NGS), which is also known as RNA sequencing (RNA-seq), there is a reliable and powerful tool to identify global miRNAs that are associated with diseases due to some methodological advantages such as higher throughput, less RNA input, increased detection depth, reduced background noise, greater reproducibility, and discovery of novel miRNAs.13,14
In this study, we collected serum samples from healthy controls and CKD stage 1 (CKD1) and CKD stage 5 (CKD5) patients with primary glomerulonephritis (GN), which is the leading cause of CKD in China.15 A NGS platform (Solexa sequencing) and qRT-PCR methods were used to identify differentially expressed miRNAs (DEMs) in serum as non-invasive diagnostic biomarkers for CKD.
Materials and methods
Ethics statement
The present study was approved by the Institutional Review Board of the Fourth Hospital of Hebei Medical University (Shijiazhuang, China). Written informed consent was obtained from all participants who were involved in this study.
Patients and sample collection
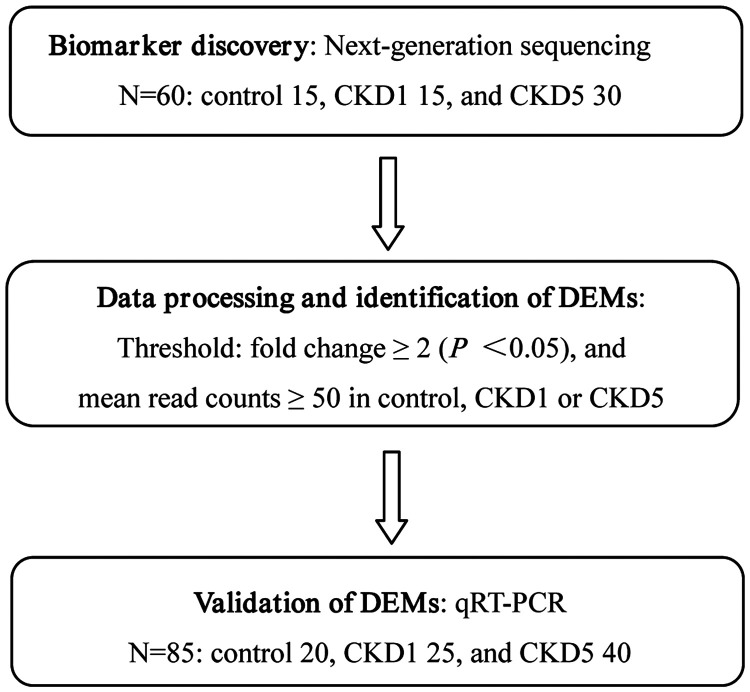
A flow diagram of this study design is presented in Figure 1. We collected serum samples from 40 patients with CKD1, 70 patients with CKD5 before hemodialysis, and 35 healthy controls at the Fourth Hospital of Hebei Medical University (Shijiazhuang, China) from March 2016 to August 2017. The patients were diagnosed with primary GN by renal biopsy or clinical history. The exclusion criteria were as follows: acute infection, and concurrent cancer, heart, brain, liver, and systemic diseases such as diabetes mellitus. Healthy controls did not have any known kidney disease history, and the same exclusion criteria applied. Written informed consent was obtained from all participants who were involved in this study.
Figure 1.
Flow chart of the study design. NGS was performed for biomarker discovery, and DEMs were validated using qRT-PCR.
NGS, next-generation sequencing; DEMs, differentially expressed miRNAs; miRNAs, microRNAs; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Demographic data from the CKD1 and CKD5 patients and healthy controls are summarized in Table 1. All the blood samples were drawn within 24 hours after admission. The coagulated blood samples were collected in tubes and centrifuged at 1500 ×g for 15 minutes at 4°C. The supernatant serum was stored at −80°C until use.
Table 1.
The demographic data from CKD1 and CKD5 patients and healthy controls.
| Variable | CKD1 patients | CKD5 patients | Healthy controls |
|---|---|---|---|
| Biomarker discovery set | |||
| Number | 15 | 30 | 15 |
| Age (years, mean ± SD) | 51.40±10.12 | 53.80±12.75 | 52.40±7.75 |
| Gender (M/F) | 9/6 | 18/12 | 9/6 |
| CRP (mg/L) | 4.56±2.78 | 20.52±16.96 | 2.82±1.89 |
| eGFR (mL/minute/1.73 m2) | 106.32±10.24 | 11.71±2.04 | — |
| Biomarker validation set | |||
| Number | 25 | 40 | 20 |
| Age (years, mean ± SD) | 51.16±9.75 | 50.70±14.75 | 49.39±11.10 |
| Gender (M/F) | 16/9 | 25/15 | 12/8 |
| CRP (mg/L) | 5.27±4.66 | 18.58±17.81 | 3.22±2.07 |
| eGFR (mL/minute/1.73 m2) | 104.87±12.31 | 12.23±2.22 | — |
CKD, chronic kidney disease; SD, standard deviation; M, male; F, female; CRP, C-reactive protein; eGFR, estimated glomerular filtration rate; —, no data.
NGS and bioinformatics analysis
In the initial biomarker discovery stage, we analyzed serum samples from 15 CKD1 individuals, 30 CKD5 individuals, and 15 age- and gender-matched healthy controls (Table 1). Among the 15 serum samples from CKD1 patients, every five samples from five individuals (gender: male/female = 3/2) were pooled together in an equal volume (0.2 mL each) as one replicate to reduce individual variation, resulting in three replicates for CKD1. The CKD5 samples were treated in the same manner as those of CKD1. Total RNA from each pooled sample was isolated using the RNeasy Mini Kit (Qiagen, Shanghai, China), in accordance with the manufacturer’s protocol. RNA quality and quantity were measured using a Nanodrop spectrophotometer (ND-1000, Nanodrop Technologies, Wilmington, DE, USA), and RNA integrity was assessed by gel electrophoresis. Then, the RNA was used to prepare the miRNA sequencing library, which included the following steps: 3ʹ-adaptor ligation, 5ʹ-adaptor ligation, cDNA synthesis, PCR amplification, and size selection of approximately 135 to 155 bp PCR amplified fragments (corresponding to approximately 15 to 35 nt small RNAs). The libraries were denatured as single-stranded DNA molecules, which were captured on Illumina flow cells, amplified in in situ as clusters, and finally sequenced for 50 cycles on Illumina NextSeq (Illumina Inc, San Diego, CA, USA), per the manufacturer’s instructions.
After sequencing, the Solexa CHASTITY quality filtered reads were harvested as Clean Reads. The adaptor sequences were trimmed and the adaptor-trimmed reads (≥15 nt) were left. The trimmed reads were aligned to merged pre-miRNA databases (known pre-miRNA from miRBase v21 plus the newly predicted pre-miRNAs) using Novoalign software (v2.07.11; Novocraft Technologies, Selangor, Malaysia) with at most one mismatch. Reads (counts < 2) were discarded when calculating the miRNA expression. To characterize the isomiR variability, sequences that matched the miRNA precursors in the mature miRNAs region ± 4 nt (no more than one mismatch) were accepted as mature miRNA isomiRs, which were grouped according to the 5ʹ (5p) or 3ʹ (3p) arm of the precursor hairpin. The numbers of mapped tags were defined as the raw expression levels of that miRNA. To correct for the difference in tag counts among different samples, the tag counts were scaled to TPM (the copy number of transcripts per million) based on the total number of tags that were aligned. Choosing a different isomiR sequence for measuring miRNA expression can affect the ability to detect differential miRNA expression. We used the most abundant isomiR, mature miRNA annotated in miRbase, and all miRNA isoforms (5p or 3p) to calculate the miRNA expression. When comparing the DEM profiles between two groups, the fold change and P-values were calculated and used to identify significant DEMs (based on ALL_Isoform value).
DEMs between two groups were filtered through miRNA fold-change and mean read counts. Hierarchical clustering was performed based on DEMs. miRNA target prediction was performed by TargetScan (http://www.targetscan.org/vert_71/) and mirdbV5 (http://mirdb.org/miRDB/), and the overlapping results of two databases were generally accepted. All of the above-mentioned procedures were performed by KangChen Bio-tech (Shanghai, China).
miRNA quantification by qRT-PCR
Among the DEMs, miR-483-5p and miR-363-3p were selected to be further validated in serum samples from 25 CKD1, 40 CKD5 patients, and 20 healthy controls using qRT-PCR. We analyzed the predicted target genes related to kidney diseases for miR-483-5p and miR-363-3p using the GeneCards Database (https://www.genecards.org/). Because of the extremely low miRNA concentration in serum, developing a proper method to enrich the concentration and for normalization is challenging. In this study, we extracted total RNA from each individual serum sample using a miRcute serum/plasma miRNA isolation kit (Tiangen Biotech, Beijing, China) in accordance with the manufacturer’s instructions. Because U6 and 5S rRNA are degraded in serum, and there is no current consensus on the use of housekeeping miRNAs for qRT-PCR analysis.8 Therefore, synthetic Caenorhabditis elegans miR-39 (cel-miR-39) was spiked into all samples before RNA extraction as an exogenous reference that has been recommended previously.16 The mature miRNA was transcribed into first-strand cDNA using a miRcute Plus miRNA first-strand cDNA synthesis kit (Tiangen Biotech, Beijing, China), followed by qRT-PCR with the SYBR Green miRcute Plus miRNA qPCR Detection Kit (Tiangen Biotech, Beijing, China). Specific forward primers of cel-miR-39 (Cat. No. CR100-01), miR-483-5p (Cat. No. CD201-0181), and miR-363-3p (Cat. No. CD201-0106) were purchased from Tiangen Biotech (Beijing, China). Amplification was conducted under the following conditions: 95°C for 15 minutes, followed by 40 cycles of 94°C for 20 s, and 60°C for 34 s. All reactions were performed in triplicate. The relative miRNA expression levels compared with cel-miR-39 were calculated using the 2−ΔΔCt method.17 Data are presented as the mean ± standard error of mean (SEM). Statistical analysis was performed using the Statistical Package for Social Sciences (SPSS) 22.0 for Windows (IBM Corp., Armonk, NY, USA). miRNA expression differences between different groups were analyzed using a one-way analysis of variance (ANOVA) test, and statistical significance was considered when P < 0.05.
Results
Overall, there were 145 participants enrolled into this study. In the initial biomarker discovery stage, 60 participants were enrolled into the following groups: CKD1 (n = 15); CKD5 (n = 30); and control (n = 15). There were 36 men and 24 women, with nine men and six women in the CKD1 group, 18 men and 12 women in the CKD5 group, and nine men and six women in the control group. The average age of the participants in the CKD1, CKD5, and control groups was 51.40 ± 10.12 years, 53.80 ± 12.75 years, and 52.40 ± 7.75 years, respectively. In the following biomarker validation stage, 85 participants were enrolled: CKD1 (n = 25); CKD5 (n = 40); and control (n = 20). There were 53 men and 32 women, with 16 men and nine women in the CKD1 group, 25 men and 15 women in the CKD5 group, and 12 men and eight women in the control group. The average age of participants in the CKD1, CKD5, and control groups was 51.16 ± 9.75 years, 50.70 ± 14.75 years, and 49.39 ± 11.10 years, respectively (Table 1).
miRNA expression analysis by NGS
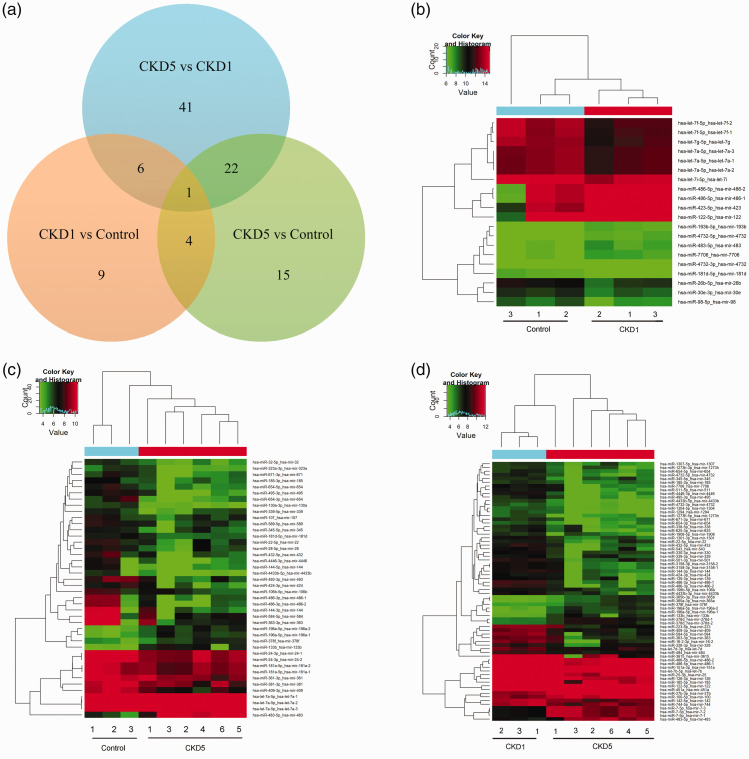
To select serum miRNA biomarkers for CKD patients, we first performed initial genome-wide miRNA expression profiling of serum samples that were derived from CKD1 or CKD5 patients and control subjects using Solexa sequencing. In this study, significant DEMs were identified as those with a fold change ≥2.0 (P < 0.05) and mean read counts ≥50. Compared with the control group, the CKD1 and CKD5 groups displayed 20 DEMs (nine up-regulated, 11 down-regulated) and 42 DEMs (five up-regulated, 37 down-regulated), respectively, and 70 dysregulated miRNAs (14 up-regulated and 56 down-regulated) were also observed in the CKD5 group compared with the CKD1 group. The expression patterns of the DEMs in the pairwise comparisons among the control, CKD1, and CKD5 groups are shown in a Venn diagram and hierarchical clustering heatmaps (Figure 2). The fold changes of all the DEMs are listed in Tables 2 to 4.
Figure 2.
DEM expression patterns in the pairwise comparisons among the control, CKD1, and CKD5 groups are shown in a Venn diagram and hierarchical clustering heatmaps. a, Venn diagram showing the DEMs and overlap. b, Hierarchical cluster analysis of DEMs in the CKD1 group compared with the control group. c, Hierarchical cluster analysis of DEMs in the CKD5 group compared with the control group. d, Hierarchical cluster analysis of DEMs in the CKD5 group compared with the CKD1 group. Red pixels correspond to an increased abundance of miRNA in the serum, whereas green pixels indicate decreased miRNA levels.
DEMs, differentially expressed miRNAs; miRNAs, microRNAs; CKD, chronic kidney disease.
Table 2.
Differentially expressed miRNAs in CKD1 patients compared with the healthy controls.
| Mature miRNAs | pre-miRNA | Fold change | Mature miRNAs | pre-miRNA | Fold change |
|---|---|---|---|---|---|
| Up-regulated miRNAs | |||||
| miR-122-5p | mir-122 | 5.63 | miR-486-5p | mir-486-1 | 3.46 |
| miR-486-5p | mir-486-2 | 3.44 | miR-423-5p | mir-423 | 3.07 |
| miR-193b-5p | mir-193b | 2.99 | miR-483-5p | mir-483 | 2.90 |
| miR-4732-5p | mir-4732 | 2.83 | miR-4732-3p | mir-4732 | 2.42 |
| miR-7706-3p | mir-7706 | 2.24 | |||
| Down-regulated miRNAs | |||||
| let-7f-5p | let-7f-1 | 0.26 | let-7f-5p | let-7f-2 | 0.28 |
| let-7g-5p | let-7g | 0.30 | miR-26b-5p | mir-26b | 0.32 |
| miR-181d-5p | mir-181d | 0.34 | let-7i-5p | let-7i | 0.40 |
| miR-98-5p | mir-98 | 0.40 | let-7a-5p | let-7a-2 | 0.47 |
| let-7a-5p | let-7a-3 | 0.47 | let-7a-5p | let-7a-1 | 0.47 |
| miR-30e-3p | mir-30e | 0.47 | |||
miRNAs, microRNAs; CKD, chronic kidney disease.
Table 3.
Differentially expressed miRNAs in CKD5 patients compared with the healthy controls.
| Mature miRNAs | pre-miRNA | Fold change | Mature miRNAs | pre-miRNA | Fold change |
|---|---|---|---|---|---|
| Up-regulated miRNAs | |||||
| miR-483-5p | mir-483 | 25.20 | miR-133b-3p | mir-133b | 8.24 |
| miR-196a-5p | mir-196a-2 | 6.00 | miR-196a-5p | mir-196a-1 | 4.83 |
| miR-378f-3p | mir-378f | 2.97 | |||
| Down-regulated miRNAs | |||||
| miR-144-3p | mir-144 | 0.09 | miR-584-5p | mir-584 | 0.10 |
| miR-486-3p | mir-486-2 | 0.10 | miR-363-3p | mir-363 | 0.11 |
| miR-486-3p | mir-486-1 | 0.11 | miR-654-3p | mir-654 | 0.11 |
| miR-409-3p | mir-409 | 0.13 | miR-495-3p | mir-495 | 0.16 |
| miR-493-3p | mir-493 | 0.18 | miR-144-5p | mir-144 | 0.19 |
| miR-4446-3p | mir-4446 | 0.19 | miR-130a-3p | mir-130a | 0.19 |
| miR-381-3p | mir-381 | 0.20 | miR-654-5p | mir-654 | 0.20 |
| miR-432-5p | mir-432 | 0.22 | miR-4433b-5p | mir-4433b | 0.24 |
| miR-671-3p | mir-671 | 0.26 | miR-424-3p | mir-424 | 0.28 |
| miR-32-5p | mir-32 | 0.33 | miR-22-5p | mir-22 | 0.33 |
| miR-589-5p | mir-589 | 0.34 | miR-24-3p | mir-24-2 | 0.35 |
| miR-185-3p | mir-185 | 0.35 | miR-24-3p | mir-24-1 | 0.35 |
| miR-106b-5p | mir-106b | 0.36 | miR-28-5p | mir-28 | 0.37 |
| miR-323a-3p | mir-323a | 0.38 | let-7a-5p | let-7a-2 | 0.40 |
| let-7a-5p | let-7a-3 | 0.40 | let-7a-5p | let-7a-1 | 0.40 |
| miR-181d-5p | mir-181d | 0.42 | miR-339-5p | mir-339 | 0.43 |
| miR-107-3p | mir-107 | 0.44 | miR-361-3p | mir-361 | 0.46 |
| miR-345-5p | mir-345 | 0.47 | miR-181a-5p | mir-181a-1 | 0.50 |
| miR-181a-5p | mir-181a-2 | 0.50 | |||
miRNAs, microRNAs; CKD, chronic kidney disease.
Table 4.
Differentially expressed miRNAs in the CKD5 patients compared with the CKD1 patients.
| Mature miRNAs | pre-miRNA | Fold change | Mature miRNAs | pre-miRNA | Fold change |
|---|---|---|---|---|---|
| Up-regulated miRNAs | |||||
| miR-483-5p | mir-483 | 8.70 | miR-7-5p | mir-7-1 | 8.41 |
| miR-7-5p | mir-7-3 | 8.26 | miR-7-5p | mir-7-2 | 8.26 |
| miR-133b-3p | mir-133b | 7.56 | miR-196a-5p | mir-196a-2 | 6.50 |
| miR-196a-5p | mir-196a-1 | 6.44 | miR-365a-3p | mir-365a | 6.23 |
| miR-365b-3p | mir-365b | 6.23 | miR-378d-3p | mir-378d-1 | 6.18 |
| miR-378d-5p | mir-378d-2 | 5.94 | miR-27b-3p | mir-27b | 4.93 |
| miR-100-5p | mir-100 | 3.48 | miR-378f-3p | mir-378f | 3.33 |
| Down-regulated miRNAs | |||||
| miR-486-3p | mir-486-1 | 0.09 | miR-486-3p | mir-486-2 | 0.09 |
| miR-4446-3p | mir-4446 | 0.10 | miR-584-5p | mir-584 | 0.11 |
| miR-486-5p | mir-486-1 | 0.14 | miR-486-5p | mir-486-2 | 0.15 |
| miR-432-5p | mir-432 | 0.15 | miR-16-2-3p | mir-16-2 | 0.16 |
| miR-4433b-5p | mir-4433b | 0.17 | miR-25-3p | mir-25 | 0.18 |
| miR-122-5p | mir-122 | 0.19 | miR-409-3p | mir-409 | 0.19 |
| miR-363-3p | mir-363 | 0.19 | miR-451a-5p | mir-451a | 0.20 |
| miR-223-5p | mir-223 | 0.21 | miR-495-3p | mir-495 | 0.21 |
| miR-3158-3p | mir-3158-1 | 0.21 | miR-3158-3p | mir-3158-2 | 0.21 |
| miR-185-5p | mir-185 | 0.22 | miR-4732-5p | mir-4732 | 0.23 |
| miR-1301-3p | mir-1301 | 0.24 | miR-4433b-3p | mir-4433b | 0.26 |
| miR-144-3p | mir-144 | 0.26 | miR-654-5p | mir-654 | 0.26 |
| miR-7706-3p | mir-7706 | 0.26 | miR-4732-3p | mir-4732 | 0.26 |
| miR-501-3p | mir-501 | 0.28 | miR-424-3p | mir-424 | 0.29 |
| miR-511-5p | mir-511 | 0.30 | miR-1273h-5p | mir-1273h | 0.30 |
| miR-330-3p | mir-330 | 0.30 | miR-1304-5p | mir-1304 | 0.30 |
| miR-671-3p | mir-671 | 0.31 | miR-151a-3p | mir-151a | 0.31 |
| miR-654-3p | mir-654 | 0.31 | miR-1294-5p | mir-1294 | 0.31 |
| miR-126-3p | mir-126 | 0.32 | miR-543-3p | mir-543 | 0.32 |
| miR-625-3p | mir-625 | 0.33 | miR-338-5p | mir-338 | 0.33 |
| miR-139-3p | mir-139 | 0.33 | miR-328-3p | mir-328 | 0.34 |
| let-7b-5p | let-7b | 0.35 | miR-3615-3p | mir-3615 | 0.35 |
| miR-1908-5p | mir-1908 | 0.37 | miR-185-3p | mir-185 | 0.39 |
| miR-484-5p | mir-484 | 0.40 | let-7d-3p-3p | let-7d | 0.40 |
| miR-142-5p | mir-142 | 0.41 | miR-1273h-3p | mir-1273h | 0.42 |
| miR-1307-5p | mir-1307 | 0.43 | miR-744-5p | mir-744 | 0.44 |
| miR-339-3p | mir-339 | 0.46 | miR-345-5p | mir-345 | 0.48 |
| miR-22-5p | mir-22 | 0.49 | miR-106b-5p | mir-106b | 0.50 |
miRNAs, microRNAs; CKD, chronic kidney disease.
Predicted DEM targets
Identification of predicted targets may help to understand the biological role of the DEMs. We used TargetScan and mirdbV5 to identify predicted target mRNAs for the DEMs. The overlapping results of two databases were considered to be the predicted targets for each miRNA. Thus, one miRNA can target many genes and one gene can be targeted by different miRNAs, which suggests that the miRNAs may participate in a series of biological processes by regulating multiple target genes.
Validation of DEMs by qRT-PCR
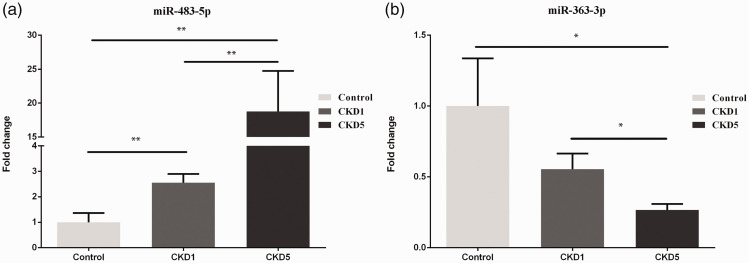
Based on the sequencing data and related literature reports, miR-483-5p and miR-363-3p were chosen to be validated by qRT-PCR. The results showed that miR-483-5p was up-regulated in CKD1 (fold change = 2.56, P < 0.01) and CKD5 (fold change = 18.77, P < 0.01) patients compared with the control subjects, and it also increased in the CKD5 compared with CKD1 patients (fold change = 7.34, P < 0.01; Figure 3a). miR-363-3p was down-regulated in the CKD5 patients compared with the control subjects (fold change = 0.27, P < 0.05) and CKD1 patients (fold change = 0.48, P < 0.05; Figure 3b).
Figure 3.
Validation of DEMs in serum samples from healthy controls, CKD1, and CKD5 patients by qRT-PCR. a, validation of miR-483-5p; b, validation of miR-363-3p. All samples were normalized to cel-miR-39. The data are expressed as the mean SEM. The significance of differences for miRNA expression was calculated using a one-way ANOVA. *, P<0.05; **, P<0.01.
DEMs, differentially expressed miRNAs; miRNAs, microRNAs; CKD, chronic kidney disease; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; SEM, standard error of the mean; ANOVA, analysis of variance.
Discussion
The increased incidence of CKD has led to higher morbidity and mortality rates worldwide. Early identification of CKD patients is of paramount importance to perform prompt interventions, reduce progression to kidney failure, and improve the prognosis.18 Thus, it is significant to identify valid, sensitive, non-invasive, and specific biomarkers that correlate well with kidney pathology and disease progression. However, existing measurements of kidney function such as SCr, blood urea nitrogen (BUN), and proteinuria seem to be insufficient.1,18
Over the past several years, miRNAs have been shown to play an important regulatory role in renal physiology, such as normal renal development, homeostasis, vascular calcification, and osmoregulation.19,20 It was estimated that there are more than 300 miRNAs in kidneys,21 and miRNA dysregulation has been observed in various renal diseases, which indicates that miRNAs can be examined for their potential role as diagnostic, prognostic, and even therapeutic biomarkers for renal disease.22 To identify miRNAs as diagnostic biomarkers for CKD, we performed NGS to determine the genome-wide serum miRNA expression profiling of CKD5, CKD1, and healthy controls. There were 20 and 42 DEMs in the CKD1 and CKD5 groups compared with the control group, respectively, and 70 DEMs in the CKD5 group compared with the CKD1 group.
miR-483-5p was the only miRNA that was found to be differentially expressed across all three groups in the Venn diagram, and it was further validated using qRT-PCR. The human miR-483-5p gene was reported to be located at the second intron of its host gene insulin-like growth factor 2 (IGF2) on chromosome 11p15.5.23 It was recently shown to be involved in cancers,24 fibrosis,25 and kidney diseases.26–28 For example, Dai et al.26 reported that miR-483-5p was up-regulated in the renal biopsies of primary immunoglobulin A nephropathy (IgAN) patients using microarray analysis. Another study reported that miR-483-5p was also up-regulated in peripheral blood mononuclear cells (PBMCs) of lupus nephritis patients.27 In addition, serum miR-483-5p could serve as a new diagnostic biomarker for kidney transplantation.28 In this study, serum miR-483-5p was shown to be significantly up-regulated in CKD1 and CKD5 groups based on sequencing and qRT-PCR results. However, the reason is not clear. The predicted target genes that were related to kidney diseases for miR-483-5p were analyzed using the GeneCards Database. As one of seven target genes (Table 5), cystathionine beta synthase (CBS) was reported to be an inhibitor in renal fibrosis.29 Because renal fibrosis is one of the histopathological hallmarks for CKD, miR-483-5p was predicted to regulate target genes that were enriched in pathways that are related to fibrosis.25 However, whether miR-483-5p plays a role in the development of CKD via regulating CBS or other fibrosis-related genes remains unknown.
Table 5.
Predicted target genes related to kidney diseases for miR-483-5p and miR-363-3p.
| miRNAs | Predicted target genes related to kidney diseases |
|---|---|
| miR-483-5p | ACBD6, CBS, CDK15, MSC, PIP5K1A, SRSF4, STK40 |
| miR-363-3p | ADAM10, AIDA, ANKIB1, ANKRD44, ANP32E, APPL1, B3GALT2, BCL2L11, BSDC1, BTG2, C11orf24, CAMK2A, CBLN4, CCNC, CD69, CDCA7L, CHST7, CIC, CLDN11, CNEP1R1, COL1A2, CPEB2, CPEB3, DCAF6, DNAJB9, DNAJC30, DOCK9, DPY30, DSC2, DSCAML1, DUS2, DUSP10, DUSP5, EFR3A, ELOVL4, ERGIC2, FAM110B, FAM135A, FAM20C, FAR1, FBXW7, FHL2, FMR1, FNDC3B, FNIP1, FOXN2, FRY, FZD10, G3BP2, GFPT2, GLRA1, GLYR1, GNAQ, GOLGA3, GOLGA4, GOLGA8A, GPR180, GRHL1, HAND1, HAND2, HERPUD2, HIPK3, IDH1, IQGAP2, ITGA5, ITGA6, ITGAV, JOSD1, KAT2B, KBTBD8, KIAA1109, KLF4, KLF6, KLHL14, LATS2, LHFPL2, LRCH1, MAN2A1, MAP2K4, MIA3, MMD, MOAP1, MORC3, MPP1, MTMR9, MYCBP2, MYLIP, MYO1B, NECAP1, NEFH, NEFM, NOVA1, NSG1, NSMF, OTUD3, PAX9, PCDH11Y, PCMTD1, PCOLCE2, PDE10A, PDZD2, PER2, PHTF2, PIK3CB, PIKFYVE, PKDCC, PLEKHA1, PLEKHB2, PPCS, PPP1R12A, PPP1R12C, PPP1R37, PTEN, PTGER4, RAB23, RAD21, RASSF3, RBM47, REV3L, RGL1, RGS17, RHPN2, RNF180, RPS6KA4, RSBN1, SERTAD3, SESN3, SGPP1, SLC12A5, SLC17A6, SLC24A3, SLC25A32, SLC32A1, SNAPC1, SNN, SOCS5, SOSTDC1, SPRYD4, SRPR, ST6GAL2, SYNDIG1, TACC2, TAGAP, TCF21, TEF, TGIF1, TMEM229A, TMEM87A, TMF1, TOB1, TOB2, UBE2Z, UBXN4, UGP2, USP28, WASL, WWP2, ZDHHC5, ZFC3H1, ZFYVE21 |
miRNAs, microRNAs.
miR-363-3p is a component of the oncogenic miR-17 to 92 family of miRNA clusters,30 and it has been reported to play a role as a tumor suppressor or oncogene in cancers.31 Moreover, aberrant expression of miR-363-3p has also been seen in some renal diseases. For example, Tan et al.32 reported that miR-363-3p was down-regulated in renal biopsies from primary IgAN patients. Urinary miR-363-3p could predict the development of microalbuminuria in patients with diabetic nephropathy.33 In this study, miR-363-3p was down-regulated in the CKD5 patients, and its predicted target genes related to kidney diseases were also analyzed using the GeneCards Database (Table 5). As one target gene, integrin α5 (ITGA5) was reported to be down-regulated in healthy renal cells by miR-363-3p, miR-328-3p, and miR-25-3p.31 ITGA5 was also shown to be a well-established activator of transforming growth factor-β (TGF-β), which is known as a central mediator of renal fibrosis in CKD.19 Serum miR-363-3p, miR-328-3p, and miR-25-3p were all decreased in the CKD5 group in the present study. Therefore, we suggest that these decreased miRNAs synergistically up-regulate ITGA5, and ITGA5 may cause up-regulation of TGF-β, thereby participating in the development of CKD. However, the hypothesis should be verified experimentally in future studies.
It is widely believed that cellular miRNAs are actively secreted from donor cells or passively released from damaged cells into the extracellular microenvironment or body fluids via microvesicles, exosomes, lipoproteins, or RNA-binding proteins.13,19 Extracellular miRNAs may transmit biological information from donor cells to recipient cells, or carry alarm signals from apoptotic cells to other cells.19,34 Therefore, extracellular miRNAs, such as serum miRNAs, act as disease biomarkers and they also function as critical regulators of cellular crosstalk.10 Generally, serum miRNAs were mostly derived from peripheral blood mononuclear cells and other blood cells under normal conditions.8 Once the body is abnormal, such as in CKD, certain miRNAs may be released from damaged cells or blood cells and lead to increased serum miRNA expressions. However, most DEMs in our results were down-regulated. The reason may be partly associated with renal dysfunction. Neal et al.35 reported that the total circulating miRNA concentration was significantly reduced as CKD progresses because of dialysis or increased RNAse activity. CKD pathophysiology is complicated, and excess comorbidity is present in almost all CKD patients. Because the CKD population is heterogeneous and a single miRNA is insufficient to meet all the expectations, miRNA panels will be necessary to allow CKD patients to be distinguished from controls or determine patient prognosis. This suggests that there is an advantage to using miRNAs that are involved in different pathophysiological pathways to obtain a complete picture.
In conclusion, to the best of our knowledge, this is the first identification of a genome-wide serum microRNA expression profile in CKD patients using NGS. This study served as a preliminary investigation and the results should be further confirmed in future large-scale studies.
Footnotes
Author contributions: JSX and YLB are responsible for study design. XYL and WJW are responsible for performing the study, data analysis, and manuscript writing. HRZ, SLZ, LH, WZ, and DXZ are responsible for patient recruitment and preparation of laboratory specimens.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This work was supported by the Hebei Science and Technology Planning Project (16397733D), the Hebei Province Medical Technology Tracking project (G2018050), and the Hebei Province Key Research and Development Project (20377704D), as well as the Health and Family Planning Commission of Hebei Province (No. 20181499).
ORCID iD: Jinsheng Xu https://orcid.org/0000-0001-6735-6579
References
- 1.Rysz J, Gluba-Brzozka A, Franczyk B, et al. Novel biomarkers in the diagnosis of chronic kidney disease and the prediction of its outcome. Int J Mol Sci 2017; 18: 1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senanayake S, Gunawardena N, Palihawadana P, et al. Validity and reliability of the Sri Lankan version of the kidney disease quality of life questionnaire (KDQOL-SF). Health Qual Life Outcomes 2017; 15: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016; 388: 1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Wang F, Wang L, et al. Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet 2012; 379: 815–822. [DOI] [PubMed] [Google Scholar]
- 5.Zhou M, Wang H, Zhu J, et al. Cause-specific mortality for 240 causes in China during 1990–2013: a systematic subnational analysis for the Global Burden of Disease Study 2013. Lancet 2016; 387: 251–272. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 7.Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011; 8: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- 9.Wang J Chen J andSen S.. MicroRNA as biomarkers and diagnostics. J Cell Physiol 2016; 231: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenzen JM andThum T.. Circulating and urinary microRNAs in kidney disease. Clin J Am Soc Nephrol 2012; 7: 1528–1533. [DOI] [PubMed] [Google Scholar]
- 11.Konta T, Ichikawa K, Suzuki K, et al. A microarray analysis of urinary microRNAs in renal diseases. Clin Exp Nephrol 2014; 18: 711–717. [DOI] [PubMed] [Google Scholar]
- 12.Gholaminejad A Abdul Tehrani H andGholami Fesharaki M.. Identification of candidate microRNA biomarkers in diabetic nephropathy: a meta-analysis of profiling studies. J Nephrol 2018; 31: 813–831. [DOI] [PubMed] [Google Scholar]
- 13.Lopez JP, Diallo A, Cruceanu C, et al. Biomarker discovery: quantification of microRNAs and other small non-coding RNAs using next generation sequencing. BMC Med Genomics 2015; 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z Gerstein M andSnyder M.. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang L, Yao J, Kong X, et al. Increasing prevalence of membranous nephropathy in patients with primary glomerular diseases: a cross-sectional study in China. Nephrology 2017; 22: 168–173. [DOI] [PubMed] [Google Scholar]
- 16.Kroh EM, Parkin RK, Mitchell PS, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010; 50: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmittgen TD andLivak KJ.. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 18.Wasung ME Chawla LS andMadero M.. Biomarkers of renal function, which and when? Clin Chim Acta 2015; 438: 350–357. [DOI] [PubMed] [Google Scholar]
- 19.Trionfini P Benigni A andRemuzzi G.. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 2015; 11: 23–33. [DOI] [PubMed] [Google Scholar]
- 20.Lee CT, Lee YT, Tain YL, et al. Circulating microRNAs and vascular calcification in hemodialysis patients. J Int Med Res 2019; 47: 2929–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCall MN, Kim MS, Adil M, et al. Toward the human cellular microRNAome. Genome Res 2017; 27: 1769–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul P, Chakraborty A, Sarkar D, et al. Interplay between miRNAs and human diseases. J Cell Physiol 2018; 233: 2007–2018. [DOI] [PubMed] [Google Scholar]
- 23.Ma N, Wang X, Qiao Y, et al. Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol Cell Endocrinol 2011; 333: 96–101. [DOI] [PubMed] [Google Scholar]
- 24.Agosta C, Laugier J, Guyon L, et al. MiR-483-5p and miR-139-5p promote aggressiveness by targeting N-myc downstream-regulated gene family members in adrenocortical cancer. Int J Cancer 2018; 143: 944–957. [DOI] [PubMed] [Google Scholar]
- 25.Chouri E, Servaas NH, Bekker CPJ, et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J Autoimmun 2018; 89: 162–170. [DOI] [PubMed] [Google Scholar]
- 26.Dai Y, Sui W, Lan H, et al. Microarray analysis of micro-ribonucleic acid expression in primary immunoglobulin A nephropathy. Saudi Med J 2008; 29: 1388–1393. [PubMed] [Google Scholar]
- 27.Te JL, Dozmorov IM, Guthridge JM, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One 2010; 5: e10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sui W, Yang M, Li F, et al. Serum microRNAs as new diagnostic biomarkers for pre- and post-kidney transplantation. Transplant Proc 2014; 46: 3358–3362. [DOI] [PubMed] [Google Scholar]
- 29.Yuan X, Zhang J, Xie F, et al. Loss of the protein cystathionine beta-synthase during kidney injury promotes renal tubulointerstitial fibrosis. Kidney Blood Press Res 2017; 42: 428–443. [DOI] [PubMed] [Google Scholar]
- 30.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008; 132: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boguslawska J, Rodzik K, Poplawski P, et al. TGF-beta1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. Cancer Lett 2018; 412: 155–169. [DOI] [PubMed] [Google Scholar]
- 32.Tan K, Chen J, Li W, et al. Genome-wide analysis of microRNAs expression profiling in patients with primary IgA nephropathy. Genome 2013; 56: 161–169. [DOI] [PubMed] [Google Scholar]
- 33.Argyropoulos C, Wang K, Bernardo J, et al. Urinary microRNA profiling predicts the development of microalbuminuria in patients with type 1 diabetes. J Clin Med 2015; 4: 1498–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayraktar R Van Roosbroeck K andCalin GA.. Cell-to-cell communication: microRNAs as hormones. Mol Oncol 2017; 11: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neal CS, Michael MZ, Pimlott LK, et al. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant 2011; 26: 3794–3802. [DOI] [PubMed] [Google Scholar]