Abstract
The lateral habenula (LHb) integrates critical information regarding aversive stimuli that shapes decision making and behavioral responses. The three major LHb outputs innervate dorsal raphe nucleus (DRN), ventral tegmental area (VTA), and the rostromedial tegmental nucleus (RMTg). LHb neurons that project to these targets are segregated and nonoverlapping, and this led us to consider whether they have distinct molecular phenotypes and adaptations to stress exposure. In order to capture a time-locked profile of gene expression after repeated forced swim stress, we used intersectional expression of RiboTag in rat LHb neurons and next-gen RNA sequencing to interrogate the RNAs actively undergoing translation from each of these pathways. The “translatome” in the neurons comprising these pathways was similar at baseline, but diverged after stress, especially in the neurons projecting to the RMTg. Using weighted gene co-expression network analysis, we found one module, which had an overrepresentation of genes associated with phosphoinositide 3 kinase (PI3K) signaling, comprising genes downregulated after stress in the RMTg-projecting LHb neurons. Reduced PI3K signaling in RMTg-projecting LHb neurons may be a compensatory adaptation that alters the functional balance of LHb outputs to GABAergic vs. monoaminergic neurons following repeated stress exposure.
Keywords: Lateral habenula, Gene expression, RiboTag, rnaSEQ, PI3K
1. Introduction
Stress disorders, anxiety, and depression are associated with altered functional connectivity of key brain regions and are critical determinants of associated symptoms (Williams, 2016), including the lateral habenula (LHb), a small nucleus in the epithalamus that acts as an anti-reward nucleus. While it receives inputs from diverse regions throughout the brain, it has three main efferent pathways – to the ventral tegmental area (VTA), the rostromedial tegmental nucleus (RMTg), and the dorsal raphe nucleus (DRN) (Brinschwitz et al., 2010; Geisler and Trimble, 2008)—these three outputs contribute to different elements of adaptive responses to stress (Brinschwitz et al., 2010; Coffey et al., 2020b). The neurons comprising these three main output pathways appear to be quite segregated as they do not send collaterals to more than one of these targets (Bernard and Veh, 2012; Goncalves et al., 2012; Kalen et al., 1989; Quina et al., 2015). LHb neurons are mainly glutamatergic, contributing excitatory synapses onto both GABAergic and monoaminergic neurons in the DRN and the VTA at roughly equivalent rates (Brown and Shepard, 2016; Lammel et al., 2012; Ogawa et al., 2014; Omelchenko et al., 2009; Stamatakis and Stuber, 2012; Weissbourd et al., 2014). LHb afferents to RMTg are also excitatory, and in turn RMTg inhibits both the DRN and VTA through GABAergic projection neurons. Both the DRN and VTA send reciprocal projections to each other and back to the LHb, and serotonin and dopamine presumably released from these projections modulate LHb excitability (Gruber et al., 2007; Han et al., 2015; Kowski et al., 2009; Li et al., 1993; Shen et al., 2012; Xie et al., 2016; Zhang et al., 2018). Interestingly, norepinephrine from the VTA may also activate D4 receptors in the LHb (Root et al., 2015). Thus, there is a complex interplay of excitation and inhibition of DRN and VTA that is controlled by the relative activation of these LHb output pathways.
The LHb may be an important target for treating stress disorders (Browne et al., 2018; Geisler and Trimble, 2008; Lee and Huang, 1988). Stress exposure activates LHb neurons intensely and induces c-Fos, a marker of neuronal activity (Wirtshafter et al., 1994). Stimulation of LHb neurons promotes passive avoidance (Ootsuka and Mohammed, 2015; Stamatakis and Stuber, 2012) while inhibition of the LHb, and its connection to both serotonergic and dopaminergic regions, reduces anxiety- and depression-like behaviors (Gill et al., 2013; Luo et al., 2015; Zhang et al., 2016). Inhibition of the LHb via the inhibitory DREADD receptor hM4Di decreases passive coping in the forced swim test (FST), a measure of behavioral despair (Nair et al., 2013). LHb projections to the DRN are thought to mediate this effect (Coffey et al., 2020b).
Recently, a surge in transcriptomic data emphasizes the heterogeneity of LHb neurons. Wagner et al. (2016) used in situ hybridization images from the Allen Brain Atlas to map transcript expression throughout the LHb and its subregions, finding ten distinct anatomical patterns of expression. Recent single cell RNA-seq profiles of the habenular nuclei identified several types of distinct neurons (Hashikawa et al., 2020; Wallace et al., 2020), although none of these appeared to be pathway-specific. However, these studies did not explicitly examine gene expression between the three major LHb outputs. Cerniauskas et al. (2019) identified a depression-like phenotype which gave rise to differential gene expression in mouse LHb neurons projecting to the VTA. Given the heterogeneity of the LHb transcriptome and the functional differences between its anatomically segregated output pathways, it seems plausible that gene expression in these pathways respond differentially to stress. Additionally, while Hashikawa et al. (2020), Wallace et al. (2020) and Cerniauskas et al. (2019) studied the mouse habenula, this is the first study examining the genetic profile of LHb neurons in the rat.
To investigate this, we used next-gen sequencing after intersectional expression of RiboTag in neurons from each of these pathways to isolate ribosome-associated RNAs actively undergoing translation (Sanz et al., 2009), a method that reveals evolving changes in gene expression in response to stress (Lesiak et al., 2020). This is the first such application of intersectional translatome analysis in rat neurons and by capturing ribosome-associated RNA from the soma and dendrites, may be sensitive to rapid changes in translation following stress. We found that LHb neurons projecting to RMTg are differentially regulated by stress in comparison to those projecting to DRN or VTA.
2. Materials and Methods
2.1. Animals
For all experiments, male Sprague-Dawley rats (n = 43, Charles River, Raleigh, NC) weighing 251–275 g were used. Rats were double-housed in a temperature- and humidity-controlled vivarium with a 14-10 light-dark cycle and allowed to acclimate to the facility for two weeks prior to surgery. All experiments were carried out during the light period. Food and water were freely available at all times. All experimental procedures were approved by the University of Washington Institutional Animal Care and Use Committee and were conducted in accordance to the guidelines of the ‘Principles of Laboratory Animal Care’ (NIH publication no. 86–23, 1996). 9 animals were excluded because viral-mediated gene expression of RiboTag was below the threshold. A second cohort of 13 rats was used for a replication of differentially expressed genes in the RMTg-projected LHb neurons using the same procedures as the RNAsequencing cohort.
2.2. Experimental design
Our overarching experimental strategy was to use an intersectional viral vector approach to express RiboTag (Sanz et al., 2009) in LHb neurons selectively projecting to VTA, RMTg or DRN based using our recently published procedures (Coffey et al., 2020b). This was achieved through injection of Cre-dependent adeno-associated viral vectors (AAV8-hSyn-DIO-RiboTag) into LHb and a retrogradely transported canine adenovirus-2 expressing Cre (CAV2-Cre) into one of the three output regions. Separate groups of animals were used for each pathway to evaluate the effect of repeated swim stress on gene expression. All experimental procedures were approved by the University of Washington Institutional Animal Care and Use Committee and were conducted in accordance with National Institutes of Health (NIH) guidelines.
2.3. Surgical methods
For intersectional surgeries, anesthesia was induced with 5% isoflurane/95% oxygen and maintained at 1–3% isoflurane during the surgical procedure. Using a custom robotic stereotaxic instrument (Coffey et al., 2013), 27 animals received AAV8-hSyn-DIO-RiboTag injected into LHb. Blunt 28 g needles were inserted bilaterally at a 10° angle terminating at A/P −3.2, M/L ±0.7, and D/V −5.25 and 1 μl of AAV8-hSyn-DIO-RiboTag was injected at a rate of 0.2 μl/min. Nine animals received bilateral 1 μl injections of CAV2-Cre into the DRN at a 15° angle terminating at A/P −7.8, M/L ±0.23, and D/V −6.85. Nine animals received bilateral 1 μl injections of CAV2-Cre into the VTA at a 10° angle terminating at A/P −5.8, M/L ±0.6 and D/V −8.6. Nine animals received bilateral 1 μl injections of CAV2-Cre into the RMTg at a 10° angle terminating at A/P −7.6, M/L ±0.62 and D/V −8.5. For the replication experiment, an additional 13 rats received bilateral injections of AAV8-hSyn-DIO-RiboTag into the LHb and CAV2-Cre into the RMTg. After surgeries, rats were given meloxicam (0.2 mg/kg, s. c.) for pain management and monitored for at least 3 days. Accuracy of injection coordinates was confirmed by RTqPCR detection of RiboTag and Cre RNAs from LHb homogenate; these injection volumes and coordinates were optimized to produce selective transduction of LHb neurons with minimal expression adjacent regions. Rats recovered for three weeks post-surgery to give time for viral expression to occur. For “sham” control samples, rats were anesthetized and placed in the robotic stereotaxic instrument; however, no needle was inserted, nor any virus injected.
2.4. Behavioral methods
After three weeks of handling post-surgery, rats in the stressed grouping underwent a two-day forced swim protocol. Rats were placed in an inescapable 40 cm tall x 20 cm diameter Plexiglas cylinder filled with water (23±2C) to 30 cm, a level deep enough to prevent them from standing on the bottom (Clark et al., 2002). Rats were stressed for 15 min on day one; 24 h later, they were placed into the same chamber for 5 min. Rats in the unstressed group were handled as normal. Decapitation and tissue extraction occurred 3 h following this protocol.
2.5. RiboTag extraction
RiboTag-associated RNA extraction as previously described (Lesiak et al., 2020; Lesiak and Neumaier, 2016). The LHb was extracted using a 4 mm punch and homogenized in 2 mL of supplemented homogenizing buffer [S-HB, 50 mM Tris-HCl, 100 mM KCl, 12 mM MgCl2, 1% NP40, 1 mM DTT, 1 × Protease inhibitor cocktail (Sigma-Aldrich), 200 U/mL RNasin (Promega, Madison, WI), 100 μg/mL cyclohexamide (Sigma-Aldrich), 1 mg/mL heparin (APP Pharmaceuticals, Lake Zurich, IL)]. Samples were centrifuged at 4 °C at 11,934×g for 10 min, and supernatant was collected, reserving 50 μL (10%) as an input fraction. Mouse monoclonal HA-specific antibody (2.5 μL) (HA.11, ascites fluid; Covance, Princeton, NJ) was added to the remaining supernatant, and RiboTag-IP fractions were rotated at 4 °C for 4 h. Protein A/G magnetic beads (200 μL) (Pierce) were washed with Homogenizing Buffer (HB 50 mM Tris-HCl, 100 mM KCl, 12 mM MgCl2, 1% NP40) prior to addition to the RiboTag-IP fraction and were rotated at 4 °C overnight. The next day, RiboTag-IP fractions were placed on DynaMag-2 magnet (Life Technologies), and the bead pellet was washed 3 times for 15 min with high salt buffer (HSB; 50 mM Tris, 300 mM KCl, 12 mM MgCl2, 1% NP40, 1 mM DTT, and 100 μg/mL cyclohexamide) and placed on a rotator. After the final wash, HSB was removed and beads were re-suspended in 400 μL supplemented RLT buffer (10 μL β-mercaptoethanol/10 mL RLT Buffer) from the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany) and vortexed vigorously. These samples were then placed back on the magnet and the RLT buffer was removed from the magnetic beads prior to RNA extraction. 350 μL supplemented RLT buffer was added to the Input Fraction prior to RNA extraction. RNA was extracted using Qiagen RNeasy Plus Micro kit according to package directions. RNA from both IP and input fractions were isolated using RNeasy Plus Micro Kit and eluted with 14–16 μl of water. RNA concentration was measured using Quant-iT RiboGreen RNA Assay (ThermoFisher Cat. R11490, Waltham, MA). Total RNA yield for the RiboTag-IP fraction 36 ng (sham controls 12 ng) and for the input fraction 196 ng (sham controls 188 ng).
2.6. RNAseq library preparation
RNAseq libraries were prepared using SMARTer Stranded Total RNA-Seq Kit v2 – Pico Input Mammalian (Takara Bio USA, Inc. Cat. 635,007, Mountain View, CA). 10 ng of RNA or average equivalent volumes of “sham” control samples were used to generate the negative control samples. RNAseq libraries were submitted to Northwest Genomics Center at University of Washington (Seattle, WA) where library quality control was measured using a BioAnalyzer, library concentrations were measured using Qubit dsDNA HS Assay Kit (ThermoFisher), and then samples were normalized and pooled prior to cluster generation on HiSeq High Output for Paired-end reads. RNAseq libraries were sequenced on the HiSeq 4000, Paired-end 75bp to sufficient read depth with PhiX spike-in controls (7%) (Illumina San Diego, CA).
2.7. RT-qPCR analysis
The remaining RNA was reverse transcribed to create cDNA libraries for qPCR using Superscript VILO Master Mix (ThermoFisher Cat. 11,754,050, Waltham, MA), and then cDNA libraries were diluted to a standard concentration before running the qPCR assay using Power Sybr Green on ViiA 7 Real-Time PCR System (Thermo Fisher) or QuantStudio 5 Real-Time PCR System (Thermo Fisher). The normalized relative starting quantities (NRStQ) was determined by qPCR analysis conducted using the standard curve method and normalized to four housekeeping genes (Gapdh, Ppia, Hprt, and Actinb) (Lesiak et al., 2015). NRStQ data was analyzed using ANOVA with Bonferroni Post-Hoc.
For the replication experiment, several RNA transcripts identified in the WGCNA analysis were quantified on QuantStudio 7 Pro Real-Time PCR System (Thermo Fisher); RiboTag expression was checked for surgical misses. NRStQ data of Kdr, Tek, and Ptpn13 was analyzed using unpaired t-tests.
2.8. Bioinformatics
RNA sequencing was analyzed as previously described (Coffey et al., 2020a; Lesiak et al., 2020) with modification as noted below.
2.8.1. Transcript quantification and quality control
Raw fastq files were processed using multiple tools through the Galaxy platform 26. Fastq files were inspected for quality using FastQC (Galaxy Version 0.7.0), and then passed to Salmon 27 (Galaxy Version 0.8.2) for quantification of transcripts. The Salmon index was built using the protein coding transcriptome GRCm38-mm10.
2.8.2. Non-specific immunoprecipitated RNA subtraction
Due to the inherent issue of non-specific precipitation of immunoprecipitation (IP) procedures, we used a novel computational method for subtraction of non-specific RNA counts from the pathway specific RiboTag sample (Coffey et al., 2020a). Briefly, an IP was performed on an equivalent tissue sample from animals that did not receive the RiboTag virus (“sham” controls). RNA quantity from these samples was then quantified (mean ∼12 ng) along with the RiboTag-expressing experimental IP samples (mean ∼36 ng). Non-specific RNA contamination is considered to be a random sampling of the Input RNA, and the proportion of the IP sample that is contaminated is calculated from the true RNA quantity calculated for each sample. We capped the contamination level at 30% to prevent over-correction. All IP data presented in main manuscript have undergone this adjustment, but we also performed all bioinformatics on the raw IP files as well, which are available in the data archive. The adjusted bioinformatics are labeled “IP Minus Noise” while the raw files are labeled “IP”.
2.8.3. Differential expression analysis
Differential gene expression was calculated using DESeq228 (Galaxy Version 2.11.39). All Salmon and DESeq2 settings were left default and our analysis pipeline is archived on our Galaxy server. Differential expression analysis was performed on the IP and Input samples. To determine pathway specific gene enrichment, all IP samples were compared to pooled Input samples. For all other test of differential expression, IP samples were compared to IP samples. To determine the pathway expression at baseline, the unstressed DRN pathway was compared to the unstressed RMTg or VTA pathways and the unstressed RMTg pathway was compared to the unstressed VTA pathway. To determine the effects of stress between each pathway, the stressed DRN pathway was compared to the stressed RMTg or VTA pathways and the stressed RMTg pathway was compared to the stressed VTA pathway. To determine the effects of stress within each pathway, the stressed DRN pathway was compared to the unstressed DRN pathway, stressed RMTg to unstressed RMTg, and stressed VTA to unstressed VTA. For all comparisons, a positive Wald statistic means that a gene is expressed more in the first group as compared to the second group. An FDR of q = 0.10 was used as the threshold for differential expression throughout the manuscript.
2.8.4. Gene set enrichment analysis (GSEA)
Wald statics generated by DESeq2 were used as the ranking variable for gene set enrichment analysis (GSEA). The Wald statistic has a benefit over log 2 (fold change) as it incorporates an estimate of variance, and ultimately is used to determine significance in differential expression analysis. All genes with reliable statistical comparisons (those not filtered by DeSeq2) were entered into WebGestalt 201,929 (Liao et al., 2019) and we performed GSEA on all pertinent comparisons (Stressed vs Unstressed for each pathway, pairwise comparisons between the three stressed pathways, and pairwise comparisons between the three unstressed pathways). Gene sets used include three Gene Ontology (GO) classes: Biological Process, Molecular Function, and Cellular Component, as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Panther, Transcription Factor Targets, and MicroRNA Targets. All advanced parameters were left default except for significance level, which was set to FDR = 0.1.
2.8.5. Weighted gene Co-expression network analysis (WGCNA)
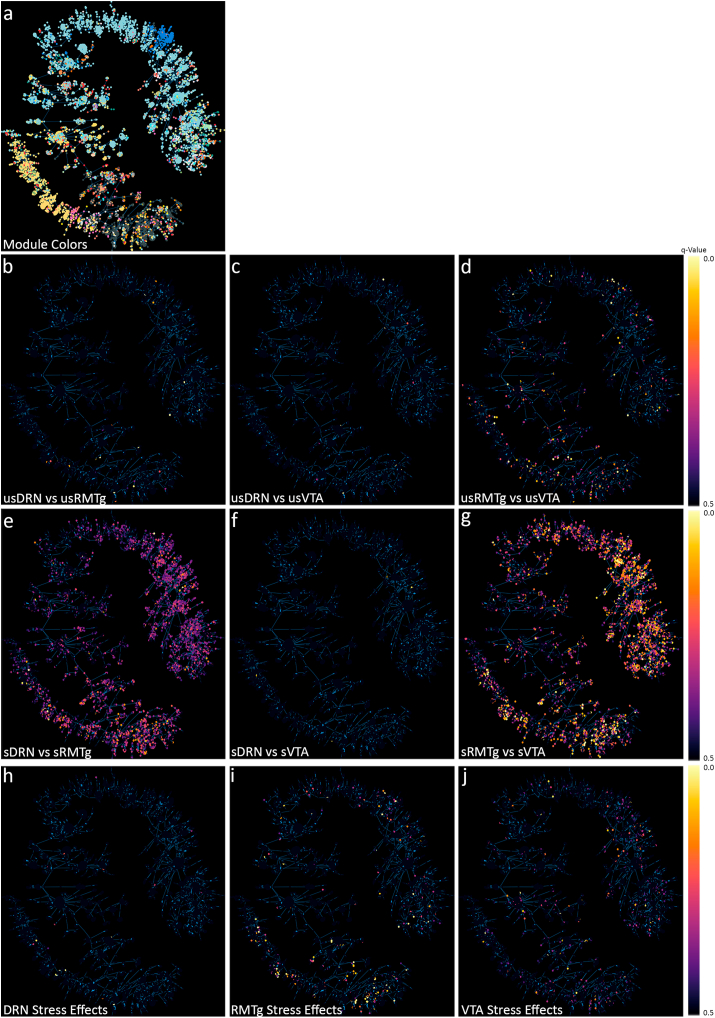
Topological overlap matrix for IP samples were generated, and module clustering was accomplished using the WGCNA (Langfelder and Horvath, 2008) package for R (R Development Core Team, 2020). Briefly, the TPM matrix for each group was filtered to remove zero-variance genes, and a signed adjacency matrix was generated using “bicor” as the correlation function. From this a signed topological overlap matrix was generated, followed by a dissimilarity topological overlap matrix. Finally, module membership was assigned using a dynamic tree cut. The complete code used to run WGCNA are available in the data archive. Post processing of the topological overlap matrix was completed with a custom object based Matlab class structure that we released previously (Coffey et al., 2020a). The WGCNA class structure can be accessed on https://github.com/levinsmr/WGCNA and the code used to generate all of the WGCNA figures in this manuscript is available in the Supplementary Code and Data for use as an example. When the average Wald score of a module was >2.0, we performed a secondary overrepresentation analysis of the component genes to evaluate for overrepresentation of genes associated with particular biological functions with WebGestalt.
3. Results
3.1. Read depth
Three weeks after viral vector injections, we compared handled but unstressed rats to rats subjected to a two-day forced swim stress and then all animals were sacrificed 3 h after completion of the stress procedure. This differs from the conventional Porsolt Forced Swim Test, where the behavior is measured during the second swim session. RNA was purified, sequencing libraries were prepared, and samples were sequenced as described in the Materials and Methods section (Fig. 1). The IP samples were sequenced to a depth of 7.4 ± 0.4 × 106 double stranded reads whereas the Input RNA samples were sequenced to a read depth of 1.1 ± 0.1 × 107 double stranded reads (Table 1). We sequenced Input RNA from pooled samples including each animal from that treatment group rather than each individual; this allowed us to estimate and subtract the small amount of contaminating input RNA that is carried forward during RiboTag immunoprecipitation (Coffey et al., 2020a).
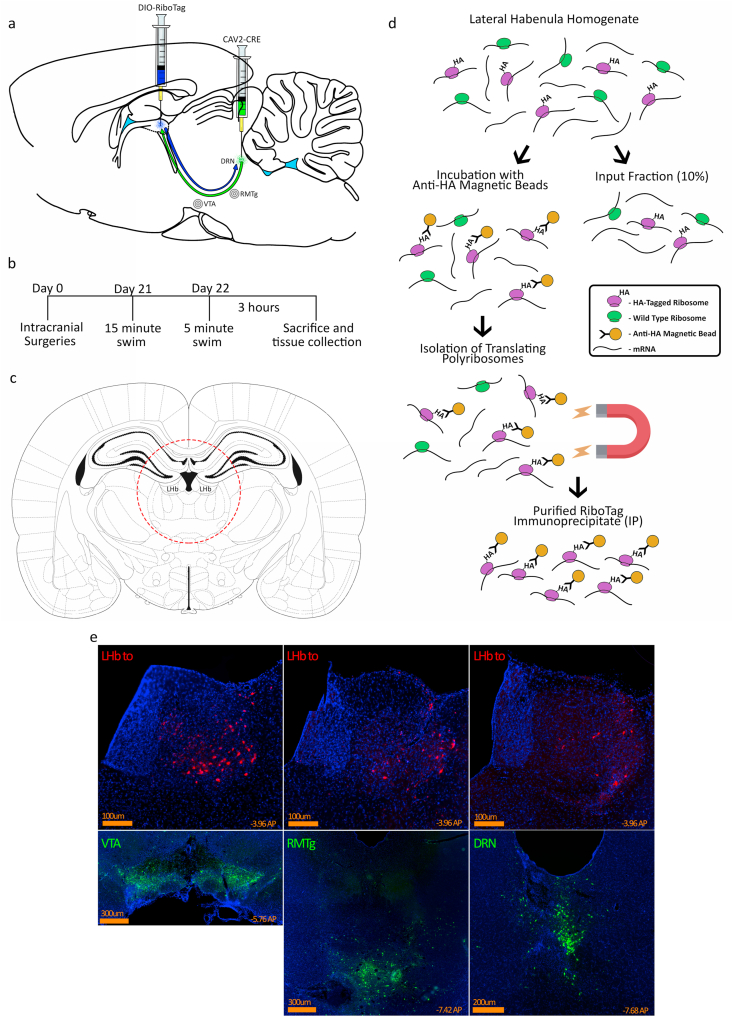
Fig. 1.
Intersectional viral mediated gene transfer and RiboTag strategy. a) Rats were injected with AAV8-DIO-RiboTag into the LHb and CAV2-Cre injected into one of the three target regions, either the DRN, RMTg or VTA. b) Experimental timeline. c) Location of tissue punch for LHb extraction. d) RiboTag immunoprecipitation protocol. e) Representative images from animals injected with the same AAV8-DIO vector expressing hM4Di-mCherry (injected into LHb) and CAV2-Cre mixed with 5% CAV2-ZsGreen (injected into VTA, DRN, or RMTg). Of note, ZsGreen expression marks the target area that was injected very brightly whereas mCherry is only expressed in neurons that project from LHb to the site of the target where CAV2-Cre was injected and retrogradely infected the LHb neurons.
Table 1.
Read depth.
| Sample Type | Pathway | Stress | n | Read Depth (mean ± SEM) |
|---|---|---|---|---|
| IP | DRN | Stressed | 3 | 6.624 × 106 (±1.742 × 106) |
| IP | DRN | Unstressed | 3 | 8.601 × 106 (±0.958 × 106) |
| IP | RMTg | Stressed | 3 | 7.957 × 106 (±0.077 × 106) |
| IP | RMTg | Unstressed | 3 | 7.001 × 106 (±0.079 × 105) |
| IP | VTA | Stressed | 3 | 7.055 × 106 (±0.077 × 106) |
| IP | VTA | Unstressed | 3 | 7.217 × 106 (±0.202 × 106) |
| Input | DRN, RMTg, VTA | Stressed | 3 (pooled each pathway) | 11.84 × 106 (±1.667 × 106) |
| Input | DRN, RMTg, VTA | Unstressed | 3 (pooled each pathway) | 10.01 × 107 (±1.371 × 106) |
| IP | Sham | Unstressed | 3 | 5.299 × 106 (±0.773 × 106) |
| Input | Sham | Unstressed | 3 | 8.05 × 106 (±0.813 × 106) |
3.2. Stress induced divergent gene expression in LHb Output pathways
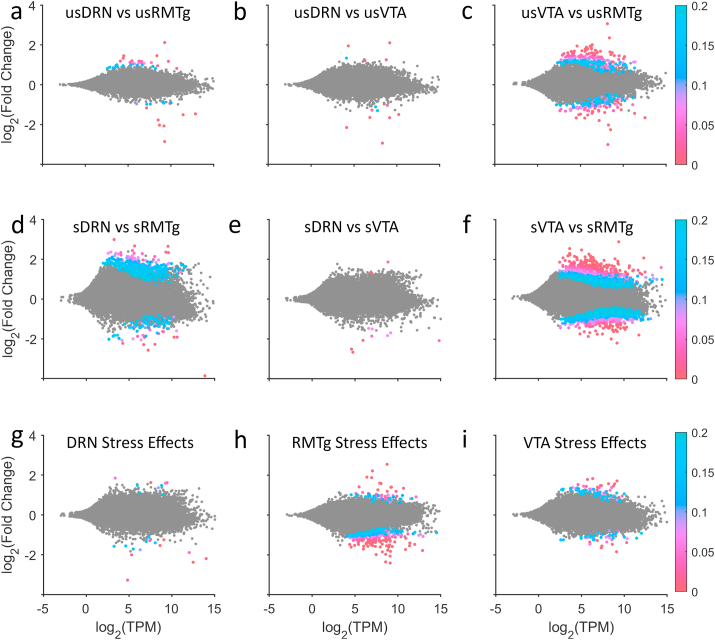
Initially, we compared differential gene expression in the three pathways using pairwise comparisons among the unstressed conditions (usDRN, usVTA, usRMTg), then the stressed conditions (sDRN, sVTA, sRMTg), and then between stressed and unstressed conditions within each pathway using DESeq2 (Fig. 2). In all cases, if the gene was more highly expressed in the first group, the Wald score is positive (upregulated) and if it was more highly expressed in the second group, the Wald score is negative (downregulated). We used an FDR of q = 0.1 for these initial pair-wise comparisons between pathways. Gene expression profiles in the LHb projection neurons were relatively similar among the three pathways under the basal, unstressed condition (Fig. 2a–c). Thirty-one genes were differentially expressed between the unstressed DRN pathway and unstressed RMTg pathway (Fig. 2a). Of these, 21 genes were more highly expressed in the DRN pathway. Eleven genes were differentially expressed between DRN and VTA-projecting LHb neurons (Fig. 2b). The VTA and RMTg pathways showed the greatest difference at baseline, with 236 Differentially Expressed Genes (DEGs) (Fig. 2c). Most of these genes (141) were expressed at greater levels in the VTA pathway compared to the RMTg pathway.
Fig. 2.
Pairwise DESeq2 comparisons. The top row shows comparisons between the unstressed pathways (a–c); there were few differences between LHb neurons projecting to the DRN and neurons projecting to the RMTg or VTA, although several hundred genes differed between the RMTg and VTA pathways at baseline. The middle row shows comparisons between the stressed pathways (d–f). There were only a few differences between the DRN and the VTA pathway, but stress exposure caused dramatic gene expression changes in the RMTg-projecting neurons relative to either the DRN or VTA pathways. The bottom row shows stress effects within each pathway (g–i). As is evident, the neurons projecting to the RMTg had the most differentially expressed genes, and most of those were downregulated.
Interestingly, gene expression in stressed rats showed striking differences between the pathways (Fig. 2d–f). 125 genes were differentially expressed between the stressed DRN and RMTg pathways, and 95 of those genes were more highly expressed in the DRN pathway (Fig. 2d). As with the unstressed comparisons, the stressed DRN and VTA pathways remained similar – only 11 genes were differentially expressed (Fig. 2e). Further, 1,078 genes were differentially expressed between the stressed VTA and RMTg pathways (Fig. 2f). The majority of these genes (657) were expressed at greater levels in the VTA pathway.
We next determined the effects of stress within the neurons of each pathway (Fig. 2g–i). Stress had the smallest impact on gene expression within the DRN-projecting LHb neurons–only 14 genes were different with 5 being upregulated and 9 being downregulated following stress (Fig. 2g). In the RMTg-projecting neurons, 204 genes were changed by stress; with 38 being upregulated and 166 being downregulated (Fig. 2h). 63 genes were changed by stress in the VTA pathway – 40 were upregulated and 23 were downregulated (Fig. 2i).
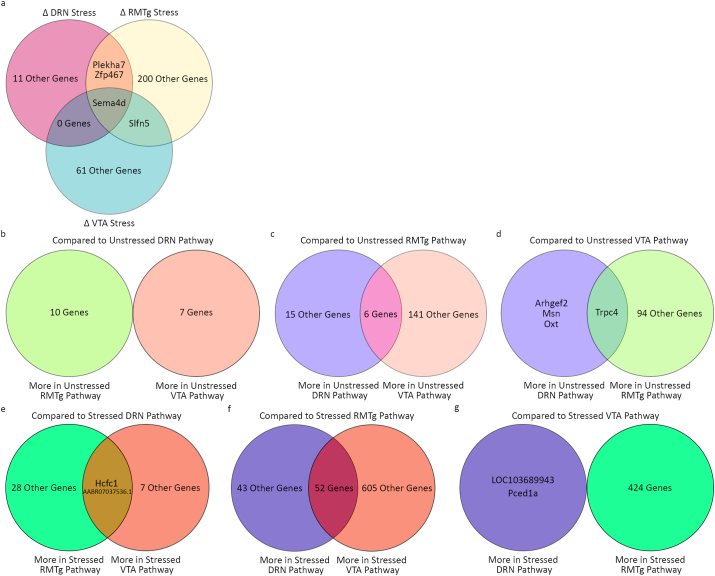
Fig. 3 illustrates overlapping DEGs between and within the pathways. When comparing the effects of stress on each pathway, there were only a small number of gene expression differences that overlapped across pathways. Sema4d was consistently downregulated with stress in all three pathways (Wald scores = -5.2 to −8, q score <0.001). Several other genes (Plekha7, Zfp467, and Slfn5) were significantly differentially expressed in some but not all cases (q < 0.001–0.09) (Fig. 3a). All specific genes mentioned throughout this manuscript can be located in Table 3. When comparing the unstressed pathways, only a few genes overlapped. Compared to usDRN, no genes were more heavily expressed in both the usRMTg and usVTA (Fig. 3b). When compared to the usRMTg pathway, 6 were more highly expressed in both the usDRN and usVTA (Fig. 3c). When comparing to the usVTA pathway, Trpc4 was more highly expressed in both usDRN and usRMTg pathways (usDRN vs usVTA Wald = 4.7, q = 0.006; usVTA vs usRMTg Wald = -3.4, q = 0.06) (Fig. 3d). Oxytocin was positively differentially expressed in the DRN in both of these comparisons (usDRN vs usRMTg Wald = 4.93, q = 0.0011; usDRN vs usVTA Wald = 5.63, q < 0.0001) (Fig. 3c and d). We confirmed the oxytocin expression results with RTqPCR on RiboTag RNA samples from the same brains (Supplemental Figs. 2 and 4). Compared to the sDRN pathway, Hcfc1 (sDRN vs sRMTg Wald = -8.4, q < 0.0001; sDRN vs sVTA Wald = -4.4, q = 0.04) and AABR07037536.1 (sDRN vs sRMTg Wald = -3.4, q = 0.09; sDRN vs sVTA Wald = -5.7, q = 0.0002) were more heavily expressed in the sRMTg and sVTA pathways (Fig. 3e). While there were only a handful of overlapping DEGs between the pairwise comparisons within stressed and unstressed cases, 52 genes were more heavily expressed in both the stressed DRN and VTA pathways than in the stressed RMTg pathway (Fig. 3f). Finally, when compared to the sVTA pathway, no genes were more heavily expressed in both the sDRN and sRMTg pathways (Fig. 3g).
Fig. 3.
Venn diagrams of differentially expressed genes (q < 0.1). a) The intersection of DEGs in stressed vs. unstressed neurons in each pathway were few; Sema4d was the only gene to be downregulated after stress in all pathways. b-d) Pairwise comparisons between RNA from the unstressed conditions revealed few differences between the pathways although the DRN and VTA pathways differed the least from each other and shared the most DEGs relative to RMTg. e-g) RNA in the RMTg-projecting neurons from stressed animals diverged from the other pathways. While the RNA from the DRN and VTA pathways of stressed animals were similar to one another, RNA in the RMTg-projecting neurons had ∼4.5 fold more DEGs than in the comparisons from unstressed animals.
Table 3.
Genes named.
| Gene ID | Context |
|---|---|
| Sema4d | Consistently downregulated after stress across pathways |
| Plekha7 | Upregulated after stress in DRN and RMTg pathways |
| Zfp467 | Downregulated after stress in DRN and RMTg pathways |
| Slfn5 | Downregulated after stress in RMTg and VTA pathways |
| Trpc4 | Less expressed in usVTA than usDRN or usRMTg |
| Oxt | More expressed in usDRN than usVTA or usRMTg |
| Hcfc1 | Less expressed in sDRN than sVTA or sRMTg |
| AABR07037536.1 | Less expressed in sDRN than sVTA or sRMTg |
| Kdr | Highlighted in PI3K signaling pathway |
| Pld2 | Highlighted in PI3K signaling pathway |
| Plxnb1 | Highlighted in PI3K signaling pathway |
| Ptpn13 | Highlighted in PI3K signaling pathway |
| Tek | Highlighted in PI3K signaling pathway and ICSBP transcription factor |
| Col4a1 | Highlighted in ICSBP transcription factor |
| Fryl | Highlighted in ICSBP transcription factor |
| Rrbp1 | Highlighted in ICSBP transcription factor |
| Slc12a4 | Highlighted in ICSBP transcription factor |
| Tnfrsf19 | Highlighted in ICSBP transcription factor |
3.3. Stress produced opposite patterns of gene set enrichment in DRN and VTA pathways as compared to the RMTg pathway
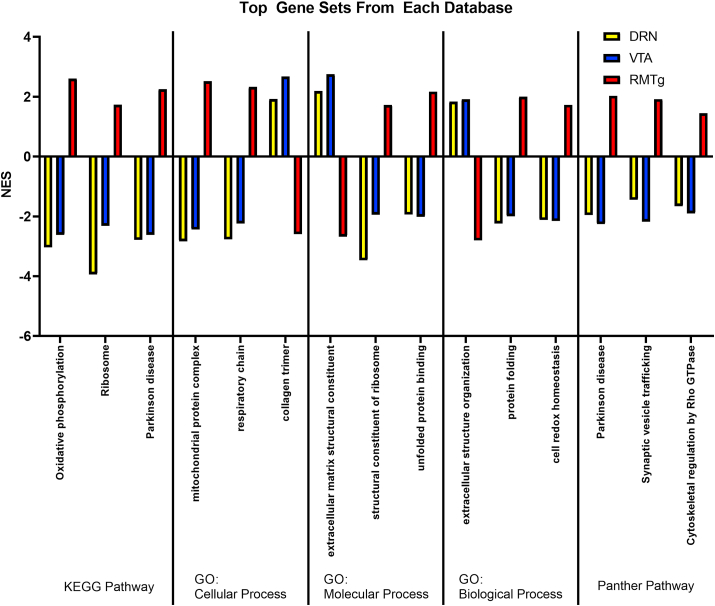
Next we used a curated gene set strategy to evaluate whether there were patterns of changes in sets of related genes using WebGestalt to perform Gene Set Enrichment Analysis (GSEA). The significantly enriched gene sets were similar across different databases and showed a clear pattern: in most cases stress led to a reduction in expression of a particular gene set within the DRN (sDRN vs usDRN) and VTA (sVTA vs usVTA) pathways, but an increase within RMTg (sRMTg vs usRMTg); in the few cases where this pattern was not replicated, stress induced changes in RMTg were always in the opposite direction than in VTA or DRN (Fig. 4). While the function-defined gene sets shared this consistent pattern, gene sets relating to microRNA or transcription factor targets were not prominent nor shared across pathways and are not presented.
Fig. 4.
Gene set enrichment analysis. The average normalized enrichment scores (NES) for stressed vs. unstressed animals for each pathway of the three gene sets found in all three pairwise comparisons with the largest differences are illustrated for each data base queried. Of note, in each gene set, the direction of RMTg changes (i.e. upregulation or downregulation of DEGs) was in the opposite direction as for the DRN and VTA pathways. Unlike other analyses in this paper, GSEA mostly detected gene sets that were upregulated by stress in the RMTg pathway. GO – Gene Ontology.
3.4. Stress downregulated a PI3-Kinase related gene network in the RMTg pathway
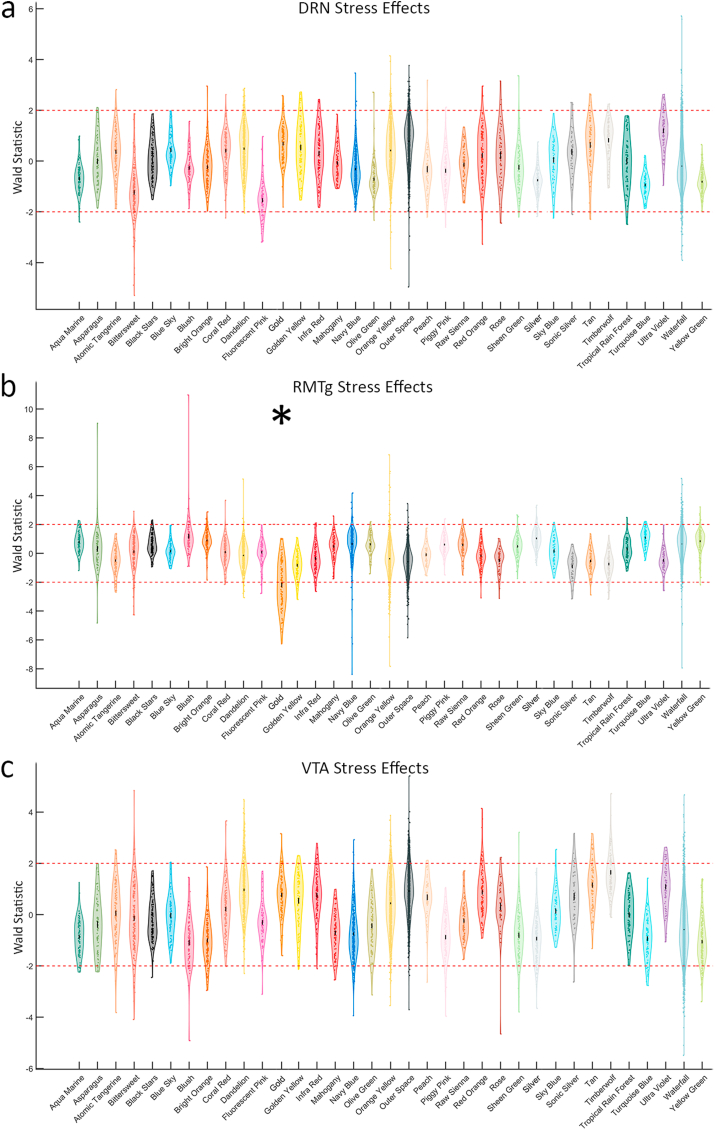
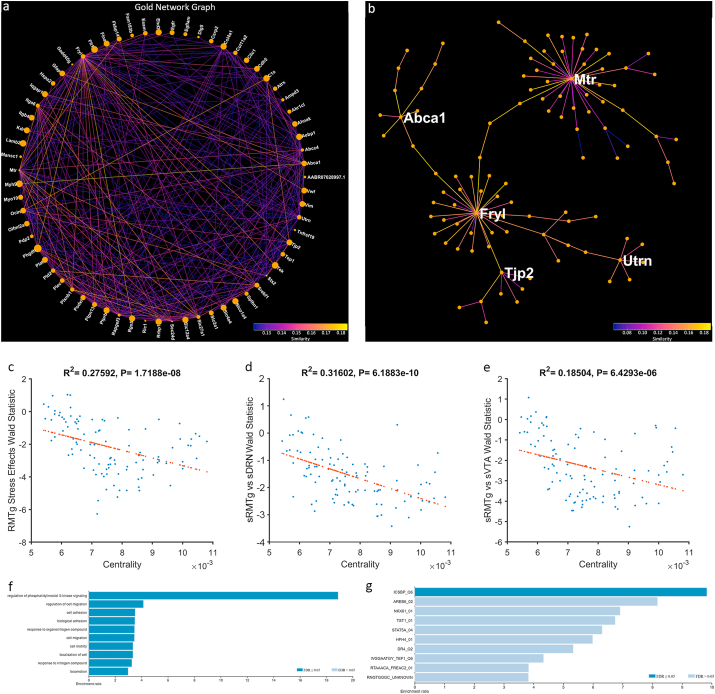
Lastly, we performed Weighted Gene Co-expression Network Analysis (WGCNA), an unguided method for identifying modules of genes with correlated expression. WGCNA uses a special type of correlation matrix called a topological overlap matrix to represent the network connectedness of gene expression across all samples. This matrix was transformed into a hierarchical tree and clustered with the dynamic tree cut library for R (Langfelder and Horvath, 2008). WGCNA modules are randomly re-named using the “Crayola Color Palette” in order to combat attribution of meaning or importance to numbered modules. Initially, WGCNA identified 96 modules; however, many of the module eigengenes were highly correlated. Modules were merged down to 35 using hierarchical clustering with a tree cut height of 1.5. The module colors (Fig. 5a) and q-scores for every gene for each of the nine pairwise comparisons are mapped onto minimum spanning trees in Fig. 5. Notably, while the unstressed pathway comparisons (Fig. 5b–d), and even the within-pathway stress comparisons (Fig. 5h–j) have relatively few low q-values, the stressed DRN vs stressed RMTg (Fig. 5e) and stressed RMTg vs stressed VTA (Fig. 5g) contained many genes with low q-values as illustrated in the pseudocolor scale. Many of these DEGs map to the “Gold” module. Additionally, the distribution of Wald scores for each pair-wise comparison in each module are presented as violin plots in Fig. 6. The Gold module stood out with an average Wald score >±2 in multiple comparisons. The Gold module reached this threshold in the Stressed vs. Unstressed RMTg pathway (average Wald = −2.192, Fig. 6b) and the Stressed RMTg vs. Stressed VTA pathway (average Wald = −2.317, not shown). Additionally, this module reached a less stringent threshold of >±1.5 in the Stressed RMTg vs. Stressed DRN pathway comparison (average Wald = −1.584, not shown). Fig. 7 depicts each gene in this module's importance within the network using a centrality metric (the more central genes have many strong bright lines); the genes most central to this module topologically are analogous to “hub” genes in other schemes. The centrality of individual genes in the Gold module correlated significantly with the Wald score in pairwise comparisons of differential expression for stressed vs. unstressed RMTg (Fig. 7c), stressed RMTg vs stressed DRN (Fig. 7d), and stressed RMTg vs stressed VTA (Fig. 7e). Further, the Timberwolf, Turquoise Blue, and Silver modules reached a threshold of >1.5 in the Stressed DRN vs. Stressed RMTg and Stressed VTA vs. Stressed RMTg pathway comparisons (see Supplemental Code and Data). However, only the Gold and Silver modules demonstrated significant correlations between each gene's centrality in the module and that gene's Wald score. Interestingly, the Gold network genes tended to be decreased after stress in the RMTg pathway (i.e. downregulated) and were similarly downregulated in stressed RMTg compared to either the stressed DRN pathway or the stressed VTA pathway; we interpret this to mean that this gene module as a whole was downregulated in the stressed RMTg pathway.
Fig. 5.
Minimum spanning trees of WGCNA results. a) Two-dimensional minimum spanning trees illustrate the 35 gene modules identified by weighted gene coexpression network analysis (WGCNA). Using the original DESeq2 analysis, the q-values for each individual gene is shown for pairwise comparisons of unstressed DRN to unstressed RMTg (b), unstressed DRN to unstressed VTA (c), and unstressed RMTg to unstressed VTA (d). There were relatively few changes apparent between the pathways in unstressed animals, but after stress, there were larger differences apparent in in the stressed DRN vs. stressed RMTg (e), stressed DRN vs. stressed VTA (f), and stressed RMTg vs. stressed VTA (g). The RMTg vs. VTA showed the most significant differences. (h–j) shows the pairwise comparisons for stressed vs. unstressed DRN (h), RMTg (i), and VTA (j).
Fig. 6.
The Wald score for each gene within the 35 modules assigned by WGCNA. a) Stress effects in the DRN pathway. b) Stress effects in the RMTg pathway. The Gold module is downregulated with stress in the RMTg pathway (average Wald = −2.192) c) Stress effect in the VTA pathway. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 7.
The Gold Network. a) Gold Gene Network projected onto a two-dimensional space using a circle layout. Line color represents pairwise gene expression similarity as calculated by WGCNA. Node size represents centrality to the network. b) Gold Network projected as a minimum spanning tree. The most central genes are labeled. c)Genes which are more downregulated with stress in the RMTg pathway are more central to the network (r2 = 0.276, p < 0.001). d) Genes which are less highly expressed in the stressed RMTg than the stressed DRN pathway are more central to the network (r2 = 0.316, p < 0.001). e) Genes which are less highly expressed in the stressed RMTg than the stressed VTA pathway are more central to the network (r2 = 0.185, p < 0.001). f) Overrepresentation analysis of GO: Biological Process of the entire network. g) Overrepresentation analysis of transcription factor targets for the 25 most central genes to the network. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We next performed an overrepresentation analysis to determine if the genes in the Gold module were associated with identified gene sets using WebGestalt. While there was significant overrepresentation of genes from several pathways, the regulation of PI3K signaling gene set was markedly overrepresented (enrichment ratio = 18.89; Fig. 7f and Table 2). This gene set included Kdr, Pld2, Plxnb1, Ptpn13, and Tek. Furthermore, these genes are downregulated in the stressed RMTg pathway compared to the unstressed RMTg, the stressed VTA or the stressed DRN pathways. Table 2 shows the Wald statistic and q score for each gene in this set for each comparison. Additionally, when investigating the top 25 most central genes in this network, 6 of them are regulated by the transcription factor ICSBP (IRF8) (enrichment ratio = 9.8, FDR = 0.01) (Fig. 7g). This gene set included Col4a1, Fryl, Rrbp1, Slc12a4, Tek, and Tnfrsf19. All specific genes mentioned in this article are located in Table 3.
Table 2.
Phosphoinositide 3 kinase signaling genes in the gold networ.
| Gene Symbol | Gene name | Relative to sRMTg Pathway |
|||||
|---|---|---|---|---|---|---|---|
| usRMTg |
sDRN |
sVTA |
|||||
| Wald | Q | Wald | Q | Wald | Q | ||
| Kdr | kinase insert domain receptor | 4.829 | <0.001 | 2.133 | 0.301 | 3.871 | 0.008 |
| Pld2 | phospholipase D2 | 2.096 | 0.543 | 2.577 | 0.234 | 2.569 | 0.121 |
| Plxnb1 | plexin B1 | 2.512 | 0.331 | 2.139 | 0.301 | 2.426 | 0.147 |
| Ptpn13 | protein tyrosine phosphatase, non-receptor type 13 | 3.339 | 0.068 | 2.532 | 0.237 | 3.060 | 0.052 |
| Tek | TEK receptor tyrosine kinase | 4.067 | 0.008 | 2.128 | 0.303 | 4.389 | 0.002 |
3.5. Validation of downregulation of PI3-Kinase related genes in the stressed RMTg pathway
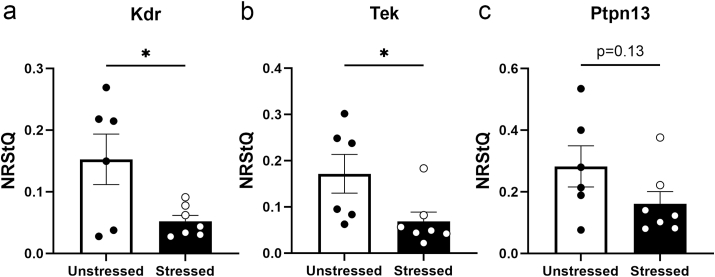
In order to validate the RNAseq results, we repeated the stress procedure in a new cohort of rats that were first injected with AAV-DIO-RiboTag in LHb and CAV2-Cre into RMTg. RiboTag-purified RNA was prepared from RMTg-projecting LHb neurons and used to quantify the expression level of three DEGs from the Gold network identified by WGCNA that are associated with PI3K signaling (Table 2). Kdr and Tek RNAs were significantly reduced in the ribosome-associated RNA from the sRMTg pathway compared to the usRMTg pathway (t (11) = 2.570, p = 0.0260; t (11) = 2.323, p = 0.0404, respectively; Fig. 8) while there was a nonsignificant trend for Ptpn13 to be reduced (t (11) = 1.614, p = 0.1348). These changes replicated the results of the WGCNA analysis of the RNAseq data in a separate cohort of animals indicating that forced swim stress reduced the expression of several genes associated with PI3K signaling in RMTg-projecting LHb neurons.
Fig. 8.
RTqPCR validates downregulation of PI3-Kinase related genes in RMTg-projecting LHb neurons. Based on the RNAseq analysis, we expressed RiboTag in the pathway from LHb to RMTg in a new cohort of control rats (n = 6) and rats subjected to forced swim stress (n = 7). RTqPCR of the ribosome-associated RNA showed that Kdr (a) and Tek (b) were significantly reduced after stress; there was a trend for stress to reduce Ptpn13 (c). ∗p < 0.05.
4. Discussion
In this study we investigated the impact of stress on RNAs actively undergoing translation in LHb neurons comprising the three major output pathways to VTA, DRN, and RMTg. We compared expression of individual genes between pathways and between unstressed and stressed conditions in a pairwise fashion. We found that GSEA and WGCNA revealed patterns in gene expression changes with stress that differed between these efferent LHb pathways. Whereas GSEA is based upon previously identified sets of functionally related genes, WGCNA uses an unbiased method of clustering genes into “modules” based on patterns of expression levels across the entire data set without assumptions about gene function. When we examined the average expression levels of all of the modules that were identified, just one repeatedly demonstrated significant differential expression between experimental groups. In most cases, the gene sets or modules were downregulated in RMTg-projecting LHb neurons after stress but upregulated in DRN- and VTA-projecting neurons after stress.
When we compared the pathways to one another in a pairwise fashion in unstressed rats, there were relatively few differences in RNA expression; in particular, there were no dramatic phenotype-defining differences observed. A recent single cell RNAseq study in mice reached similar conclusions (Wallace et al., 2020). While some LHb neurons express neuropeptides and perhaps GABA, nearly every neuron in the LHb is glutamatergic (Aizawa et al., 2012; Stamatakis and Stuber, 2012; Zuo et al., 2017). Therefore, it is less surprising that the pathways are homogeneous at baseline and only change with stress. Our a priori hypothesis was that the neurons comprising these three pathways might be quite different because the pathways are highly segregated with few collaterals, but this was rejected; however, some differences between these pathways emerged following stress exposure.
Neurons projecting to different target regions are intermixed within subregions of the LHb yet have minimal collateralization between DRN, VTA, and RMTg. Given that the LHb neurons targeting these areas have similar phenotypes at baseline, perhaps the differences become apparent after stress because each pathway receives different input information during stress. The LHb receives inputs from myriad sources throughout the brain. Perhaps, stress-sensitive inputs preferentially synapse onto neurons projecting to specific targets, and differential activation leads to changes in their subsequent gene expression. Indeed, a recent paper found that CaMKII-expressing neurons projecting from the entopeduncular nucleus tended to synapse onto LHb neurons projecting to the VTA; whereas, those from the lateral hypothalamus and VTA tended to synapse onto neurons projecting to the DRN (Cerniauskas et al., 2019).
We found that RMTg-projecting neurons changed the most following stress. This was unexpected since we previously found that the pathway to the DRN was the most involved in adaptations to forced swim immobility (Coffey et al., 2020b). Chemogenetically inhibiting DRN-projecting neurons in between the first and second swim sessions decreased immobility and decreased perseverative seeking in the reward omission task. Conversely, activating these neurons increased perseverative seeking but did not further change immobility in the forced swim test. Inhibiting either of the other two pathways did not change behavior in any of the tests performed. Thus, we initially expected the pathway to the DRN to be the most affected by stress. One possible interpretation of these results is that inhibiting the LHb-RMTg pathway had no effect because the plasticity in this pathway already served to reduce its excitability, and inhibition with hM4Di had no additional impact. Supporting this interpretation, Proulx and colleagues found that acute optogenetic stimulation of RMTg-projecting LHb neurons transiently increased immobility during a single FST session (Proulx et al., 2018), suggesting that it indeed is not maximally activated during forced swim alone. It is interesting to consider that, if the pathway from LHb to RMTg becomes less excitable due to changes in gene expression following forced swim stress, perhaps this leads to a relative increased activation of VTA and DRN by LHb.
The circuitry of the LHb and its major targets, RMTg, DRN, and VTA, is complex. LHb stimulation generally decreases dopaminergic activity within the VTA; although there is also evidence of direct excitation of dopamine neurons (Brown and Shepard, 2016). Similarly, activation of the LHb has complex effects on the output of serotonin from raphe neurons; only high frequency stimulation increased serotonin release into striatum; this was further augmented by bicuculline injection into DRN (Kalen et al., 1989). Indeed, lesions of the habenula ablate the increase in serotonin in the DRN during and after stress (Amat et al., 2001). LHb neurons project to both serotonergic and GABAergic neurons in DRN at roughly equal rates (Weissbourd et al., 2014). While direct projections to VTA or DRN may either activate monoaminergic or GABAergic neurons, the projection to RMTg innervates only GABAergic neurons which in turn inhibit VTA and DRN (Yang et al., 2018). In this report we observed differential gene expression plasticity between neurons projecting to VTA and DRN on the one hand and RMTg on the other. Since the changes in RMTg-projecting neurons involved reduced expression of genes associated with excitability (such as in the PI3K signaling pathway), perhaps the overall balance of LHb stimulation of downstream GABAergic tone is reduced after repeated forced swim stress.
There is some diversity in the responses of LHb neurons to aversive stimuli. Both inescapable shock and reward omission increase firing in subpopulations of the LHb, exciting 30–50% of these neurons (Lecca et al., 2017; Shabel et al., 2019). Furthermore, roughly 10% of LHb neurons are actually inhibited by foot-shocks (Congiu et al., 2019). Further, RMTg-projecting LHb neurons have shown increased excitability 24 h to 14 days after cocaine exposure, but VTA-projecting neurons did not (Maroteaux and Mameli, 2012; Meye et al., 2015). Thus, it is possible that the observed stress-induced reduction in the excitability of LHb projections to RMTg disinhibits the VTA and DRN monoaminergic outputs. On the contrary, activation of LHb-RMTg neurons optogenetically interferes with reward motivated behavior and reduces activity in the FST (Proulx et al., 2018). Perhaps the pathway to the RMTg becomes less active following an inescapable stressor via an allostatic adaptation in neuronal excitability.
Cerniauskas et al. (2019) characterized the effect of chronic mild stress on LHb neurons projecting to either the VTA/RMTg or the DRN. They observed greater excitability in neurons projecting to the VTA/RMTg in mice with the greatest composite depression-like behaviors. No such pattern was observed in the pathway to the DRN. Further, chemogenetic activation of LHb-VTA/RMTg neurons produced a depression-like phenotype in stress-naïve mice. Conversely, DREADD-mediated inhibition of this pathway in stressed mice reduced their depression-like behaviors. Their data suggested that entopeduncular nucleus projections to LHb neurons that in turn project to VTA may be a critical functional circuit. Surgical targeting of VTA without hitting RMTg is difficult in mice, while these target areas can reliably be discriminated with retrograde strategies in rats (Coffey et al., 2020b; Fu et al., 2017; Goncalves et al., 2012), as we did in this study, so it is difficult to directly compare our results and theirs. Additionally, this study included pathway-specific scRNAseq investigating transcriptional differences between the LHb neurons projecting to the DRN and those to the VTA/RMTg as well as differences with the LHb-VTA/RMTg neurons from mice which displayed prolonged immobility in the Tail Suspension Test and those which did not. When comparing our data to theirs, a few caveats must be considered. The transcriptome and translatome represent distinct RNA pools and will inevitably be different. Additionally, Cerniauskas et al. (2019) probed the cells electrophysiologically prior to collection; whereas, we homogenized tissue and extracted actively translating ribosomes. Each of these methods may result in different artifacts within the data by altering transcription or translation. However, several of the gene expression changes between pathways were similar to changes we observed. While none of the more highly-expressed genes in the DRN-projecting neurons, nor any genes which differed in relation to the tail suspension test criteria were differentially expressed in our study, several of the genes which they found that were more heavily expressed in the pathway to the VTA/RMTg approached significance in our data. Specifically, Grid 2 and Sytl2 trended towards increased expression in the sRMTg versus sVTA comparison (q = 0.126 and 0.18, respectively). Interestingly, both genes were more heavily expressed in the sRMTg pathway.
While our GSEA results suggest that gene expression in these pathways diverge after stress, we focused on the unbiased topological analysis with WGCNA, which also indicated that the RMTg-projecting neurons diverged from the VTA- and DRN-projecting neurons. WGCNA clusters genes with no a priori knowledge of function; however, it is not uncommon for modules to have overrepresentations of biologically relevant sets of genes. This was exemplified by the Gold module, which had an overrepresentation of downregulated genes associated with PI3K signaling after stress, found both via GO: Biological Process and the KEGG database. In order to validate these results, we performed RTqPCR from ribosome-associated RNA isolated from the RMTg-projecting neurons in a new cohort of control and stressed rats. We measured the three differentially expressed PI3K-associated genes identified by WGCNA and confirmed the downregulation of Kdr and Tek while Ptpn13 exhibited a similar trend. We propose that this reduction in PI3K-associated genes may reduce excitability of RMTg-projecting LHb neurons. PI3K signaling regulates anxiety-like behavior, may be involved in the antidepressant effects of deep brain stimulation, and is implicated in the mood-stabilizing effects of lithium in post-mortem analyses of patients with major depressive disorder (Voleti and Duman, 2012). Phosphorylation of PI3K is implicated in the antidepressant effects of baicalin in a mouse model of depression (Guo et al., 2019). We predict that downregulation of PI3K in the RMTg-projecting neurons after stress would be a compensatory adaptation that reduces their excitability, thereby disinhibiting the monoaminergic neurons in the DRN and VTA. Since we examined RNA actively undergoing translation in this study, the impact of these changes on protein production might take additional time to manifest. Future studies should examine the role of PI3K signaling in this pathway on resilience after stress exposure.
5. Conclusions
In summary, RNA expression in the major efferent pathways from the LHb was more similar than anticipated at baseline; however, stress led to divergent patterns of gene expression. In particular, the RMTg-projecting neurons had a downregulation of PI3K signaling genes, which might lead to an altered balance of activity of these pathways during the response to stress.
Funding
This work was supported by MH106532 (JFN), NS099578 (MRL), and DA007278 (KRC).
CRediT authorship contribution statement
Marjorie R. Levinstein: Conceptualization, Methodology, Formal analysis, Investigation, Writing - original draft, Visualization. Kevin R. Coffey: Methodology, Software, Visualization, Writing - review & editing. Russell G. Marx: Methodology, Software. Atom J. Lesiak: Conceptualization, Methodology. John F. Neumaier: Conceptualization, Resources, Supervision, Funding acquisition, Project administration, Writing - review & editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2020.100268.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aizawa H., Kobayashi M., Tanaka S., Fukai T., Okamoto H. Molecular characterization of the subnuclei in rat habenula. J. Comp. Neurol. 2012;520:4051–4066. doi: 10.1002/cne.23167. [DOI] [PubMed] [Google Scholar]
- Amat J., Sparks P.D., Matus-Amat P., Griggs J., Watkins L.R., Maier S.F. The role of the habenular complex in the elevation of dorsal raphe nucleus serotonin and the changes in the behavioral responses produced by uncontrollable stress. Brain Res. 2001;917:118–126. doi: 10.1016/s0006-8993(01)02934-1. [DOI] [PubMed] [Google Scholar]
- Bernard R., Veh R.W. Individual neurons in the rat lateral habenular complex project mostly to the dopaminergic ventral tegmental area or to the serotonergic raphe nuclei. J. Comp. Neurol. 2012;520:2545–2558. doi: 10.1002/cne.23080. [DOI] [PubMed] [Google Scholar]
- Brinschwitz K., Dittgen A., Madai V.I., Lommel R., Geisler S., Veh R.W. Glutamatergic axons from the lateral habenula mainly terminate on gabaergic neurons of the ventral midbrain. Neuroscience. 2010;168:463–476. doi: 10.1016/j.neuroscience.2010.03.050. [DOI] [PubMed] [Google Scholar]
- Brown P.L., Shepard P.D. Functional evidence for a direct excitatory projection from the lateral habenula to the ventral tegmental area in the rat. J. Neurophysiol. 2016;116:1161–1174. doi: 10.1152/jn.00305.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne C.A., Hammack R., Lucki I. Dysregulation of the lateral habenula in major depressive disorder. Front. Synaptic Neurosci. 2018;10:46. doi: 10.3389/fnsyn.2018.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerniauskas I., Winterer J., de Jong J.W., Lukacsovich D., Yang H., Khan F., Peck J.R., Obayashi S.K., Lilascharoen V., Lim B.K., Foldy C., Lammel S. Chronic stress induces activity, synaptic, and transcriptional remodeling of the lateral habenula associated with deficits in motivated behaviors. Neuron. 2019;104:899–915. doi: 10.1016/j.neuron.2019.09.005. e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.S., Sexton T.J., McClain M., Root D., Kohen R., Neumaier J.F. Overexpression of 5-ht1b receptor in dorsal raphe nucleus using herpes simplex virus gene transfer increases anxiety behavior after inescapable stress. J. Neurosci. 2002;22:4550–4562. doi: 10.1523/JNEUROSCI.22-11-04550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey K.R., Barker D.J., Ma S., West M.O. Building an open-source robotic stereotaxic instrument. J Vis Exp. e51006. 2013 doi: 10.3791/51006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey K.R., Lesiak A.J., Marx R.G., Vo E.K., Garden G.A., Neumaier J.F. Ribotag-seq reveals a compensatory camp responsive gene network in striatal microglia induced by morphine withdrawal. 2020. bioRxiv. [DOI]
- Coffey K.R., Marx R.G., Vo E.K., Nair S.G., Neumaier J.F. Chemogenetic inhibition of lateral habenula projections to the dorsal raphe nucleus reduces passive coping and perseverative reward seeking in rats. Neuropsychopharmacology. 2020 doi: 10.1038/s41386-020-0616-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congiu M., Trusel M., Pistis M., Mameli M., Lecca S. Opposite responses to aversive stimuli in lateral habenula neurons. Eur. J. Neurosci. 2019;50:2921–2930. doi: 10.1111/ejn.14400. [DOI] [PubMed] [Google Scholar]
- Fu R., Mei Q., Zuo W., Li J., Gregor D., Bekker A., Ye J. Low-dose ethanol excites lateral habenula neurons projecting to vta, rmtg, and raphe. Int J Physiol Pathophysiol Pharmacol. 2017;9:217–230. [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Trimble M. The lateral habenula: No longer neglected. CNS Spectr. 2008;13:484–489. doi: 10.1017/s1092852900016710. [DOI] [PubMed] [Google Scholar]
- Gill M.J., Ghee S.M., Harper S.M., See R.E. Inactivation of the lateral habenula reduces anxiogenic behavior and cocaine seeking under conditions of heightened stress. Pharmacol. Biochem. Behav. 2013;111:24–29. doi: 10.1016/j.pbb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves L., Sego C., Metzger M. Differential projections from the lateral habenula to the rostromedial tegmental nucleus and ventral tegmental area in the rat. J. Comp. Neurol. 2012;520:1278–1300. doi: 10.1002/cne.22787. [DOI] [PubMed] [Google Scholar]
- Gruber C., Kahl A., Lebenheim L., Kowski A., Dittgen A., Veh R.W. Dopaminergic projections from the vta substantially contribute to the mesohabenular pathway in the rat. Neurosci. Lett. 2007;427:165–170. doi: 10.1016/j.neulet.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Guo L.T., Wang S.Q., Su J., Xu L.X., Ji Z.Y., Zhang R.Y., Zhao Q.W., Ma Z.Q., Deng X.Y., Ma S.P. Baicalin ameliorates neuroinflammation-induced depressive-like behavior through inhibition of toll-like receptor 4 expression via the pi3k/akt/foxo 1 pathway. J. Neuroinflammation. 2019;16:95. doi: 10.1186/s12974-019-1474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L.N., Zhang L., Li L.B., Sun Y.N., Wang Y., Chen L., Guo Y., Zhang Y.M., Zhang Q.J., Liu J. Activation of serotonin(2c) receptors in the lateral habenular nucleus increases the expression of depression-related behaviors in the hemiparkinsonian rat. Neuropharmacology. 2015;93:68–79. doi: 10.1016/j.neuropharm.2015.01.024. [DOI] [PubMed] [Google Scholar]
- Hashikawa Y., Hashikawa K., Rossi M.A., Basiri M.L., Liu Y., Johnston N.L., Ahmad O.R., Stuber G.D. Transcriptional and spatial resolution of cell types in the mammalian habenula. Neuron. 2020;106:1–16. doi: 10.1016/j.neuron.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalen P., Strecker R.E., Rosengren E., Bjorklund A. Regulation of striatal serotonin release by the lateral habenula-dorsal raphe pathway in the rat as demonstrated by in vivo microdialysis: role of excitatory amino acids and gaba. Brain Res. 1989;492:187–202. doi: 10.1016/0006-8993(89)90901-3. [DOI] [PubMed] [Google Scholar]
- Kowski A.B., Veh R.W., Weiss T. Dopaminergic activation excites rat lateral habenular neurons in vivo. Neuroscience. 2009;161:1154–1165. doi: 10.1016/j.neuroscience.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Lammel S., Lim B.K., Ran C., Huang K.W., Betley M.J., Tye K.M., Deisseroth K., Malenka R.C. Input-specific control of reward and aversion in the ventral tegmental area. Nature. 2012;491:212–217. doi: 10.1038/nature11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. Wgcna: an r package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecca S., Meye F.J., Trusel M., Tchenio A., Harris J., Schwarz M.K., Burdakov D., Georges F., Mameli M. Aversive stimuli drive hypothalamus-to-habenula excitation to promote escape behavior. Elife. 2017;6 doi: 10.7554/eLife.30697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.H., Huang S.L. Role of lateral habenula in the regulation of exploratory behavior and its relationship to stress in rats. Behav. Brain Res. 1988;30:265–271. doi: 10.1016/0166-4328(88)90169-6. [DOI] [PubMed] [Google Scholar]
- Lesiak A.J., Brodsky M., Neumaier J.F. Ribotag is a flexible tool for measuring the translational state of targeted cells in heterogeneous cell cultures. Biotechniques. 2015;58:308–317. doi: 10.2144/000114299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak A.J., Coffey K., Cohen J., Liang K.J., Chavkin C., Neumaier J.F. Sequencing the serotonergic neuron translatome reveals a new role for fkbp5 in stress. Mol. Psychiatry. 2020 doi: 10.1038/s41380-020-0750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak A.J., Neumaier J.F. Ribotag: not lost in translation. Neuropsychopharmacology. 2016;41:374–376. doi: 10.1038/npp.2015.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Q., Takada M., Shinonaga Y., Mizuno N. The sites of origin of dopaminergic afferent fibers to the lateral habenular nucleus in the rat. J. Comp. Neurol. 1993;333:118–133. doi: 10.1002/cne.903330110. [DOI] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. Webgestalt 2019: gene set analysis toolkit with revamped uis and apis. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X.F., Zhang B.L., Li J.C., Yang Y.Y., Sun Y.F., Zhao H. Lateral habenula as a link between dopaminergic and serotonergic systems contributes to depressive symptoms in Parkinson's disease. Brain Res. Bull. 2015;110:40–46. doi: 10.1016/j.brainresbull.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Maroteaux M., Mameli M. Cocaine evokes projection-specific synaptic plasticity of lateral habenula neurons. J. Neurosci. 2012;32:12641–12646. doi: 10.1523/JNEUROSCI.2405-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meye F.J., Valentinova K., Lecca S., Marion-Poll L., Maroteaux M.J., Musardo S., Moutkine I., Gardoni F., Huganir R.L., Georges F., Mameli M. Cocaine-evoked negative symptoms require ampa receptor trafficking in the lateral habenula. Nat. Neurosci. 2015;18:376–378. doi: 10.1038/nn.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S.G., Strand N.S., Neumaier J.F. Dreadding the lateral habenula: a review of methodological approaches for studying lateral habenula function. Brain Res. 2013;1511:93–101. doi: 10.1016/j.brainres.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S.K., Cohen J.Y., Hwang D., Uchida N., Watabe-Uchida M. Organization of monosynaptic inputs to the serotonin and dopamine neuromodulatory systems. Cell Rep. 2014;8:1105–1118. doi: 10.1016/j.celrep.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko N., Bell R., Sesack S.R. Lateral habenula projections to dopamine and gaba neurons in the rat ventral tegmental area. Eur. J. Neurosci. 2009;30:1239–1250. doi: 10.1111/j.1460-9568.2009.06924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootsuka Y., Mohammed M. Activation of the habenula complex evokes autonomic physiological responses similar to those associated with emotional stress. Phys. Rep. 2015;3 doi: 10.14814/phy2.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx C.D., Aronson S., Milivojevic D., Molina C., Loi A., Monk B., Shabel S.J., Malinow R. A neural pathway controlling motivation to exert effort. Proc. Natl. Acad. Sci. U. S. A. 2018;115:5792–5797. doi: 10.1073/pnas.1801837115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quina L.A., Tempest L., Ng L., Harris J.A., Ferguson S., Jhou T.C., Turner E.E. Efferent pathways of the mouse lateral habenula. J. Comp. Neurol. 2015;523:32–60. doi: 10.1002/cne.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: a language and environment for statistical computing. [Google Scholar]
- Root D.H., Hoffman A.F., Good C.H., Zhang S., Gigante E., Lupica C.R., Morales M. Norepinephrine activates dopamine d4 receptors in the rat lateral habenula. J. Neurosci. 2015;35:3460–3469. doi: 10.1523/JNEUROSCI.4525-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E., Yang L., Su T., Morris D.R., McKnight G.S., Amieux P.S. Cell-type-specific isolation of ribosome-associated mrna from complex tissues. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel S.J., Wang C., Monk B., Aronson S., Malinow R. Stress transforms lateral habenula reward responses into punishment signals. Proc. Natl. Acad. Sci. U. S. A. 2019;116:12488–12493. doi: 10.1073/pnas.1903334116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Ruan X., Zhao H. Stimulation of midbrain dopaminergic structures modifies firing rates of rat lateral habenula neurons. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.M., Stuber G.D. Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nat. Neurosci. 2012;15:1105–1107. doi: 10.1038/nn.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voleti B., Duman R.S. The roles of neurotrophic factor and wnt signaling in depression. Clin. Pharmacol. Ther. 2012;91:333–338. doi: 10.1038/clpt.2011.296. [DOI] [PubMed] [Google Scholar]
- Wagner F., French L., Veh R.W. Transcriptomic-anatomic analysis of the mouse habenula uncovers a high molecular heterogeneity among neurons in the lateral complex, while gene expression in the medial complex largely obeys subnuclear boundaries. Brain Struct. Funct. 2016;221:39–58. doi: 10.1007/s00429-014-0891-9. [DOI] [PubMed] [Google Scholar]
- Wallace M.L., Huang K.W., Hochbaum D., Hyun M., Radeljic G., Sabatini B.L. Anatomical and single-cell transcriptional profiling of the murine habenular complex. Elife. 2020;9 doi: 10.7554/eLife.51271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbourd B., Ren J., DeLoach K.E., Guenthner C.J., Miyamichi K., Luo L. Presynaptic partners of dorsal raphe serotonergic and gabaergic neurons. Neuron. 2014;83:645–662. doi: 10.1016/j.neuron.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.M. Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry. 2016;3:472–480. doi: 10.1016/S2215-0366(15)00579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtshafter D., Asin K.E., Pitzer M.R. Dopamine agonists and stress produce different patterns of fos-like immunoreactivity in the lateral habenula. Brain Res. 1994;633:21–26. doi: 10.1016/0006-8993(94)91517-2. [DOI] [PubMed] [Google Scholar]
- Xie G., Zuo W., Wu L., Li W., Wu W., Bekker A., Ye J.H. Serotonin modulates glutamatergic transmission to neurons in the lateral habenula. Sci. Rep. 2016;6:23798. doi: 10.1038/srep23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Wang H., Hu J., Hu H. Lateral habenula in the pathophysiology of depression. Curr. Opin. Neurobiol. 2018;48:90–96. doi: 10.1016/j.conb.2017.10.024. [DOI] [PubMed] [Google Scholar]
- Zhang H., Li K., Chen H.S., Gao S.Q., Xia Z.X., Zhang J.T., Wang F., Chen J.G. Dorsal raphe projection inhibits the excitatory inputs on lateral habenula and alleviates depressive behaviors in rats. Brain Struct. Funct. 2018;223:2243–2258. doi: 10.1007/s00429-018-1623-3. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Feng J.J., Yang S., Liu X.F., Li J.C., Zhao H. Lateral habenula as a link between thyroid and serotoninergic system modiates depressive symptoms in hypothyroidism rats. Brain Res. Bull. 2016;124:198–205. doi: 10.1016/j.brainresbull.2016.05.007. [DOI] [PubMed] [Google Scholar]
- Zuo W., Wang L., Chen L., Krnjevic K., Fu R., Feng X., He W., Kang S., Shah A., Bekker A., Ye J.H. Ethanol potentiates both gabaergic and glutamatergic signaling in the lateral habenula. Neuropharmacology. 2017;113:178–187. doi: 10.1016/j.neuropharm.2016.09.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.