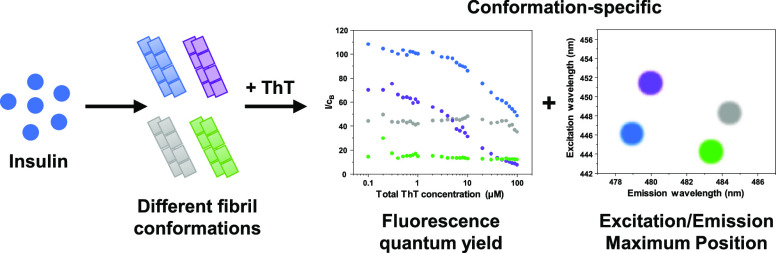
Abstract
Amyloidogenic protein aggregation into highly structured fibrils is linked to more than 30 amyloidoses, including several neurodegenerative disorders. Despite significant progress in trying to understand the process of amyloid formation, there is still no cure or effective treatment available. A number of studies involving potential anti-amyloid compounds rely on the use of a fluorescent probe—thioflavin-T—to track the appearance, growth, or disassembly of these cytotoxic aggregates. Despite the wide application of this dye molecule, its interaction with amyloid fibrils is still poorly understood. Recent reports have shown it may possess distinct binding modes and fluorescence intensities based on the conformation of the examined fibrils. In this work, we generate insulin fibrils under four different conditions and attempt to identify distinct conformations using both classic methods, such as atomic force microscopy and Fourier-transform infrared spectroscopy, as well as their ThT binding ability and fluorescence quantum yield. We show that there is a significant variance of ThT fluorescence quantum yields, excitation/emission maxima positions, and binding modes between distinct insulin fibril conformations.
Introduction
Protein misfolding and association into amyloid fibrils is linked with more than 30 amyloidoses, which lead to organ dysfunction, including neurodegenerative disorders, such as Alzheimer’s or Parkinson’s disease.1−3 These afflictions affect millions of people worldwide,4 and the number is projected to increase further as the average human lifespan continues to increase.5,6 Currently, both the mechanism7,8 and the resulting structure9,10 of such aggregates are still not fully understood, which is one of the main reasons why there is still no effective cure or treatment available.11
To better understand the process of how these fibrils form, what their resulting structure is, and how potential drug molecules are able to alter the aggregation process, countless experiments are conducted in vitro using amyloidogenic proteins.12−16 To track this process and determine whether there are amyloids present in the sample, multiple techniques are used. These include atomic force microscopy (AFM),17,18 light scattering,19 Fourier-transform infrared (FTIR) spectroscopy,20 and fluorescence spectroscopy.21 One common fluorescent probe used to investigate the kinetics of fibrilization, as well as to quantify the concentration of fibrils, is thioflavin-T (ThT).22,23 Upon binding to β-sheet grooves located on the surface of amyloids, the conformation of ThT becomes locked, which results in both a red shift of its excitation/emission wavelengths and a significant increase in its fluorescence quantum yield.24
Despite the relatively simple structure and specific affinity toward amyloid fibrils, this dye molecule has been proven to be far more complex than initially perceived. One interesting aspect of ThT is its ability to bind to fibrils in more than one mode,25 with distinct modes having specific excitation and emission wavelength maximum positions and fluorescence quantum yields.26,27 Multiple experimental and computational reports have shown that the number of binding modes for ThT can vary from one to six,25,28−32 depending on the protein that the fibril is composed of. It was also demonstrated that α-synuclein33 and prion proteins34 can form two distinct conformations that differ by their ThT fluorescence intensity. In addition to this, we have also recently observed distinct fluorescence intensities for two insulin conformations generated at different protein concentrations,35 as well as a massive increase in ThT fluorescence when EGCG was used to alter the aggregation reaction.36
Insulin is a peptide hormone that regulates carbohydrate, fat, and protein metabolism. Its aggregation is associated with insulin-derived amyloidosis, localized at injection sites in patients with diabetes.37 Considering that insulin is also a model protein used to study fibril formation,38 as well as to screen potential anti-amyloid drugs,39−41 such conformation-specific variation in the ThT fluorescence intensity could significantly alter the experimental results. If a compound is capable of slightly altering the structure of fibrils, which results in a different or additional ThT binding mode, this would, in turn, cause an increase or decrease in the signal’s intensity, leading to a conclusion that the molecule worked as an aggregation enhancer or inhibitor. The occurrence of such an event is not unlikely, as it is known that variations in pH42 or concentration35 can have a sizeable impact on the resulting structure of insulin fibrils. On the other hand, if the differences in ThT signal intensities or excitation/emission maxima positions are conformation-specific, then this factor could also be used as a quick and simple way of distinguishing between insulin fibril conformations and may be applied to other amyloidogenic proteins, such as the previously mentioned α-synuclein33 and prion proteins.34
Materials and Methods
Human recombinant insulin (Sigma-Aldrich, Cat. No. 91077C) was dissolved in either 20% acetic acid solution (AC), containing 100 mM NaCl, 100 mM sodium phosphate buffer (pH 2.0), 100 mM sodium phosphate buffer, containing 100 mM NaCl (pH 2.4) or phosphate buffer saline (pH 7.4) to a final protein concentration of 200 μM and placed into 1.5 mL test tubes (Fisher, Cat. No. 15432545) to a final volume of 1 mL (Table 1). The samples were incubated at 60 °C for 24 h in a Ditabis Thermomixer. The phosphate-buffered saline (PBS) samples also contained two glass beads each (Merck, Cat. No. 104015) and were agitated at 600 rpm throughout the incubation period.
Table 1. Insulin Sample Aggregation Reaction Conditions.
| sample name | reaction solution | agitation |
|---|---|---|
| AC | 20% acetic acid | none |
| 100 mM NaCl | ||
| PH20 | 100 mM sodium phosphate buffer (pH 2.0) | none |
| PH24 | 100 mM sodium phosphate buffer (pH 2.4) | none |
| 100 mM NaCl | ||
| PH74 | phosphate buffer saline (pH 7.4) | 600 rpm + 2 glass beads |
Atomic Force Microscopy
Each sample (30 μL) was deposited on mica disks, left to adsorb for 1 min, gently washed with 1 mL of H2O, and dried using airflow. AFM images were scanned using a Dimension Icon (Bruker) atomic force microscope (tapping mode), equipped with a silicon cantilever (40 N/m, Budget Sensors). The 1024 × 1024 pixel resolution images were acquired using a scan rate of 0.5 Hz. AFM images were analyzed using Gwyddion 2.5.5 software. The fibril height and width were determined by tracing a line perpendicular to the fibril axis, while the fibril length was determined by tracing parallel to the fibril axis. The height, width, and length distributions were calculated from 50 traces for each sample.
Fourier-Transform Infrared Spectroscopy
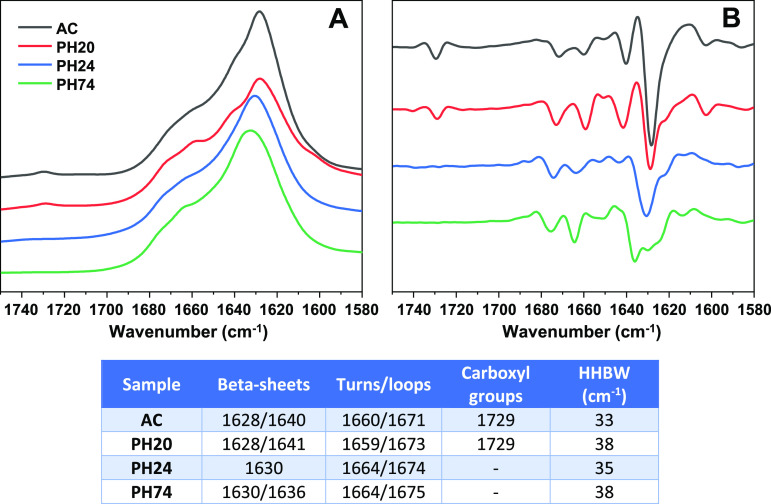
Each sample was centrifuged at 10 000g for 30 min, after which the supernatant was removed and fibrils were resuspended in D2O. The centrifugation and resuspension steps were repeated four times. Finally, the fibrils were resuspended in a small volume of D2O (final fibril concentration ∼10 mg/mL). The fibril samples were sonicated for 1 min using a Bandelin Sonopuls ultrasonic homogenizer with an MS 73 tip (40% total power). The FTIR spectra were recorded using a Vertex 80v (Bruker) IR spectrometer with a mercury cadmium telluride detector at room temperature under near-vacuum conditions. A total of 256 interferograms with 2 cm–1 resolution were averaged. The spectrum of D2O was subtracted from each sample’s spectrum. All spectra were normalized to the same area of amide I/I′ band (1700–1580 cm–1) using GRAMS software. To calculate the amide I/I′ band’s width at its half-height (HHBW),43 the spectra were baseline-corrected between 1700 and 1580 cm–1 before normalization.
Sample Preparation for ThT Binding Assays
Each sample was centrifuged at 10 000g for 30 min, after which the supernatant was removed and the fibrils were resuspended in MilliQ H2O. The centrifugation and resuspension procedure was repeated five times. Each sample was then sonicated for 1 min using a Bandelin Sonopuls ultrasonic homogenizer with an MS 73 tip (40% total power). The samples were then combined to a final protein concentration of 200 μM. The combined solution was then further sonicated for 10 min using the previously described method with 30 s sonication/rest intervals. The resuspension and sonication procedure resulted in highly dispersed and fragmented aggregates (Figure S1A–D), with a greatly reduced average fibril length (Figure S2A,B). The four fibril samples had similar width, and the height distribution remained comparable to the untreated fibrils (Figure S1E,F).
ThT (Sigma-Aldrich, Cat. No. T3516) was dissolved in MilliQ water to a final concentration of 10 mM. The exact concentration was determined by taking an aliquot of the dye solution, diluting it 200 times with MilliQ water and scanning its absorbance at 412 nm (ε412 = 23 250 M–1 cm–1). The ThT solution was then diluted to prepare 200, 20, and 2 μM stock solutions. The sonicated fibrils were mixed with ThT stock solutions and H2O to a range of ThT concentrations (final protein concentration was 100 μM in all cases).
ThT Excitation–Emission Matrix (EEM) Analysis
Each fibril–ThT solution (100 μL) was placed in a 3 mm pathlength cuvette, and its excitation–emission matrix was scanned using a Varian Cary Eclipse fluorescence spectrophotometer (excitation range was 435–465 nm with 1 nm steps and 5 nm slit width, emission range was 460–500 nm with 1 nm steps and 2.5 nm slit width; all other device parameters were kept the same for all sample scans). Three EEMs were scanned for every sample, the background spectrum was subtracted, and the resulting matrices were averaged. Due to slightly different fibril cross-interactions in the presence of ThT, the EEM areas where light scattering is prevalent cannot be accurately subtracted. Therefore, for further data analysis, the EEM region present 7 nm or less away from the excitation wavelength was not taken into account.
To correct for the inner filter effect caused by ThT, the absorbance spectra of each sample were scanned from 300 to 600 nm using 1 nm steps (each spectrum was the average of three repeats). The spectrum of fibrils without ThT was subtracted from each fibril–ThT spectrum. Because of ThT-induced differences in fibril association, the fibril spectrum could not be subtracted by a factor of 1 in certain cases (light absorbance differences induced by fibril clumping). The fibril spectrum was therefore subtracted by multiplying it by a certain factor (usually between 0.9 and 1.1) until there was no slope observed in the 550–600 nm range in the resulting spectrum.
The inner filter effect was corrected for every EEM point by using the following equation
| 1 |
where AEx is the sample’s absorbance at the excitation wavelength, AEm is the sample’s absorbance at the emission wavelength, Im is the signal intensity observed during measurement, and Ic is the corrected signal intensity.
The EEM intensity “center of mass” was calculated by selecting the top 10% intensity values and using the following equation
| 2 |
where λ is the wavelength of either the excitation or emission center of mass, λn is the excitation or emission wavelength, ∑In is the sum of all signal intensities at λn, and ∑Ia is the sum of all signal intensities.
This method helps avoiding the discrepancy caused by variations in light scattering at emission wavelengths close to the excitation wavelength, as well as EEM maxima position deviations due to background noise.
Bound ThT Concentration Determination
Two methods used to determine the concentration of fibril-bound ThT include sample centrifugation26 or sample dialysis.29 Since each sample is sonicated, certain fibril types become difficult to separate from solution by centrifugation. Microdialysis techniques can lack accuracy at low ThT concentrations due to dye–membrane association and the relatively long time needed that can result in fibril clumping. To determine the concentration of bound ThT, a different method was devised based on two ThT–fibril interaction factors. First, based on the reported dye binding constants,29 when the sample contains 100 μM insulin aggregates and 1 μM or less ThT, the majority of it should be bound to fibrils. Second, the difference between the absorbance spectra of free and bound ThT is much greater than the difference between spectra of ThT bound in distinct binding modes.
In this case, the absorbance spectra of ThT in the range from 0.1 to 1.0 μM should be the result of bound ThT molecules. By taking the absorbance values at 412 nm (free ThT spectrum maximum position) and 450 nm (bound ThT absorbance spectrum shifts toward higher wavelengths), we can calculate the extinction coefficients of bound ThT at these two wavelengths (ε412 and ε450) for all four fibril samples. Since the ε412 and ε450 values are known for free ThT (23 250 and 5880 M–1 cm–1 respectively), the concentration of bound ThT can be calculated using the following system of equations
| 3 |
| 4 |
where cF is the concentration of free ThT, cB is the concentration of bound ThT, ε412 and ε450 are the extinction coefficients of bound ThT, and A412 and A450 are sample absorbance values at 412 and 450 nm, determined by subtracting the fibril absorbance spectrum from the fibril–ThT spectrum, as described previously.
The concentration of bound ThT determined using this method may lose accuracy if distinct binding modes have vastly different ε412 and ε450 values; however, the calculated total ThT concentration has a linear dependence on total ThT present in the sample (Figure S3), which indicates that there are no drastic changes to bound ThT extinction coefficients.
Results
Four distinct conformation insulin amyloid fibrils were generated by aggregating the protein under four different environmental conditions: 20% acetic acid solution (further referred to as AC),35 pH 2.0 and pH 2.4 sodium phosphate buffers (PH20 and PH24, respectively),42 as well as pH 7.4 phosphate buffer saline (PH74).44 To evaluate the effectiveness of identifying insulin amyloid fibrils with different conformations, control experiments had to be carried out using well-established methods, such as Fourier-transform infrared spectroscopy and atomic force microscopy.
Atomic Force Microscopy
The four fibril samples were first evaluated based on their AFM images, as well as by comparing the height and width of aggregates. The AC (Figure 1A), PH20 (Figure 1B), and PH24 (Figure 1C) samples contain long, linear fibrils, with some self-association observed in the case of AC and PH24 conditions. The PH74 sample (Figure 1D), on the other hand, contains highly fragmented and short aggregates. When considering the height distribution (Figure 1E), PH20 and PH74 aggregates have the smallest height (average value is 4 nm). The AC fibril height is slightly bigger—5 nm—and the pH 2.4 sample contains the highest fibrils—7 nm. Based on the width of aggregates (Figure 1F), all four samples have different values, with PH74 having the lowest (13 nm), PH20 (14 nm), pH 2.4 (16 nm), and AC (18 nm). By combining the height and width measurements and the visual inspection of AFM images, the PH20 and PH74 samples are easy to differentiate from the rest, while AC and PH24 are quite similar to one another.
Figure 1.
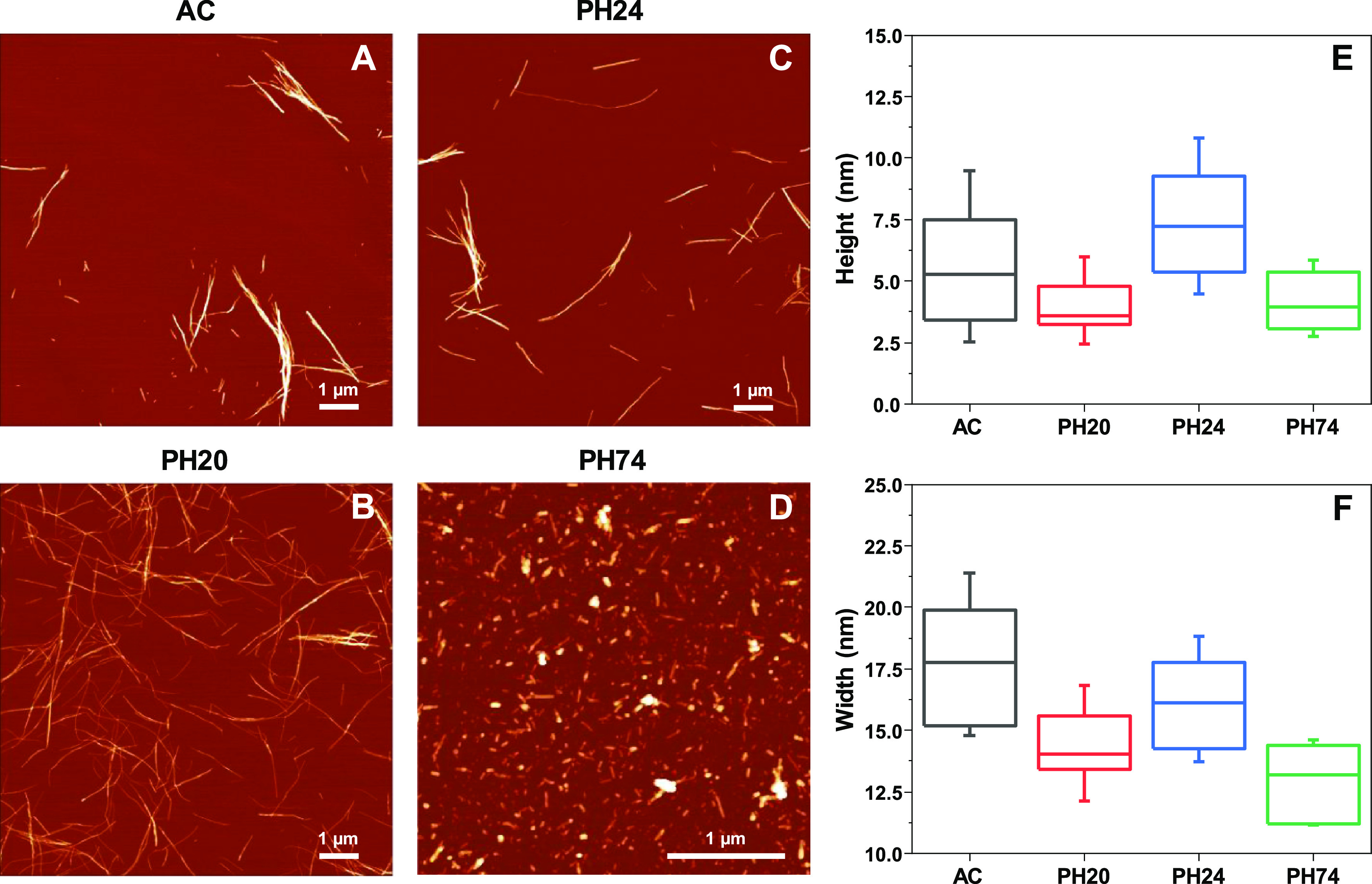
Atomic force microscopy images and fibril height and width distributions of insulin samples prepared under different conditions. Insulin fibrils were prepared under AC (A), PH20 (B), PH24 (C), and PH74 (D) conditions. Fibril height (E) and width (F) distribution (n = 50), where box plots indicate the interquartile range and error bars are 1 standard deviation.
Fibril length was not used as a means of distinguishing between insulin fibril conformations due to fragmentation having a substantial effect on this factor, as can be seen in the case of PH74, where the sample was agitated during aggregation (Figure S2A,B). There were also no clear periodicity patterns observed for any of the fibrils, where such a parameter could be determined (Figure S4). Prior to further experiments, the samples were sonicated, which resulted in all four aggregates having a comparable length distribution (Figure S2B). The width distribution experienced a slight increase (Figure S1F), likely due to the lateral association of small fibril fragments.45 Sonication had a very minimal influence on all four aggregate type height distributions (Figure S1E).
Fibril Secondary Structure
FTIR was used to detect differences in the fibril secondary structure, relying on the amide I/I′ region (Figure 2). The AC and PH20 samples share similarities in the fact that they both contain a small peak at 1729 cm–1, which is associated with deuterated carboxyl groups. The appearance of this peak can be associated to the disappearance of salt bridge interactions and subsequent deuteration of the resulting carboxyl group.46 They also both have the main minima in the second derivative at 1628 cm–1, which can be associated with stronger hydrogen bonds in the β-sheet structure, while the bands at 1640 cm–1 for AC and 1641 cm–1 for PH20 are related to weaker hydrogen bonds. The difference between AC and PH20 fibrils lays in more expressed bands at 1659 and 1673 cm–1 (associated with turns and/or loops) in the second derivative spectrum of PH20. There is also a considerable difference between half-height band widths43 of AC and PH20 amide I/I′ bands (Figure 2). The PH24 sample displays the only main minimum at 1630 cm–1, which suggests a single hydrogen bonding profile in the β-sheet structure. This spectrum also has no peak associated with deuterated carboxyl groups. The most distinct FTIR spectrum belongs to the PH74 sample, which has the main minimum at 1636 cm–1, with smaller bands at 1628–1630 cm–1, indicating a dominant presence of weaker and a smaller fraction of stronger hydrogen bonds in the β-sheet structure. Based on these observations, it is not difficult to separate the PH24 and PH74 samples from the rest; however, the AC and PH20 FTIR spectra have quite a few similarities and can only be accurately distinguished based on different HHBW values. All four types of fibrils contain parallel β-sheets, and there is no clear indication of the presence of antiparallel ones.47
Figure 2.
Insulin sample’s FTIR spectra (A), second derivatives (B), and spectrum positions associated with β-sheets, turns, loops, deuterated carboxyl groups, and each spectrum’s band’s width at its half-height (table inserted).
Bound ThT Intensities
Before conducting experiments with ThT binding, all fibril samples were repeatedly resuspended into MilliQ water and sonicated to negate any effect that the solution’s ionic strength, additives, or fibril superstructural organization may have on dye binding or fluorescence.
After measuring each sample’s fluorescence EEMs and absorbance spectra under a range of ThT concentrations, the maximum fluorescence intensities, as well as bound ThT concentrations, were determined. Since fibrils may possess different dye binding modes or have parts of their surface covered due to cluster formation, the absolute fluorescence intensity could not be used to differentiate between samples. Instead, to accurately compare each sample, the ratios between ThT fluorescence intensities and bound dye concentrations (I/cB) were calculated by dividing the fluorescence intensity with the concentration of bound dye molecules at each examined ThT concentration.
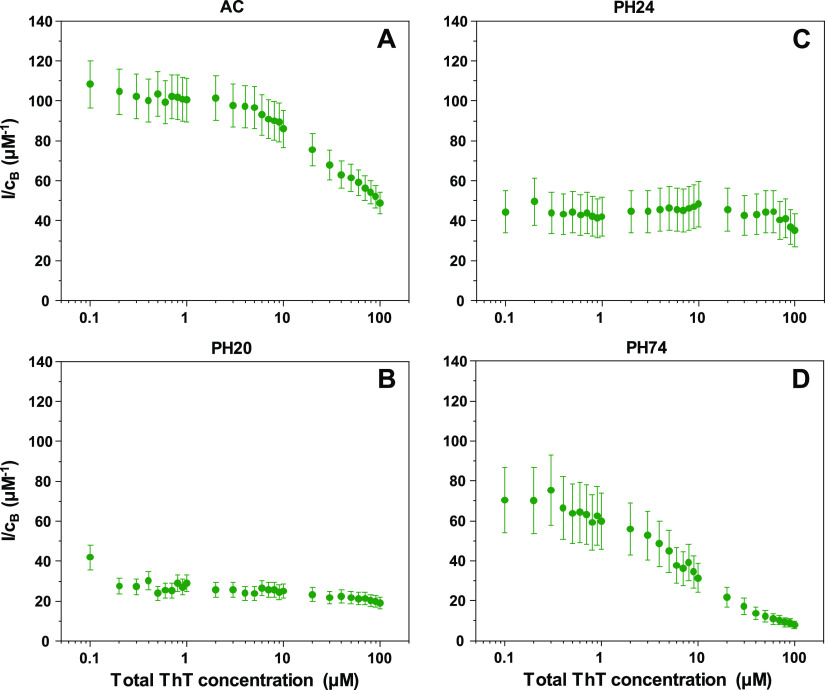
In the case of AC (Figure 3A), the I/cB ratio is around 110 μM–1 when the total concentration of ThT is low. Once the dye’s concentration reaches 3–4 μM, the ratio begins to decrease, eventually reaching 50 μM–1. Such reductions in the fluorescence quantum yield are associated with self-quenching of dye molecules due to binding in close proximity on the fibril’s surface.48−50 For PH20 (Figure 3B), both the initial (30–40 μM–1) and final (20–30 μM–1) ratio values are quite low in comparison and they do not experience such a drastic drop upon an increase of ThT concentrations. The PH24 sample (Figure 3C) initial and final ratios are within the margin of error (35–50 μM–1), and there is almost no reduction in values throughout the entire ThT concentration range. The PH74 (Figure 3D) ratio dependence on total ThT concentration has a sigmoidal shape, with initial values being 60–80 μM–1, which then drop to 10 μM–1. Based on I/cB ratios, all four samples have distinct dependencies on the total ThT concentration. AC and PH74 have similar shapes but different initial and final values, as do PH20 and PH24 samples.
Figure 3.
Insulin sample’s ThT fluorescence intensity and bound ThT concentration ratios (I/cB) at different total dye concentrations. Insulin fibrils were prepared under AC (A), PH20 (B), PH24 (C) and PH74 (D) conditions.
To verify the distinct ways of dye–fibril association, the bound ThT absorbance29 and fluorescence spectra at 1 μM ThT (where most of the dye molecules are in their bound state) were compared (Figure S5A,B). The PH20 sample had the lowest absorbance values, as well as the lowest maximum absorbance wavelength (420 nm). PH24 had a wider peak, but the absorbance intensity was similar to PH20. Both AC and PH74 had the highest absorbance peaks, which were also shifted toward 445 nm. This partial similarity between PH20 and PH24, as well as between AC and PH74, is in line with the observed I/cB ratio dependencies on total ThT concentration. Under these conditions, the fluorescence intensities correlate with the absorbance intensities (Figure S5B).
EEM Maxima Positions
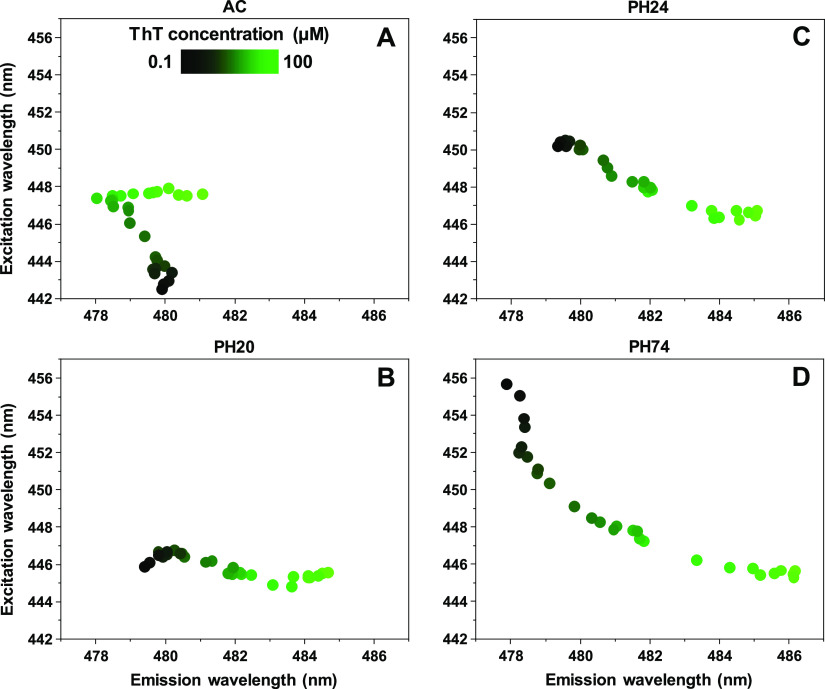
Different modes of ThT binding may possess specific excitation and emission wavelength maximum positions, which was also used to differentiate between the four samples. In the case of AC (Figure 4A), at low ThT concentrations, the maximum positions are located at 443/480 nm excitation/emission wavelengths. As the dye’s concentration increases, the excitation wavelength shifts toward 448 nm and the emission wavelength shifts to 478 nm. At higher ThT concentrations, the emission wavelength remains stable, but the excitation wavelength shifts toward 481 nm. This type of maximum position movement is a likely indicator of three ThT binding modes. For PH20 (Figure 4B), there is minimal variation in the excitation wavelength, which remains at 445–447 nm; however, there is a shift in the emission wavelength—from 479 to 485 nm. The PH24 sample’s (Figure 4C) EEM maximum position moves from 450/479 to a similar position to the PH20 sample (446/485). This means that PH20 and PH24 fibrils likely have one different and one similar ThT binding mode. The PBS sample’s (Figure 4D) EEM maximum position experiences the most significant movement, forming an arc-shape from 456/478 to 445/486. It has partial overlap with PH20 and PH24 sample positions at higher ThT concentrations, suggesting the existence of a similar binding mode for all three cases. The position at low ThT values, however, is different from all of the other samples. Such a significant movement and the arc-shape is also a likely indicator of more than two modes of ThT binding.
Figure 4.
Insulin sample’s ThT fluorescence EEM intensity center of mass positions at different total ThT concentrations. Insulin fibrils were prepared under AC (A), PH20 (B), PH24 (C), and PH74 (D) conditions.
Discussion
Using the two classical methods (AFM and FTIR) to compare insulin fibrils formed at different conditions, it can be identified that the four samples possess distinct morphologies and secondary structures. In some instances, there were similarities between AFM images or fibril heights and widths, and in others, there were similarities between FTIR spectra. Combining both methods, however, allows us to very clearly differentiate between the four insulin conformations.
When we examine the sample fluorescence intensity and bound ThT concentration ratios (I/cB), we see that there are clear distinctions between the four samples. Besides the fact that this allows us to differentiate between the distinct conformations, there is also an interesting I/cB ratio dependence on total ThT concentrations. In two of the cases, namely, AC and PH74, the ratio decreases with increasing ThT concentrations. Such an event is to be expected, as ThT is known to induce a self-quenching effect.48 However, PH20 and PH24 samples do not experience such a significant decrease, even at the highest dye concentrations. It is possible that ThT binds in such a manner that it does not allow for a self-quenching effect to occur. The absorbance spectra of 1 μM bound ThT displays significant distinctions between all four samples, further supporting the idea that these four fibril conformations have specific ThT binding characteristics.
Comparing each sample’s excitation–emission matrix and their intensity center of mass also leads to four different EEM maximum position distributions. At low ThT concentrations, all four samples have clearly distinct positions. In three of the cases, namely, PH20, PH24, and PH74, increasing the total ThT concentration leads to a maximum position convergence at 446/485 nm. This means that, while each fibril conformation possibly has unique ThT binding modes, some conformations may share a similar binding mode.
Since these ThT binding/fluorescence characteristics stem from either morphological or secondary structure variations, we have to examine which of them could be responsible for such dye–fibril association. In the case of morphology, the only real difference present between all four samples after sonication is their height distribution (Figure S1E). There does not seem to be any correlation between the I/cB ratio and fibril height, as PH20 and PH24 samples have the largest height difference, yet similar shape I/cB ratio distribution dependencies on the total ThT concentration. No correlation is also present in the case of fluorescence EEM positions. This leads to the idea that fibril morphology is not the main determining factor for ThT binding characteristics.
In the case of secondary structure, the relation between each structural motif and dye binding/fluorescence has to be considered. The deuterated carboxyl group, related to the loss of salt bridges46 during insulin aggregation, does not appear to be a significant factor, as there is a massive difference in both I/cB and EEM parameters between the two insulin conformations that have a peak at 1729 cm–1 in FTIR spectra. The turn/loop motives do not pertain to significant variations for them to be a factor, which leaves hydrogen bond strength in the β-sheet structure as the main suspect. The highest I/cB ratio at low ThT concentrations is observed in the AC sample, where fibrils have the largest amount of stronger hydrogen bonds (as indicated by the major 1628 cm–1 band in the second derivative spectrum). The PH20 sample also has a peak at 1628 cm–1, but in this case, the 1641 cm–1 band is relatively larger than in the case of AC. This sample has a significantly lower I/cB ratio than AC, however, which may indicate that the weaker hydrogen bonds in the fibril’s structure create binding positions with higher ThT binding affinity and lower fluorescence quantum yield. The PH24 FTIR spectrum has a single β-sheet-related band, and, coincidentally, the I/cB ratio experiences the least amount of variation throughout the entire ThT concentration range. The PH74 FTIR spectrum is the most peculiar of all, displaying mostly weaker hydrogen bonds within the β-sheet structure but also some stronger ones. Its corresponding I/cB ratio and EEM positions also experience the most drastic changes throughout the whole ThT concentration range. These observations hint at a possibility that weaker hydrogen bonds in the fibril’s structure lead to binding positions with higher ThT binding affinity and lower fluorescence quantum yield, while stronger ones result in binding position with lower ThT binding affinity and higher quantum yield.
When we combine AFM with FTIR and I/cB with EEM positions, it is clear that both pairs of methods are highly efficient at differentiating insulin fibrils prepared under different environmental conditions. Considering that a ThT binding examination has comparable efficiency to both AFM and FTIR, we have to discuss the advantages that this method has. In the case of FTIR, to acquire a high-quality spectrum and detect minor differences, a high concentration of fibrils has to be used. For AFM, the method of sample deposition can influence the overall fibril distribution (fibril clump formation, washing away smaller fibrils/oligomers, different aggregate/mica association propensities). Conversely, fibril–ThT fluorescence I/cB ratio and EEM position profile outlines, as well as bound ThT absorbance spectra, can be acquired by scanning relatively low fibril concentration samples at four ThT concentrations (0.1, 1, 10, and 100 μM). This method also uses fibril resuspension into MilliQ water, which negates the effect that the initial solution’s ionic strength or additives may have on ThT binding. The aggregates are also sonicated to both homogenize the sample, as well as disrupt any superstructural organization that may form during aggregation.
Despite insulin being a model for the study of amyloid aggregation and not considered as a neurodegenerative disease-associated protein, its ability to form multiple distinct fibril conformations serves as a means to display this method’s effectiveness. As of this time, there have been reports showing that different strains of disease-related proteins, such as α-synuclein33 or prion proteins,34 also possess different ThT binding modes, resulting in distinct fluorescence profiles. This means that ThT binding could serve in conjunction with other commonly used methods to both identify and differentiate the large variety of conformations that amyloid fibrils can obtain.
Conclusions
ThT molecule interactions with insulin amyloid fibrils, prepared under different conditions, display a wide variety of bound ThT quantum yields, as well as distinct binding modes. Such interactions can be used to differentiate between fibrils to a similar extent as other widely used methods, such as AFM or FTIR. As this ThT binding assay requires minimal amounts of fibrils and is not affected by the initial aggregation solution or sample deposition, it can be applied as either an alternative or supplemental method in amyloid fibril research.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biomac.0c01178.
Atomic force microscopy images of insulin fibrils, prepared in AC, PH20, PH24, and PH74 conditions after multiple rounds of resuspension into MilliQ H2O and sonication (Figure S1); AC, PH20, PH24, and PH74 fibril length distributions before and after resuspension into MilliQ H2O and sonication (Figure S2); comparison of total (bound + free) ThT concentration calculated from sample absorbance data and total ThT added to the sample (Figure S3); AC, PH20, and PH24 fibril surface height along the fibril axis (Figure S4); and absorbance spectra of free and fibril-bound ThT and fluorescence intensity of all four types of fibrils in the presence of 1 μM ThT (Figure S5) (PDF)
Author Contributions
M.Z. and V.S. designed the studies. M.Z. and A.S. undertook the experimental work. M.Z. and V.S. analyzed the data and prepared the manuscript.
This research was funded by Vilnius University, Grant No. MSF-JM-3.
The authors declare no competing financial interest.
Supplementary Material
References
- Chiti F.; Dobson C. M. Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 2006, 75, 333–366. 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Knowles T. P. J.; Vendruscolo M.; Dobson C. M. The Amyloid State and Its Association with Protein Misfolding Diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Baker K. R.; Rice L. The Amyloidoses: Clinical Features, Diagnosis And Treatment. Methodist DeBakey Cardiovasc. J. 2012, 8, 3 10.14797/mdcj-8-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isik A. T. Late Onset Alzheimer’s Disease in Older People. Clin. Interventions Aging 2010, 5, 307. 10.2147/CIA.S11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert L. E.; Weuve J.; Scherr P. A.; Evans D. A. Alzheimer Disease in the United States (2010–2050) Estimated Using the 2010 Census. Neurology 2013, 80, 1778–1783. 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur K. C.; Calvo A.; Price T. R.; Geiger J. T.; Chiò A.; Traynor B. J. Projected Increase in Amyotrophic Lateral Sclerosis from 2015 to 2040. Nat. Commun. 2016, 7, 12408 10.1038/ncomms12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatani E.; Yamamoto N. Recent Progress on Understanding the Mechanisms of Amyloid Nucleation. Biophys. Rev. 2018, 10, 527–534. 10.1007/s12551-017-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisl G.; Kirkegaard J. B.; Arosio P.; Michaels T. C. T.; Vendruscolo M.; Dobson C. M.; Linse S.; Knowles T. P. J. Molecular Mechanisms of Protein Aggregation from Global Fitting of Kinetic Models. Nat. Protoc. 2016, 11, 252–272. 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick A. W. P.; Debelouchina G. T.; Bayro M. J.; Clare D. K.; Caporini M. A.; Bajaj V. S.; Jaroniec C. P.; Wang L.; Ladizhansky V.; Muller S. A.; MacPhee C. E.; Waudby C. A.; Mott H. R.; De Simone A.; Knowles T. P. J.; Saibil H. R.; Vendruscolo M.; Orlova E. V.; Griffin R. G.; Dobson C. M. Atomic Structure and Hierarchical Assembly of a Cross-β Amyloid Fibril. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 5468–5473. 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fändrich M.; Nyström S.; Nilsson K. P. R.; Böckmann A.; LeVine H.; Hammarström P. Amyloid Fibril Polymorphism: A Challenge for Molecular Imaging and Therapy. J. Intern. Med. 2018, 283, 218–237. 10.1111/joim.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J.; Lee G.; Ritter A.; Sabbagh M.; Zhong K. Alzheimer’s Disease Drug Development Pipeline: 2019. Alzheimer’s Dementia: Transl. Res. Clin. Interventions 2019, 5, 272–293. 10.1016/j.trci.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šneideris T.; Baranauskienė L.; Cannon J. G.; Rutkienė R.; Meškys R.; Smirnovas V. Looking for a Generic Inhibitor of Amyloid-like Fibril Formation among Flavone Derivatives. PeerJ 2015, 3, e1271 10.7717/peerj.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan E.; Rajasekaran R. Probing the Inhibitory Activity of Epigallocatechin-Gallate on Toxic Aggregates of Mutant (L84F) SOD1 Protein through Geometry Based Sampling and Steered Molecular Dynamics. J. Mol. Graphics Model. 2017, 74, 288–295. 10.1016/j.jmgm.2017.04.019. [DOI] [PubMed] [Google Scholar]
- Goyal D.; Shuaib S.; Mann S.; Goyal B. Rationally Designed Peptides and Peptidomimetics as Inhibitors of Amyloid-β (Aβ) Aggregation: Potential Therapeutics of Alzheimer’s Disease. ACS Comb. Sci. 2017, 19, 55–80. 10.1021/acscombsci.6b00116. [DOI] [PubMed] [Google Scholar]
- Byeon S. R.; Lee J. H.; Sohn J.-H.; Kim D. C.; Shin K. J.; Yoo K. H.; Mook-Jung I.; Lee W. K.; Kim D. J. Bis-Styrylpyridine and Bis-Styrylbenzene Derivatives as Inhibitors for Aβ Fibril Formation. Bioorg. Med. Chem. Lett. 2007, 17, 1466–1470. 10.1016/j.bmcl.2006.10.090. [DOI] [PubMed] [Google Scholar]
- Konar M.; Bag S.; Roy P.; Dasgupta S. Gallic Acid Induced Dose Dependent Inhibition of Lysozyme Fibrillation. Int. J. Biol. Macromol. 2017, 103, 1224–1231. 10.1016/j.ijbiomac.2017.05.158. [DOI] [PubMed] [Google Scholar]
- Ruggeri F. S.; Šneideris T.; Vendruscolo M.; Knowles T. P. J. Atomic Force Microscopy for Single Molecule Characterisation of Protein Aggregation. Arch. Biochem. Biophys. 2019, 664, 134–148. 10.1016/j.abb.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podestà A.; Tiana G.; Milani P.; Manno M. Early Events in Insulin Fibrillization Studied by Time-Lapse Atomic Force Microscopy. Biophys. J. 2006, 90, 589–597. 10.1529/biophysj.105.068833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streets A. M.; Sourigues Y.; Kopito R. R.; Melki R.; Quake S. R. Simultaneous Measurement of Amyloid Fibril Formation by Dynamic Light Scattering and Fluorescence Reveals Complex Aggregation Kinetics. PLoS One 2013, 8, e54541 10.1371/journal.pone.0054541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandomeneghi G.; Krebs M. R. H.; McCammon M. G.; Fändrich M. FTIR Reveals Structural Differences between Native β-Sheet Proteins and Amyloid Fibrils. Protein Sci. 2004, 13, 3314–3321. 10.1110/ps.041024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmos K. G.; Blancas-Mejia L. M.; Weber B.; Buchner J.; Ramirez-Alvarado M.; Naiki H.; Otzen D. ThT 101: A Primer on the Use of Thioflavin T to Investigate Amyloid Formation. Amyloid 2017, 24, 1–16. 10.1080/13506129.2017.1304905. [DOI] [PubMed] [Google Scholar]
- Picken M. M.; Herrera G. A.. Thioflavin T Stain: An Easier and More Sensitive Method for Amyloid Detection. In Amyloid and Related Disorders; Picken M. M.; Herrera G. A.; Dogan A., Eds.; Humana Press: Totowa, NJ, 2012; pp 187–189. [Google Scholar]
- Wetzel R.; Chemuru S.; Misra P.; Kodali R.; Mukherjee S.; Kar K. An Aggregate Weight-Normalized Thioflavin-T Measurement Scale for Characterizing Polymorphic Amyloids and Assembly Intermediates. Methods Mol. Biol. 2018, 1777, 121–144. 10.1007/978-1-4939-7811-3_6. [DOI] [PubMed] [Google Scholar]
- Groenning M. Binding Mode of Thioflavin T and Other Molecular Probes in the Context of Amyloid Fibrils—Current Status. J. Chem. Biol. 2010, 3, 1–18. 10.1007/s12154-009-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart A.; Ye L.; Judd D. B.; Merritt A. T.; Lowe P. N.; Morgenstern J. L.; Hong G.; Gee A. D.; Brown J. Evidence for the Presence of Three Distinct Binding Sites for the Thioflavin T Class of Alzheimer’s Disease PET Imaging Agents on β-Amyloid Peptide Fibrils. J. Biol. Chem. 2005, 280, 7677–7684. 10.1074/jbc.M412056200. [DOI] [PubMed] [Google Scholar]
- Ziaunys M.; Smirnovas V. Additional Thioflavin-T Binding Mode in Insulin Fibril Inner Core Region. J. Phys. Chem. B 2019, 123, 8727–8732. 10.1021/acs.jpcb.9b08652. [DOI] [PubMed] [Google Scholar]
- Sulatskaya A. I.; Kuznetsova I. M.; Belousov M. V.; Bondarev S. A.; Zhouravleva G. A.; Turoverov K. K. Stoichiometry and Affinity of Thioflavin T Binding to Sup35p Amyloid Fibrils. PLoS One 2016, 11, e0156314 10.1371/journal.pone.0156314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenning M.; Norrman M.; Flink J. M.; van de Weert M.; Bukrinsky J. T.; Schluckebier G.; Frokjaer S. Binding Mode of Thioflavin T in Insulin Amyloid Fibrils. J. Struct. Biol. 2007, 159, 483–497. 10.1016/j.jsb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Kuznetsova I. M.; Sulatskaya A. I.; Uversky V. N.; Turoverov K. K. Analyzing Thioflavin T Binding to Amyloid Fibrils by an Equilibrium Microdialysis-Based Technique. PLoS One 2012, 7, e30724 10.1371/journal.pone.0030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai R.; Araki M.; Yoshimura M.; Kamiya N.; Ono M.; Saji H.; Okuno Y. Core Binding Site of a Thioflavin-T-Derived Imaging Probe on Amyloid β Fibrils Predicted by Computational Methods. ACS Chem. Neurosci. 2018, 9, 957–966. 10.1021/acschemneuro.7b00389. [DOI] [PubMed] [Google Scholar]
- Ivancic V. A.; Ekanayake O.; Lazo N. D. Binding Modes of Thioflavin T on the Surface of Amyloid Fibrils Studied by NMR. ChemPhysChem 2016, 17, 2461–2464. 10.1002/cphc.201600246. [DOI] [PubMed] [Google Scholar]
- Mao X.; Guo Y.; Wang C.; Zhang M.; Ma X.; Liu L.; Niu L.; Zeng Q.; Yang Y.; Wang C. Binding Modes of Thioflavin T Molecules to Prion Peptide Assemblies Identified by Using Scanning Tunneling Microscopy. ACS Chem. Neurosci. 2011, 2, 281–287. 10.1021/cn200006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu A.; Vaneyck J.; Blum C.; Segers-Nolten I.; Subramaniam V. Polymorph-Specific Distribution of Binding Sites Determines Thioflavin-T Fluorescence Intensity in α-Synuclein Fibrils. Amyloid 2018, 25, 189–196. 10.1080/13506129.2018.1517736. [DOI] [PubMed] [Google Scholar]
- Ziaunys M.; Sneideris T.; Smirnovas V. Formation of Distinct Prion Protein Amyloid Fibrils under Identical Experimental Conditions. Sci. Rep. 2020, 10, 4572 10.1038/s41598-020-61663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakalauskas A.; Ziaunys M.; Smirnovas V. Concentration-Dependent Polymorphism of Insulin Amyloid Fibrils. PeerJ 2019, 7, e8208 10.7717/peerj.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneideris T.; Sakalauskas A.; Sternke-Hoffmann R.; Peduzzo A.; Ziaunys M.; Buell A. K.; Smirnovas V. The Environment Is a Key Factor in Determining the Anti-Amyloid Efficacy of EGCG. Biomolecules 2019, 9, 855 10.3390/biom9120855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta Y.; Singla G.; Singla R. Insulin-Derived Amyloidosis. Indian J. Endocrinol. Metab. 2015, 19, 174–177. 10.4103/2230-8210.146879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foderà V.; Cataldo S.; Librizzi F.; Pignataro B.; Spiccia P.; Leone M. Self-Organization Pathways and Spatial Heterogeneity in Insulin Amyloid Fibril Formation. J. Phys. Chem. B 2009, 113, 10830–10837. 10.1021/jp810972y. [DOI] [PubMed] [Google Scholar]
- Gong H.; He Z.; Peng A.; Zhang X.; Cheng B.; Sun Y.; Zheng L.; Huang K. Effects of Several Quinones on Insulin Aggregation. Sci. Rep. 2015, 4, 5648 10.1038/srep05648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-B.; Wang Y.-M.; Zeng C.-M. Quercetin Inhibits Amyloid Fibrillation of Bovine Insulin and Destabilizes Preformed Fibrils. Biochem. Biophys. Res. Commun. 2011, 415, 675–679. 10.1016/j.bbrc.2011.10.135. [DOI] [PubMed] [Google Scholar]
- Jayamani J.; Shanmugam G. Gallic Acid, One of the Components in Many Plant Tissues, Is a Potential Inhibitor for Insulin Amyloid Fibril Formation. Eur. J. Med. Chem. 2014, 85, 352–358. 10.1016/j.ejmech.2014.07.111. [DOI] [PubMed] [Google Scholar]
- Sneideris T.; Darguzis D.; Botyriute A.; Grigaliunas M.; Winter R.; Smirnovas V. PH-Driven Polymorphism of Insulin Amyloid-Like Fibrils. PLoS One 2015, 10, e0136602 10.1371/journal.pone.0136602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzwolak W.; Smirnovas V.; Jansen R.; Winter R. Insulin Forms Amyloid in a Strain-Dependent Manner: An FT-IR Spectroscopic Study. Protein Sci. 2004, 13, 1927–1932. 10.1110/ps.03607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannuzzi C.; Borriello M.; Portaccio M.; Irace G.; Sirangelo I. Insights into Insulin Fibril Assembly at Physiological and Acidic Ph and Related Amyloid Intrinsic Fluorescence. Int. J. Mol. Sci. 2017, 18, 2551 10.3390/ijms18122551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhiet P.-E.; Yamamoto D.; Berthoumieu O.; Dosset P.; Le Grimellec C.; Verdier J.-M.; Marchal S.; Ando T. Deciphering the Structure, Growth and Assembly of Amyloid-Like Fibrils Using High-Speed Atomic Force Microscopy. PLoS One 2010, 5, e13240 10.1371/journal.pone.0013240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmacz-Chwedoruk W.; Nieznańska H.; Wójcik S.; Dzwolak W. Cross-Seeding of Fibrils from Two Types of Insulin Induces New Amyloid Strains. Biochemistry 2012, 51, 9460–9469. 10.1021/bi301144d. [DOI] [PubMed] [Google Scholar]
- Barth A. Infrared Spectroscopy of Proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Lindberg D. J.; Wenger A.; Sundin E.; Wesén E.; Westerlund F.; Esbjörner E. K. Binding of Thioflavin-T to Amyloid Fibrils Leads to Fluorescence Self-Quenching and Fibril Compaction. Biochemistry 2017, 56, 2170–2174. 10.1021/acs.biochem.7b00035. [DOI] [PubMed] [Google Scholar]
- Ran C.; Zhao W.; Moir R. D.; Moore A. Non-Conjugated Small Molecule FRET for Differentiating Monomers from Higher Molecular Weight Amyloid Beta Species. PLoS One 2011, 6, e19362 10.1371/journal.pone.0019362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziaunys M.; Mikalauskaite K.; Smirnovas V. Amyloidophilic Molecule Interactions on the Surface of Insulin Fibrils: Cooperative Binding and Fluorescence Quenching. Sci. Rep. 2019, 9, 20303 10.1038/s41598-019-56788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.