Abstract
The repeated emergence of similar morphologies in the dental elements of Permian Sweetognathus conodonts has been a hypothesized example of parallel evolution. To test if morphological parallelisms occur between isolated Sweetognathus lineages, this study uses two-dimensional-based geometric morphometrics combined with a revised and expanded phylogeny of Permian Sweetognathus conodonts to quantify dental element trait distributions and compare the phenotypic trajectories between lineages. A hierarchical clustering method was used to identify recurrent species pairs based on principal component scores describing their morphological variation, with the further incorporation of widely used ecological metrics such as limiting similarity and morphological overlap. Our research implies that a major contributor to conodont diversity in Palaeozoic marine trophic networks is the emergence of recurrent parallel morphologies via disruptive and directional selection. This study illustrates the mechanisms through which conodonts achieved their status as hyper-diverse predators and scavengers, contributing substantially to the complexity of Palaeozoic marine communities.
Keywords: parallel evolution, conodont trophic networks, Sweetognathus
1. Introduction
Conodonts are an extinct group of early marine vertebrates (approx. 510–201 Ma) that were the first to develop a biomineralized phosphatic dental skeleton, which consisted of tooth-like elements forming a complex feeding apparatus [1–3]. Phylogenetically, their position remains unresolved between cyclostomes and stem gnathostomes, with their closest living relative being lampreys [3,4]. Based on wear and fracture patterns in their dental elements [5,6], biomechanics [7], growth allometry [8] and microstructural adaptation [9], filter-feeding or parasitic habit can be excluded for most conodonts. They were most likely a free-swimming animal capable of actively pursuing their prey and of mechanical mastication of its tissues.
The Sweetognathus lineage evolved near the beginning of the Permian Period (298.9 Ma) within near-equatorial, shallow-water seas. Reconstructions of its evolutionary history are based on their P1 elements, which were located in the pharynx of the animal and, as in many conodonts, developed food-processing adaptations analogous with mammalian molars [10]. Their adaptive radiation and dispersions followed numerous significant relative sea-level fluctuations controlled by the waxing and waning of continental ice-sheets during the Late Palaeozoic ice age, with derived populations becoming isolated during sea-level lowstands when Gondwana ice volume was at its greatest (figure 1) [12].
Figure 1.
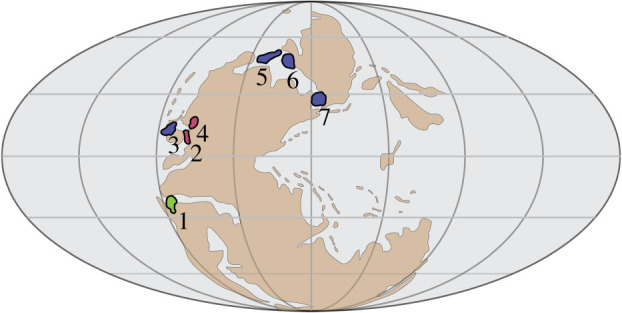
Palaeogeographic map of sampled Sweetognathus populations during sea-level lowstands [11]. Colours (and numbers) correspond to time of diversification; green (1): Early Asselian, red (2,4): Late Asselian, blue (3,5,6,7): Sakmarian and Artinskian. The numbered localities refer to the locations of Sweetognathus populations used in this study: 1: Apillapampa Section, Bolivia; 2: Florence Limestone, Kansas; 3: Carlin Canyon, Nevada; 4: Tensleep, Wyoming; 5: Tingmissut Gorge Section, Melville Island; 6: East Blind Fiord, Ellesmere Island; 7: Dalny Tulkas Section, Russia. Species of Sweetognathus have a wider palaeogeographic range during sea-level highstands. (Online version in colour.)
In this study, we test whether repeated episodes of morphological radiations in Sweetognathus conodonts [11,13,14] satisfy the definition of parallel evolution. With the advent of modern genomic techniques, multiple definitions of parallel evolution have been proposed, which attempt to address the underlying genetic mechanisms that lead to parallelisms, as well as the affect environmental selective pressures have on phenotypic expression (reviewed in [15]). Because the underlying genetic mechanisms of parallel evolution cannot be tested in deep-time studies [16], we adopt a definition that enables us to directly address morphological parallelisms. Parallel evolution will thus be defined as the similarity in phenotypic trajectories between two closely related lineages ([17], reviewed in [18]).
The present study has two objectives. The first is to quantify the morphological variation in the oral surface of Sweetognathus species, including morphological overlap and limiting similarity, using geometric morphometric (GM) techniques [19,20]. This provides a measure of intra- and interspecific morphological variation between populations and allows us to recognize potential species pairs. The second objective is to test if parallel evolution between Sweetognathus lineages has occurred by computing and comparing their phenotypic trajectories [16,21]. If significant similarities in the phenotypic trajectories among Sweetognathus populations exist, our goal will then be to evaluate the drivers of the parallel evolution by comparing our observations with the causes documented in extant fish species demonstrating the same phenomenon.
2. Methods
Three-dimensional (3D) surface models of Sweetognathus P1 dental elements were created at Friedrich-Alexander University Erlangen-Nürnberg, GeoZentrum Nordbayern, Germany using a phoenix v|tome|xs research edition computer tomographic scanner at a resolution of 1.362 µm3. Beam voltage was set to 80 kV, current at 100 µA, resulting in an 8 W beam. Images were taken with 400 ms exposure, averaging over five images, and skipping the initial image. A total of 1500 images were taken from the 0° starting position. Image stacks as well as meshes are stored through the open source repository MorphoBank [22,23] (http://morphobank.org/permalink/?P3736), while R analysis code is stored through the open source repository GitHub [24] (https://github.com/WyattSP/Evidence-of-Parallel-Evolution-in-the-Dental-Elements-of-Sweetognathus-Conodonts). A total of 59 Sweetognathus P1 dental elements were scanned for this study. They originated from seven locations (electronic supplementary material, Locality Information; figure 1). Sweetognathus elements are curated at the University of Calgary.
(a). Model generation, landmark selection and general procrustes analysis
3D models were rendered in VG Studio Max from the acquired image stacks. Elements not displaying dextral curvature (right-curving chirality) were inverted in the open source software IDAV Landmark ([25], v. 3.0).
Discrete cross sections (n = 401) were computed through individual denticles on the dorsal platform in the open source software Meshlab ([26], 64bit v. 2016.12] and imported into R Studio ([27], v. 1.1.423) running R Software ([28], v. 3.6.2) using the R package Momocs ([29], v. 1.2.9). For example, a specimen with nine dorsal platform denticles would yield nine discrete cross sections describing their individual shapes (figure 2a).
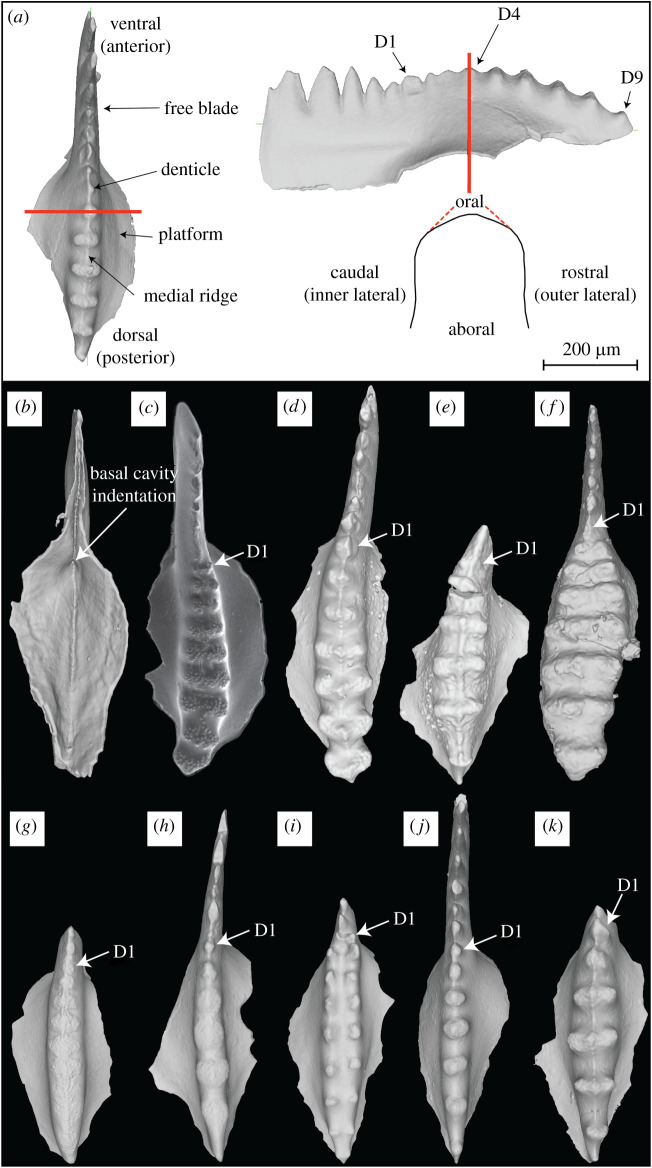
Figure 2.
Sweetognathus P1 elements. (a) Terminology used to describe Sweetognathus P1 morphology (for more information on element orientation refer to [30]). The red line is an example of a cross section taken through the denticle. The oral surface of the element contains the denticles. (b) 3D rendered model of aboral surface illustrating the basal cavity indentation. (c) Scanning electron microscopy image of Sw. whitei from the Tensleep Section in Wyoming, small nodes cover the surface of the denticles; the element displays left curvature. 3D rendered models of Sw. whitei (d), Sw. asymmetricus (e), Sw. clarki (f), Sw. cf. adenticulatus (g), Sw. aff. binodosus (h), Sw. obliquidentatus (i), Sw. binodosus (j) and Sw. anceps (k). (Online version in colour.)
One crucial benefit in using 3D rendered models is for their ability to show subtle morphological characteristics because models can be easily reoriented. This is usually difficult when using an optical microscope or a scanning electron microscope as conodont elements tend to be quite small and difficult to physically manipulate. In the 3D models, we were able to identify a small anteriorly (ventrally) directed indentation (inside the basal cavity) on the aboral side of the conodont element that points to the first carinal denticle (figure 2b); compressed ventral denticles are on the free blade. For reference, all Sweetognathus P1 elements essentially lack a true cusp that is clearly distinguishable from the other denticles, as it is not clear where the apex of the basal cavity points. With this discovery, the cusp can be consistently and confidently diagnosed between taxa, assuring correspondence in the assignment of denticle position in this study, as well as establishing a homologous character with species outside the Sweetognathus genus (figure 2c–k).
Shape variation in the denticle cross sections was then computed using two-dimensional (2D) GM methods [18,19] (reviewed in [31,32]) including the analysis of phenotypic trajectories between identified species pairs [21]. A 2D GM methodology was implemented in this paper for two reasons. The Sweetognathus genus has very few homologous anatomical structures that would allow for the assignment of landmarks, and as such an insufficient number of landmarks exist to define the boundaries of 3D semi-landmark patches. Additionally, differences in element taphonomy between specimens further limits the availability of homologous anatomical landmarks. Based upon this fact, a 2D approach could assure positional and functional homology between all of the specimens was followed, especially as this is the first attempt to quantify shape variation in Sweetognathus conodonts. Second, the usage of histological sections identical in orientation and morphology to the cross sections used in this study, and of which are being used in finite-element analysis for other conodont taxa [5], make the results obtained between these different methods comparable and accommodating to a unified interpretation on functional adaptations.
A qualitative assessment of this method's ability to differentiate between different morphotypes as well as its ability to accommodate user error in cross-section selection was assessed beforehand in the electronic supplementary material, Denticle Cross Section Sensitivity Analysis. Type 1 (homology-based) landmarks could not be consistently defined on the elements to accurately recreate its shape [33], therefore, 50 equally spaced semi-landmarks were placed around the circumference of the median vertical section through each denticle, with two landmarks positioned on either extremes of the cross section (start of the curve and end of the curve) (figure 2).
Conodont elements are known to record wear (e.g. scratches, flaking or spalling) as a result of their dental function [7,10,34], but their shape is periodically restored through a mechanism unique to conodonts [6]. Here, some degree of wear has probably changed the overall profile of the denticles (see D1 in figure 2d). To mitigate the possible affects wear has had on the analysis methods, species denticle averages (electronic supplementary material, figure S12) were computed and used as input parameters in the hierarchical clustering (HC) and in the other quantitative metrics (see below sections; denticle asymmetries seen in figure 2d are no longer present when species denticle means are taken). Increased within-species variance in shape may be caused by sampling individuals in different phases of the wear-repair cycle, as well as by the species-specific distribution of wear across the denticles. The inclusion of an occlusal model to help deduce the changes in denticle outlines incurred by wear would be beneficial, unfortunately only a handful of reconstructed occlusal models exist, as fossil preservation needs to be exceptional [5,7,34].
The assignment of landmarks along each cross section was done in the R package geomorph [35] (see R analysis code for detailed methods [24]). Semi-landmarks were placed at equidistant intervals along the curve, between a start and endpoint, of the denticle outline. Finally, a generalized Procrustes analysis (GPA) [36,37] was completed to calculate the shape differences between the 50 sliding semi-landmarks and two landmarks that defined the shape of each cross section [35]. For the GPA, bending energy was minimized as the optimization criterion during Procrustes superimposition [36,37].
(b). Sweetognathus phylogenetic hypothesis
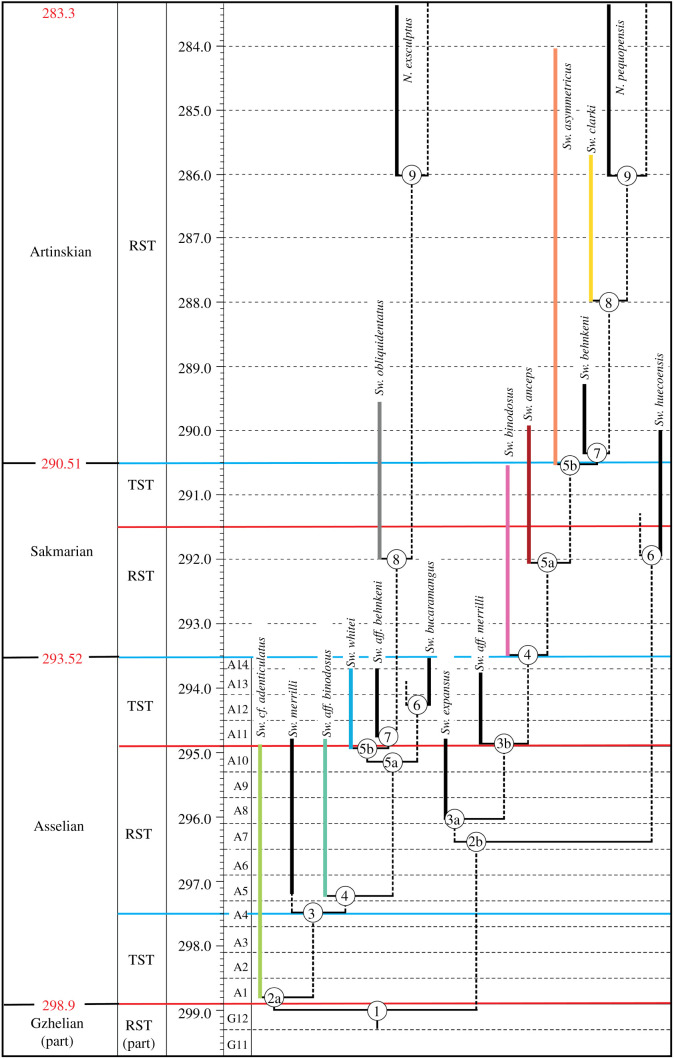
The evolutionary relationships of the Sweetognathus phylogenetic hypothesis (figure 4) presented in this paper were formulated using a non-quantitative cladistic approach and considers the timing of introduction of interpreted shared derived characters, characters that have long been used to identify species of Sweetognathus (similar approaches can be found in [11] and [12]). This approach is possible thanks to abundant data on Sweetognathus occurrences in the fossil record, with consideration to the systematic distribution of gaps, owing to its biostratigraphic significance. The ranges of species are well constrained using numerous methods including radiometric age dating [38,39], and astronomical tuning of 405 thousand-year cyclothems in the Canadian Arctic and mid-continent USA [12,40]. The topological relationships (branch pattern) are constrained by the relative timing of key character introductions and a pattern of two distinct lineages was considered most likely given the average 4 Myr separation of first occurrences of given characters.
Figure 4.
Sweetognathus phylogenetic hypothesis. The numbered nodes refer to character transitions (electronic supplementary material, Phylogenetic Hypothesis). Sea-level fluctuations are indicated by the blue and red lines which represent transgressive system tracts (TST) and regressive system tracts (RST) [40]. Colours correspond to species in figure 3. Evolutionary lineages examined in this study are early Asselian taxa, Sw. cf. adenticulatus and Sw. aff. binodosus, late Asselian taxa, Sw. whitei and Sw. obliquidentatus, Sakmarian taxa, Sw. binodosus and Sw. anceps and finally Artinskian taxa, Sw. asymmetricus and Sw. clarki. (Online version in colour.)
(c). Quantification of morphological variation in Sweetognathus specimens
To visualize the morphological differences between specimens, Procrustes-aligned coordinates were then subjected to principal component analysis (PCA) [32]. Mean principal component (PC) values were then calculated for each denticle grouped by species and subjected to HC via a multiscale bootstrap resampling method as implemented by the R package pvclust ([41], v. 2.2; 1000 bootstrap replications) to identify clusters of morphologically similar taxa. The mean PC values were used for HC as some specimens had fewer than eight denticles and missing values cannot be accommodated in the character matrix (first nine PC values included in HC and weighted by per cent variance, this accounted for 99% of the overall variance). Denticle position was assessed based on an assumed functional homology as outlined above (i.e. the cusp on the platform will have a homologous function between all taxa). An additional assumption made during analysis is that all conodont specimens are adults, based on the comparable size between the specimens and their similar number of carinal denticles. A Euclidean distance measure and Ward's method was used for HC. The cutoff value for clustering was α = 0.05.
A multivariate approach was then taken to assess if the identified clusters were statistically different from each other. First, a multivariate analysis of variance (MANOVA) was run to test for differences between the clustered groups, this was completed for both the overall species means as well as for each individual denticle (electronic supplementary material, table S5). Procrustes-aligned coordinates [18,42,43] were used in the analysis with the grouping term set to the clustered groups (trait spaces). If the results calculated for Pillai's trace test statistic in the MANOVA were significant, the same inputs were subjected to a pairwise comparison. All multivariate analysis was completed in the RRPP package [44,45] (see R Analysis Code [24]).
Two other metrics were also calculated to quantify the intra- and interspecific variation between species. The ratio of the distance between trait means (µMV for PCs; mean) to their trait widths (σMV for PCs; standard deviation) was calculated for each species ([46], reviewed in [47]). This ratio is equivalent to MacArthur & Levins [46] quotient of interspecific niche means to interspecific niche widths (limiting similarity). Limiting similarity is an idealized case of competitive exclusion whereby competitive selective pressures lead to the extinction of a species [46]. This can be understood in terms of the ‘competitive exclusion principle’, which states that species cannot coexist if they occupy identical niche spaces and are in complete competition with each other [48]. The coexistence of these closely related species will then be dependent on the degree of trait displacement between species that enable individuals to use different resources [49,50]. By calculating the degree of similarity between closely related species we can get a quantitative metric on the degree of trait overlap. This can help us to understand the potential evolutionary pathways whereby morphological innovations could have arisen. This ratio is used as a metric to quantify niche similarity between species. Next, the species-wide variance () relative to the total variance of the genus () was assessed for each species [47,51] to determine the degree of morphological overlap across the Sweetognathus phylogeny.
(d). Analysis of phenotypic change between Sweetognathus lineages
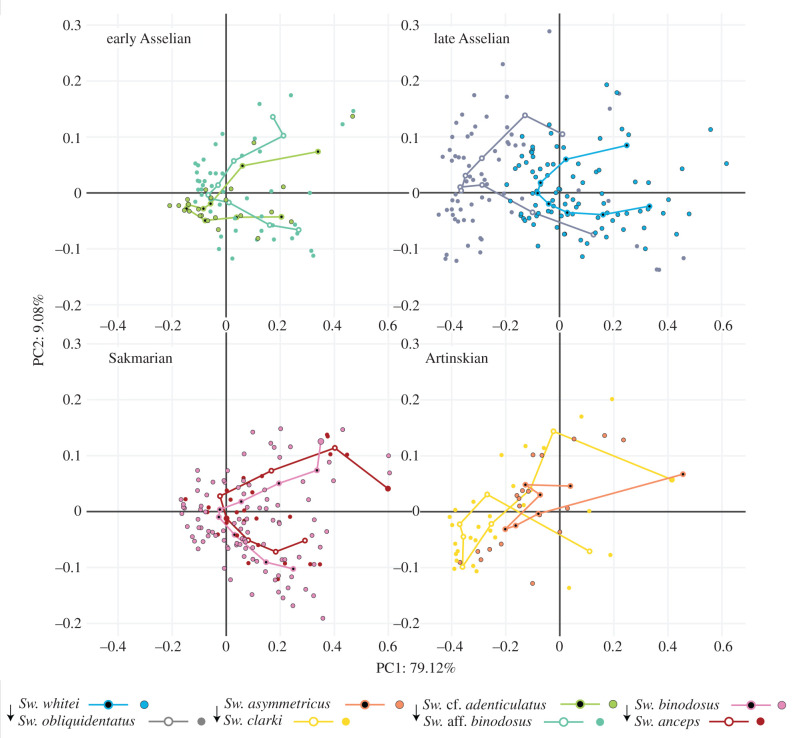
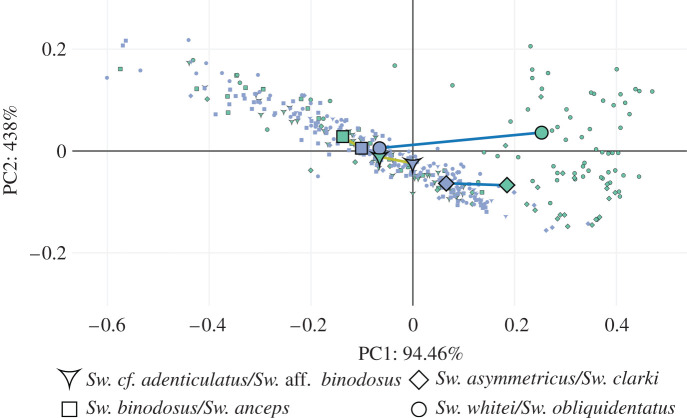
To test the evolutionary mechanisms at play in Sweetognathus diversification, phenotypic evolution needed to be quantified and compared between the different lineages in the Sweetognathus genus. Adams & Collyer [21] define phenotypic evolution trajectories as an ordered sequence of estimated phenotypes along an evolutionary path. As such, the magnitude and direction of these phenotypic trajectories can be quantified and compared to examine the evolution of phenotypes through evolutionary time between species. Phenotypic trajectories were calculated and compared using the R package RRPP [44,45] (trajectories calculated using Procrustes-aligned coordinates; see R analysis code for detailed methods [24]). The input requires an ordered sequence of phenotypes through evolutionary time as well as species groupings. Taking into account the phenotypic groupings found from the HC, species were split into four groups based on the hypothesized Sweetognathus phylogeny. Group one consisted of early Asselian taxa, Sw. cf. adenticulatus and Sw. aff. binodosus. Group two consisted of Sakmarian taxa, Sw. binodosus and Sw. anceps. Group three consisted of late Asselian taxa, Sw. whitei and Sw. obliquidentatus. Finally, group four consisted of Artinskian taxa, Sw. asymmetricus and Sw. clarki. Based on the results obtained from HC, the Sweetognathus phylogeny, and relationship in morphospace (figure 3), it was hypothesized that group one and two as well as group three and four contained parallel species pairs, respectively. Phenotypic trajectories were then plotted and compared to evaluate the potential drivers of evolutionary change between the identified parallel species pairs (figure 5).
Figure 3.
Principal component plots of Sweetognathus species grouped by time of diversification (evolutionary lineage; figure 4). In each of the plots, the initial phenotype expressed in the evolutionary lineage is indicated by the arrow. The early Asselian and Sakmarian taxa are hypothesized to follow parallel phenotypic trajectories, as well as the late Asselian and Artinskian taxa. Parallel species pairs (recurrent morphotypes) between the two hypothesized parallel lineages can be found in table 1. Open points represent the average values per species denticle, while closed (solid colour) points represent individual values for each denticle. The most likely parallel species pairs identified by cluster analysis are Sw. aff. binodosus, Sw. binodosus, and Sw. whitei (trait space 2), and Sw. obliquidentatus and Sw. clarki (trait space 5). The other species fall along trajectories of phenotypic evolution (trait spaces 1, 3, 4). (Online version in colour.)
Figure 5.
Phenotypic trajectories between species of Sweetognathus grouped by time of diversification and geographical location (evolutionary lineages). Principal components (PCs) were calculated from the covariation matrix of mean shapes using the RRPP package [44,45]. The blue points represent the initial phenotype expressed by a population in an evolutionary sequence, while the green points (outlined in black) represent the end phenotype expressed. Species were grouped into evolutionary sequences based on their degree of relatedness (figure 4), time and location of diversification, and HC results. Sweetognathus cf. adenticulatus and Sw. aff. binodosus were tested against Sw. binodosus and Sw. anceps as having parallel evolutionary sequences (gold lines). Sweetognathus whitei and Sw. obliquidentatus were tested against Sw. asymmetricus and Sw. clarki as having parallel evolutionary sequences (dark blue lines). (Online version in colour.)
3. Results
(a). Element morphospace
The first and second principal components (PC1 and PC2) described 79% and 9% of the denticle shape variance, respectively, encompassing a total 88% of the total variance (electronic supplementary material, figures and tables). Most of the species in the morphospace, grouped by denticle position, plot closely with each other and have a large amount of overlap, with Sw. obliquidentatus and Sw. clarki having the highest degree of separation from the other species (figure 3; electronic supplementary material, figures S9–S11).
To aid in the characterization and description of the shape differences between the denticle morphologies, thin-plate spline deformation grids of the mean denticle shape warped from the global mean (consensus configuration) in each species were generated (electronic supplementary material, figures S8 and S12; [19]; reviewed in [32]).
Sweetognathus whitei, Sw. aff. binodosus, Sw. binodosus and Sw. anceps have the first and second denticles elongated in the oral direction, with an overall triangular appearance. Denticles 3–5 are all fairly equidimensional, with a slight asymmetry in the denticle apex (medial ridge) depending on the species. Denticles 6 and 7 for Sw. whitei and Sw. aff. binodosus maintain the equidimensional denticle form, while Sw. binodosus and Sw. anceps begin to elongate orally and form a rounded/hemi-spherical upper surface. Denticles 8 and 9 for Sw. whitei, Sw. aff. binodosus, Sw. binodosus and Sw. anceps continue with the trend of oral elongation, but to varying degrees. Sweetognathus clarki, Sw. obliquidentatus and Sw. cf. adenticulatus have a more equidimensional first denticle, though still orally elongated, as compared to Sw. whitei, Sw. aff. binodosus, Sw. binodosus and Sw. anceps. Sweetognathus asymmetricus has an orally elongated first denticle constricted at the base, with a rounded/hemi-spherical upper surface. Denticles 2, 3, 4 and 5 of Sw. clarki and Sw. obliquidentatus are laterally elongated (orally compressed). A prominent axial groove is present in Sw. obliquidentatus and weakly in Sw. clarki. In denticles 6 and 7 of Sw. clarki, the denticle upper surface becomes rounded/hemi-spherical. Denticle 8 in Sw. clarki and denticle 9 in Sw. obliquidentatus are equidimensional, as denticle 9 in Sw. clarki is elongated orally, medially constricted, and has a rounded/hemi-spherical upper surface, while denticle 8 in Sw. obliquidentatus retains the axial groove. Sweetognathus asymmetricus and Sw. cf. adenticulatus tend to have slightly orally compressed denticles. A faint medial ridge can be seen on the upper surface of the denticles, but all are much more rounded/hemi-spherical than the other species. Sweetognathus asymmetricus diverges in morphology from Sw. cf. adenticulatus as the posterior denticles tend to be slightly more rounded/hemi-spherical on the upper surface.
Along the positive direction of the first PC axis, denticles tend to elongate orally (electronic supplementary material, figure S8). Along the negative direction of the first PC axis, denticles are elongated laterally, having a prominent axial groove. Along the positive direction of the second PC axis, denticles tend to be orally constricted at their base, be laterally elongated at the top of the denticle, and have a flat upper oral surface. Finally, along the negative direction of the second PC axis, denticles tend to be triangular.
(b). Cluster analysis
HC using a multiscale bootstrap resampling method yielded two statistically significant clusters using the first nine PC scores (accounting for 99% of the variance) weighted using per cent variance (electronic supplementary material, figure S9). Clustered species consisted of Sw. binodosus, Sw. whitei, and Sw. aff. binodosus (p-value = 0.05), and Sw. clarki and Sw. obliquidentatus (p-value < 0.01). Sweetognathus asymmetricus and Sw. cf. adenticulatus (p-value = 0.19) were associated together, but not significantly. Sweetognathus anceps was least associated with Sw. clarki and Sw. obliquidentatus.
(c). Analysis of Sweetognathus species in trait space
PC plots of the first and second PCs are presented in figure 3, grouped by potential evolutionary sequences as hypothesized by the Sweetognathus phylogeny and the cluster analysis results. Species were classified a priori into five groups based on the HC results, referred to as trait spaces as they encompass the full range of trait variation and as such allow for the identification of recurrent morphotypes (species pairs) (trait space 1: Sw. cf. adenticulatus; trait space 2: Sw. aff. binodosus, Sw. binodosus, Sw. whitei; trait space 3: Sw. anceps; trait space 4: Sw. asymmetricus; trait space 5: Sw. clarki, Sw. obliquidentatus).
When trait spaces were compared for the entire dataset (not filtering data based on denticle position) MANOVA revealed significant variation among the trait spaces (Phillai's trace = 0.62, Z = 7.73, p = 0.0001). Pairwise comparisons were then made between each trait space revealing a continuum of trait spaces in morphospace, with trait space 5 being different from all the others (electronic supplementary material, table S5). The same multivariate analysis was also completed on trait spaces filtered by denticle position (shape data for each denticle position run independently) with similar results, apart from slight denticle specific variations (electronic supplementary material, table S5). For example, trait spaces were less defined for anterior carinal denticles then posterior carinal denticles.
Next, the ratio of the distance between species trait means to trait widths was calculated (limiting similarity). In all cases, the ratio of limiting similarity was smallest for species that inhabited similar trait spaces (electronic supplementary material, table S3). Finally, species-wide variance () relative to the total variance of the genus () was calculated for each species () (table 1). Trait spaces all maintained low values, that gradually increased from trait space 1 to trait space 5 (table 1).
Table 1.
Values of the first principal component score trait means, variance, community-wide variance (), overlap (species trait width/total trait width), trait spaces found from hierarchical clustering, and lineage.
| PC 1 | variance | mean | overlap | trait space | lineage | |
|---|---|---|---|---|---|---|
| Sw. cf. adenticulatus | 0.03 | −0.01 | 0.16 | 0.64 | 1 | early Asselian |
| Sw. aff. binodosus | 0.02 | 0.06 | 0.16 | 0.55 | 2 | early Asselian |
| Sw. binodosus | 0.03 | 0.1 | 0.17 | 0.72 | 2 | Sakmarian |
| Sw. whitei | 0.03 | 0.06 | 0.18 | 0.76 | 2 | late Asselian |
| Sw. anceps | 0.03 | 0.14 | 0.18 | 0.7 | 3 | Sakmarian |
| Sw. asymmetricus | 0.04 | −0.1 | 0.2 | 0.77 | 4 | Artinskian |
| Sw. clarki | 0.05 | −0.19 | 0.22 | 0.77 | 5 | Artinskian |
| Sw. obliquidentatus | 0.05 | −0.24 | 0.21 | 0.85 | 5 | late Asselian |
Based on the above results, it appears that the trait spaces inhabit different regions in morphospace and probably represent recurrent morphotypes or species pairs between the Sweetognathus lineages.
(e). Evolutionary trajectories between Sweetognathus lineages
Morphological parallelisms were assessed by examining species in morphospace (figure 3) and comparing phenotypic trajectories between identified species pairs in the Sweetognathus lineages (figure 5). The null hypothesis in the model assumed the angle theta between the phenotypic trajectories is 0°, indicating parallel phenotypic trajectories. Between the early Asselian and Sakmarian evolutionary sequences, the angle theta between the two phenotypic trajectories was 28.42° (p-value = 0.18) and differences in the magnitude of 0.02 (p-value = 0.71). This indicates that the direction between the two phenotypic trajectories was similar and that the magnitude of phenotypic evolution between the two lineages was similar. In the case of the late Asselian and Artinskian evolutionary sequences, the angle theta between the two phenotypic trajectories was 24.65° (p-value = 0.25) and differences in the magnitude of 0.20 (p-value = 0.0002). This indicates that the direction between the two phenotypic trajectories was similar but the magnitude of phenotypic evolution between the two lineages was significantly different.
4. Discussion
(a). Parallel species pairs and evolutionary trajectories in Sweetognathus conodonts
In our study, we demonstrated the occurrence of recurrent parallel species pairs that follow similar phenotypic trajectories (figures 3 and 5). The strongest evidence for morphological parallelisms is observed in the analysis of phenotypic trajectories that found two instances of parallel evolution in Sweetognathus populations (figure 5). The first example is observed between the early Asselian and Sakmarian populations, where trajectories indicated a shift towards more symmetrical anterior carinal denticles and orally elongated posterior carinal denticles, laterally constricted at their base. The second example is observed between the late Asselian and Artinskian populations, where trajectories indicated the repeated tendency of the denticles to elongate laterally and compress orally, as well as to form a distinct axial groove, losing the denticle apex. The catalyst for the emergence of these recurrent parallel morphologies in both examples of parallel phenotypic evolution is probably some form of repeated directional selection brought on by similarities in environmental selective pressures.
The differences in the trajectory direction between the early Asselian and Sakmarian populations and the late Asselian and Artinskian populations could be the result of strong environmental selective pressures leading to disruptive and directional selection on extreme phenotypes [52,53]. The selective regimes occurring between these geographically isolated populations might have been an important mechanism in trophic diversification, which contributed to Sweetognathus species adapting to divergent niches in terms of their food base.
No direct interpretation of functional morphology exists for these morphological trends as only a few conodont taxa have been found in preservation that allow for the reconstruction of their occlusion (e.g. [11]). Broad, laterally expanded P1 elements have been reconstructed to have repeatedly evolved to allow higher loads generated during mechanical food digestion [7], whereas sharp, laterally flattened elements were adapted to have a cutting function [34]. The morphological variation has been in all cases associated with expansion to new trophic niches [7,54].
It appears that the ancestral type morphology in this study is represented by Sw. cf. adenticulatus, as it occupies the initial trait space in the phenotypic evolutionary sequence between the early Asselian and Sakmarian populations (figure 3, figures 4 and 5). Sweetognathus binodosus and Sw. aff. binodosus probably represent intermediary morphologies in the phenotypic evolutionary sequence, before the appearance of Sw. anceps (figure 5). Based on prior biostratigraphic studies (see [11]), Sw. expansus, Sw. merrilli and Sw. aff. merrilli probably represent equivalent ancestral type morphologies in the Sweetognathus lineage to that of Sw. cf. adenticulatus, though they are not included in this study. Not sampled in this study but recognized in previous biostratigraphic studies is Sw. aff. obliquidentatus, which probably represents trait space 5 missing from the early Asselian lineage. Other species such as Sw. huecoensis and Sw. bucaramangus also probably represent an undefined trait space for each lineage (figure 4).
(b). Potential causes of recurrent parallel species pairs in Sweetognathus conodonts
Empirical neontological studies have recognized parallel species pairs to emerge as a result of similar selective pressures [55,56]. Threespine sticklebacks inhabiting coastal lakes in British Columbia [57–63] provide an analogue, as do Nicaraguan cichlid fishes [64–66], to the emergence of parallel species pairs in Sweetognathus conodonts. In each case, isolated populations derived from a single ancestral population repeatedly radiated in lake environments to optimize feeding ecology, limit competition and increase fitness, demonstrating the importance of natural selection in shaping morphological parallelisms [64]. The calculated values for limiting similarity (electronic supplementary material, table S3) further support this idea, as species inhabiting equivalent niches (values approaching 0) tend not to coexist in the same lineage or region. Instead, adaptive responses (directional or disruptive selection) to new environmental pressures resulted in phenotypic shifts in the direction of a new phenotypic optima [52,53], facilitating the diversification of new species to new trophic positions, resulting in recurrent morphologies under similar selective pressures.
The extent of deviations in the magnitude and direction (figure 5) of the evolutionary trajectories between the Sweetognathus lineages from strict parallelism might reflect local environmental differences [17], developmental constraints [67] or differences in time available for diversification that changed both the order of occurrence in morphotypes and the rate at which they emerged. The incorporation of T-statistics (reviewed in [60]) and other measures of variation when comparing the intra- and interspecific variation in morphospace has provided more basis to infer on the selective pressures that may have led to deviations in evolutionary trajectories.
Community-wide variance () is used as a measure of the strength of external filters on a population, with values that approach 0 representing strong environmental selective pressures, and values that approach 1 representing high degrees of internal selective pressures (i.e. competition) [60]. In all the cases of trait spaces presented, environmental selective pressures appear to be exerting the greatest force on community assembly (table 1). For species inhabiting trait spaces 1, 2 and 3 it appears that environmental selective pressures exerted the highest degree of pressure on community assembly (table 1; ranging from 0.16 to 0.18), while more derived forms experienced higher degrees of internal selective pressures (competition) (table 1; ranging from 0.20 to 0.22). When taken in context with the ratio of trait overlap, it appears that species inhabiting initial trait spaces quickly specialized to new environmental conditions. Later, trait spaces then began to become more generalist retaining both ancestral traits as well as evolving new traits. Comparable analogues of this phenomenon are observed in Nicaraguan cichlid fishes [64,65]. Furthermore, recent studies examining Gondolellid evolution have found evidence for parallel evolution between lineages that experienced similar environmental selective pressures [67]. It, therefore, seems that the occurrence of recurrent parallel species pairs is the product of similar environmental selective pressures on dental morphologies.
5. Conclusion
Quantification of Sweetognathus dental element morphologies using GMs and analysed in association with phylogeny revealed morphological parallelisms and recurrent parallel species pairs in multiple isolated populations. The outstanding fossil record and rapid morphological radiation of Sweetognathus during the early Permian provide a long-term evolutionary perspective on parallel evolution and its role in shaping trophic diversifications in late Palaeozoic shallow marine ecosystems. Based on comparisons with cases analysing morphological variability in extant species, both intraspecific competition arising from repeated colonization of similar environments and strong environmental selective pressures leading to disruptive and directional selection drove parallelisms in Sweetognathus. Deviations in the magnitude and direction of evolutionary trajectories as well as differences in trait overlap, community-wide variance and limiting similarity between species indicate the strong role both environmental selective pressures and community assembly play in shaping Sweetognathus diversity.
Supplementary Material
Acknowledgements
We are grateful to the reviewers, Carlos Martínez-Pérez and one anonymous, for their comments. The authors are responsible for the final content, but reviewer feedback resulted in a much better formulated paper. Discussion with Mary Jane West-Eberhard on aspects of past versions was also invaluable. Finally, a thanks to NSERC (Discovery Grant to C.M.H.), the University of Calgary and Friedrich-Alexander University for supporting this work.
Data accessibility
Uncompiled computer tomographic scans and mesh files for this study, including specimen identification numbers, have been deposited in a Morphobank repository (http://morphobank.org/permalink/?P3736). The R analysis code used during data analysis and for figure creation in this study have been deposited in a GitHub repository (https://github.com/WyattSP/Evidence-of-Parallel-Evolution-in-the-Dental-Elements-of-Sweetognathus-Conodonts).
Authors' contributions
C.M.H. created the initial idea to explore the occurrence of Sweetognathus morphotypes. C.M.H. further provided valuable insight into the taxonomic notes and compiled the new Sweetognathus phylogeny. C.M.H. additionally commented on and edited the manuscript. W.P. designed and carried out the methods for this study as well as completed the analysis and interpretations for this study. W.P. wrote the manuscript for this study. E.J. aided in the scanning and processing of scanned specimens. E.J. further assisted in the development of methods and interpretations for the study. E.J. additionally commented and edited the manuscript. K.D.B. provided valuable input on the interpretations for this study and commented on the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by Dr Hertha and Helmut Schmauser-Stiftung at Friedrich-Alexander University Erlangen-Nuremberg, NSERC Discovery Grant to C.M.H. and Deutsche Forschungsgemeinschaft (project number: JA 2718/3-1).
References
- 1.Donoghue PCJ. 1998. Growth and patterning in the conodont skeleton. Phil. Trans. R. Soc. Lond. B 353, 633–666. ( 10.1098/rstb.1998.0231) [DOI] [Google Scholar]
- 2.Murdock DJE, Dong XP, Repetski JE, Marone F, Stamponi M, Donoghue PCJ. 2013. The origin of conodonts and of vertebrate mineralized skeletons. Nature 502, 546–549. ( 10.1038/nature12645) [DOI] [PubMed] [Google Scholar]
- 3.Terrill DF, Henderson CM, Anderson JS. 2018. New applications of spectroscopy techniques reveal phylogenetically significant soft tissue residue in Paleozoic conodonts. J. Anal. At. Spectrom. 33, 992–1002. ( 10.1039/c7ja00386b) [DOI] [Google Scholar]
- 4.Donoghue PCJ, Keating JN. 2014. Early vertebrate evolution. Palaeontology 57, 879–893. (doi:10.1111.pala.12125) [Google Scholar]
- 5.Martínez-Pérez C, Rayfield EJ, Purnell MA, Donoghue PCJ. 2014. Finite element, occlusal, microwear and microstructural analyses indicate that conodont microstructure is adapted to dental function. Palaeontology 57, 1059–1066. ( 10.1111/pala.12102) [DOI] [Google Scholar]
- 6.Shirley B, Grohganz M, Bestmann M, Jarochowska E. 2018. Wear, tear and systematic repair: testing models of growth dynamics in conodonts with high-resolution imaging. Proc. R. Soc. B 285, 20181614 ( 10.1098/rspb.2018.1614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez-Pérez C, Rayfield EJ, Botella H, Donoghue PCJ. 2016. Translating taxonomy into the evolution of conodont feeding ecology. Geology 44, 247–250. ( 10.1130/G37547.1) [DOI] [Google Scholar]
- 8.Purnell MA. 1993. Feeding mechanisms in conodonts and the function of the earliest vertebrate hard tissues. Geology 21, 375–377. () [DOI] [Google Scholar]
- 9.Donoghue PCJ, Sansom JJ. 2002. Origin and early evolution of vertebrate skeletonization. Microsc. Rec. Tech. 59, 352–372. ( 10.1002/jemt.10217) [DOI] [PubMed] [Google Scholar]
- 10.Donoghue PCJ, Purnell MA. 1999. Mammal-like occlusion in conodonts. Paleobiology 25, 58–74. [Google Scholar]
- 11.Mei S, Henderson CM, Wardlaw BR. 2002. Evolution and distribution of the conodonts Sweetognathus and Iranognathus and related genera during the Permian, and their implications for climate change. Palaeogeogr. Palaeoclimatol. Palaeoecol. 180, 57–91. ( 10.1016/S0031-0182(01)00423-0) [DOI] [Google Scholar]
- 12.Henderson CM. 2018. Permian conodont biostratigraphy. In The Permian timescale, vol. 450, (eds Lucas SG, Shen SZ), pp. 119–142, London, UK: Geological Society; ( 10.1144/SP450.9) [DOI] [Google Scholar]
- 13.Wang CY, Ritter SM, Clark DL. 1987. The Sweetognathus complex in the Permian of China: implications for evolution and homeomorphy. J. Paleontol. 61, 1047–1057. ( 10.1017/S0022336000029395) [DOI] [Google Scholar]
- 14.Sweet WC. 1988. The Conodonta: morphology, taxonomy, paleoecology, and evolutionary history of a long-extinct animal phylum. Oxford Monographs on Geology and Geophysics, no. 10, 212 p New York, NY: Clarendon Press [Google Scholar]
- 15.Elmer KR, Meyer A. 2011. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol. Evol. 26, 298–306. ( 10.1016/j.tree.2011.02.008) [DOI] [PubMed] [Google Scholar]
- 16.Monnet C, De Baets K, Klug C. 2011. Parallel evolution controlled by adaptation and covariation in ammonoid cephalopods. BMC Evol. Biol. 11, 115 ( 10.1186/1471-2148-11-115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaster J, Berger J. 1977. Convergent and parallel evolution: a model illustrating selection, phylogeny and phenetic similarity. Biosystems 9, 195–200. ( 10.1016/0303-2647(77)90003-X) [DOI] [PubMed] [Google Scholar]
- 18.Oke KB, Bukhari M, Kaueffer R, Rolshausen G, Räsänen K, Bolnick DI, Peichel CL, Hendry AP. 2016. Does plasticity enhance or dampen phenotypic parallelism? A test with three lake-stream stickleback pairs. J. Evol. Biol. 29, 126–143. ( 10.1111/jeb.12767) [DOI] [PubMed] [Google Scholar]
- 19.Bookstein FL. 1991. Morphometric tools for landmark data: geometry and biology. New York, NY: Cambridge University Press. [Google Scholar]
- 20.Rohlf FJ, Marcus LF. 1993. A revolution in morphometrics. Trends Ecol. Evol. 8, 129–132. ( 10.1016/0169-5347(93)90024-J) [DOI] [PubMed] [Google Scholar]
- 21.Adams DC, Collyer ML. 2009. A general framework for the analysis of phenotypic trajectories in evolutionary studies. Evolution 63, 1143–1154. ( 10.1111/j.1558-5646.2009.00649.x) [DOI] [PubMed] [Google Scholar]
- 22.O'Leary MA, Kaufman SG. 2012. MorphoBank 3.0: Web application for morphological phylogenetics and taxonomy See http://www.morphobank.org.
- 23.Petryshen W, Henderson C, De Baets K, Jarochowska E. 2020. Data from: Evidence of parallel evolution in the dental elements of Sweetognathus conodonts MorphoBank (http://morphobank.org/permalink/?P3736) [DOI] [PMC free article] [PubMed]
- 24.Petryshen W, Henderson C, De Baets K, Jarochowska E.. 2020. R Analysis Code from: Evidence of parallel evolution in the dental elements of Sweetognathus conodonts. v. 1.0.0. Zenodo. ( 10.5281/zenoda.4117371). (https://github.com/WyattSP/Evidence-of-Parallel-Evolution-in-the-Dental-Elements-of-Sweetognathus-Conodonts) [DOI] [PMC free article] [PubMed]
- 25.Wiley DF, et al. 2005. Evolutionary morphing. IEEE. Vis 05. In Visulization Conf., IEEE, Minneapolis, Minnesota, pp. 431–438. ( 10.1109/VISUAL.2005.1532826) [DOI] [Google Scholar]
- 26.Cignoni P, Callieri M, Corsini M, Dellepiane M, Ganovelli F, Ranzuglia G. 2008. Meshlab: an open-source mesh processing tool. In Sixth Eurographics Italian Chapter Conf., 2008, pp. 129–136. Geneva, Switzerland: Eurographics Association. [Google Scholar]
- 27.RStudio Team. 2020. RStudio: integrated development for R. Boston, MA: RStudio, PBC; See http://www.rstudio.com/. [Google Scholar]
- 28.R Core Team. 2019. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See https://www.R-project.org/. [Google Scholar]
- 29.Bonhomme V, Picq S, Gaucherel C, Claude J. 2014. Momocs: outline analysis using R. J. Stat. Softw. 56, 1–24. ( 10.18637/jss.v056.i13) [DOI] [Google Scholar]
- 30.Purnell MA, Donoghue PCJ, Aldridge RJ. 2000. Orientation and anatomical notation in conodonts. J. Paleontol. 75, 113–122. ( 10.1017/S0022336000031292) [DOI] [Google Scholar]
- 31.Adams DC, Rohlf FL, Slice DE. 2013. A field comes of age: geometric morphometrics in the 21st century. Hystrix It. J. Mamm. 24, 7–14. ( 10.4404/hystrix-24.1-6283) [DOI] [Google Scholar]
- 32.Mitteroecker P, Gunz P. 2009. Advances in geometric morphometrics. Evol. Biol. 36, 235–247. ( 10.1007/s11692-009-9055-x) [DOI] [Google Scholar]
- 33.Zelditch ML, Swiderski DL, Sheets HD. 2012. Geometric morphometrics for biologists: a primer. Amsterdam, The Netherlands: Academic Press. [Google Scholar]
- 34.Jones D, Evans AR, Siu KK, Rayfield EJ, Donoghue PC. 2012. The sharpest tools in the box? Quantitative analysis of conodont element functional morphology. Proc. R. Soc. B 279, 2849–2854. ( 10.1098/rspb.2012.0147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams DC, Collyer ML, Kaliontzopoulou A.. 2018. Geomorph: software for geometric morphometric analyses. R package version 3.0.6 See https://cran.r-project.org/package=geomorph.
- 36.Gower JC. 1975. Generalized Procrustes analysis. Psychometrika 40, 33–51. ( 10.1007/BF02291478) [DOI] [Google Scholar]
- 37.Rohlf FL, Slice DE. 1990. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Zool. 39, 40–59. ( 10.2307/2992207) [DOI] [Google Scholar]
- 38.Schmitz MD, Davydov VI. 2012. Quantitative radiometric and biostratigraphic calibration of the global Pennsylvanian – Early Permian time scale. Geol. Soc. Am. Bull. 124, 549–577. ( 10.1130/B30385.1) [DOI] [Google Scholar]
- 39.Henderson CM, Schmitz M, Crowley J, Davydov VI. 2009. Evolution and geochronology of the Sweetognathus lineage from Bolivia and the Urals of Russia: Biostratigraphic problems and implications for Global Stratotype Section and Point (GSSP) definition. Permophiles (ICOS Abstracts) 53, 20. [Google Scholar]
- 40.Beauchamp B, Calvo Gonzalez D, Henderson CM, Baranova DV, Wang HY, Pelletier E. 2020. Late Pennsylvanian–early Permian tectonically-driven stratigraphic sequences and carbonate sedimentation along northern margin of Sverdrup Basin (Otto Fiord Depression), Arctic Canada. In Late Paleozoic tectonostratigraphy and biostratigraphy of western Pangea (eds Henderson CM, Ritter S, Snyder WS). SEPM Special Publication No. 113; Broken Arrow, OK: SEPM. [Google Scholar]
- 41.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. ( 10.1093/bioinformatics/btl117) [DOI] [PubMed] [Google Scholar]
- 42.Dryden IL, Mardia KV. 1998. Statistical shape analysis. New York, NY: John Wiley & Sons. [Google Scholar]
- 43.Rohlf FL. 1999. Shape statistics: Procrustes superimpositions and tangent spaces. J. Classif. 16, 197–223. ( 10.1007/s003579900054) [DOI] [Google Scholar]
- 44.Collyer M, Adams DC. 2019. RRPP: linear model evaluation with randomized residuals in a permutation. R package v. 0.5.1. Procedure. See https://CRAN.R-project.org/package=RRPP.
- 45.Collyer ML, Adams DC. 2018. RRPP: an R package for fitting linear models to high-dimensional data using residual randomization. Methods Ecol. Evol. 9, 1772–1779. ( 10.1111/2041-210X.13029) [DOI] [Google Scholar]
- 46.MacArthur R, Levins R. 1967. The limiting similarity, convergence, and divergence of coexisting species. Am. Nat. 101, 377–385. ( 10.1086/282505) [DOI] [Google Scholar]
- 47.Violle C, Enquist BJ, McGill BJ, Jiang L, Albert CH, Hulshof C, Jung V, Messier J. 2012. The return of the variance: intraspecific variability in community ecology. Cell Press 27, 244–252. ( 10.1016/j.tree.2011.11.014) [DOI] [PubMed] [Google Scholar]
- 48.Gause GF. 1934. The struggle for existence. Baltimore, MD: Williams & Wilkins. [Google Scholar]
- 49.Turcotte MM, Levine JM. 2016. Phenotypic plasticity and species coexistence. Trends Ecol. Evol. 31, 803–813. ( 10.1016/j.tree.2016.07.013) [DOI] [PubMed] [Google Scholar]
- 50.Albert CH, Thuiller W, Yoccoz NG, Douzet R, Aubert S, Lavorel S. 2010. A multi-trait approach reveals the structure and the relative importance of intra- vs. interspecific variability in plant traits. Methods Ecol. Evol. 24, 1192–1201. ( 10.1111/j.1365-2435.2010.01727.x) [DOI] [Google Scholar]
- 51.Holsinger K, Weir B. 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10, 639–650. ( 10.1038/nrg2611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price TD, Qvarnström A, Irwin DE. 2003. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B 270, 1433–1440. ( 10.1098/rspb.2003.2372) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghalambor CK, McKay JK, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407. ( 10.1111/j.1365-2435.2007.01283.x) [DOI] [Google Scholar]
- 54.Ginot S, Goudemand N. 2019. Conodont size, trophic level, and the evolution of platform elements. Paleobiology 45, 458–468. ( 10.1017/pab.2019.19) [DOI] [Google Scholar]
- 55.Clark BC. 1975. The contribution of ecological genetics to evolutionary theory: detecting the direct effects of natural selection on particularly polymorphic loci. Genetics 79(Suppl.), 101–113. [PubMed] [Google Scholar]
- 56.Endler JA. 1986. Natural selection in the wild. Princeton, NJ: Princeton University Press. [Google Scholar]
- 57.McPhail JD, Lindsey CC. 1970. Freshwater fishes of northwestern Canada and Alaska. Bull. Fish. Res. Board Can. 173, 1–381. [Google Scholar]
- 58.Bell MA. 1974. Reduction and loss of the pelvic girdle in Gasterosteus (Pisces): a case of parallel evolution. Nat. Hist. Mus. Los Angeles Cty. Contrib. Sci. 257, 1–36. [Google Scholar]
- 59.Bell MA. 1976. Evolution of phenotypic diversity in Gasterosteus aculeatus superspecies on the Pacific coast of North America. Syst. Zool. 25, 211–227. ( 10.2307/2412489) [DOI] [Google Scholar]
- 60.Bell MA. 1987. Interacting evolutionary constraints in pelvic reduction of threespine sticklebacks, Gasterosteus aculeatus (Pisces, Gasterosteidea). Biol. J. Linn. Soc. 31, 347–382. ( 10.1111/j.1095-8312.1987.tb01998.x) [DOI] [Google Scholar]
- 61.Bell MA, Foster SA. 1994. The evolutionary biology of threespine stickleback. Oxford, UK: Oxford University Press. [Google Scholar]
- 62.Walker JA, Bell MA. 2000. Net evolutionary trajectories of body shape evolution within a microgeographic radiation of threespine sticklebacks (Gasterosteus aculeatus). J. Zool. 253, 293–302. ( 10.1111/j.1469-7998.2000.tb00624.x) [DOI] [Google Scholar]
- 63.Schluter D, Clifford EA, Nemethy M, McKinnon JS. 2004. Parallel evolution and inheritance of quantitative traits. Am. Nat. 163, 809–820. ( 10.1086/383621) [DOI] [PubMed] [Google Scholar]
- 64.Elmer KR, Shaohua F, Kusche H, Spreitzer ML, Kautt AF, Franchini P, Meyer A. 2014. Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat. Commun. 5, 5168 ( 10.1038/ncomms6168) [DOI] [PubMed] [Google Scholar]
- 65.Elmer KR, Kusche H, Lehtonen TK, Meyer A. 2010. Local variation and parallel evolution: morphological and genetic diversity across a species complex of neotropical crater lake cichlid fishes. Phil. Trans. R. Soc. B 365, 1763–1782. ( 10.1098/rstb.2009.0271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Villa J. 1976. Ichthyology of the lakes of Nicaragua: historical perspective. In Investigations of the ichthyology of Nicaraguan lakes (ed. Thorson TB.), pp. 101–113. Lincoln, NE: University of Nebraska Press. [Google Scholar]
- 67.Guenser P, Souquet L, Dolédec S, Mazza M, Rigo M, Goudemand N. 2019. Deciphering the roles of environment and development in the evolution of a Late Triassic assemblage of conodont elements. Paleobiology 45, 440–457. ( 10.1017/pab.2019.14) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Petryshen W, Henderson C, De Baets K, Jarochowska E. 2020. Data from: Evidence of parallel evolution in the dental elements of Sweetognathus conodonts MorphoBank (http://morphobank.org/permalink/?P3736) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Uncompiled computer tomographic scans and mesh files for this study, including specimen identification numbers, have been deposited in a Morphobank repository (http://morphobank.org/permalink/?P3736). The R analysis code used during data analysis and for figure creation in this study have been deposited in a GitHub repository (https://github.com/WyattSP/Evidence-of-Parallel-Evolution-in-the-Dental-Elements-of-Sweetognathus-Conodonts).