Abstract
How many parasites are there on Earth? Here, we use helminth parasites to highlight how little is known about parasite diversity, and how insufficient our current approach will be to describe the full scope of life on Earth. Using the largest database of host–parasite associations and one of the world’s largest parasite collections, we estimate a global total of roughly 100 000–350 000 species of helminth endoparasites of vertebrates, of which 85–95% are unknown to science. The parasites of amphibians and reptiles remain the most poorly described, but the majority of undescribed species are probably parasites of birds and bony fish. Missing species are disproportionately likely to be smaller parasites of smaller hosts in undersampled countries. At current rates, it would take centuries to comprehensively sample, collect and name vertebrate helminths. While some have suggested that macroecology can work around existing data limitations, we argue that patterns described from a small, biased sample of diversity aren’t necessarily reliable, especially as host–parasite networks are increasingly altered by global change. In the spirit of moonshots like the Human Genome Project and the Global Virome Project, we consider the idea of a Global Parasite Project: a global effort to transform parasitology and inventory parasite diversity at an unprecedented pace.
Keywords: parasites, biodiversity estimation, systematics, museum collections, tapeworms
1. Introduction
Parasitology is currently trapped between apparently insurmountable data limitations and the urgent need to understand how parasites will respond to global change. Parasitism is arguably the most species-rich mode of animal life on Earth [1–3], and parasites probably comprise a majority of the undescribed or undiscovered species left to modern science [2,4]. In recent years, the global diversity and distribution of parasite richness has become a topic of particular concern [1,5,6], both in light of the accelerating rate of disease emergence in wildlife, livestock and humans [7], and growing recognition of the ecological significance of many parasites [8]. Parasitic taxa are expected to face disproportionately high extinction rates in the coming century, causing a cascade of unknown but possibly massive ecological repercussions [5,9]. Understanding the impacts of global change relies on baseline knowledge about the richness and biogeography of parasite diversity, but some groups are better studied than others. Emerging and potentially zoonotic viruses dominate this field [10–14]; macroparasites receive comparatively less attention.
Despite the significance of parasite biodiversity, the actual richness of most macroparasitic groups remains uncertain, due to a combination of underlying statistical challenges and universal data limitations for symbiont taxa. Particularly deserving of reassessment are helminth parasites (hereafter helminths), a polyphyletic group of parasitic worms including, but not limited to, the spiny-headed worms (acanthocephalans; Phylum: Acanthocephala), tapeworms (cestodes; Phylum: Platyhelminthes, Class: Cestoda), roundworms (nematodes; Phylum: Nematoda), and flukes (trematodes; Phylum: Platyhelminthes, Class: Trematoda). Helminth parasites exhibit immense diversity [1,6], tremendous ecological and epidemiological significance [15,16], and a wide host range across vertebrates, invertebrates and plants [1,17,18]. Estimates of helminth diversity remain controversial [1,2,19], especially given uncertainties arising from the small fraction of total diversity described so far [4]. Though the task of describing parasite diversity has been called a ‘testimony to human inquisitiveness’ [1], it also has practical consequences for the global task of cataloging life; one recent study proposed there could be 80 million or more species of nematode parasites of arthropods, easily reaffirming the Nematoda as a contender for the most diverse phylum on Earth. [2]
With the advent of metagenomics and bioinformatics, and the increasing digitization of natural history collections, funders are becoming interested in massive ‘moonshot’ endeavours to catalogue global diversity. Last year, the Global Virome Project was established with the stated purpose of cataloging 85% of viral diversity within vertebrates (particularly mammals and birds, which host almost all emerging zoonoses), with an investment of $1.2 billion over 10 years. Whereas the Global Virome Project is ultimately an endeavour to prevent the future emergence of the highest-risk-potential zoonoses—the natural evolution of decades of pandemic-oriented work at the edge of ecology, virology and epidemiology—we suggest parasitologists have the opportunity to set a more inclusive goal. Between a quarter and half of named virus species can infect humans [14], while human helminthiases are a small, almost negligible fraction of total parasite diversity despite their massive global health burden. The need to understand global parasite diversity reflects a more basic set of questions about the world we live in, and the breadth of life within it.
Here we ask, what it would take to completely describe global helminth diversity in vertebrates? The answer is just as dependent on how many helminth species exist as it is on the rate and efficiency of parasite taxonomic description efforts. We set out to address three questions:
-
I.
What do we know about the global process of describing and documenting parasite biodiversity, and how will it change in the future?
-
II.
How many helminth species should we expect globally, and how much of that diversity is described?
-
III.
How many years are we from describing all of global parasite diversity, and what can (and cannot) we do with what we have?
From there, we make recommendations about where the next decade of parasite systematics and ecology might take us.
2. The data
To answer all three questions, we take advantage of two collections-based datasets that have been made available in the last decade (figure 1). The biological collections housed at museums, academic research institutions, and various private locations around the world are one of the most significant ‘big data’ sources for biodiversity research [20], especially for parasites [21,22]. The Natural History Museum in London (NHM) curates the Host–Parasite Database, which includes regional lists of helminth-host associations, including full taxonomic citations for helminth species [23,24]. By species counts alone, the NHM dataset is perhaps the largest species interaction dataset published so far in ecological literature. [6] In our updated scrape of the web interface, which will be the most detailed version of the dataset ever made public, there are a raw total of 109 060 associations recorded between 25 740 helminth species (including monogeneans, which we exclude to focus on endoparasites) and 19 097 hosts (vertebrate and invertebrate).
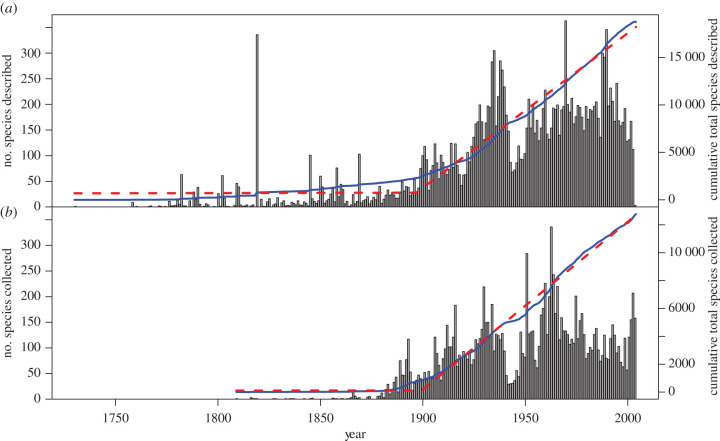
Figure 1.
Rates of helminth (a) descriptions (from NHM data) and (b) collections (from the US National Parasite Collection (USNPC)). Solid lines (blue) indicate cumulative totals, and dashed lines (red) give a breakpoint regression with a single breakpoint (1912 for the NHM data, 1903 for the USNPC data). Although the current trend appears to be levelling off, it is unlikely this indicates a saturating process (as comparably illustrated by the drop in sampling during the Second World War, 1940–1945). (Online version in colour.)
The US National Parasite Collection (USNPC) is one of the largest parasite collections in the world, and is one of the most significant resources used by systematists to discover, describe and document new species [21,25]. The published records constitute the largest open museum collection database for helminths, especially in terms of georeferenced data availability [5]. Here, we use a recent copy of the USNPC database comprising 89 580 specimen records, including 13 426 species recorded in the groups Acanthocephala, Nematoda and Platyhelminthes (of these, the vast majority are vertebrate parasites [26]). In combination, the two datasets represent the growing availability of big data in parasitology, and allow us to characterize parasite diversity much more precisely than we could have a decade ago.
3. How does parasite biodiversity data accumulate?
Describing the global diversity of parasites involves two major processes: documenting and describing diversity through species descriptions, geographic distributions, host associations, etc.; and consolidating and digitizing lists of valid taxonomic names and synonyms (e.g. ITIS, Catalogue of Life, WoRMS). Both efforts are important, time-consuming, and appear especially difficult for parasites.
(a). Why has helminth diversity been so difficult to catalogue?
The most obvious reason is the hyperdiversity of groups like the Nematoda, but this only tells part of the story. Other hyperdiverse groups, like the sunflower family (Asteraceae), have far more certain richness estimates (and higher description rates) despite being comparably speciose. Several hypotheses are plausible: surveys could be poorly optimized for the geographical and phylogenetic distribution of helminth richness, or remaining species might be objectively harder to discover and describe than known ones were. Perhaps the most popular explanation is that taxonomists’ and systematists’ availability might be the limiting factor [27,28]; the process of describing helminth diversity relies on the dedicated work of systematic biologists, and the availability and maintenance of long-term natural history collections. However, Costello et al. [29] observed that the number of systematists describing parasites has increased steadily since the 1960s, with apparently diminishing returns. Costello posited that this was evidence the effort to describe parasites has reached the ‘inflection point’, with more than half of all parasites described; this assessment disagrees with many others in the literature. [28]
(b). Have we actually passed the inflection point?
No, probably not. We show this by building species accumulation curves over time, from two different sources: the dates given in taxonomic authority citations in the NHM data, and the date of first accession in the USNPC data, for each species in the dataset (figure 1). Both are a representation of total taxonomic effort, and vary substantially between years. Some historical influences are obvious, such as a drop during the Second World War (1939–1945). Recently, the number of parasites accessioned has dropped slightly, but it seems unlikely (especially given historical parallels) that this reflects a real inflection point in parasite sampling, and is probably instead reflects a limitation of the data structure; the NHM data, in particular, have not been updated since 2013. Despite interannual variation, the accumulation curves both demonstrate a clear cut pattern: sometime around the turn of the twentieth century, they turn upward and increase linearly. Since 1897, an average of 163 helminth species have been described annually (R2 = 0.991, p < 0.001), while an average of 120 species are added to collections every year since 1899 (R2 = 0.998, p < 0.001). The lack of slowing down in those linear trends is a strong indicator that we remain a long way from a complete catalogue of helminth diversity.
(c). Are we looking in the wrong places?
An alternate explanation for the slow rate of parasite discovery is that the majority of parasite diversity is in countries where sampling effort is lower, and vice versa most sampling effort and research institutions are in places with more described parasite fauna [30]. Recent evidence suggests species discovery efforts so far have been poorly optimized for the underlying—but mostly hypothetical—richness patterns of different helminth groups [30,31]. Ecologists have started to ask questions that could help optimize sampling: do parasites follow the conventional latitudinal diversity gradient? Are there unique hotspots of parasite diversity, or does parasite diversity peak in host biodiversity hotspots [1,6,30,32–34]? But our ability to answer these types of questions is predicated on our confidence that observed macroecological patterns in a small (and uncertain) percentage of the world’s helminths are representative of the whole.
(d). Are species described later qualitatively different?
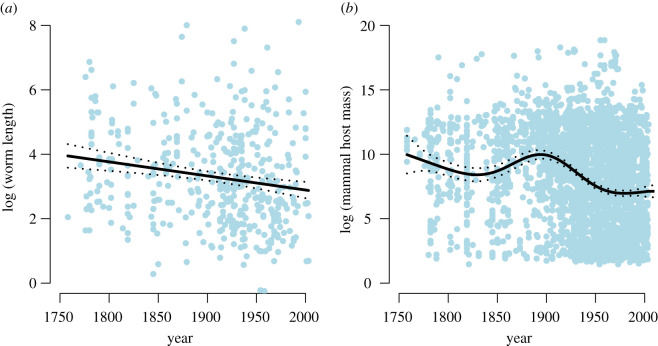
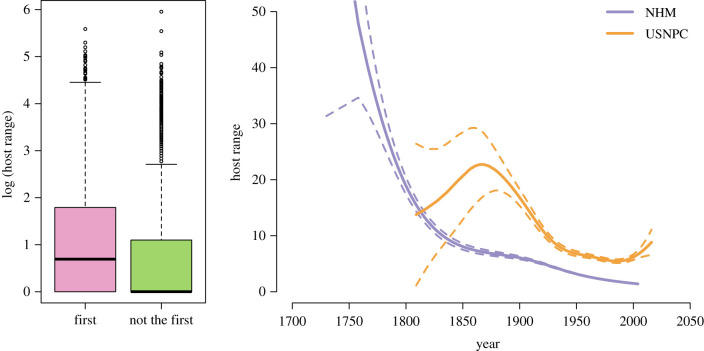
If helminth descriptions have been significantly biased by species’ ecology, this should produce quantitative differences between the species that have and have not yet been described. We examine two easily intuited sources of bias: body size (larger hosts and parasites are better sampled) and host specificity (generalist parasites should be detected and described sooner). We found a small but highly significant trend of decreasing body size over time for both hosts and parasites, suggesting the existence of a sampling bias, but not necessarily suggesting unsampled species should be massively different (figure 2). For host specificity, measured as the total host range (number of hosts), we find an obvious pattern relative to description rates, though less so for collections data (figure 3). The inflection point around 1840 is probably a by-product of the history of taxonomy, as the Series of Propositions for Rendering the Nomenclature of Zoology Uniform and Permanent—now the International Code for Zoological Nomenclature—was first proposed in 1842, leading to a standardization of host nomenclature and consolidation of the proliferation of multiple names for single species.
Figure 2.
We found evidence of weak but highly significant declines over time in (a) parasite adult body length (smooth term p = 0.0003) and (b) host body size across known host associations (smooth term p < 0.0001). This confirms a mild description bias for larger parasites in larger hosts. (Online version in colour.)
Figure 3.
The type species (the first described in a genus) has a statistically significantly higher average host range than those that follow. Parasites described earlier typically have a higher degree of generalism (greater number of recorded hosts), especially prior to the 1840s; specimens collected after roughly the 1870s also apparently tend towards more host-specific species than those from older collections. (Curves are generalized additive models fit assuming a negative binominal distribution, with dashed lines for the 95% confidence bounds.) (Online version in colour.)
The temporal trend also likely reflects the history of taxonomic revisions, as the first species reported in a genus tends to have a higher range of hosts, morphology and geography, while subsequent revisions parse these out into more appropriate, narrower descriptions. Using the NHM data, we can easily show that the first species reported in every genus (usually the type species but not always, given incomplete sampling) generally has significantly higher reported numbers of hosts (Wilcoxon rank sum test: W = 22 390 629, p < 0.001; figure 3). This is because type species often become umbrella descriptors that are subsequently split into more species after further investigation, each with only a subset of the initial total host range. Based on our results, we can expect undescribed species of helminths to be disproportionately host-specific.
4. How many helminths?
(a). How do we count parasites?
For many groups of parasites, the number of species known to science is still growing exponentially, preventing estimation based on the asymptote of sampling curves [35]. In some cases, there are workarounds: for example, the diversity of parasitoid wasps (Hymenoptera: Braconidae) has been estimated based on the distribution of taxonomic revisions rather than descriptions [36]. But for helminths, every major estimate of diversity is based on the scaling between host and parasite richness, a near-universal pattern across spatial scales and taxonomic groups [6,37,38]. The scaling of hosts and fully host-specific parasites can be assumed to be linear: for example, every arthropod is estimated to have at least one host-specific nematode [2]. Poulin & Morand [35] proposed an intuitive correction for generalists, where parasite richness P can be estimated () as a linear function of host richness H, using estimates:
| 4.1 |
Poulin & Morand [35] compiled independently sourced estimates of host specificity and per-species richness, and the resulting estimate of approximately 75 000–300 000 helminth species was canon for a decade [1].
(b). What do we know now that we didn’t before?
A previous study by Strona & Fattorini [19] showed that the linear method of estimating parasite diversity is inconsistent with the properties of real data. Using the NHM dataset (as we do here), they showed that subsampling a host–parasite network approximately generates power law scaling, not linear scaling, which reduced estimates by of helminth diversity (in helminth and vertebrate taxon pairs) by an average of 58%. However, they made no overall corrected estimate of helminth diversity in vertebrates.
Examining bipartite host–affiliate networks across several types of symbiosis, including the vertebrate–helminth network (from the NHM data), we previously found approximate power law behaviour in every scaling curve [14]. The underlying reasons for this pattern are difficult to ascertain, and may or may not be connected to approximate power-law degree distributions in the networks. Regardless, the method seems to work as a tool for estimating richness; using the new R package codependent [39], we used these tools to show that viral diversity in mammals is probably only about 2–3% of the estimates generated with linear extrapolation by the Global Virome Project [14].
Here, we build on this work by adding confidence intervals using the codependent package. Moreover, we show that association data can be used to estimate the proportion of overlap among groups, and thereby correct the total when adding together parasite richness sub-totals. (See Material and methods.) This allows us to extend Strona and Fattorini’s analysis to produce a total corrected estimate of the diversity of helminth endoparasites of vertebrates.
(c). How many species are there?
Building on previous studies [1,19], we used the power law method to re-estimate global helminth diversity. We derived these estimates using codependent, a taxonomically cleaned version of the NHM dataset, and a new formula for combining parasite richness across groups (table 1). In total, we estimated 103 078 species of helminth parasites of vertebrates, most strongly represented by trematodes (44 262), followed by nematodes (28 844), cestodes (23 749) and acanthocephalans (6223). Using an updated estimate of bony fish richness significantly increased these estimates from previous ones, with over 37 000 helminth species in this clade alone. Birds and fish were estimated to harbour the most helminth richness, but reptiles and amphibians had the highest proportion of undescribed diversity. The best-described groups were nematode parasites of mammals (possibly because so many are zoonotic and livestock diseases) and cestode parasites of the cartilaginous fishes (perhaps due to the expertise of a strong collaborative research community, including the participants in the Planetary Biodiversity Inventory project on cestode systematics) [40].
Table 1.
Helminth diversity, re-estimated: How many helminth species (top), and what percentage of species have been described (bottom)?
| Chondrichthyes | Osteichthyes | Amphibia | Reptilia | Aves | Mammalia | total | |
|---|---|---|---|---|---|---|---|
| Acanthocephala | 169 | 3572 | 765 | 785 | 1184 | 886 | 6223 |
| (4%) | (13%) | (3%) | (4%) | (14%) | (12%) | (11%) | |
| Cestoda | 2108 | 5875 | 637 | 2153 | 10 257 | 4061 | 23 749 |
| (28%) | (12%) | (5%) | (5%) | (14%) | (26%) | (16%) | |
| Nematoda | 566 | 10 712 | 2148 | 4537 | 3925 | 7902 | 28 844 |
| (14%) | (11%) | (10%) | (12%) | (19%) | (30%) | (17%) | |
| Trematoda | 391 | 17 745 | 3700 | 12 153 | 8778 | 4550 | 44 262 |
| (16%) | (19%) | (6%) | (4%) | (17%) | (23%) | (14%) | |
| total | 3234 | 37 904 | 7250 | 19 628 | 24 144 | 17 399 | 103 078 |
| (23%) | (15%) | (7%) | (6%) | (16%) | (26%) | (15%) |
(d). Do we trust these estimates?
Although estimates from a decade ago were surprisingly close given methodological differences [1], we now have a much greater degree of confidence in our overall estimate of vertebrate helminth richness. However, some points of remaining bias are immediately obvious. The largest is methodological: by fitting power law curves over host richness, we assumed all hosts had at least one parasite from any given helminth group. While this assumption worked well for mammal viruses, it may be more suspect especially for the less-speciose groups like Acanthocephala. On the other hand, the power-law method is prone to overestimation in several ways enumerated in [14]. Furthermore, Dallas et al. [41] estimated that 20–40% of the host range of parasites is underdocumented in the Global Mammal Parasite Database, a sparser but comparable dataset. If these links were recorded in our data, they would substantially expand the level of host-sharing and cause a reduction of the scaling exponent of power laws, causing lower estimates. On the other hand, if we know that the majority of undescribed parasite diversity is far more host specific than known species, our estimates would severely underestimate in this regard. At present, it is essentially impossible to estimate the sign of the these errors once compounded together.
(e). What about cryptic diversity?
One major outstanding problem is cryptic diversity, the fraction of undescribed species that are genetically distinct but morphologically indistinguishable, or at least so subtly different that their description poses a challenge. Many of the undescribed species could fall in this category, and splitting them out might decrease the apparent host range of most species, further increasing estimates of total diversity. Dobson et al. [1] addressed this problem by assuming that the true diversity of helminths might be double and double again their estimate; while this makes sense conceptually, it lacks any data-driven support. The diversity of cryptic species is unlikely to be distributed equally among all groups; for example, long-standing evidence suggests it may be disproportionately higher for trematodes than cestodes or nematodes [42].
We can loosely correct our overall richness estimates for cryptic diversity. A recently compiled meta-analysis suggests an average of 2.6 cryptic species per species of acanthocephalan, 2.4 per species of cestode, 1.2 per species of nematode, and 3.1 per species of digenean. [43] Using these numbers, we could push our total estimates to at most 22 404 acanthocephalan species, 80 747 cestodes, 63 457 nematodes and a whopping 181 474 species of trematodes, with a total of 348 082 species of helminths. However, there may be publication bias that favours higher cryptic species rates (or at least, zeros may be artificially rare), making these likely to be overestimates. Increased sampling will push estimates higher for many species, and eventually will allow a more statistically certain estimate of the cryptic species ‘multiplication factor’ needed to update the estimates we present here.
5. Could we describe the world’s parasite diversity?
(a). How long would it take to catalogue global helminth diversity?
We estimated 103 079 total helminth species on Earth, of which 13 426 (13.0%) are in the USNPC and 15 817 (15.3%) are in the NHM Database. At the current rates, we estimated, it would take 536 years to describe global helminth diversity and catalogue at least some host associations (based on the NHM data as a taxonomic reference), and 745 years to add every species to the collection (based on the USNPC). Including the full range of possible cryptic species would push the total richness to 348 082 helminth species (95% undescribed), which would require 2040 years to describe and 2779 years to collect.
Even with hypothetical overcorrections, these are daunting numbers: for example, if the NHM only captures one-tenth of known helminth diversity, and thereby underestimates the rate of description by an order of magnitude, it would still take two centuries to describe remaining diversity. These estimates are also conservative in several ways: the majority of remaining species will be more host-specific and therefore harder to discover, and the process would almost certainly undergo an asymptote or at least a mild saturating process. Moreover, many of the 13 426 unique identifiers in the USNPC are either currently or may be synonyms of valid names and may be corrected through taxonomic revision and redetermination; previous estimates suggest invalid names may outnumber valid ones, in some data [29].
(b). Where is the undescribed diversity?
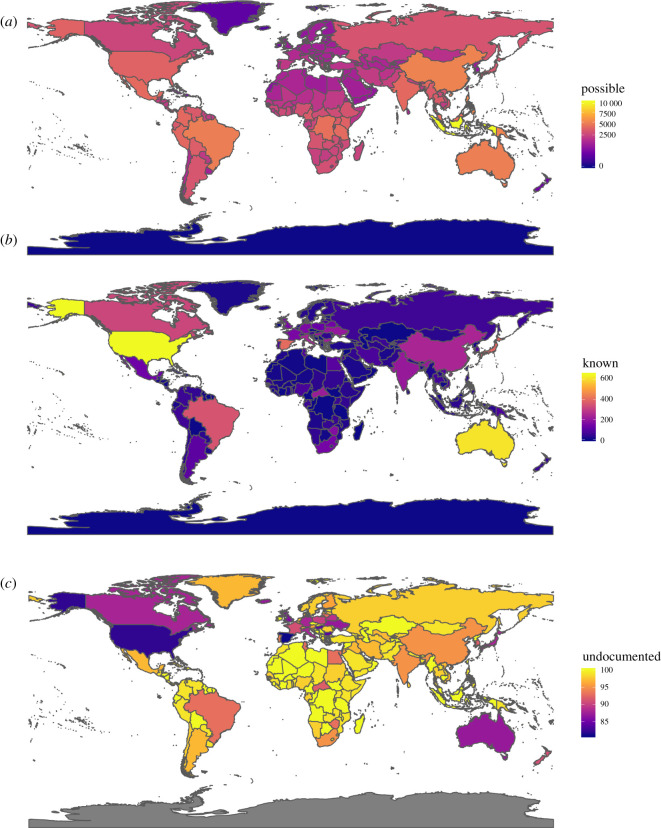
Previous work has argued that current patterns of helminth description are poorly matched to underlying richness patterns, though those patterns are also unknown and assumed to broadly correspond to host biodiversity [30]. Here, we used the scaling between host and parasite diversity to predict the ‘maximum possible’ number of parasites expected for a country’s mammal fauna, and compared that to known helminths described from mammals in the NHM dataset (figure 4). While these estimates are liberal in the sense that they include the global range of parasite fauna associated with given hosts, they are also conservative in that they are uncorrected for cryptic diversity, or the possibility of higher host specificity in the tropics.
Figure 4.
The distribution of (a) maximum possible helminth richness in mammals, (b) the number of known helminth parasites of mammals as recorded by country in the NHM data and (c) the maximum percentage of undocumented helminth fauna by country. (Online version in colour.)
We found that helminths were best known in the handful of countries that dominate parasite systematics work (the USA, Australia, Brazil, Canada, China and some European countries). But even in these places, most species are probably undescribed; many countries have no records at all, including large countries like the Democratic Republic of the Congo that are mammal diversity hotspots. Between 80% and 100% of possible parasite diversity could be locally undescribed for most of the world—high estimates, but plausible given a global undescribed rate of 85–95%. This spatial pattern probably reflects a combination of language and access barriers (data in Chinese and Russian collections, for example, are known to be substantial, but inaccessible to our present work), and a broader inequity arising from the concentration of institutions and researchers in wealthy countries, and the corresponding disproportionate geographical focus of research [44]. Previous research has noted that African parasitology has been especially dominated by foreign researchers [45], and African parasitologists remain particularly underrepresented in Western research societies [46].
(c). How much can we do with what we have?
Or, to put the question another way: with such a small fraction of parasite diversity described, how confident can we be in macroecological patterns? A parallel problem was encountered by Quicke [47] as part of a longer-term effort to estimate global parasitoid wasp diversity [36,48]. Only a year after publishing a paper [49] exploring similar macroecological patterns to those we have previously explored [6,50], Quicke concluded ‘we know too little’ to make conclusions about macroecological patterns like latitudinal trends [47]. For parasitoid wasps, the problem is attributable to a similar set of systemic biases, like underdescription of tropical fauna, or a bias in species description rates towards larger species first.
Given that almost 90% of helminth diversity is undescribed (and closer to 100% is undescribed in many places), parasite ecologists need to approach work with ‘big data’ with a similar degree of caution. Working at the level of ecosystems or narrowly defined taxonomic groups may help sidestep some of these issues [33]. But at the global level, patterns like a latitudinal diversity gradient could be the consequence of real underlying trends, or just as easily be the consequence of extreme spatial sampling bias in collections and taxonomic descriptions and revisions.
It will take decades or even centuries before datasets improve substantially enough to change our degree of confidence in existing macroecological hypotheses. Given this problem, Poulin [28] recommended abandoning the task of estimating parasite diversity, and assuming parasite richness is determined ‘simply [by] local host species richness’. However, at global scales, this is not necessarily supported [51]; Dallas et al. [6] showed that the per-host richness of parasite fauna varied over an order of magnitude across different countries in the NHM data, a spatial pattern with little correlation to mammal biodiversity gradients. Even this result is nearly impossible to disentangle from sampling incompleteness and sampling bias. Moreover, even at mesoscales where ‘host diversity begets parasite diversity’ is usually a reliable pattern, anthropogenic impacts are already starting to decouple these patterns [52]. At the present moment, helminth richness patterns could be functionally unknowable at the global scale. The same is likely to be true of many other groups of metazoan parasites that are far more poorly described.
6. The case for a Global Parasite Project
Given the extensive diversity of helminths, some researchers have argued in favour of abandoning the goal of ever fully measuring or cataloging parasite diversity, focusing instead on more ‘practical’ problems [28]. At current rates of description, this is a reasonable outlook; even with several sources of unquantifiable error built into our estimates, it might seem impossible to make a dent within a generation. However, we dispute the idea that nothing can be done to accelerate parasite discovery. Funding and support for most scientific endeavours are at an unprecedented high in the twenty-first century. Other scientific moonshots, from the Human Genome Project to the Event Horizon Telescope image of the M87 black hole, would have seemed impossible within living memory.
For parasitology, the nature of the problem might call for a similarly unprecedented effort. For some purposes, the 5–15% of diversity described may be adequate to form and test ecoevolutionary hypotheses. But the reliability and accuracy of these data will become more uncertain in the face of global change, which will re-assemble host–parasite interactions on a scale that is nearly impossible to predict today. Shifting environmental suitability will drive range shifts in many parasites, or change their transmission intensity; some may go extinct, while others may become epizootics [5,53,54]. Already, some parasites have been observed disappearing in ecosystems undergoing biodiversity loss [55,56]. Others will jump into new host species as hosts undergo range shifts, and encounter new parasites in local fauna, leading to new evolutionary opportunities [57–59]. As climate change progresses, an increasing amount of our time and energy will be spent attempting to differentiate ecological signals from noise and anthropogenic signals. Though some consider the task of cataloging parasite diversity a ‘testimony to human inquisitiveness’ [1], it is also a critical baseline for understanding biological interactions in a world on the brink of ecological collapse.
Along the same lines of the Global Virome Project, we suggest that parasitology could be transformed by a ‘Global Parasite Project’: an internationally coordinated, bottom-up effort to accelerate parasite description, and catalogue half the parasite diversity on Earth (as proposed in the global parasite conservation plan [16]). No such effort currently exists, or has been proposed, and this study is not an announcement. Instead, we consider it as a hypothetical example of how international coordination and targeted investment could change the status quo we identified: parasite taxonomy and collections have grown at a steady but funding-limited pace over the last century; much of the remaining parasite diversity is in undersampled host groups and undersampled biodiversity hotspots, which may pose an increasing challenge; and within the current limits of scientific infrastructure, sifting through this undescribed diversity would take hundreds of years. None of these are likely to change on their own, and—given funding shortages, limited incentives for careers in taxonomy, and the growing challenges of international cooperation—these challenges may only become more entrenched.
In practice, many different strategies could be used to address these challenges. However, our analysis highlights several key points about how a Global Parasite Project could be defined, and what might help it succeed. First, modern methods of estimating parasite diversity make it possible to set realistic and tangible targets for sampling, and budget accordingly. Recently, the global parasite conservation plan [16] proposed an ambitious goal of describing 50% of parasite diversity in the next decade. From the bipartite rarefaction curves, we used above [14,19], we can back-estimate how many hosts we expect to randomly sample before we reach that target. For example, describing 50% of terrestrial nematode parasites would require sampling 3215 new reptile host species, 2560 birds, 2325 amphibians and only 995 mammals. These estimates assume diversity accumulates randomly, and hosts are sampled in an uninformed way. In practice, with knowledge about existing ecological and geographical biases, we can target sampling to accelerate species discovery, just as previous programmes like the Planetary Biodiversity Inventory tapeworm project have, to great success [40].
Second, any moonshot effort to describe parasite diversity would have to start with museums and collections. Systematics is the backbone of biodiversity science [60,61], and especially in parasitology, collections are the backbone of systematics [26,27]. Our analyses show how valuable these collections can be, not just as a hotbed of parasite taxonomic research, but as a source of foundational data to track trends and challenges in parasite discovery. They are also some of the most vulnerable research institutions in modern science: collections are chronically underfunded and understaffed, sometimes to the point of dissolving. Even well-funded collections are still mostly undigitized, ungeoreferenced and unsequenced [21], and massive volumes of ‘grey data’ are unaccounted for in collections that are isolated from the global research community, or fall on opposite sides of deep historical divides (e.g. between Soviet and American science). In all likelihood, hundreds or thousands of parasite species have already been identified and are waiting to be described from museum backlogues, or their descriptions have been recorded in sources inaccessible due to digital access, language barriers and paywalls. Technological advances in the coming decade—like faster bioinformatic pipelines for digitization, easier DNA extraction from formalin-fixed samples, or cryostorage of genomic-grade samples—will expand the possibilities of collections-based work, but are insufficient to fix many of the structural problems in the field.
Whereas viral discovery efforts have mostly focused on capacity building for field sampling and laboratory work, a Global Parasite Project could probably accelerate parasite description the most by focusing on collections science. If the existing research and funding model continues into the next decade, most ‘available’ parasite data will be collected by Western scientists running field trips or long-term ecological monitoring programmes that mostly feed into collections at their home institutions. Building out American and European parasite collections with globally sourced specimens would only perpetuate existing data gaps and research inefficiencies, and the structural inequities and injustices they reflect. Increasingly, biomedical research is under legitimate scrutiny for parachute research—Western-driven research ‘partnerships’ that leverage international project design for exploitative and extractive sampling, with little benefit to partners in the Global South [62–64]. Though our hypothetical Global Parasite Project would be focused primarily on biodiversity and ecology, rather than biomedical or global health priorities, systematics and conservation are no exception to these conversations.
A Global Parasite Project, and its governance principles, would need to focus on supporting collections work and strengthening infrastructure around the world, with explicit priority on equity and local leadership. Recent developments in international law are particularly relevant to this end [65]. The Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity establishes a regime to ensure that access to genetic resources—which some countries may define to include parasites—is coupled with the equitable sharing of benefits from their use. While implementation of the Nagoya Protocol varies between countries, it codifies important norms addressing injustices in obtaining parasites for collections, and inequities in the benefits arising directly or indirectly from their use, which may include capacity building, technology transfer and recognition in scientific publications.
Done right, a Global Parasite Project would build resilient capacities for local priorities, through financial and technical support that empowers local researchers in resource-constrained settings. The support provided could include a combination of training, funding, conferences and meetings, and technology transfer. These can be identified on a case-by-case basis to meet local priorities, which could include formalizing parasite collections, in cases where the component collections are distributed across departments; improving or modernizing specimen preservation methods or physical infrastructure; and digitizing and sequencing collections [40,66]. Following these steps could fill major data gaps, and make collections around the world more resistant to damage, disasters, and gaps in research support. In turn, there is a wealth of local technical knowledge and expertise in countries where parasite collections are underserved. This can be an opportunity for multilateral capacity-building, and, where appropriate, dissemination of local knowledge to the broader scientific community with clear principles for locally led publications and clear attribution. In particular, this work should prioritize expanding avenues for parasitologists in the Global South to be recognized and engaged as active participants in the global research community.
Third, a Global Parasite Project would need to focus not just on completeness in parasite descriptions, but in host–parasite interaction data. Our analysis and several recent others [6,14,50] highlight how many uses these data can have, especially for estimating parasite biodiversity. However, the sparseness of existing network datasets can add an order of magnitude to the uncertainty of these estimates [14], and describing new parasites as fast as possible might make this problem more pronounced if novel parasites are only identified in one host at a time. An active effort needs to be made to fill in the 20–40% of missing links in association matrices, potentially using model-predicted links to optimize sampling [41]. Better characterizing the full host–parasite network would have major benefits for actionable science, ranging from the triage process for parasite conservation assessments [16], to work exploring the apparently emerging sylvatic niche of Guinea worm and its implications for disease eradication [67].
This is where ecologists fit best into a parasite moonshot. Rather than establishing an entirely novel global infrastructure for field research, we can expand parasitology in existing biodiversity inventories. The vast majority of animals already collected by field biologists have easily documented symbionts, which are nevertheless neglected or discarded during sampling. In response, recent work has suggested widespread adoption of integrative protocols for how to collect and document the entire symbiont fauna of animal specimens [68,69]. Building these protocols into more biodiversity inventories will help capture several groups of arthropod, helminth, protozoan and fungal parasites, without unique or redundant sampling programs for each. In cases where destructive sampling is challenging (rare or elusive species) or prohibitive (endangered or protected species), nanopore sequencing and metagenomics may increasingly be used to fill sampling gaps. Collecting data these ways will improve detection of parasites’ full host range, and allow researchers to explore emerging questions about how parasite metacommunities form and interact [70]. As novel biotic interactions form and are detected in real time, this could become a major building block of global change research [16].
Despite decades of work calling out the shortage of parasitologists and the ‘death’ of systematics [27,71], the vast diversity of undescribed parasites has never stopped the thousands of taxonomists and systematists who compiled our datasets over the last century—mostly without access to modern luxuries like digital collections or nanopore sequencing. A testimony to persistence and resourcefulness, these data provide the roadmap for a new transformative effort to describe life on Earth. In an era of massive scientific endeavours, a coordinated effort to describe the world’s parasite diversity seems more possible than ever. There may never be a Global Parasite Project per se, but the current moment may be the closest we’ve ever been to the ‘right time’ to try for one. If biologists want to understand how the entire biosphere is responding to a period of unprecedented change, there is simply no alternative.
7. Material and methods
(a). Data assembly and cleaning
The data we use in this study come from two sources: the USNPC and the NHM Host–Parasite Database. We describe the cleaning process for both of these sources in turn. All data, and all code, are available on Github at github.com/cjcarlson/helminths.
The USNPC has been housed at the Smithsonian National Museum of Natural History since 2013, and is one of the largest parasite collections in the world. The collection is largely digitized and has previously been used for global ecological studies [5]. We downloaded the collections database from EMu in September 2017. The collection includes several major parasitic groups, not just helminths, and so we filtered data down to Acanthocephala, Nematoda, and Platyhelminthes. Metadata associated with the collection has variable quality, and host information is mostly unstandardized, so we minimize its use here.
The NHM Host–Parasite Database is an association list for helminths and their host associations, dating back to the Host–Parasite Catalogue compiled by H. A. Baylis starting in 1922. The database itself is around 250 000 unique, mostly location-specific association records digitized from a reported 28 000 scientific studies. The NHM dataset has been used for ecological analysis in previous publications [6,72,73], but here we used an updated scrape of the online interface to the database. Whereas previous work has scraped association data by locality, we scraped by parasite species list from previous scrapes, allowing records without locality data to be included, and therefore including a more complete sample of hosts. The total raw dataset comprised 100 370 host–parasite associations (no duplication by locality or other metadata), including 17 725 hosts and 21 115 parasites.
We subset the data to the four focal groups, and excluded monogeneans (which are recorded separately from the Trematoda in the NHM database), given our interest in helminth endoparasites. We cleaned the NHM data with a handful of validation steps. First, we removed all host and parasite species with no epithet (recorded as ‘sp.’), and removed all pre-revision name parentheticals. We then ran host taxonomy through ITIS with the help of the taxize package in R, and updated names where possible. This also allowed us to manually re-classify host names by taxonomic grouping. Parasite names were not validated because most parasitic groups are severely under-represented (or outdated) in taxonomic repositories like WORMS and ITIS. At present, no universal, reliable dataset exists for validating parasite taxonomy. After cleaning, there were a total of 13 162 host species and 20 016 parasite species with a total of 73 273 unique interactions; this is compared to, in older scrapes, what would have been a processed total of 61 397 interactions among 18 583 parasites and 11 749 hosts. We finally validated all terrestrial localities by updating to ISO3 standard, including island territories of countries like the UK; many localities stored in the NHM data predate the fall of the USSR or are have similar anachronisms.
(b). Trends over time
(i). Description rates
In the NHM data, we assigned dates of description by extracting year from the full taxonomic record of any given species (e.g. Ascaris lumbricoides Linnaeus, 1758) using regular expressions; in the USNPC data, we extracted year from the accession date recorded for a given specimen. We added together the total number of species described (NHM) and collected (USNPC), and fitted a break-point regression using the segmented package for R [74].
(ii). Body size
We examined trends in body size of hosts and parasites over time using the date of description given in the NHM dataset. For parasite body size, we used a recently published database of trait information for acanthocephalans, cestodes and nematodes [75], and recorded the adult stage body length for all species present in the NHM dataset. For host body size, we subsetted associations to mammals with body mass information in PanTHERIA [76]. We examined trends in worm length and host mass over time using generalized additive models (GAMs) with a smoothed fixed effect for year, using the mgcv package in R [77].
(iii). Host specificity
To test for a description bias in host specificity, we identified the year of description from every species in the NHM data, and coded for each species whether or not they were the first species recorded in the genus. We compared host range for first and non-first taxa and tested for a difference with a Wilcoxon test (chosen given the non-normal distribution of host specificity). To test for temporal trends in host specificity, we fit two GAM models with host specificity regressed against a single smoothed fixed effect for time. In the first, we used the year of species description in the NHM data; in the second, we recorded the year of first accession in the USNPC.
(c). Estimating species richness
Strona & Fattorini [19] discovered that subsampling the host-helminth network produces an approximately power-law scaling pattern, leading to massively reduced richness estimates compared to Dobson et al. [1]. This pattern was recently found by Carlson et al. [14] to be general across large bipartite networks, who developed the R package codependent [39] as a tool for fitting these curves and extrapolating symbiont richness.
We use this approach to re-estimate the total diversity of helminth parasites, repeating the same analysis as Strona & Fattorini [34]. As they did, we mostly ignore questions about species definitions (which are problematic for many parasite clades), and simply use the same definition of ‘species’ operationalized in the available datasets. We used the cleaned host-helminth network and codependent to fit curves for each of 20 groups, and extrapolate to independent richness estimates for all host groups. We sourced the estimate of every terrestrial group’s diversity from the 2014 IUCN Red List estimates. Fish were split into bony and cartilaginous fish in the same style as Dobson et al. [1], but because they have much poorer consolidated species lists, we used estimates of known richness from a fish biology textbook [78].
The software also allows generation of 95% confidence intervals generated procedurally from the fitting of the networks, and while we have used these in previous work [14], here we elected not to. In our assessment, the epistemic uncertainty around cryptic species, the per cent of documented links, and even basic choices like the number of bony fish far outweigh the uncertainty of the model fit for the power-law curves.
One major methodological difference between Carlson et al. [14] and our study is that in their study, they back-corrected estimates by the proportion of viruses described for the hosts in their network (via validation on independent metagenomic datasets). We have no confident way to evaluate how comprehensive the NHM dataset is, as it is certainly the largest dataset available describing host–helminth interactions, and widely believed to be one of the most thorough [6]. Consequently, our estimates account for the proportion of undescribed diversity due only to unsampled hosts, and underestimates by assuming all recorded hosts have no undescribed parasites. This error is likely overcorrected by the back of the envelope correction we perform for cryptic richness.
(d). Estimating total richness across host groups
The overall number of parasites for all orders considered is smaller than the sum of estimates for each order, as some parasites would be expected to infect vertebrates from more than one order. Here we present a new mathematical approach to correcting richness estimates for affiliates across multiple groups, based on the inclusion–exclusion principle.
(i). Inclusion–exclusion principle
The inclusion–exclusion principle from set theory allows us to count the number of elements in the union of two or more sets, ensuring that each element is counted only once. For two sets, it is expressed as follows:
where is the number of elements in the union of the set, |A| and |B| are the number of elements in A and B, respectively, and is number of elements in both A and B. For three sets, it is expressed as follows:
For a greater number of sets, the pattern continues, with elements overlapping an even number of sets subtracted, and elements overlapping an odd number of sets added.
(ii). Inclusion–exclusion and parasite estimates
The overall estimated number of parasites of two groups, , is given as the expected size of . Adapting the inclusion-exclusion principle, we can assume that the overlap between groups N1 and N2 in collections is similar to the overlap of not yet discovered parasites:
We average the estimated number in both groups over and , rather than just scaling by , because we cannot be sure that and scale with N1 and N2 roughly proportionally. (For example, we estimated that the description rate of mammal trematodes is almost an order of magnitude higher than in reptiles.) Instead of estimating the average overlap for a given total number, we estimate the number of multi-order parasites for a given order’s count, and average that across the groups.
For h orders, this can be generalized as follows:
We provide a new implementation of this approach with the multigroup function in an update to the R package codependent.
(e). Mapping potential richness
To map species richness, we used the IUCN range maps for mammals, and counted the number of mammals overlapping each country. Using mammal richness for each country, we predicted the expected number of parasitic associations those species should have globally, running models separately by parasite group (acanthocephalans, cestodes, nematodes and trematodes), and totalled these. We call these ‘possible’ associations and not expected richness, for two reasons: (1) most macroparasites, especially helminths, are not found everywhere their hosts are found. (2) Host specificity may vary globally [79], but as we stress in the main text, it is difficult to disentangle our knowledge of macroecological patterns from the massive undersampling of parasites in most countries. We compared patterns of possible richness against known helminth associations recorded in a given country, the grounds on which parasite richness has previously been mapped [6]. Finally, we mapped the percentage of total possible unrecorded interactions (an upper bound for high values, except when 100% is reported, indicating that no parasites have been recorded in the NHM data from a country). All maps were generated in R.
Supplementary Material
Acknowledgements
Thanks to Shweta Bansal, Phil Staniczenko and Joy Vaz for formative conversations, and to the Georgetown Environment Initiative for funding.
Data accessibility
All data are available at https://github.com/cjcarlson/helminths.
Authors' contributions
C.J.C., T.A.D. and L.W.A. performed analyses. All authors contributed to the conception of the paper, the design of analyses and the writing of the paper.
Competing interests
We declare we have no competing interests.
Funding
No funding has been received for this article.
References
- 1.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. 2008. Homage to Linnaeus: how many parasites? How many hosts? Proc. Natl Acad. Sci. USA 105, 11482–11489. ( 10.1073/pnas.0803232105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen BB, Miller EC, Rhodes MK, Wiens JJ. 2017. Inordinate fondness multiplied and redistributed: the number of species on earth and the new pie of life. Q Rev. Biol. 92, 229–265. ( 10.1086/693564) [DOI] [Google Scholar]
- 3.Rohde K. 1982. Ecology of marine parasites. St Lucia, Australia: University of Queensland Press. [Google Scholar]
- 4.Okamura B, Hartigan A, Naldoni J. 2018. Extensive uncharted biodiversity: the parasite dimension. Integr. Comp. Biol. 58, 1132–1145. ( 10.1093/icb/icy039) [DOI] [PubMed] [Google Scholar]
- 5.Carlson CJ. et al. 2017. Parasite biodiversity faces extinction and redistribution in a changing climate. Sci. Adv. 3, e1602422 ( 10.1126/sciadv.1602422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallas TA, Aguirre AA, Budischak S, Carlson C, Ezenwa V, Han B, Huang S, Stephens PR. 2018. Gauging support for macroecological patterns in helminth parasites. Global Ecol. Biogeogr. 27, 1437–1447. ( 10.1111/geb.12819) [DOI] [Google Scholar]
- 7.Han BA, Kramer AM, Drake JM. 2016. Global patterns of zoonotic disease in mammals. Trends Parasitol. 32, 565–577. ( 10.1016/j.pt.2016.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty ER, Carlson CJ, Bueno VM, Burgio KR, Cizauskas CA, Clements CF, Seidel DP, Harris NC. 2016. Paradigms for parasite conservation. Conserv. Biol. 30, 724–733. ( 10.1111/cobi.12634) [DOI] [PubMed] [Google Scholar]
- 9.Cizauskas CA, Carlson CJ, Burgio KR, Clements CF, Dougherty ER, Harris NC, Phillips AJ. 2017. Parasite vulnerability to climate change: an evidence-based functional trait approach. R. Soc. Open Sci. 4, 160535 ( 10.1098/rsos.160535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han BA, Schmidt JP, Bowden SE, Drake JM. 2015. Rodent reservoirs of future zoonotic diseases. Proc. Natl Acad. Sci. USA 112, 7039–7044. ( 10.1073/pnas.1501598112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han BA, Schmidt JP, Alexander LW, Bowden SE, Hayman DT, Drake JM. 2016. Undiscovered bat hosts of filoviruses. PLoS Negl. Trop. Dis. 10, e0004815 ( 10.1371/journal.pntd.0004815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650. ( 10.1038/nature22975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll D, Daszak P, Wolfe ND, Gao GF, Morel CM, Morzaria S, Pablos-Méndez A, Tomori O, Mazet JA. 2018. The global virome project. Science 359, 872–874. ( 10.1126/science.aap7463) [DOI] [PubMed] [Google Scholar]
- 14.Carlson CJ, Zipfel CM, Garnier R, Bansal S. 2019. Global estimates of mammalian viral biodiversity accounting for host sharing. Nat. Ecol. Evol. 3, 1070–1075. ( 10.1038/s41559-019-0910-6) [DOI] [PubMed] [Google Scholar]
- 15.Brooker S, Clements AC, Bundy DA. 2006. Global epidemiology, ecology and control of soil-transmitted helminth infections. Adv. Parasitol. 62, 221–261. ( 10.1016/S0065-308X(05)62007-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson CJ. et al. 2020. A global plan for parasite conservation. Biol. Conserv. 250, 108596 ( 10.1016/j.biocon.2020.108596) [DOI] [Google Scholar]
- 17.Hugot J-P, Baujard P, Morand S. 2001. Biodiversity in helminths and nematodes as a field of study: an overview. Nematology 3, 199–208. ( 10.1163/156854101750413270) [DOI] [Google Scholar]
- 18.Holterman M. et al. 2017. Disparate gain and loss of parasitic abilities among nematode lineages. PLoS ONE 12, e0185445 ( 10.1371/journal.pone.0185445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strona G, Fattorini S. 2014. Parasitic worms: how many really? Int. J. Parasitol. 44, 269–272. ( 10.1016/j.ijpara.2014.01.002) [DOI] [PubMed] [Google Scholar]
- 20.Schilthuizen M, Vairappan CS, Slade EM, Mann DJ, Miller JA. 2015. Specimens as primary data: museums and ‘open science’. Trends Ecol. Evol. 30, 237–238. ( 10.1016/j.tree.2015.03.002) [DOI] [PubMed] [Google Scholar]
- 21.Bell KC, Carlson CJ, Phillips AJ. 2018. Parasite collections: overlooked resources for integrative research and conservation. Trends Parasitol. 34, 637–639. ( 10.1016/j.pt.2018.04.004) [DOI] [PubMed] [Google Scholar]
- 22.DiEuliis D, Johnson KR, Morse SS, Schindel DE. 2016. Opinion: specimen collections should have a much bigger role in infectious disease research and response. Proc. Natl Acad. Sci. USA 113, 4–7. ( 10.1073/pnas.1522680112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson D, Bray R, Harris E. 2005. Host–parasite database of the Natural History Museum, London See www.nhm.ac.uk/research-curation/scientific-resources/taxonomy-systematics/host-parasites/database/index.jsp.
- 24.Dallas T. 2016. helminthR: an R interface to the London Natural History Museum’s host–parasite database. Ecography 39, 391–393. ( 10.1111/ecog.02131) [DOI] [Google Scholar]
- 25.Lichtenfels J. 1984. Methods for conserving, storing, and studying helminths in the US National Parasite Collection. Syst. Parasitol. 6, 250–251. ( 10.1007/BF00012199) [DOI] [Google Scholar]
- 26.Hoberg EP. 2002. Foundations for an integrative parasitology: collections, archives, and biodiversity informatics. Comp. Parasitol. 69, 124–132. ( 10.1654/1525-2647(2002)069[0124:FFAIPC]2.0.CO;2) [DOI] [Google Scholar]
- 27.Brooks DR, Hoberg EP. 2001. Parasite systematics in the 21st century: opportunities and obstacles. Trends Parasitol. 17, 273–275. ( 10.1016/S1471-4922(01)01894-3) [DOI] [PubMed] [Google Scholar]
- 28.Poulin R. 2014. Parasite biodiversity revisited: frontiers and constraints. Int. J. Parasitol. 44, 581–589. ( 10.1016/j.ijpara.2014.02.003) [DOI] [PubMed] [Google Scholar]
- 29.Costello MJ. 2016. Parasite rates of discovery, global species richness and host specificity. Integr. Comp. Biol. 56, 588–599. ( 10.1093/icb/icw084) [DOI] [PubMed] [Google Scholar]
- 30.Jorge F, Poulin R. 2018. Poor geographical match between the distributions of host diversity and parasite discovery effort. Proc. R. Soc. B 285, 20180072 ( 10.1098/rspb.2018.0072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulin R, Jorge F. 2019. The geography of parasite discovery across taxa and over time. Parasitology 146, 168–175. ( 10.1017/S003118201800118X) [DOI] [PubMed] [Google Scholar]
- 32.Clark NJ. 2018. Phylogenetic uniqueness, not latitude, explains the diversity of avian blood parasite communities worldwide. Global Ecol. Biogeogr. 27, 744–755. ( 10.1111/geb.12741) [DOI] [Google Scholar]
- 33.Preisser W. 2019. Latitudinal gradients of parasite richness: a review and new insights from helminths of cricetid rodents. Ecography 42, 1315–1330. ( 10.1111/ecog.04254) [DOI] [Google Scholar]
- 34.Strona G, Fattorini S. 2014. A few good reasons why species-area relationships do not work for parasites. BioMed Res. Int. 2014, 271680 ( 10.1155/2014/271680) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulin R, Morand S. 2004. Parasite biodiversity. Washington, DC: Smithsonian Institution. [Google Scholar]
- 36.Jones OR, Purvis A, Baumgart E, Quicke DL. 2009. Using taxonomic revision data to estimate the geographic and taxonomic distribution of undescribed species richness in the braconidae (Hymenoptera: Ichneumonoidea). Insect Conserv. Divers. 2, 204–212. ( 10.1111/j.1752-4598.2009.00057.x) [DOI] [Google Scholar]
- 37.Kamiya T, O’wyer K, Nakagawa S, Poulin R. 2014. Host diversity drives parasite diversity: meta-analytical insights into patterns and causal mechanisms. Ecography 37, 689–697. ( 10.1111/j.1600-0587.2013.00571.x) [DOI] [Google Scholar]
- 38.Kamiya T, O’dwyer K, Nakagawa S, Poulin R. 2014. What determines species richness of parasitic organisms? a meta-analysis across animal, plant and fungal hosts. Biol. Rev. 89, 123–134. ( 10.1111/brv.12046) [DOI] [PubMed] [Google Scholar]
- 39.Carlson CJ. 2019. codependent R package. version 1.1. See https://github.com/cjcarlson/codependent.
- 40.Caira JN, Jensen K. 2017. Planetary biodiversity inventory (2008–2017): Tapeworms from vertebrate bowels of the earth. Natural History Museum, University of Kansas.
- 41.Dallas T, Huang S, Nunn C, Park AW, Drake JM. 2017. Estimating parasite host range. Proc. R. Soc. B 284, 20171250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulin R. 2011. Uneven distribution of cryptic diversity among higher taxa of parasitic worms. Biol. Lett. 7, 241–244. ( 10.1098/rsbl.2010.0640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de León GP-P, Poulin R. 2018. An updated look at the uneven distribution of cryptic diversity among parasitic helminths. J. Helminthol. 92, 197–202. ( 10.1017/S0022149X17000189) [DOI] [PubMed] [Google Scholar]
- 44.Martin LJ, Blossey B, Ellis E. 2012. Mapping where ecologists work: biases in the global distribution of terrestrial ecological observations. Front. Ecol. Environ. 10, 195–201. ( 10.1890/110154) [DOI] [Google Scholar]
- 45.Smit N, Basson L, Vanhove MP, Scholz T. 2018. History of fish parasitology in Africa. In A guide to the parasites of African freshwater fishes (eds T Scholz, MPM Vanhove, N Smit, Z Jayasundera, M Gelnareds), pp. 15–30. Brussels, Belgium: RBINS.
- 46.Caira J. 2011. The American Society of Parasitologists: who are we now?. J. Parasitol. 97, 967–973. ( 10.1645/GE-2980.1) [DOI] [PubMed] [Google Scholar]
- 47.Quicke DL. 2012. We know too little about parasitoid wasp distributions to draw any conclusions about latitudinal trends in species richness, body size and biology. PLoS ONE 7, e32101 ( 10.1371/journal.pone.0032101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolphin K, Quicke DL. 2001. Estimating the global species richness of an incompletely described taxon: an example using parasitoid wasps (Hymenoptera: Braconidae). Biol. J. Linnean Soc. 73, 279–286. ( 10.1111/j.1095-8312.2001.tb01363.x) [DOI] [Google Scholar]
- 49.Santos AM, Quicke DL. 2011. Large-scale diversity patterns of parasitoid insects. Entomol. Sci. 14, 371–382. ( 10.1111/j.1479-8298.2011.00481.x) [DOI] [Google Scholar]
- 50.Stephens PR. et al. 2016. The macroecology of infectious diseases: a new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 19, 1159–1171. ( 10.1111/ele.12644) [DOI] [PubMed] [Google Scholar]
- 51.Nunn CL, Altizer SM, Sechrest W, Cunningham AA. 2005. Latitudinal gradients of parasite species richness in primates. Divers. Distributions 11, 249–256. ( 10.1111/j.1366-9516.2005.00160.x) [DOI] [Google Scholar]
- 52.Wood CL, Zgliczynski BJ, Haupt AJ, Guerra AS, Micheli F, Sandin SA. 2018. Human impacts decouple a fundamental ecological relationship—the positive association between host diversity and parasite diversity. Glob. Change Biol. 24, 3666–3679. ( 10.1111/gcb.14159) [DOI] [PubMed] [Google Scholar]
- 53.Hoberg EP, Brooks DR. 2015. Evolution in action: climate change, biodiversity dynamics and emerging infectious disease. Phil. Trans. R. Soc. B 370, 20130553 ( 10.1098/rstb.2013.0553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ostfeld RS. 2009. Climate change and the distribution and intensity of infectious diseases. Ecology 90, 903–905. ( 10.1890/08-0659.1) [DOI] [PubMed] [Google Scholar]
- 55.Esser HJ, Herre EA, Kays R, Liefting Y, Jansen PA. 2019. Local host-tick coextinction in neotropical forest fragments. Int. J. Parasitol. 49, 225–233. ( 10.1016/j.ijpara.2018.08.008) [DOI] [PubMed] [Google Scholar]
- 56.Sitko J, Heneberg P. 2020. Systemic collapse of a host-parasite trematode network associated with wetland birds in europe. Parasitol. Res. 119, 935–945. ( 10.1007/s00436-020-06624-4) [DOI] [PubMed] [Google Scholar]
- 57.Brooks DR, Hoberg EP. 2007. How will global climate change affect parasite–host assemblages?. Trends Parasitol. 23, 571–574. ( 10.1016/j.pt.2007.08.016) [DOI] [PubMed] [Google Scholar]
- 58.Carlson CJ, Albery GF, Merow C, Trisos CH, Zipfel CM, Eskew EA, Olival KJ, Ross N, Bansal S. 2020. Climate change will drive novel cross-species viral transmission. bioRxiv. ( 10.1101/2020.01.24.918755) [DOI] [PubMed] [Google Scholar]
- 59.Hoberg EP, Brooks DR. 2008. A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host–parasite systems. J. Biogeogr. 35, 1533–1550. ( 10.1111/j.1365-2699.2008.01951.x) [DOI] [Google Scholar]
- 60.Littlewood D. 2011. Systematics as a cornerstone of parasitology: overview and preface. Parasitology 138, 1633–1637. ( 10.1017/S0031182011001533) [DOI] [PubMed] [Google Scholar]
- 61.Monis P. 1999. Invited review the importance of systematics in parasitological research. Int. J. Parasitol. 29, 381–388. ( 10.1016/S0020-7519(98)00216-1) [DOI] [PubMed] [Google Scholar]
- 62.Yozwiak NL, Happi CT, Grant DS, Schieffelin JS, Garry RF, Sabeti PC, Andersen KG. 2016. Roots, not parachutes: research collaborations combat outbreaks. Cell 166, 5–8. ( 10.1016/j.cell.2016.06.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Health TLG. 2018. Closing the door on parachutes and parasites. The Lancet Global health 6, e593 ( 10.1016/S2214-109X(18)30239-0) [DOI] [PubMed] [Google Scholar]
- 64.Serwadda D, Ndebele P, Grabowski MK, Bajunirwe F, Wanyenze RK. 2018. Open data sharing and the global south–who benefits? Science 359, 642–643. ( 10.1126/science.aap8395) [DOI] [PubMed] [Google Scholar]
- 65.Prathapan K, Rajan PD. 2020. Advancing taxonomy in the global south and completing the grand linnaean enterprise. Megataxa 1, 73–77. ( 10.11646/megataxa.1.1.15) [DOI] [Google Scholar]
- 66.Janzen DH. 2004. Now is the time. Phil. Trans. R. Soc. Lond. B 359, 731–732. ( 10.1098/rstb.2003.1444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiele EA, Eberhard ML, Cotton JA, Durrant C, Berg J, Hamm K, Ruiz-Tiben E. 2018. Population genetic analysis of chadian guinea worms reveals that human and non-human hosts share common parasite populations. PLoS Negl. Trop. Dis. 12, e0006747 ( 10.1371/journal.pntd.0006747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cook JA. et al. 2016. Transformational principles for neon sampling of mammalian parasites and pathogens: a response to springer and colleagues. BioScience 66, 917–919. ( 10.1093/biosci/biw123) [DOI] [Google Scholar]
- 69.Galbreath KE. et al. 2019. Building an integrated infrastructure for exploring biodiversity: field collections and archives of mammals and parasites. J. Mammal. 100, 382–393. ( 10.1093/jmammal/gyz048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dallas TA, Laine A-L, Ovaskainen O. 2019. Detecting parasite associations within multi-species host and parasite communities. Proc. R. Soc. B 286, 20191109 ( 10.1098/rspb.2019.1109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mariaux J. 1996. Cestode systematics: any progress? Int. J. Parasitol. 26, 231–243. ( 10.1016/0020-7519(95)00129-8) [DOI] [PubMed] [Google Scholar]
- 72.Dallas T, Park AW, Drake JM. 2017. Predictability of helminth parasite host range using information on geography, host traits and parasite community structure. Parasitology 144, 200–205. ( 10.1017/S0031182016001608) [DOI] [PubMed] [Google Scholar]
- 73.Dallas T, Gehman ALM, Aguirre AA, Budischak SA, Drake JM, Farrell MJ, Ghai R, Huang S, Morales-Castilla I. 2019. Contrasting latitudinal gradients of body size in helminth parasites and their hosts. Global Ecol. Biogeogr. 28, 804–813. ( 10.1111/geb.12894) [DOI] [Google Scholar]
- 74.Muggeo VM, Muggeo MVM. 2017. Package ‘segmented’. Biometrika 58, 516. [Google Scholar]
- 75.Benesh DP, Lafferty KD, Kuris A. 2017. A life cycle database for parasitic acanthocephalans, cestodes, and nematodes. Ecology 98, 882–882 ( 10.1002/ecy.1680) [DOI] [PubMed] [Google Scholar]
- 76.Jones KE. et al. 2009. Pantheria: a species-level database of life history, ecology, and geography of extant and recently extinct mammals: Ecological archives e090-184. Ecology 90, 2648–2648 ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 77.Wood SN. 2001. mgcv: GAMs and generalized ridge regression for R. R news 1, 20–25. [Google Scholar]
- 78.Nelson JS. 2006. Fishes of the world, 4th edn New York, NY: John Wiley & Sons. [Google Scholar]
- 79.Wells K, Gibson DI, Clark NJ. 2019. Global patterns in helminth host specificity: phylogenetic and functional diversity of regional host species pools matter. Ecography 42, 416–427. ( 10.1111/ecog.03886) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available at https://github.com/cjcarlson/helminths.