Abstract
Positive-sense single-stranded RNA viruses, such as coronaviruses, flaviviruses and alphaviruses, carry out transcription and replication inside virus-induced membranous organelles within host cells1–7. The remodelling of the host-cell membranes for the formation of these organelles is coupled to the membrane association of viral replication complexes and to RNA synthesis. These viral niches allow for the concentration of metabolites and proteins for the synthesis of viral RNA, and prevent the detection of this RNA by the cellular innate immune system8. Here we present the cryo-electron microscopy structure of non-structural protein 1 (nsP1) of the alphavirus chikungunya virus, which is responsible for RNA capping and membrane binding of the viral replication machinery. The structure shows the enzyme in its active form, assembled in a monotopic membrane-associated dodecameric ring. The structure reveals the structural basis of the coupling between membrane binding, oligomerization and allosteric activation of the capping enzyme. The stoichiometry—with 12 active sites in a single complex—redefines viral replication complexes as RNA synthesis reactors. The ring shape of the complex implies it has a role in controlling access to the viral organelle and ensuring the exit of properly capped viral RNA. Our results provide high-resolution information about the membrane association of the replication machinery of positive-sense single-stranded RNA viruses, and open up avenues for the further characterization of viral replication on cell membranes and the generation of antiviral agents.
Subject terms: Membrane proteins, Viral proteins, Alphaviruses, Cryoelectron microscopy
Cryo-electron microscopy structures of non-structural protein 1 (nsP1) of chikungunya virus reveal the mechanisms that underpin the association of viral replication machinery with virus-induced membranous organelles within host cells.
Main
Alphaviruses generate invaginations in the plasma membrane or cytoplasmic vacuoles of host cells, creating small (around 50 nm in diameter) lipid baskets known as spherules2. Spherules have a small aperture that connects with the external cytosol, which is the only way out for the newly synthetized viral RNA and the only way in for the metabolites and proteins that are required for RNA synthesis. The extruded viral RNAs are capped and polyadenylated, ready for translation of the viral proteins by the host. This is achieved by viral replication complexes that assemble onto cellular membranes, promoting spherule formation and synthesizing viral RNAs in a coordinated fashion.
Alphaviruses have a replication complex that contains four non-structural proteins (nsP1, nsP2, nsP3 and nsP4), which are produced following cleavage of the viral polyprotein precursor by a viral protease in nsP2. nsP1, nsP2, nsP3 and nsP4 are essential for replication9,10. nsP1 is the only one of these proteins that is known to interact directly with the membrane, and it displays two activities that are necessary for the capping of viral RNA: S-adenosyl-l-methionine (SAM)-dependent methyltransferase (MTase) and m7GTP transferase (GTase) activities11–14, both of which are dependent on membrane binding15. Here we show the structure of the nsP1 complex, which provides a structural basis for these phenomena and challenges biochemical data that have accumulated over decades on positive-sense single-stranded RNA viruses replicating in membrane-associated viral factories.
Assembly of nsP1 into active dodecameric pores
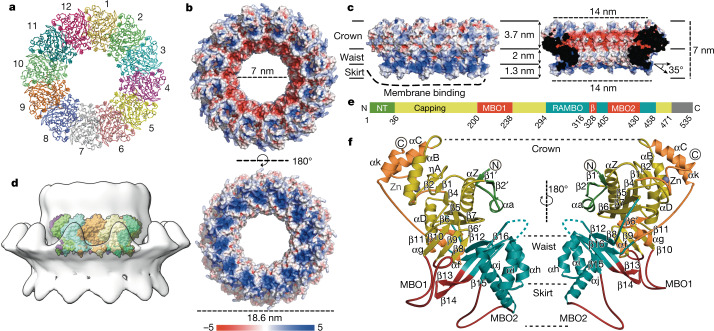
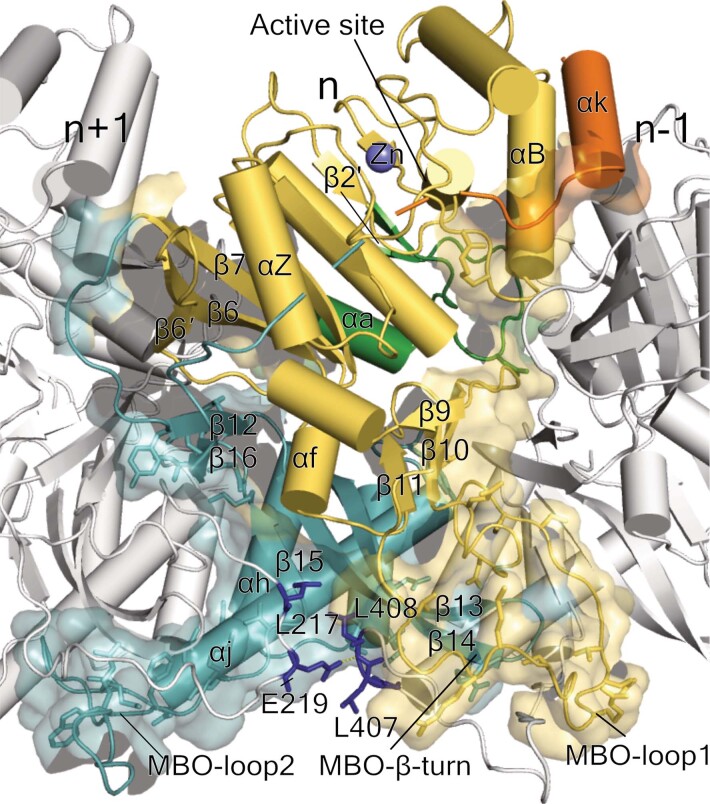
We expressed nsP1 recombinantly in Escherichia coli and in insect cells16, and recovered inactive monomeric and active membrane-associated nsP1, respectively. Detergent screening enabled us to solubilize and purify active ring-shaped complexes of nsP1 from insect cells and inactive monomeric protein, which indicates that nsP1 of chikungunya virus (CHIKV) needs to be assembled as oligomers for activity (Extended Data Fig. 1, Methods). We determined the structure of the complex using single-particle cryo-electron microscopy (cryo-EM). The sorting of particles into two-dimensional (2D) and three-dimensional (3D) class averages was consistent with two volumes, which correspond to dodecameric single and double rings of nsP1: the structures of these rings were determined at a resolution of 2.6 Å and 2.9 Å, respectively (Extended Data Figs. 2, 3, Extended Data Table 1). The absence of contacts between the interface of the double rings confirms that the single ring is the biological assembly (Extended Data Fig. 4). Twelve molecules of nsP1 assemble in the ring with C12 symmetry (Fig. 1a). The rings are 18.6 nm in diameter and 7 nm in height, and have a central aperture that is 7 nm wide (Fig. 1b, c). Three regions are well-defined in the complex: the crown, the waist and the membrane-binding skirt, which together confer a bodice-like shape to the complex (Fig. 1c). The crown is formed from the capping domains of the 12 subunits (as discussed in ‘Atomic structure of CHIKV nsP1 protein’), generating a cone-shaped inner chamber that is negatively charged, about 3.7 nm in depth and has a diameter of 14 nm at the top that narrows to 7 nm at the bottom. The top of the crown is lined with positively charged pockets that correspond to the active sites of the capping domain. Under the crown, the 7-nm-wide waist region defines a pore that is 2 nm in depth as well as a positively charged region outside the complex. The inner walls of the pore have a neutral charge and would allow for the passage of globular proteins smaller than about 70–90 kDa in size. Under the waist, spikes from the membrane-binding skirt project into the detergent micelles at an angle of around 35° from the equatorial plane of the pore, generating a concave and positively charged surface that extends from the bottom of the complex to the lower external waist (with the exception of the hydrophobic tips of the spikes). Tomographic reconstructions of spherule necks, performed in a nodavirus (Flock House virus (FHV)) at low resolution, maintain dimensions and architecture similar to those shown by the nsP1 complex9 (Fig. 1d, Extended Data Fig. 5). In conclusion, the nsP1 complex forms enzymatically active rings that are monotopically associated with the membrane within the spherule necks, and which gate access to the organelle. This is the foundation of the generation of spherule-like viral replication factories.
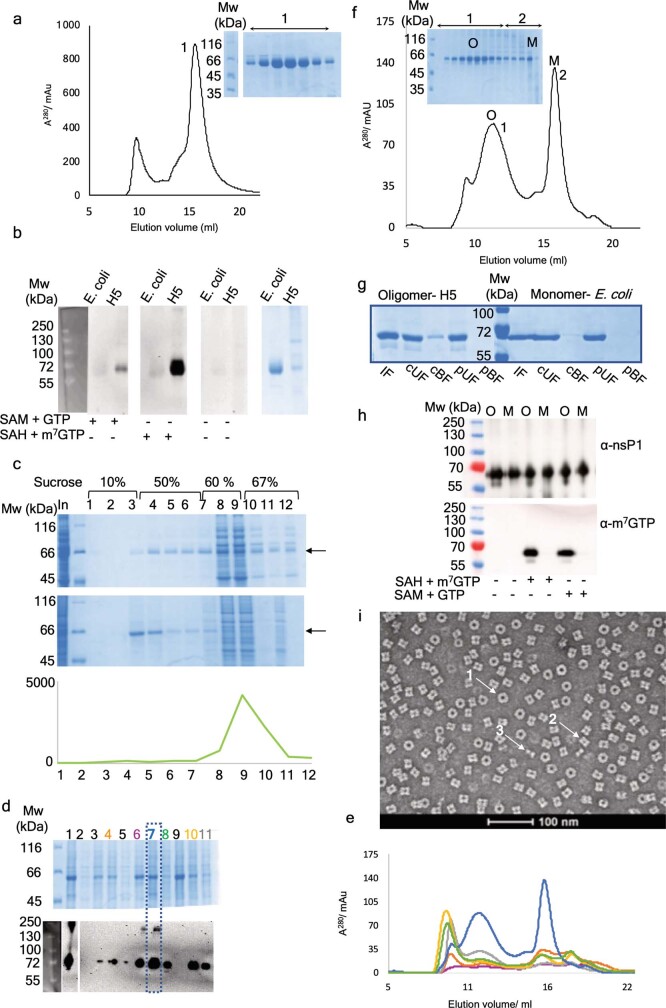
Extended Data Fig. 1. Isolation and biochemical characterization of nsP1 ring complexes.
a, Size-exclusion profile of monomeric CHIKV nsP1 expressed in E. coli cells after purification by affinity chromatography. The elution volume corresponds to a molecular weight of approximately 61 kDa, corresponding to a nsP1 monomer. Fractions corresponding to the peak of the monomeric nsP1are shown in the offset SDS–PAGE with the molecular weight markers on the left. b, Western-blot-based activity test with CHIKV nsP1 purified from insect cells (H5) and E. coli with an antibody specific for m3G and m7G. The capping intermediate of the methylated GMP covalently bound to nsP1 is detected at 61 kDa for samples purified from insect cells when SAM and GTP are added in the reaction, and substantially more when m7GTP is added as substrate instead of GTP. Conversely, the monomeric nsP1 produced in E. coli shows no activity despite a substantially higher amount of protein used in the experiments. This is shown in the SDS–PAGE gel on the right. c, SDS–PAGE gels of fractions derived from a sucrose gradient flotation assay (Methods), in which extracts from E. coli cells (top gel) and Hi5 insect cells (bottom gel) expressing CHIKV nsP1 were subjected to flotation analysis. Total samples (In) were loaded in the 60% sucrose layer of the gradient (fractions 8 and 9), and during centrifugation membrane-associated proteins float to the 10–50% sucrose interface (fractions 3 and 4) and soluble proteins remain in the 60% layer. nsP1 expressed in Hi5 cells floats and that expressed in E. coli is mainly soluble (arrows). The sucrose concentration along the gradient is indicated, and molecular weight markers are in the second lane from the left. The fluorescence emission of soluble YFP, used for following insect cell expression with the MultiBac system16, was monitored as an internal control (graph below). As expected, the YFP migrates to the soluble fraction. d, Screening to identify a detergent suitable for solubilization of CHIKV nsP1 from Hi5 membranes. Isolated membranes were incubated with a range of detergents at 1% concentration and subjected to ultracentrifugation. The resulting soluble fractions were analysed by SDS–PAGE (top-right panel) and for nsP1 guanyltransferase activity in the presence of SAH and m7GMP by western blotting with the m3G and m7G antibody (bottom-right panel). Lanes are as follows: 1, crude membrane fraction; 2–11, soluble fractions with no detergent (2), anzergent 3-10 (3), cymal 5 (orange) (4), DG (5), β-OG (purple) (6), fos-choline 12 (blue) (7), DM (green) (8), SDS (9), DMNG (yellow) (10), DDM (grey) (11). e, Size-exclusion chromatograms of a subset of promising detergents that were used in scaled-up purifications (same colour scheme used as in d). f, Size-exclusion chromatogram of CHIKV nsP1 purified from insect cells with SDS–PAGE of elution fractions with molecular weight markers shown on the left. Fractions corresponding to the oligomeric peak (1) and monomeric peak (2) are indicated, as well as the fractions used for subsequent biochemical experiments with the oligomeric (O) and the monomeric (M) nsP1. g, Acyl capture experiment to assess palmitoylation of oligomeric and monomeric nsP1. After blocking of free thiols and cleavage of acyl thioester bonds, palmitoylated samples are retained by a resin that binds free thiols (cBF, cleaved bound fraction) and non-palmitoylated samples are washed away (cUF, cleaved unbound fraction). Paired negative controls in which thiols were not cleaved are also shown (pBF, protected bound fraction; pUF, protected unbound fraction). The input for the resin is labelled IF (input fraction). Only a very small proportion of oligomeric nsP1 is palmitoylated. h, Guanyltransferase activity tests of the oligomeric (O) and monomeric (M) species that were pooled after size exclusion. The top panels show a western blot with a nsP1 antibody. The lower panel shows the protein activity as monitored in b. Only the oligomers produce the intermediate of the guanyltransferase reaction with both GTP and m7GTP as substrate. Molecular weight markers are shown on the left. i, Negative-stain electron micrograph of the central fraction from the oligomeric peak shown in f. The micrograph shows top views of the ring (1), side views of single rings (2) and side views of double rings (3), all indicated by arrows. The magnification is indicated by the 100-nm-long bar below. All original gels can be found in Supplementary Fig. 1. All results from activity assays (b, d, h), acyl-capture experiments (g) and negative-stain electron-microscopy experiments (i) were successfully reproduced at least three times. Gels for sucrose flotation assays (c) are representative of two repeats.
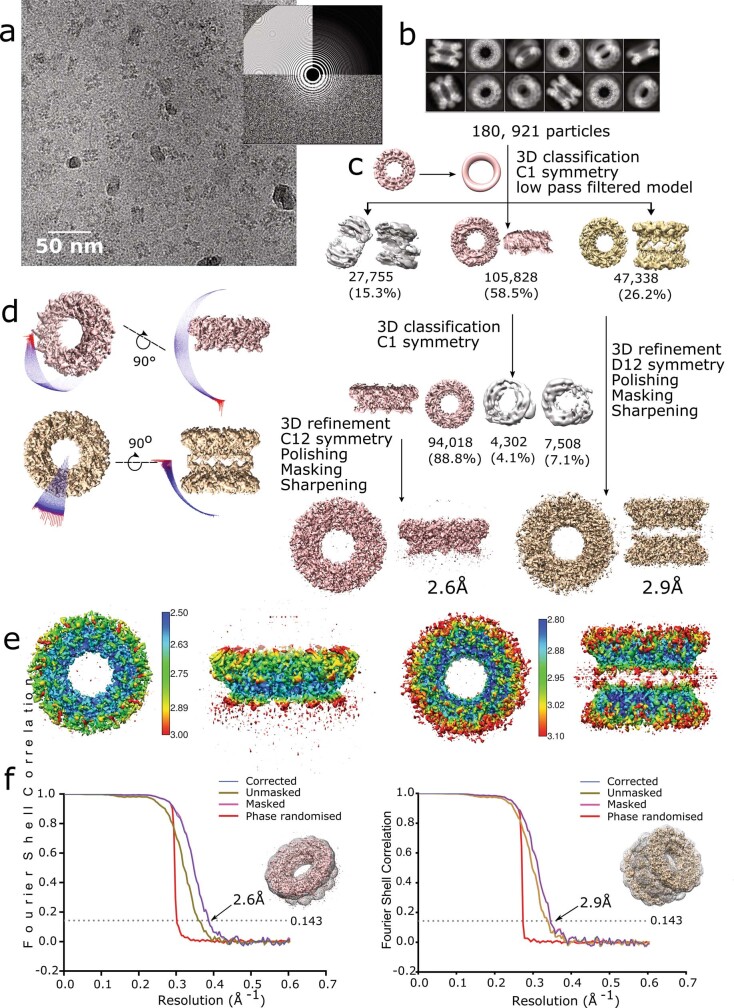
Extended Data Fig. 2. Cryo-EM reconstruction of nsP1 single and double dodecameric rings.
a, Cryo-EM micrograph of nsP1 oligomers acquired with a Titan Krios microscope (ESRF-CM01), representative of 4,948 micrographs collected. Top views and side views of the single and double rings are visible. The power spectrum of the micrograph is shown in the offset. b, Two-dimensional class averages of nsP1. Classes correspond to single or double rings in a range of orientations. c, The cryo-EM image processing workflow. The 180,981 particles selected following 2D classification and image pruning were subjected to 3D classification using an ab initio model low-pass-filtered to 60 Å. Classification with different numbers of classes consistently yielded two main classes, corresponding to single rings (pink) and double rings (gold). For the single rings, a second round of 3D classification improved homogeneity and resolution of the final reconstruction. Final 3D reconstructions were performed with masking and imposing C12 and D12 symmetry for single and double rings, respectively, following a single round of particle polishing and per-particle CTF refinement. d, Angular distribution plot of all particles included in the final C12 symmetry reconstruction of the single rings (pink) and D12 symmetry reconstruction of the double rings (gold). The length and colour of the cylinders correspond to the number of particles at a given Euler angle. The plots demonstrate a higher distribution of particles in side orientations in the case of the double rings, and top orientations for single rings, but sufficient coverage of the symmetry space to avoid this being detrimental to the reconstruction. e, Final 3D reconstructions coloured according to local resolution for single rings (left) and double (right) rings. The resolution range is relatively narrow for both reconstructions (scale indicated), where resolution is highest in the core of the complexes around the pore aperture. Resolution is lower in the top and bottom of the rings, corresponding to the region interacting with the FC12 micelles, and the C-terminal helix (αk) respectively. f, Fourier shell correlation curves for the single ring (left) and double ring (right) reconstructions. Curves for the unmasked (gold), masked (pink), phase-randomized (red) and cryo-EM density maps corrected for mask convolution effects (blue) are indicated, and the inset shows the final atomic model and mask used for the reconstructions. The resolutions estimated according to the 0.143 threshold are indicated, corresponding to 2.6 Å for single rings and 2.9 Å for double rings.
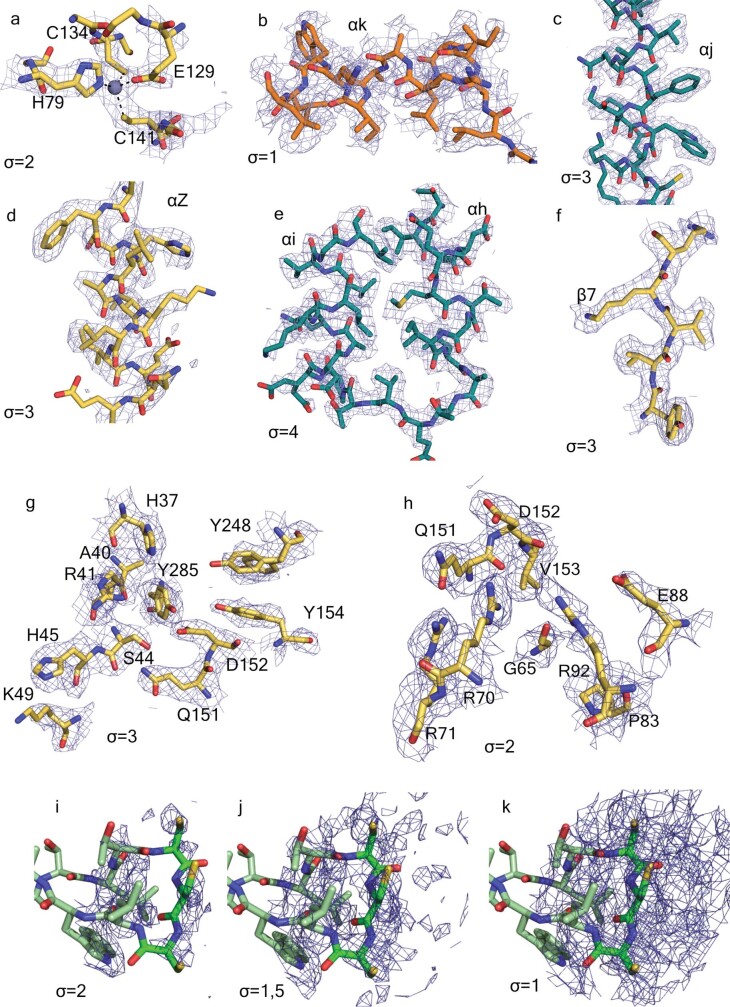
Extended Data Fig. 3. Details of the density maps of the C12 nsP1 volume reconstruction.
The panels were created in PyMOL with residues represented as sticks and density as blue mesh at the indicated σ. The carbon backbone is coloured according to domains, following Figs. 1, 2. a–h, Electron densities. a, The Zn-binding site. Dashed black lines indicate the tetragonal coordination of the Zn atom. The amino acid side chains coordinating the Zn atom are shown as sticks. b, The weak electron density for the C-terminal α-helix αk. c, The long helix αj. d, Helix αZ, e, The α-turn of the pore-wall helices αh–αi. f, The β-sheet β7. g,The GTP-binding site. h, The SAM-binding site. i–k, Three decreasing contour levels of the electron density observed around the cysteine triad at the edge of the membrane-binding spikes.
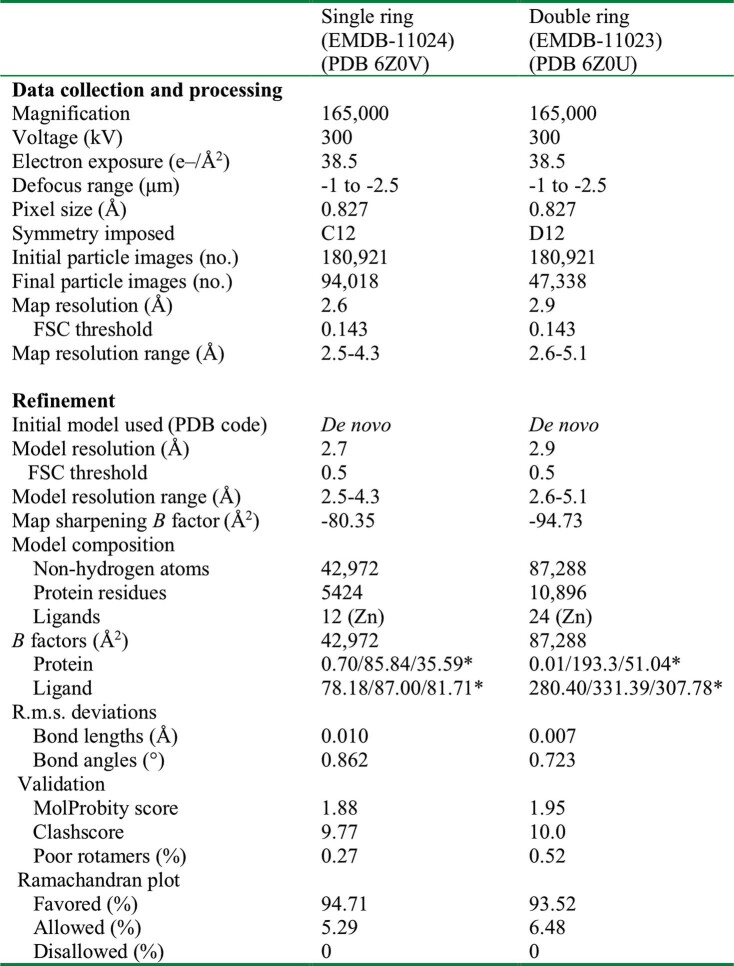
Extended Data Table 1.
Cryo-EM data collection, refinement and validation statistics
*Minimum/maximum/mean.
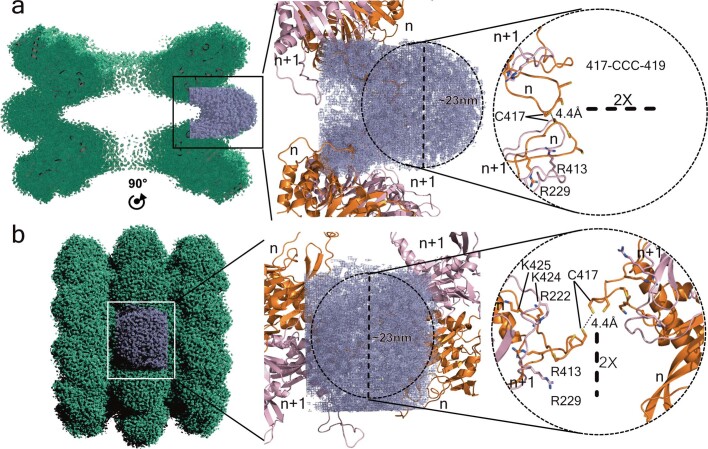
Extended Data Fig. 4. nsP1–detergent interactions.
Micelle arrangement and membrane-binding regions of the double-ring structure. a, Lateral section of the double-ring map represented by a green mesh. A section of the map including one detergent micelle is highlighted in blue (left). The central panel shows two symmetry-related nsP1 dimers (n and n + 1) interacting with the FC12 micelle, as a cartoon model coloured in pink and orange. Right, the same as in central panel without the blue mesh, showing the interaction between the two neighbouring nsP1s from each ring and the three cysteines at the tip of the spike. The closest residue between the two rings is C417 from each dimer, with an approximate distance of 4.4 Å between their sulfur atoms. b, Same representation as in a of the entire map, with the view rotated by 90° about the indicated two-fold axis.
Fig. 1. Cryo-EM structure of the spherule–pore complex of CHIKV nsP1.
a, Ribbon representation of the nsP1 complex from its cytosolic (top) view. The 12-fold symmetry is indicated by numbering; each protomer is shown in a different colour. b, Surface representation of the top cytoplasmic view (top) and membrane-bound side (bottom). The surface electrostatics were calculated using APBS (https://www.poissonboltzmann.org/) from −5 kT/e to 5 kT/e, and are represented by the colour code of the bottom bar. The diameters of the ring and the pore aperture are indicated. c, Same representation as in b, of the lateral view of the complex (left) and a section through the broader part (right). The three regions (crown, waist and skirt) are delimited by black lines. The dimensions of the height, the cone upper aperture, the pore height and the bottom skirt diameter are indicated. The membrane-binding spikes are visible at the bottom of the skirt. The detergent-binding interface (substituting the membrane) is indicated by a thick dashed line. The projection angle of the spikes with respect to the pore is also indicated. d, Lateral view of tomographic reconstructions of FHV spherule necks4 in which the CHIKV nsP1 complex is superposed (Extended Data Fig. 5). nsP1 is shown coloured by protomer inside the FHV volume (represented as a transparent grey surface). e, Schematic of the structural regions of nsP1, indicating the residues that delimit each region (CHIKV numbering). The green fragment (NT) is the N-terminal extension of the capping domain. The grey fragment corresponds to the C-terminal residues that are disordered in the structure (Extended Data Fig. 6). f, Two-side view of the nsP1 protomer structure, represented as a cartoon following the same colour code as in e. All secondary structures and the Zn atom are labelled, as are the corresponding regions of the crown, waist and skirt of the ring. The missing loop between αi and αh is indicated by blue dashed lines. The circled C and N represent the C and N termini, respectively.
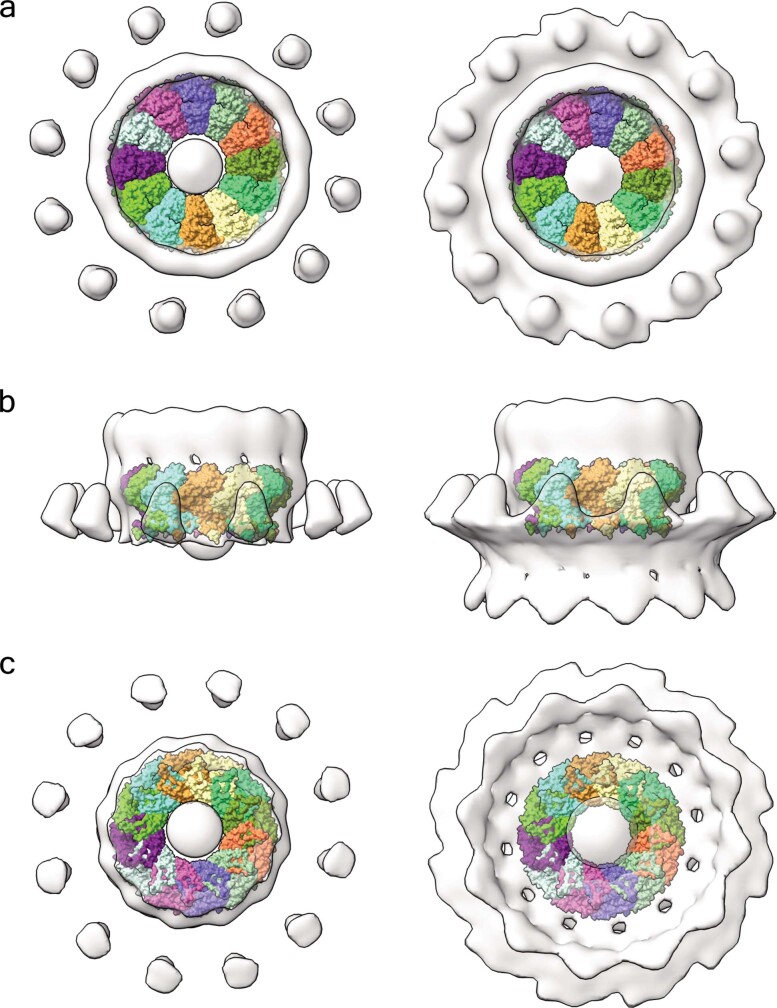
Extended Data Fig. 5. Superposition of the nsP1 complex in the FHV tomographic reconstruction of spherule necks.
a, Cytoplasmic view of the nsP1 complex (coloured by protomer) superposed into the tomography map reconstructed from FHV growing spherules (light grey surface)4. The volume is shown at two contour levels, 1.2 (left) and 0.9 (right), using ChimeraX. The superposition shows the 12-fold symmetry of both structures and that they are of a similar shape and size. The FHV central crown appears to be slightly larger. The panel on the left at lower contour level allows for visualization of the membrane around the crown. b, Lateral view of the superposition. The nsP1 fits into the lower part of the FHV crown in contact with the membrane. In the right panel, it is possible to appreciate the membrane bending carried out by the complex together with 12 external detached volumes of unidentified origin. From this view, an extension of the crown is visible at the top of the ring. This region probably corresponds to the polymerase domain of FHV protein A, which is in the same polypeptide chain as the capping enzyme that is homologous to alphavirus nsP1. c, Bottom view of the superposition. In this orientation, the inner shape of the viral spherule can be seen. The FHV volume was provided by P. Ahlquist, who has since published the same structure at higher resolution39.
Atomic structure of CHIKV nsP1 protein
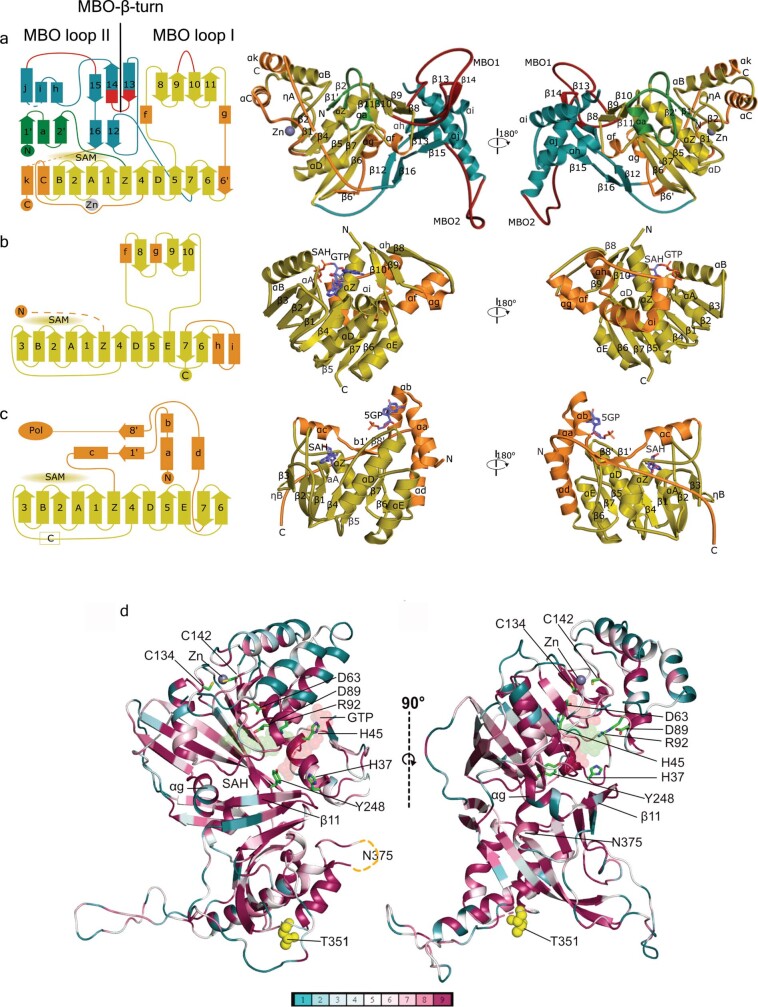
We built the atomic model of nsP1 de novo: it includes residues 3–471 out of 535, with density gaps at 364–376 and 450–458. No substantial differences were found between protomers of the double and single rings. nsP1 folds as two interconnected domains: the capping domain (located in the crown of the complex) and the ring-aperture membrane-binding and oligomerization (RAMBO) domain, which defines the waist and skirt regions of the complex (Fig. 1e, f). The capping domain (residues 1–294 and 459–472) exhibits high structural similarity with other SAM-dependent MTases: the DALI protein-structure comparison server17—using the complete Protein Data Bank (PDB) database—identifies MTases of Encephalitozoon cuniculi (PDB code 1RI1), vaccinia virus (PDB code 6RFL) and human (PDB code 5E8J) (among others) as being similar to CHIKV nsP1, with respective Z-scores of 10.1, 10.0 and 9.6, and root mean square deviations of 3.2 Å for all of them, including—respectively—188, 190 and 192 residues in the structural alignments (Extended Data Fig. 6). All of these MTases exhibit variations of a canonical SAM-dependent fold that comprises a core β-sheet of seven β-strands (β1 to β7) flanked by α-helices. In nsP1, the core β-sheet is modified through substitution of β3 by the α-helix αC (which forms an α-bundle with αB and αk; upper- and lower-case letters denote conserved secondary structures among MTases and specific secondary structures of CHIKV MTase, respectively), the incorporation of an additional β-strand (β6′) at the outer edge of the sheet and the inclusion of a Zn-binding site formed by the tetrahedral arrangement of the side chains of residues H79, E129, C134 and C141 (Extended Data Fig. 3a). The nsP1 N terminus is extended by a β–α–β motif (β1′–αa–β2′). An additional, smaller β-sheet insertion (β8 to β11) is conserved in vaccinia virus, human and E. cuniculi MTases18,19, but is absent in other positive-stranded RNA viruses such as flavivirus MTases20. This insertion is involved in the binding of a GTP acceptor in the methyltransferase reaction. Within this region, nsP1 has a 35-amino-acid-long loop insertion between β9 and β10 (residues 201–236) that is critical for membrane binding and oligomerization (which we term membrane binding and oligomerization loop 1 (MBO loop 1)) (Extended Data Fig. 6a). We could assign weak density, consistent with an α-helix (αk), to residues 462–472 at the C terminus of nsP1 (Extended Data Fig. 3b). Beyond this point, density could not be resolved, which indicates that the missing C-terminal 60 residues are disordered and projected towards the top of the crown.
Extended Data Fig. 6. Structural comparison of nsP1 with E. cuniculi and dengue virus MTases.
a, Left, topology diagram of CHIKV nsP1. The MTase conserved motifs are coloured in yellow. The CHIKV-specific features of the capping domain are coloured in orange. The RAMBO domain is coloured in teal. The MBO regions are coloured in red and labelled. The Zn-binding site and the SAM-binding regions are also indicated. Middle, right, two views of nsP1 as a cartoon model rotated by 180°. The same colour code is used as in the diagram on the left, and secondary structural elements are labelled. The Zn atom is shown as a grey sphere. b, Same representation as in a, for the MTase of E. cuniculi. (PDB code 1RI1), in which the SAH and GTP ligands are labelled. c, Same representation as in b, for the MTase of dengue virus (PDB code 1L9K). In dengue virus, the MTase and RNA-dependent RNA polymerase domain are in the same protein (NS5). The connection with the polymerase domain is indicated at the N terminus. This MTase has only one central β-sheet instead of the two β-sheet folding of E. cuniculi and nsP1. d, ConSurf representation of nsP1 sequence conservation within the alphavirus family according to the alignment of Extended Data Fig. 7. Two views of nsP1 are represented as cartoons and coloured according to the conservation score calculated by ConSurf. The lower bar indicates the conservation degree of each residue from non-conserved (blue) to fully conserved residues (purple). The GTP and SAH putative ligands are shown as red and green transparent spheres. The side chains of residues mutated in previous studies are shown as green sticks (Supplementary Information). The positions of residues suspected to bind to nsP4, T351 and N375 (missing in the structure), are shown as yellow spheres. The αg and β11 secondary structures, previously43 thought to be an amphipathic membrane-associated helix, are labelled.
The RAMBO domain (residues 295–450) folds in two β-sheets of antiparallel strands (β13–β15 and β12–β16), forming the platform on which the capping domain sits. The α-bundle that follows these strands comprises three α-helices (αh, αi and αj), which form the inner walls of the pore. A loop between αh and αi (residues 365–375) is not visible, which implies that it is highly flexible and deployed towards the cone area. Infections with temperature-sensitive mutants of the Sindbis virus RNA-dependent RNA polymerase nsP4—which prevent the synthesis of negative-strand RNA under non-permissive temperatures—could be compensated by mutations in nsP1 (T349K or T349L and N374H or N374I) (CHIKV T351 and N375) that are, respectively, located at the base and in the disordered loop at the top of the α-bundle21,22 (Extended Data Fig. 6d). This suggests that nsP4 may interact with both the inner and the outer sides of the pore. Beneath the complex, a long loop (which we term the MBO loop 2) and a pair of long antiparallel strands (β13 and β14 (which we term the MBO β-turn)) are involved in membrane binding (as discussed in ‘nsP1 oligomerization defines membrane binding’).
In conclusion, the nsP1 capping enzyme is derived from a classic SAM-dependent MTase fold with modifications that allow for the capping reaction, multimerization and membrane binding. The modifications include novel insertions and extensions at the N terminus and the C terminus of the MTase fold. Sequence similarity shows that this folding is conserved among all alphaviruses (Extended Data Figs. 6d, 7).
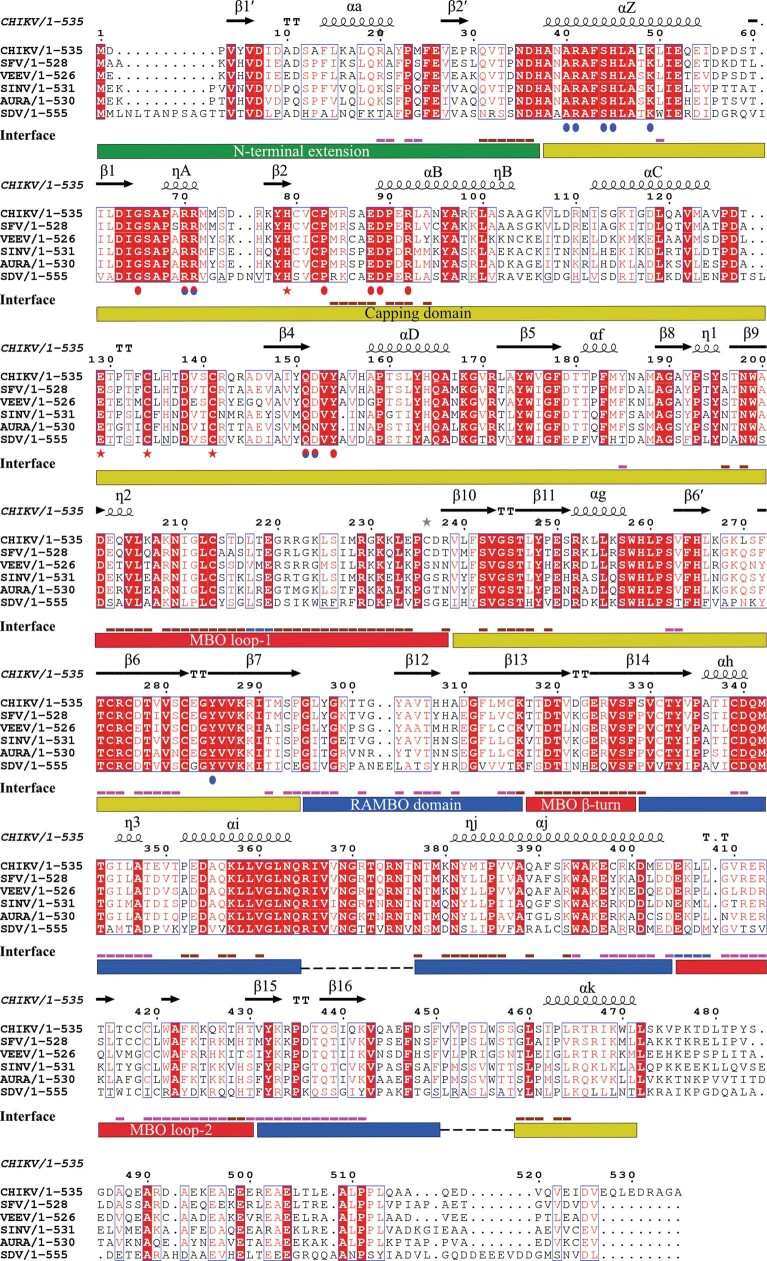
Extended Data Fig. 7. Sequence alignment of alphavirus nsP1.
The sequence alignment of nsP1 sequences of CHIKV (UniProt: Q5XXP4), Semliki Forest virus (SFV) (UniProt: P08411), Venezuelan equine encephalitis virus (VEEV) (UniProt: P27282), sindbis virus (SINV) (UniProt: P03317), aura virus (UniProt: Q86924) and salmonid sleeping disease virus (SDV) (UniProt: Q8QL53). The secondary structural elements revealed by the nsP1 structure are indicated at the top of the alignment. Below the sequences, the residues forming the GTP-binding site are highlighted by blue spots, those forming the SAM-binding site by red spots and those involved in Zn binding by red stars. Underneath, the interface line indicates the residues involved in interactions with the n + 1 nsP1 protomer in pink and with the n − 1 in brown. The blue lines indicate those residues making contacts to n − 1 and n + 1. At the bottom, rectangles indicate the domain boundaries of the nsP1 fold, coloured as described in Fig. 1e. Three motifs are conserved in the amino acid sequences of alphaviruses and similar viruses such hepatitis E virus, rubella virus and more26,27: (i) the catalytic histidine (H37) responsible for the cap transfer; (ii) the DXG motif (in which X stands for any residue) (63DIG65 in CHIKV); and (iii) and the DXXR motif (89DPER92 in CHIKV)—the last two define the SAM-binding site. This suggests that all alphavirus nsP1 have similar capping enzymes. Driven by the structural alignment of nsP1 and E. cuniculi MTase, we can more generally assign the DXG motif to the SAM-dependent MTases motif D/EX(G)XGXGXDL14,18,19, which—in alphavirus—corresponds to 63DIGSAPXRR71.
nsP1 oligomerization defines membrane binding
The structure shows that oligomerization is mediated by contacts that involve the capping and, mainly, the RAMBO domain. Our analysis of the complex interfaces using the PISA server23 reveals an extensive interface that engages 84 amino acid residues in contacts with the adjacent nsP1n − 1 and 92 residues in contacts with nsP1n + 1 subunits, burying 3,282 Å2 and 3,213 Å2 of protein surface, respectively. Each of the 12 interfaces includes 49 hydrogen bonds and 19 salt bridges, (Extended Data Figs. 7, 8, Supplementary Table 1). Altogether this network of interactions buries 29.3% of the nsP1 surface area (reaching 77,940 Å2 for the whole ring), and engages most of the nsP1 secondary structures. As a consequence, oligomerization allosterically activates the enzyme by stabilizing the conformation of the capping domain and generating a bilobular catalytic pocket in which the GTP-binding lobe faces the interior, and the SAM-binding lobe faces the exterior, of the crown (Fig. 2a).
Extended Data Fig. 8. Oligomerization interface of the nsP1 complex.
Cartoon representation of one protomer (n, coloured by the domains as outlined in Fig. 1f, Extended Data Fig. 6a) flanked by two protomers (n + 1 and n − 1, in grey) from the exterior view of the complex. The interaction interfaces are depicted as transparent surfaces. Contacts with the n − 1 protomer are maintained mainly with the capping domain, whereas the contacts with n + 1 are made mainly with the RAMBO domain. The side chains of the n protomer residues involved in n/n − 1 and n/n + 1 hydrogen bonds are represented as sticks. The Zn atom is shown as a grey sphere and labelled. The secondary structural elements involved in interactions, the active site and the MBO loops of the n protomer are labelled and indicated by arrows. MBO loop 2 folds under the n + 1 protomer and MBO loop 1 over the n − 1 protomer. The interface with nsP1n − 1 involves the MTase main β-sheet (including β6′, β6 and β7), the helix αZ and the N-terminal extension (mainly the loop between αa and β2′). The RAMBO domain is also engaged in the interaction by the strands β12, β15, β16 and the beginning of β13, the C terminus of the long helix αj and one side of αh at the pore-forming α-bundle. The interface with nsP1n − 1 is built by the capping domain small β-sheet (β9 to β11), αB and the C-terminal αk in the top of the ring and notably by the MBO loop 1, which builds up most of the interface in the membrane-binding region. A small interface between nsP1n − 1 and nsP1n + 1, which is sealing the ring, is formed by residues represented in blue sticks and labelled.
Fig. 2. nsP1 oligomerization and membrane binding are coupled processes.
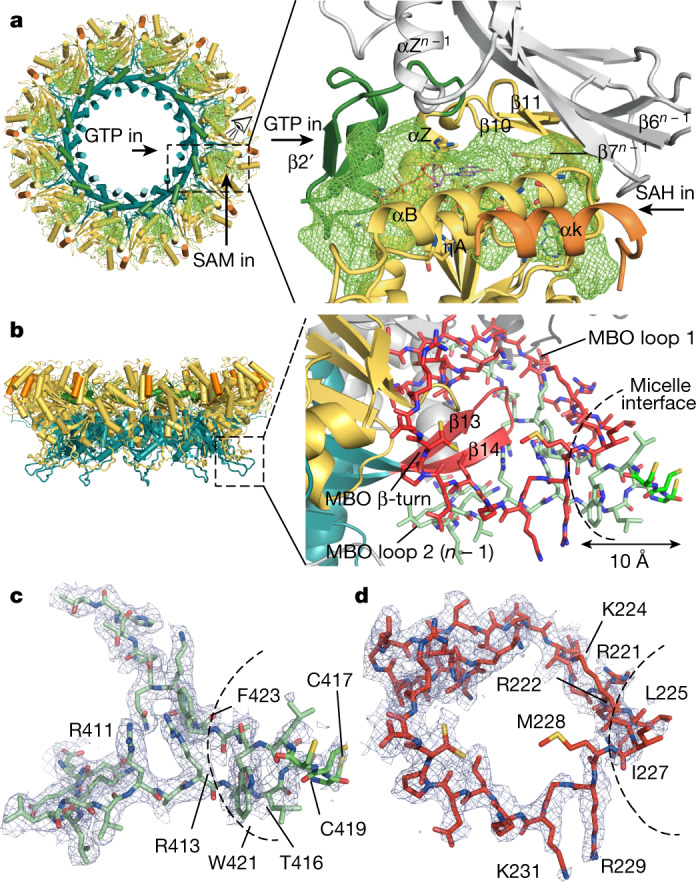
a, Left, the top view of the ring is represented as a cartoon and coloured as in Fig. 1f. The active site cavities are depicted as green meshes. Access to the GTP- and SAM-binding pockets in the active site from each side of the ring is indicated. The active site (dashed box) is magnified on the right; the view is indicated by the eye icon. The entrance of the GTP and the SAM to their respective binding pockets in the active-site cavity is indicated by arrows. The pocket is covered by the neighbouring n − 1 subunit (in grey). SAH and GTP molecules are superimposed from the structure of Ecm1 (PDB code 2RI1). b, Cartoon representation of the spike (dashed box), with magnified detail on the right. The n protomer is coloured as in a; the MBO β-turn and MBO loop 1 are coloured in red; and MBO loop 2 from the n − 1 protomer is coloured in green. The MBO loop residues are shown as sticks. The curved dashed line indicates the boundaries of the micelles found in the double-ring structure (Extended Data Fig. 4). A black arrow indicates the 10-Å length of the nsP1 fragment inserted into the FC12 micelles. c, MBO loop 2 represented as in b, with the density of the cryo-EM structure in blue mesh (drawn in PyMOL at 2σ). The FC12 detergent micelle boundary is indicated by a dashed line as in b. Details of the cysteine triad density are shown in Extended Data Fig. 3i–k. d, Representation of the MBO loop 1, as in c.
The most notable part of the interface is generated in the vicinity of the membrane by the MBO loop 2 from the RAMBO domain, which wraps under MBO loop 1 of the capping domain and the β-turn of the RAMBO domain of the nsP1n − 1 (Fig. 2b). These elements fold together, forming amphipathic membrane-binding spikes that penetrate about 10 Å into the detergent FC12 micelle. The tip of the spike is hydrophobic and includes residues 225LSIM228 from MBO loop 1 and 416TCCCLWA422 from nsPn + 1 MBO loop 2. The bases of the spikes create a positively charged belt that is continuous around the skirt, and which constitutes a platform for the binding of the negatively charged phospholipid heads. A triad of cysteines in MBO loop 2 are known to be palmitoylated24,25 in mammalian cells, but—consistent with the very low levels of palmitoylation detected in our insect-cell-expressed nsP1 (Extended Data Fig. 1g)—we do not observe acylation in the structure (Extended Data Fig. 3i–k). However, the cysteine triad appears in the structure at the end of the tip inside the micelles, consistent with the putative insertion of the palmitoyl moieties into the cellular membrane. Thus, palmitoylation is not essential for pore formation, which suggests that other eukaryotic cell factors must be required. This is consistent with previous studies that have demonstrated that palmitoylation is a determinant of nsP1 localization within the membrane, rather than of activity or membrane binding24. However, in combination, the approximately 17-Å-long palmitoyl chains and the approximately 10-Å-long inserted tip can reach a depth of around 2.7 nm into the lipid bilayer (the thickness of which ranges from about 2.5 to 3.5 nm). This represents a very strong monotopic interaction with the lipid bilayer. The well-defined electron density maps in the bases of the spikes indicates that the spikes are rigidly projected from the complex (Fig. 2c, d). The strength of the membrane binding, the rigidity of the spikes and the curvature of the positively charged skirt edges—which attract the membrane phospholipid heads—can potentially induce the marked membrane bending that is observed in spherule necks7,9. Thus, the structure of the spikes demonstrates the coupling between oligomerization, allosteric activation of the capping domain, membrane binding and membrane bending in the assembly of the nsP1 complex. This is likely to be conserved among alpha-like viruses26,27.
Structural basis of the capping reaction
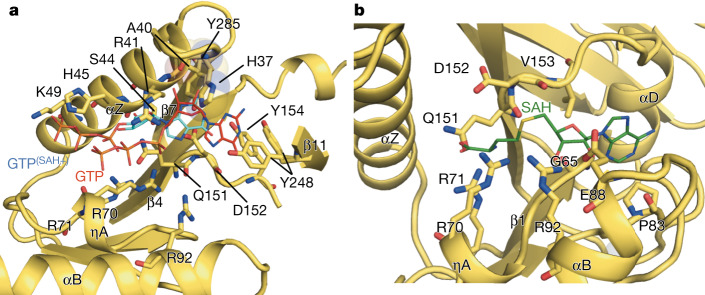
In contrast to conventional capping systems (which use sequential capping and methylation reactions to form the cap-1 structure (m7GpppRNA)), alphavirus nsP1 first methylates the GTP, covalently binds m7GMP and finally transfers m7GMP to a 5′-end diphosphate viral RNA acceptor13. Our structure shows that this is achieved by a modified SAM-dependent MTase-like domain. Superposition of the nsP1 capping domain with E. cuniculi MTase bound to S-adenysol-l-homocysteine (SAH) and GTP19 enabled us to identify residues, defining their binding sites for the guanine-N7 methylation reaction (Fig. 3, Extended Data Fig. 9a, b). The GTP-binding site is formed by residues close to the base moiety-binding position (A40, D152, Y285, Y154 and Y248) and positively charged and polar residues near the triphosphate position (K49, H45, R41, R70, R71, S44, Q151 and D152) (Fig. 3a, Extended Data Figs. 3g, 9a). The SAM-binding pocket is defined by residues shared with the GTP-binding site (R70, R71, Q151 and D152) and others that are exclusive for SAM binding (V153, G65, R92, E88, D89 and P83) (Fig. 3b, Extended Data Figs. 3h, 9b).
Fig. 3. Structure of the capping active site of nsP1.
a, Cartoon representation of the nsP1 GTP-binding-site cavity. The GTP moieties (represented as lines) are superposed in two positions, from the structures of E. cuniculi in complex with m7GpppG (PDB code 2RI2) alone (red sticks), and m7GpppG and SAH (PDB code 1RI1) (cyan sticks). Only the GTP moiety of the cap structure that was visible in these PDB entries is shown. The amino acid residues that define the cavity and form putative contacts to the GTP are shown as sticks and labelled. The secondary structures that shape the cavity are also labelled. The catalytic histidine H37 is shown as sticks and highlighted by a transparent sphere envelope. GTP(SAH+) label indicates the GTP position in the structure that includes SAH. GTP label indicates the GTP position in the structure with only m7GpppG. b, Cartoon representation of the nsP1 SAM-binding-site cavity. The cavity is shown as in a, with the carbon backbone of the SAH molecule coloured as green lines, from superposition of E. cuniculi MTase in complex with m7GpppG and SAH (PDB code 1RI1). The Zn atom is represented as a shadow sphere next to P83 and behind αB (Extended Data Fig. 9).
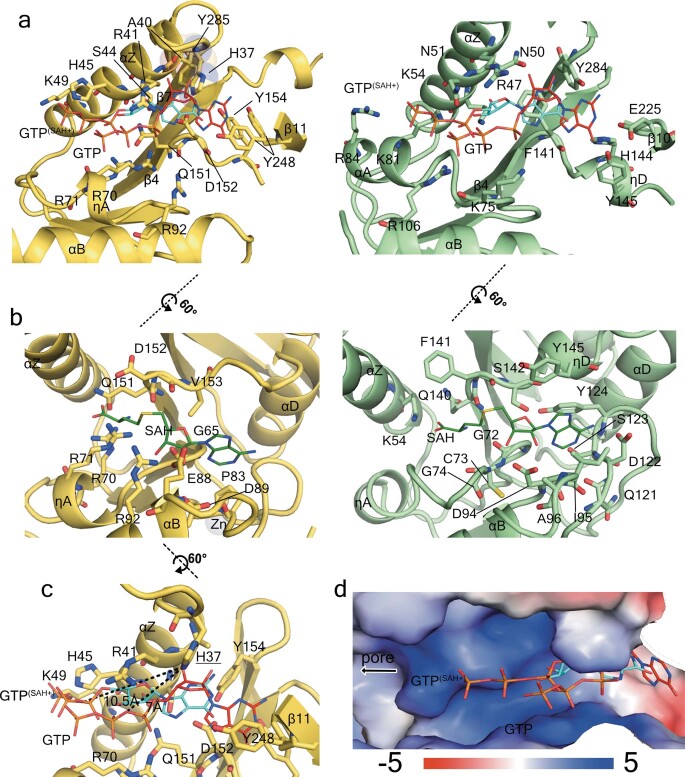
Extended Data Fig. 9. Structure of the nsP1 capping active site and comparison with E. cuniculi MTase.
a, Structural alignment of CHIKV capping domain and E. cuniculi MTase (PDB code 1RI1) showing the GTP-binding site. The structures are represented in cartoon model and coloured in yellow for CHIKV (left) and green for E. cuniculi (right). The superposed GTP visible moieties corresponding to the E. cuniculi MTase structures in complex with SAH and m7GpppG (PDB code 1RI1) and only m7GpppG, (PDB code 1RI2) are shown in both models as thin sticks with carbon atoms coloured in blue and red, respectively, and are labelled. The residues involved in GTP binding in E. cuniculi, and the residues positioned to maintain contacts with the superposed GTP in CHIKV are shown as sticks and labelled. The secondary structures are also labelled. b, Same representation as in a, showing the SAM-binding site. The SAH molecule of the E. cuniculi structure is superposed on CHIKV. c, Same representation as in a, b, focusing on the catalytic histidine H37, showing in dashed lines the distances with the GTP superposed phosphate α oxygen where covalent binding would occur for the GTase reaction. d, Surface electrostatics of the GTP-binding site by APBS are shown from −5 kT/e in red to 5 kT/e in blue as indicated in the bar below. The superposed GTP from PDB codes 1RI1 and 1RI2 are shown as in a. The direction towards the pore of the ring is indicated by a black arrow.
The nsP1–m7GMP intermediate is formed by a covalent bond between H37 and the α-phosphate of the m7GTP14 (Extended Data Fig. 9a, c). In our structure, H37 is close to the GTP-binding site but about 6–9 Å away from the superposed GTP α-phosphate position. Thus, after GTP methylation, a relocation of m7GTP in the binding pocket or a conformational change must occur to permit the formation of the covalent intermediate nsP1–m7GTP. The diphosphate viral RNA (the acceptor of the m7GMP moiety) could access the active site through a positively charged path that runs from the cone-shaped chamber of the ring to complete the cap transfer (Extended Data Fig. 9d). However, the cap transfer requires the generation of the diphosphate viral RNA by nsP2. Thus, coupling of the activities of the two proteins is required for capping viral RNA during infection. Further biochemical and structural studies will be necessary to understand the nsP1–nsP2 coupling and the precise role of the identified nsP1 residues in specificity and coordination for carrying out the three steps of the viral RNA capping reaction.
Conclusion
To our knowledge, our structure of the nsP1 complex is the first to define the structural basis of membrane binding for the replication complex of a positive-sense single-stranded RNA virus. The nsP1 pore reveals the architecture of replication complexes in the membranes, and controls the transit of molecules that enter and exit spherules that contain the double-stranded RNA viral genome, acting as a kind of viral nuclear pore complex. This markedly improves our view of the mechanisms of action of replication complexes (Extended Data Fig. 10). Moreover, the high oligomerization order and the unusual spike-mediated interaction with the membrane makes nsP1 the first example of a new class of membrane-bending, monotopic membrane proteins28,29. This class appears to be common to several positive-sense single-stranded RNA viruses. Our results thus open up avenues for the study of these viral replication complexes in the context of their membrane association.
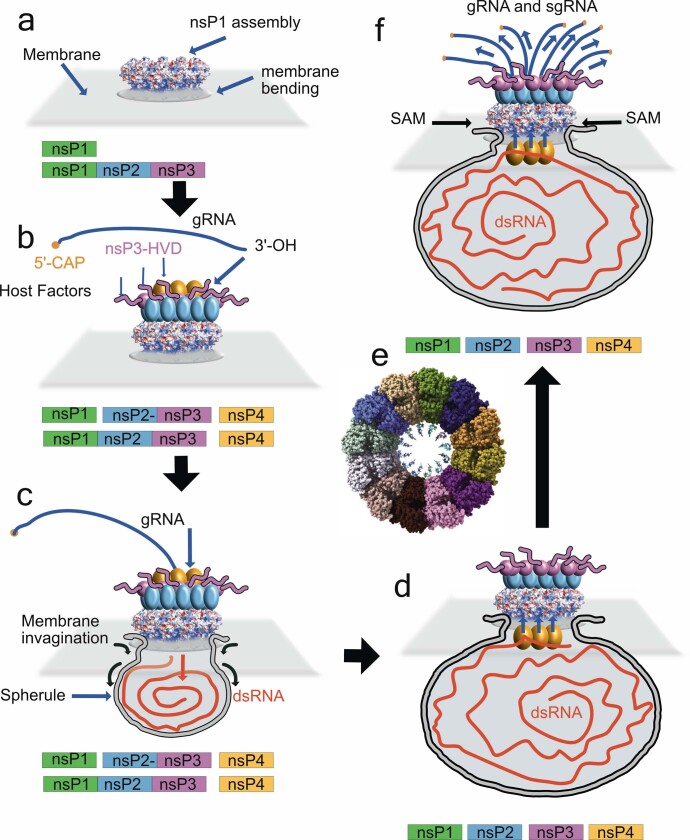
Extended Data Fig. 10. Model of replication-complex assembly, replication and spherule formation.
a, Our results demonstrate that the nsP1 active complex can assemble in the absence of other replication complex subunits. The complex could be formed by uncleaved nsP precursors as nsP12340; in this case, nsP123 would stay non-cleaved until step d. b, The nsP2, 3 and 4 subunits of the replication complex would be recruited on the spherule pore complex together with other cellular factors for the formation of active replication complexes. The negatively charged cone walls and the exposed loop between helix αi and helix αh could serve as anchoring points. c, The deep insertion of the amphipathic spikes into the membrane would have a wedging effect on the membrane, and the membrane phospholipid heads would interact with the concave positively charged patches around the waist and below the complex. Altogether, these interactions are prone to induce the first negative curvature of the membrane. Coupled with the synthesis of the negative RNA intermediate by nsP123+4, the spherule could grow below the pore, while maintaining a tight attachment to the complex. The direction of RNA synthesis into the pore is indicated by arrows, as the genomic RNA transcripts released from virions are located in the cytoplasm. d, After complete synthesis of the negative strand and growth of the spherule, transcription of capped viral RNAs may occur. The double-stranded RNA templates for viral RNA transcription must be read by the polymerase inside the spherule. A conformational change of the replication complex upon cleavage of nsP1 from nsP2 (for example, the internalization of the nsP4 proteins into the spherule) should allow the change of RNA product direction (indicated by arrows). Indeed, thermal-sensitive mutants at both sides of the pore α-bundle suggests that nsP4 interacts with both sides of the pore (see ‘Atomic structure of CHIKV nsP1 protein’). The forced traffic of the newly synthetized viral RNAs through the pore and capping active sites would ensure their capping before reaching the cytoplasm. e, The stoichiometry of the complex and size of the pore suggests that up to 12 capped viral RNA molecules could be capped simultaneously. f, After the 5′ of the viral RNA is capped the rest of the molecule will be expelled close to the MTase-domain GTP-binding site. This could facilitate the incorporation of internal methylations, a phenomenon that has previously been described for alphavirus viral RNA41,42, that confers stability to the RNA and prevents its recognition by the innate immune system.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Expression and purification of CHIKV nsP1 from insect cells
nsP1 was expressed in Hi5 cells (Thermo Fisher). The coding sequence of nsP1 (corresponding to residues 1–535 of the non-structural polyprotein sequence from the S27 African prototype, UniProt identifier: Q8JUX6) was synthesized by Gen9 as a codon-optimized gene and subcloned into a pFastBac vector (Thermo Fisher) with a C-terminal heptahistidine tag. Recombinant bacmids and viruses were produced in YFP-DH10 Bac cells16 and Sf21 cells (Thermo Fisher), respectively, according to the manufacturer’s protocols. For protein expression, Hi5 cells in suspension at a density of 0.5–1.0 × 106 ml−1 were infected with baculoviruses at 2% volume of the culture. Cells were collected at 2–3 days after cell arrest, when nsP1 expression was ascertained to be highest by western blotting.
Cells were resuspended in buffer A (35 mM Tris pH 7.6, 200 mM NaCl, 2 mM TCEP, 5% glycerol and10 mM imidazole) supplemented with 1 mM PMSF, 10 μg × ml−1 DNase, 2 μg × ml−1 RNase and 2 mM MgSO4, and lysed by sonication. Lysates were centrifuged at 15,000g to remove unbroken cells and debris, and then again at 100,000g to pellet the membrane fraction. Membranes were resuspended at 50 mg × ml−1 in buffer A containing 1 mM PMSF using a hand-held homogenizer, and then incubated with 1% fos-choline 12 detergent (Anatrace) for 2 h at 4 °C with gentle agitation for solubilization. Solubilized membranes were then recentrifuged at 100,000g, and supernatants containing nsP1 incubated with Ni-NTA resin (GE Healthcare) in batch at 4 °C for 30 min. Resin was washed with 10 column volumes of buffer A containing 40 mM imidazole and 0.13% fos-choline 12, and the protein eluted over 1.5 column volumes with buffer A containing 250 mM imidazole and 0.13% fos-choline. Elution fractions containing the protein were pooled and concentrated (Amicon 100k MW cut-off) for size-exclusion chromatography. Samples were applied to a Superose 6 10/30 column (GE Healthcare) pre-equilibrated with 25 mM tris pH 7.6, 200 mM NaCl, 2 mM TCEP and 0.13% fos-choline 12. Fractions were analysed by SDS–PAGE to assess purity, and western blotting with an nsP1 antibody and matrix-assisted laser desorption/ionization–time of flight (MALDI–TOF) mass spectrometry to confirm the presence of nsP1.
Expression and purification of CHIKV nsP1 from E. coli
The nsP1 gene was cloned into a pET28b vector in frame with a C-terminal heptahistidine tag for expression in E. coli Rosetta pLysS (DE3) cells (Novagen). Cells were grown in 2TY medium at 37 °C until reaching an optical density of 0.8 at 600 nm, when isopropyl-β-d-thiogalactopyranoside was added at 0.5 mM to induce expression at 18 °C overnight. Cells were collected by centrifugation at 4,000g and frozen until use. Cell pellets were resuspended in 35 mM Tris HCl pH 7.6, 0.3 M NaCl, 2 mM TCEP, 5% glycerol and 10 mM imidazole (buffer A) supplemented with 1 mM PMSF, 10 μg × ml−1 DNase, 2 μg × ml−1 RNase and 2 mM MgSO4 before lysis by sonication. Following centrifugation at 15,000g, the soluble fraction was applied to a 5-ml HisTrap crude FF column (GE Healthcare) pre-equilibrated in buffer A. The column was washed with buffer A containing 40 mM imidazole, and then eluted in buffer A containing 250 mM imidazole. Elution fractions containing nsP1 were concentrated (Amicon 30k MW cut-off) before application on a S200 10/30 size-exclusion column pre-equilibrated in 25 mM Tris-HCl pH 7.6, 0.2 M NaCl and 2 mM TCEP. Fractions were analysed by SDS–PAGE, MALDI–TOF and western blotting with an anti-nsP1 antibody.
Electron microscopy
The central fraction of the peak corresponding to nsP1 oligomers was selected for analysis by electron microscopy. For negative staining, 6 μl of the protein at a concentration of 60 μg × ml−1 was applied to Cu 300 mesh carbon grids (Agar Scientific), previously glow-discharged for 1 min at 15 mA using a PELCO EasiGlow cleaning system (Ted Pella). Grids were blotted, washed twice with water and stained with 6 μl of 2% (w/v) uranyl acetate before blotting and air drying. Images were acquired using a FEI Tecnai G2 Spirit microscope operating at 120 kV and equipped with a Veleta CCD camera (Olympus).
For cryo-EM, protein was diluted to 0.25 mg × ml−1 in size-exclusion buffer lacking detergent to reduce the concentration of free micelles applied to grids (to 0.5× the critical micelle concentration). Four μl of protein was applied to glow-discharged carbon-coated Cu/Rh 300 mesh Quantifoil R2/2 holey grids (Quantifoil), before manual blotting and plunge-freezing in liquid ethane using a Leica EM CPC cryo-fixation unit. Data were collected with a Titan Krios microscope operated at 300kV (ESRF-CM01)30 equipped with a post column LS/967 energy filter (Gatan) (slit width 20 eV) and K2 Summit direct electron detector (Gatan) in counting mode. Using EPU automated software (FEI), 5,626 movies were collected at a nominal magnification of 165,000× (corresponding to a sampling rate of 0.827 Å per pixel) across a defocus range of 1 to 2.5 μm. Each movie was recorded with a dose rate of 5.27 e− per pixel per s for an exposure time of 5 s distributed over 40 frames, yielding a total accumulated dose of 38.5 e− per Å2.
Cryo-EM data processing
All data were processed using RELION 3.031. Movies were corrected for drift using MotionCorr2 with dose-weighting32 and contrast transfer function (CTF) determination from the aligned non-dose-weighted micrographs was performed with CTFFind433. All frames of the movies were included for motion correction and CTF estimation. Poor-quality micrographs were discarded, yielding a subset of 5,148 images. Around 1,000 particles were selected from 100 images and subjected to 2D reference-free alignment to yield templates for the autopicking procedure of RELION. Following autopicking across the curated micrographs, 263,415 particles were extracted from the dose-weighted micrographs and subjected to 2D classification and particle sorting for removal of bad particles. Inspection of the 2D classes indicated that both single and double rings of nsP1 were present in the data, and data processing beyond this point was optimized to separate these species.
The best 2D classes (corresponding to 180,921 particles) were selected for 3D classification, using a single ring model as a template that had been generated ab initio from 2D classes obtained from a previous dataset collected from the same grid on a TALOS Arctica microscope (CNB-CIB cryo-EM facility). The model was low-pass-filtered to 60 Å, and 3D classification was performed with coarse alignment sampling (7.5°) using a circular mask of 290 Å and 3–5 classes. Ab initio model generation and all classifications were performed without imposing symmetry. Three-dimensional classification consistently yielded one major class corresponding to single rings (105,825 particles), and one class corresponding to double rings (47,338 particles). A further round of 3D classification was performed with the separated single-ring and double-ring classes to improve homogeneity, until further classification yielded no improvement. Three-dimensional classification performed with finer angular sampling did not indicate that there were conformational differences between individual protomers. A 3D reconstruction of particles from the best 3D class corresponding to single rings (94,018 particles) was performed in C1 using 3D auto-refine to a resolution of 4.3 Å (4.0 Å after map sharpening), and inspection of maps did not reveal conformational differences between protomers. The particles were thus 3D auto-refined imposing C12 symmetry to a resolution of 3.7 Å without masking, and 3.0 Å with masking, as estimated using the gold Fourier shell correlation criterion at a threshold of 0.143. Resolution improved to 3.4 Å and 2.9 Å following map sharpening, and further to 2.6 Å following a single round of particle polishing and per-particle CTF correction. Masks to perform map sharpening or refinements were generated through low-pass-filtering of the model to 15 Å and extending the map by 8 pixels and adding 6 pixels of a soft edge.
Particles from the best 3D class corresponding to double rings (47,338 particles) were auto-refined as described for single rings. Reconstructions in C1 reached a resolution of 7.1 Å (6.8 Å after map sharpening), and reconstructions performed imposing D12 symmetry reached a resolution of 3.87 Å without masking and 3.1 Å with masking. A single round of particle polishing and per-particle CTF correction finally improved the resolution to 2.9 Å. Masks were generated as outlined for single rings. Local resolution in the 3D structures was estimated using the localres implementation in RELION.
Model building
De novo model building of nsP1 was performed though fitting the protein sequence into C12- and D12-sharpened maps in Coot34. The quality of the maps allowed for unambiguous assignment of all residues with the exception of the two N-terminal residues (MG), residues 365–375, residues 451–457 and C-terminal residues 473–535, for which no density was observed. Density for residues 415–420 in MBO loop 2 within the micelle and C-terminal helix 458–472 is weaker, consistent with variations in local resolution estimates. Iterative cycles of model building and real-space refinement were performed with Coot and Phenix35 with NCS to improve the model. Validation of the model was performed using MolProbity36, and through cross-correlation analysis of the model and maps in Phenix. Pocket dimensions in the structure were calculated using POCASA37. Figures were generated using Chimera38 or PyMOL.
Sucrose gradient membrane flotation assays
Discontinuous sucrose gradients and flotation assays were adapted from a previously published method15. Small-scale cultures (10 ml) of E. coli or Hi5 cells expressing nsP1 were prepared as outlined in ‘Expression and purification of CHIKV nsP1 from insect cells’ and ‘Expression and purification of CHIKV nsP1 from E. coli’. Cells were resuspended in 2 ml of lysis buffer (35 mM Tris HCl, pH 7.6, 0.3 M NaCl and 2 mM TCEP) supplemented with 1 mM PMSF, 2 μg × ml−1 RNase and DNase and lysed by sonication. Following centrifugation at 15,000g, the soluble fraction was diluted with a 67% sucrose solution to yield a final concentration of 60%. The gradient layers (bottom to top) were as follows; 2 ml of the 67% solution, 2.5 ml of the sample, 5 ml of a 50% sucrose solution, followed by 2.5 ml of a 10% sucrose solution. All sucrose solutions were prepared in lysis buffer. Samples were centrifuged at 100,000g for 18 h in a SW40 Ti swinging-bucket rotor (Beckman Coulter) to allow the membrane fractions to float to the 10–50% interface. One-ml fractions were taken from the top of the gradient and analysed by SDS–PAGE.
Activity assays and western blotting
Five μM nsP1 was incubated with 50 μM m7GTP and 100 μM SAH or 50 μM GTP and 100 μM SAM at 30 °C for 1 h in gel filtration buffer in 20-μl reaction volumes. Two μl of the reactions were applied to 10% SDS–PAGE gels for electrophoresis at 180 mV for 1 h. Samples were transferred to nitrocellulose membranes via electroblotting at 320 mA for 1 h. Following blocking of membranes with 5% milk in TBST buffer (50 mM tris-HCl pH 7.5, 150 mM NaCl and 0.05% Tween-20) for 1 h at room temperature, membranes were incubated overnight with a primary antibody raised against nsP1 (provided by A. Merits) or an anti-m3G/m7G antibody (Synaptic Systems). Both antibodies were used at a 1/10,000 dilution in TBST buffer containing 0.5% milk. Membranes were subjected to four 10-min wash steps in TBST, before incubation with a secondary-HRP conjugated antibody for 1 h, repetition of the wash steps and revelation with Amersham Start ECL reagent (GE Healthcare). Chemiluminescence was detected with a Kodak DS Digital Science Image Station.
Detergent screening for solubilization of nsP1 from membranes
Membranes were prepared from Hi5 cells expressing nsP1 as outlined in ‘Expression and purification of CHIKV nsP1 from insect cells’. Following resuspension at 50 mg × ml−1 with a hand-held homogenizer, aliquots of the suspension were incubated with 1% detergent for 3 h at room temperature or at 4 °C overnight. Following centrifugation of samples at 100,000g for 2 h, supernatants were analysed by SDS–PAGE and assayed for guanyltransferase activity as outlined in ‘Activity assays and western blotting’. The following detergents were tested: cymal-5 (5-cyclohexyl-15-cyclohexyl-1-pentyl-β-d-maltoside), anzergent 3-10 (N-decyl-N,N-dimethyl-3-ammonio-1-propanesulfonate), DG (n-decyl-β-d-glucopyranoside), β-OG (n-octyl-β-d-glucoside), fos-choline 12 (n-dodecylphosphocholine), DM (n-decyl-β-d-maltoside), sodium dodecyl sulfate (SDS), DMNG (decyl maltose neopentyl glycol), LDAO (lauryldimethylamine-N-oxide) and DDM (n-dodecyl-β-d-maltopyranoside).
Acyl capture experiments
The palmitoylation states of purified nsP1 samples were assessed with an acyl capture kit (Badrilla). In brief, 150 μg of oligomeric or monomeric nsP1 was incubated at 60 °C for 4 h in a buffer to block free thiols before acetone precipitation. The precipitates were redissolved and incubated with a reagent designed to cleave any palmitoyl or acyl moieties, in the presence of resin that covalently bound the liberated thiols. Paired negative controls performed with a thiol protection reagent in place of the cleavage reagent yielded estimates of background binding to the resin. Bound samples were eluted from the resin through heating in the presence of Laemmli buffer. Monomeric nsP1 purified from E. coli was also included as a negative control.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-020-3036-8.
Supplementary information
This file contains Supplementary Text, Supplementary Methods, Data Availability Statement and Supplementary References.
List of atom-atom interactions across CHIKV nsP1complex protein-protein interface. List generated by the PDBsum server.
Raw data gels corresponding to Extended Data Fig. 1.
Acknowledgements
We thank the European Synchrotron Radiation Facility for provision of beam time on CM01, and E. Kandiah for assistance; J. Martin-Benito and the CNB-CIB (CSIC) cryo-EM facility for granting us access to cryo-EM equipment through a technical support contract, and J. Chichon for technical support; INSTRUCT-Eric and R. Melero for access to the computing facilities at the CNB-CSIC (PID 7046 VID 13154); A. Goulet, S. Spinelli and the electron microscopy platform at the AFMB; I. Berger and A. Aubert for supplying material and technical advice regarding eukaryotic expression; and T. Ahola, A. Merits and F. Rico for critical reading of the manuscript. The nsP1 antibody was provided by A. Merits, and the m7G cap antibody by B. Coutard. This work has been supported by the Bettencourt Shueller Fondation and an ATIP-Avenir grant (CNRS/INSERM).
Extended data figures and tables
Author contributions
R.J. performed sample production and purification, biochemical characterization and structure determination together with J.R. G.B. contributed to protein production and purification. R.A. assisted with sample preparation for cryo-EM. J.R. conceived and obtained the funding for the project. R.J. and J.R. designed the experiments and J.R. wrote the Article with the input of R.J.
Data availability
Structure coordinates are available from the PDB with accession codes 6Z0V and 6Z0U for single and double rings, respectively. The electron density maps are available from the Electron Microscopy Data Bank (EMDB) under accession codes EMD-11024 and EMD-11023 for single and double rings, respectively. All other data generated or analysed in this study are available from the corresponding author upon reasonable request.
Viral amino acid sequences used for gene synthesis and sequence alignments were retrieved from the UniProt database with the following accession numbers: CHIKV S27 African prototype (UniProt: Q8JUX6); CHIKV (UniProt: Q5XXP4), Semliki Forest virus (UniProt: P08411), Venezuelan equine encephalitis virus (UniProt: P27282), Sindbis virus (UniProt: P03317), aura virus (UniProt: Q86924) and salmonid sleeping disease virus (UniProt: Q8QL53). MTase structures for structural superpositions were retrieved from the PDB with accession codes 2RI1 and 1RI1 for E. cuniculi MTase, and 1L9K for dengue virus MTase.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature thanks Kyung Choi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s41586-020-3036-8.
Supplementary information
is available for this paper at 10.1038/s41586-020-3036-8.
References
- 1.Romero-Brey I, Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harak C, Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479-480:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egger D, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov GA, et al. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 2007;81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kujala P, et al. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T, Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 9.Pietilä MK, Hellström K, Ahola T. Alphavirus polymerase and RNA replication. Virus Res. 2017;234:44–57. doi: 10.1016/j.virusres.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomar S, Narwal M, Harms E, Smith JL, Kuhn RJ. Heterologous production, purification and characterization of enzymatically active Sindbis virus nonstructural protein nsP1. Protein Expr. Purif. 2011;79:277–284. doi: 10.1016/j.pep.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, et al. mRNA capping by Venezuelan equine encephalitis virus nsP1: functional characterization and implications for antiviral research. J. Virol. 2015;89:8292–8303. doi: 10.1128/JVI.00599-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahola T, Lampio A, Auvinen P, Kääriäinen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trowitzsch S, Bieniossek C, Nie Y, Garzoni F, Berger I. New baculovirus expression tools for recombinant protein complex production. J. Struct. Biol. 2010;172:45–54. doi: 10.1016/j.jsb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De la Peña M, Kyrieleis OJP, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 20.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirako Y, Strauss EG, Strauss JH. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology. 2000;276:148–160. doi: 10.1006/viro.2000.0544. [DOI] [PubMed] [Google Scholar]
- 22.Fata CL, Sawicki SG, Sawicki DL. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J. Virol. 2002;76:8641–8649. doi: 10.1128/JVI.76.17.8641-8649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Laakkonen P, Ahola T, Kääriäinen L. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 1996;271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 25.Karo-Astover L, Sarova O, Merits A, Zusinaite E. The infection of mammalian and insect cells with SFV bearing nsP1 palmitoylation mutations. Virus Res. 2010;153:277–287. doi: 10.1016/j.virusres.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Rozanov MN, Koonin EV, Gorbalenya AE. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 27.Ahola T. New phylogenetic grouping of positive-sense RNA viruses is concordant with replication complex morphology. MBio. 2019;10:e01402–19. doi: 10.1128/mBio.01402-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen KN, Entova S, Ray LC, Imperiali B. Monotopic membrane proteins join the fold. Trends Biochem. Sci. 2019;44:7–20. doi: 10.1016/j.tibs.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prinz WA, Hinshaw JE. Membrane-bending proteins. Crit. Rev. Biochem. Mol. Biol. 2009;44:278–291. doi: 10.1080/10409230903183472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kandiah E, et al. CM01: a facility for cryo-electron microscopy at the European synchrotron. Acta Crystallogr. D. 2019;75:528–535. doi: 10.1107/S2059798319006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams CJ, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Zhou Y, Tanaka I, Yao M. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics. 2010;26:46–52. doi: 10.1093/bioinformatics/btp599. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Unchwaniwala N, et al. Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication. Proc. Natl Acad. Sci. USA. 2020;117:18680–18691. doi: 10.1073/pnas.2006165117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellström K, et al. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J. Virol. 2017;91:e00787–17. doi: 10.1128/JVI.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frey TK, Gard DL, Strauss JH. Replication of Sindbis virus. VII. Location of 5-methyl cytidine residues in virus-specific RNA. Virology. 1978;89:450–460. doi: 10.1016/0042-6822(78)90187-3. [DOI] [PubMed] [Google Scholar]
- 42.Hsuchen CC, Dubin DT. Di-and trimethylated congeners of 7-methylguanine in Sindbis virus mRNA. Nature. 1976;264:190–191. doi: 10.1038/264190a0. [DOI] [PubMed] [Google Scholar]
- 43.Spuul P, et al. Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 2007;81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Text, Supplementary Methods, Data Availability Statement and Supplementary References.
List of atom-atom interactions across CHIKV nsP1complex protein-protein interface. List generated by the PDBsum server.
Raw data gels corresponding to Extended Data Fig. 1.
Data Availability Statement
Structure coordinates are available from the PDB with accession codes 6Z0V and 6Z0U for single and double rings, respectively. The electron density maps are available from the Electron Microscopy Data Bank (EMDB) under accession codes EMD-11024 and EMD-11023 for single and double rings, respectively. All other data generated or analysed in this study are available from the corresponding author upon reasonable request.
Viral amino acid sequences used for gene synthesis and sequence alignments were retrieved from the UniProt database with the following accession numbers: CHIKV S27 African prototype (UniProt: Q8JUX6); CHIKV (UniProt: Q5XXP4), Semliki Forest virus (UniProt: P08411), Venezuelan equine encephalitis virus (UniProt: P27282), Sindbis virus (UniProt: P03317), aura virus (UniProt: Q86924) and salmonid sleeping disease virus (UniProt: Q8QL53). MTase structures for structural superpositions were retrieved from the PDB with accession codes 2RI1 and 1RI1 for E. cuniculi MTase, and 1L9K for dengue virus MTase.