Abstract
Genetic analyses are an important contribution to wildlife reintroductions, particularly in the modern context of extirpations and ecological destruction. To address the complex historical ecology of the sea otter (Enhydra lutris) and its failed 1970s reintroduction to coastal Oregon, we compared mitochondrial genomes of pre-extirpation Oregon sea otters to extant and historical populations across the range. We sequenced, to our knowledge, the first complete ancient mitogenomes from archaeological Oregon sea otter dentine and historical sea otter dental calculus. Archaeological Oregon sea otters (n = 20) represent 10 haplotypes, which cluster with haplotypes from Alaska, Washington and British Columbia, and exhibit a clear division from California haplotypes. Our results suggest that extant northern populations are appropriate for future reintroduction efforts. This project demonstrates the feasibility of mitogenome capture and sequencing from non-human dental calculus and the diverse applications of ancient DNA analyses to pressing ecological and conservation topics and the management of at-risk/extirpated species.
Keywords: ancient DNA, mitochondrial genomes, marine mammals, applied zooarchaeology, conservation, translocation
1. Background
(a). Reintroductions and applied archaeology
The extinction and extirpation of animals and plants, and associated ecological degradation, are increasing at a rapid rate [1]. Responses to these challenges include reintroductions, translocations and other strategies used to bolster or re-establish populations of threatened or endangered species [2]. Significant challenges exist related to animal homing instincts, source population choice, predation and reproductive failures [3–6].
Genetic analyses are valuable for assessing reintroduction and translocation viability, and for documenting the impact of genetic rescue [6–8]. For example, microsatellites in desert tortoises have been used to show poor reproductive success in translocated males [5], while genomic approaches, including RADSeq and transcriptomics, highlight the importance of local adaptation in other organisms [9]. To avoid outbreeding depression and translocation failure and to account for potential local adaptation, genetic studies suggest that reintroduction efforts should maximize ecological similarity and minimize population divergence times between source and sink populations [7,8,10]. Past extirpation events may present particular challenges owing to the lack of recent genetic data on the extirpated population. Ancient DNA approaches provide a powerful method to bridge temporal gaps and provide relevant data, such as identifying appropriate source populations [11] and documenting genetic diversity before extirpation [12].
We present a novel approach integrating new methods and sources of ancient DNA to inform reintroductions of the sea otter (Enhydra lutris). The sea otter, a keystone species in kelp forest ecosystems, was hunted to the verge of extinction around the Pacific Coast during the maritime fur trade but has yet to re-occupy a large portion of its former range [13]. Our study demonstrates the importance of an applied analytical toolkit for investigating twenty-first century global extirpations and efforts to repair ecological degradation and disruption.
(b). The sea otter on the Pacific Coast
Prior to the nineteenth century, the sea otter occurred along the coast from Japan to northern Mexico [14]. Intensive hunting by Russian and Euro-American companies during the maritime fur trade (1741–1911) severely depleted sea otter populations, resulting in genetic bottlenecks [12,15–18], and triggering profound ecological and socio-cultural changes. Sea otters are an ecological priority owing to their role as a keystone species in the kelp forest ecosystem: sea otters eat urchins (benthic echinoderms) which graze on kelps, thus filling an important role in near shore community structuring [14,19]. Kelp forests protect coastlines from erosion [19], support biodiversity, and provide carbon fixation [20]. Disruptions to kelp forest ecosystems, whether through the loss of sea otters or other factors (e.g. climate change), are cause for great concern. Socio-cultural consequences of the fur trade included disruption and dismantling of Indigenous social-ecological and economic systems at the hands of colonial powers [21], and in some contexts, Indigenous peoples were coerced into hunting on behalf of fur companies [22,23]. Reviving sea otter populations to revitalize coastal ecosystems remains a conservation priority in the present, and a recent study suggests that reintroducing sea otters yields a net ecological/economic gain [20].
By the mid-twentieth century, sea otters were patchily distributed throughout their original range owing to rebound and conservation measures. Northern sea otter sub-populations (E. l. kenyoni) in the Aleutian Islands and southern sub-populations (E. l. nereis) in parts of California survived peak hunting in the late nineteenth century because of their geographical isolation [24]. In the 1960s–1970s, biologists reintroduced sea otters from southcentral and southwest Alaska to southeast Alaska, British Columbia (BC) and Washington, and successfully re-established populations in parts of their former range. However, two 1970s reintroduction attempts at Port Orford and Cape Arago in southern Oregon failed [24,25]. The ‘most plausible explanation’ for the Oregon failure was emigration (the sea otters' attempt to return to their original range/habitat) and small post-release populations that subsequently collapsed [25]. The translocated northern sea otters may also have lacked adaptations suitable for their new Oregon coast habitat [25]. Today, sea otters remain extirpated in Oregon. The species is listed as endangered on the International Union for Conservation of Nature's (IUCN) Red List [26], and the Oregon Endangered Species Act (ORS 496.171–496.192) lists the Oregon sea otter as ‘threatened’. There is growing interest in assessing whether reintroducing sea otters to Oregon is desirable and/or feasible. The Elakha Alliance non-profit is conducting a feasibility study as a step towards reintroduction, coastal ecological restoration and cultural revitalization with the partnership and support of the Confederated Tribes of Siletz and Coquille Indian Tribe [27,28].
Sea otter use and significance to tribal groups in Oregon are documented in archaeological and historical records, and tribal stories and oral histories [29–33]. Alaska Native and First Nations groups also have vested interests in sea otter conservation, use and management [21,34–36]. However, both Native and non-Native stakeholders are concerned about sea otter predation on commercially fished invertebrates [20,37,38], such as Dungeness crab in Alaska [35]. Because sea otters in Oregon were extirpated by the end of the fur trade (ca 1876, but possibly as late as 1906 [14]), there are some gaps in cultural and ecological knowledge pertaining to the species; the evidence is limited and few fur-trade era specimens or records are available for the study. As a result, available historical and archaeological Oregon sea otters represent a valuable, but often overlooked, source of data [30].
Reintroductions and subsequent management are complicated efforts involving many factors and stakeholders. Our study seeks to address a key aspect of Oregon reintroduction discussions: which post-fur trade sea otter populations are most closely related to the original (pre-fur trade) Oregon sea otter population and should serve as a source for reintroductions? We present a temporal perspective and dataset by recovering complete mitogenomes from archaeological (Late Holocene) and nineteenth-century fur trade Oregon sea otters and compare them to post-fur trade (twentieth-century and modern) sea otters to determine the relationships between populations.
(c). Previous studies
Mitochondrial DNA (mtDNA) and microsatellite analyses demonstrate that sea otter populations vary genetically along the northwest Coast [12,16,18,39–41]. Larson et al. [41] identified four modern sea otter mtDNA haplotypes on the northwest Coast. Valentine et al. [39] analysed a 222 base pair (bp) region of mtDNA from 16 archaeological Oregon sea otters and found four haplotypes: a California genotype represented by 11 Oregon individuals, an Alaska genotype represented by two Oregon individuals, and two new genotypes represented by two and one Oregon individuals, respectively. Valentine et al. [39] concluded the archaeological Oregon sea otters were, therefore, more closely related to the California sea otters, and future reintroductions using California sea otters might be more successful. Larson et al. [16] performed microsatellite analyses on pre-fur trade and modern sea otters, and found that gene flow occurred between Oregon, California and Alaska sea otters, including between Oregon and northern populations. Beichman et al. [15] identified specific aquatic adaptations and low genomic diversity in modern populations. Morphometric studies of sea otters have demonstrated some phenotypic traits vary along a latitudinal cline on the Pacific Coast, with Oregon sea otters intermediate by varying degrees [42–44]. We build on this research, presenting, to our knowledge, the first complete ancient mitogenomes for Oregon sea otters and demonstrate a new minimally destructive sampling technique using dental calculus.
(d). Current study
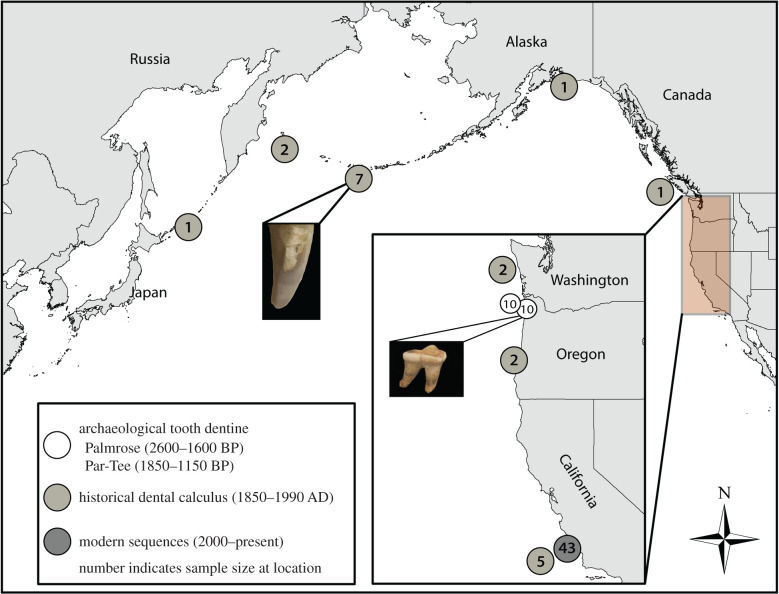
To expand upon and contribute to the current understanding of pre-fur trade Oregon sea otters, we sequenced complete mitogenomes from 20 archaeological sea otter teeth (tooth dentine). We sampled 10 right lower first molars (M1s) from the Par-Tee site (35CLT20) (1850–1150 cal BP; [45]) and 10 right M1s from the Palmrose site (35CLT47) (2600–1600 cal BP; [46,47]). Par-Tee and Palmrose are located adjacent to each other in northern Oregon (figure 1). These sites were excavated in the 1960s through to the 1970s [48] and the faunal remains are abundant and well-preserved [43,49–51]. We compared the archaeological Oregon mitogenomes to 21 historical Pacific Coast mitogenomes captured and sequenced from dental calculus for this study (table 1; figure 1) and previously published modern California mitogenomes [18]. These historical mitogenomes include sea otters from the end of the fur trade (just prior to extirpation) and the post-fur trade era, including several 1960s Amchitka Island sea otters [14,52]. Amchitka sea otters were reintroduced to southeast Alaska, BC, and Washington, and therefore probably reflect present genetic diversity in those areas [16]. Historical pre-extirpation Oregon sea otter specimens are a rare and unique data source, and dental calculus was used to minimize destructive sampling to these specimens. Sea otter mitogenomes are assumed to accurately reflect geographical origin as they are maternally inherited, and while male sea otters may travel upwards of 100 km [53,54], females tend to maintain small home ranges and geographical fidelity (extending approximately 20 km along the coast and approximately 0.3 km offshore) [55–57]. Based on previous findings [16,39,42–44], we hypothesized that the archaeological Oregon sea otters would share mitochondrial lineages with both California and northern Pacific Coast populations, but more with the latter. We also anticipated that the archaeological specimens would exhibit greater genetic diversity compared to the available modern California specimens [18] owing to past bottlenecks [15,16,58].
Figure 1.
Map showing geographical origins of sea otter archaeological dentine (2600–1600 BP; 1850–1150 BP) and historical dental calculus (1850–1990 AD), as well as published modern (2000–present) sequences. Made in ArcMap 10.0/Adobe Illustrator; data from Natural Earth and U.S. Census Bureau. (Online version in colour.)
Table 1.
Historical museum specimens from the National Museum of Natural History (NMNH) and the Santa Barbara Museum of Natural History (SBMNH).
| museum | acc no. | sex | date collected | location | haplotype |
|---|---|---|---|---|---|
| NMNH | 188636 | F | — | Kurile Islands, Japan | 22 |
| NMNH | A49492 | — | — | Copper Island, Bering Sea (Russia) | — |
| NMNH | 206458 | — | 1911 | Commander Islands (Kamchatka Peninsula) | 17 |
| NMNH | 285441 | F | 7 Apr 1949 | Amchitka Island, AK | — |
| NMNH | 285469 | M | 26 Apr 1949 | Amchitka Island, AK | 3 |
| NMNH | 285470 | — | 26 Apr 1949 | Amchitka Island, AK | 5 |
| NMNH | 396641 | M | 28 Sep 1977 | Green Island, Prince William Sound, AK | 6 |
| NMNH | 527126 | F | 17 Jun 1960 | Nagai Island, Eagle Harbor, AK | — |
| NMNH | 527134 | F | 22 Jan 1962 | Amchitka Island, AK | 1 |
| NMNH | 527162 | F | 6 Feb 1962 | Amchitka Island, AK | 2 |
| NMNH | 527170 | F | 10 Feb 1962 | Amchitka Island, AK | 4 |
| NMNH | 256971 | — | ca 1889a | Vancouver Island, BC | 21 |
| NMNH | 93954 | M | 4 Jun 1898 | Point Granville, WA | 18 |
| NMNH | 188634 | — | 1897 | Strait of Juan de Fuca, WA | — |
| NMNH | A3643 | — | ca 1859b | Port Orford, OR | 20 |
| NMNH | A13460 | — | ca 1874b | Oregon | 19 |
| SBMNH | 1922F | F | 1978 | Cooper Point, CA | 27 |
| SBMNH | 1366F | F | — | — | — |
| SBMNH | 1367M | M | 1977 | Oso Flaco Creek, CA | 26 |
| SBMNH | 3053 | M | 1983 | Sunset State Beach, CA | 25 |
| SBMNH | 3057M | M | 1983 | Point Piedras Blancas, CA | 23 |
aCollected by T. T. Minor/Dr Thomas Minor. Dr Thomas Taylor Minor was a prominent Seattle physician and the presumed collector. We assigned his year of death (1889) as an approximate date.
bBased on additional information found in Mammals and Life Zones of Oregon by Vernon Bailey [29].
2. Methods
Archaeological tooth dentine was sampled at the Laboratories of Molecular Anthropology and Microbiome Research (LMAMR) at the University of Oklahoma, Norman, in the dedicated sample preparation area following standard ancient DNA contamination protocols [59,60]. Dental calculus was sampled on location in museum research collections following a calculus-specific sampling protocol designed to reduce contamination (description in the electronic supplementary material with photos/specimen metadata). Dental calculus contains both endogenous (host) and microbial DNA [61–63] and can be removed without destruction to the specimen, preserving the integrity of rare museum collections while also addressing research questions regarding biodiversity/conservation biology. Owing to differential preservation of endogenous DNA in dental calculus [63] and documented degradation of DNA in calculus museum specimens [64], the historical DNA was extracted and sequenced following ancient DNA protocols and workflows.
Ancient and historical DNA extraction and library construction were performed in the LMAMR Ancient DNA Laboratory, a dedicated, six-room ISO-6 class cleanroom custom-built for ancient DNA and microbiome research. Detailed procedures are provided in the electronic supplementary material. In brief, DNA was extracted from dental calculus and dentine using a protocol described in Morales et al. [65]. DNA extracts were converted into dual indexed Illumina sequencing libraries and captured using a custom in-solution biotinylated RNA bait set (Arbor Biosciences). Captured libraries were sequenced on an Illumina MiSeq with 2 × 150 bp chemistry.
The raw fastq files were quality filtered using the program Adapter Removal2 (v. 2.1.7) [66] and mapped using bwa (v. 0.7.17) [67] with ancient DNA parameters to the published modern sea otter mitogenome [68] (electronic supplementary material, table S2). DNA authenticity was assessed using the program MapDamage2 [69] and fragment length plots (electronic supplementary material, figures S4 and S5). Consensus sequences were called from rescaled bam files in Geneious (v. 11.1.4) and aligned with MAFFT (v. 7.308) [70,71]. This alignment was stripped for identical sites and ambiguities and rendered into a network (figure 2) using the median-joining algorithm in PopArt [72,73]. Haplotype diversity was calculated in DnaSP (v. 6) [74]. In order to explore and visualize the temporal signal associated with haplotype diversity, we used TempNet in R (v. 3.6.3) (figure 3). We attempted estimating divergence times with BEAST (v. 1.10), but this dataset violated clocklike assumptions as tested with TempEst (v. 1.5.3). Raw sequence data are available through the NCBI Short Read Archive (SRA) under BioProject accession PRJNA550086. Consensus sequences and the alignment used for analysis (ModAlign.fa) are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.djh9w0vxz [75]).
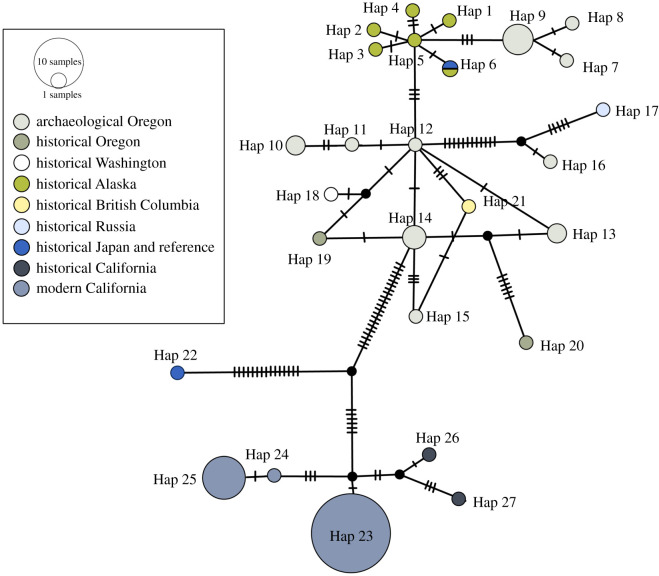
Figure 2.
Median-joining network of archaeological Oregon, historical and modern haplotypes. Node size represents haplotype frequency corresponding to table 1 and electronic supplementary material, table S3. Hash marks represent nucleotide changes between haplotypes. The reference mitogenome (Yonezawa et al. [68]) came from a sea otter in the Toba Aquarium, Mie, Japan, but shares haplotype 6 with an Alaskan sea otter. (Online version in colour.)
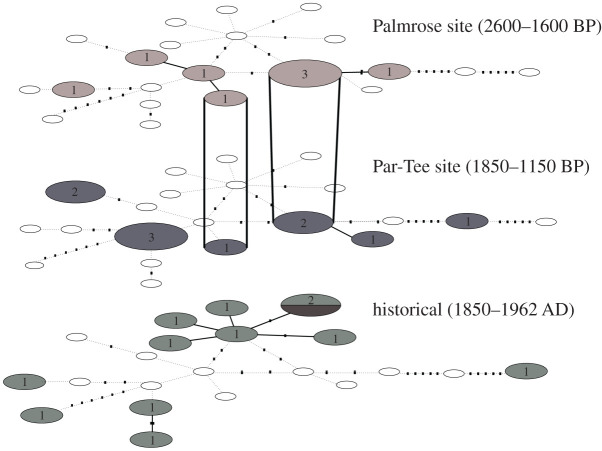
Figure 3.
TempNet analysis showing shared haplotypes between archaeological (2600–1600 BP and 1850–1150 BP, respectively) and historical sea otters (1850–1962 AD). Circles represent the haplotypes in the median-joining network; colours indicate time periods of haplotypes. Haplotype overlap (bold lines) occurs between two Palmrose and Par-Tee haplotypes (haplotypes 9 and 13 in figure 2). California samples were omitted due to divergence from archaeological and historical northwest coast haplotypes.
3. Results
Eighteen of the 20 archaeological specimens and 16 of the 21 historical specimens yielded complete mitogenomes suitable for analysis. The median-joining network analysis (figure 2) illustrates the relationships between the sea otter mitogenomes generated for this study, previously published California mitogenomes [18], and the reference mitogenome [68]. The network analysis yielded 27 haplotypes: 10 (haplotypes 7–16) represent archaeological Oregon individuals, five (haplotypes 23–27) represent historical/modern California, six (haplotypes 1–6) represent historical Alaska, two (haplotypes 19 and 20) represent historical Oregon, and haplotypes 18, 17, 21 and 22 each represent historical Washington, Russia, BC and Japan, respectively (electronic supplementary material, table S3). Of the 10 archaeological Oregon haplotypes, six are unique to single individuals (7,9,11,12,15,16), two represent five and three individuals, respectively (9 and 14), and the final two haplotypes (10 and 13) include two individuals each. The mitogenomes of the historical samples occur as expected in the network given their geographical origins, except for the historical individuals from Russia and Japan. The archaeological Oregon/northern haplotypes show substantial separation from California haplotypes. The network also demonstrates high genetic diversity in the archaeological Oregon samples.
The TempNet analysis (figure 3) shows two shared haplotypes through time. The first (haplotype 9) includes three sea otters from Palmrose and two from Par-Tee, and the second (haplotype 13) includes one individual from each site. There are no shared haplotypes between historical and archaeological individuals, despite the proximity of Oregon and northern haplotypes in the network (figure 2). The archaeological Oregon sea otters contain more overall haplotype diversity (Hd = 0.91) compared to the modern California samples (Hd = 0.44).
4. Discussion
(a). Mitogenome haplotype distributions
The mitogenome results provide new insights into archaeological/pre-extirpation Oregon sea otters. As hypothesized, the Oregon sea otter haplotypes are distinct from California haplotypes, and form several clusters with northern haplotypes in the network analysis.
Archaeological Oregon haplotypes 7, 8 and 9 (representing a total of seven individuals from both sites) are more closely related to the Alaska haplotypes (specifically haplotype 5, a 1949 Amchitka Island sea otter). Archaeological haplotypes 10–15 cluster with the historical Washington and BC haplotypes (18 and 21) and historical Oregon haplotype 19, all of which date close to extirpation (ca 1874–1898); this clustering is unsurprising given documented gene flow between northern populations prior to fur trade bottlenecks [16]. Historical Oregon haplotype 20 (ca 1859) is comparatively distant from this historical/archaeological cluster but was collected from Port Orford in southern Oregon, possibly reflecting variation on a latitudinal cline. Archaeological Oregon haplotype 16 is also distinct: it is closely associated with Russia haplotype 17 (collected 1911), and prior studies have indicated gene flow between archaeological Oregon and Russia populations occurred [16]. Interestingly, Japan (haplotype 22, no date) is separated from all other haplotypes including Russia, while the reference genome from a sea otter from the Toba Aquarium in Japan shares haplotype 6 with a 1977 historical Alaska sea otter. Overall, the distribution of haplotypes within the network analysis indicates close associations between the archaeological Oregon sea otters and pre-extirpation sea otters from northern populations, especially those immediately north of Oregon (Washington and BC), as well as the post-fur trade historical Alaska sea otters used for reintroductions.
(b). Pre-contact sea otter acquisition
Previous studies have documented Oregon sea otters sharing traits and experiencing gene flow with northern populations [16,44]. This gene flow along the coast may be responsible for the similar genetic signatures between groups, but animal and/or human behaviour may also be a factor. It is feasible sea otters travelled from southeast Alaska and were hunted in Oregon [53] (yielding the Alaska/Oregon cluster), but it seems unlikely a sea otter from populations further northwest would do so, especially in large numbers. Ethnographic data suggest that Oregon tribes (especially those in the Columbia River trading area) were the source, rather than recipients, of traded pelts [32,76], but it is possible pre-contact groups on the Pacific Rim/northwest coast may have moved animals (or in this case, their teeth or other parts) across long distances through trade networks. For example, the famous whale saddle wood carving from Ozette, WA, is inlaid with more than 700 sea otter teeth, mostly molars [77]. Such teeth could have been traded widely, perhaps as a symbol of the wealth/status associated with sea otter pelts [29,32,33]. In terms of local context, the overlap in haplotypes 9 and 13 between the two archaeological sites (figure 3) suggests the persistence of some mitochondrial lineages through time in the Seaside, OR area. Palmrose and Par-Tee are close geographically and in age so these overlaps are consistent with local sea otter hunting.
Further investigation is required to increase the archaeological, historical and modern mitogenome sample size from locations throughout their former range (especially from BC and Washington). Overall, the genomic results support our hypothesis of greater haplotype diversity in the archaeological populations.
(c). Implications for reintroduction
Prior to sea otter extirpation, the Oregon coast apparently served as a transitional zone between southern and northern phenotypes [42–44] and possibly haplotypes [16,39], and could serve a similar function in the present. The historical samples from Oregon, Washington and BC represent the end of the fur trade (approx. 1850–1900), just prior to extirpation [14,52]. The historical Amchitka Island sea otters (1949–1962 AD) were used for reintroductions to southeast Alaska, BC and Washington in the 1960s-on [25]; our results, therefore, probably reflect current populations in the northern regions. We are, therefore, able to examine the genetic landscape both before and after reintroduction and contextualize the Oregon sea otters therein.
A variety of factors may have contributed to the failed Oregon sea otter reintroductions in the 1970s [25], including the possibility that California (rather than Alaska) sea otters would have been a better stock source [39]. Our results indicate that the picture is more complicated: northern sea otters are closer to the archaeological and historical Oregon sea otters analysed in this study, probably reflecting the northern location of Palmrose and Par-Tee in Oregon. In comparison, the Oregon sea otters used by Valentine et al. [39] came from archaeological sites along the central and southern Oregon coast. Valentine et al. did find two northern haplotypes in their archaeological Oregon sea otters, while the Oregon sea otters analysed in this study did not match California haplotypes. Larson et al. [16] analysed archaeological Oregon sea otters from throughout the Oregon coast and found gene flow occurred both to the south and the north. Taken together, these results strongly point to genetic variation along a latitudinal cline.
In addition to geographical variability, there is a methodological explanation for the difference in results: Valentine et al. [39] used short D-loop sequences following standard protocols at the time, while the analysis presented here used the complete mitogenome. We trimmed the mitogenomes to the 222 bp used by Valentine et al. [39] and performed a new network analysis (electronic supplementary material, figure S1): half of the Oregon sea otters grouped with northern haplotypes, but the other half shifted to group with the modern California haplotypes. The resulting haplotype difference demonstrates the value of whole mitogenomes in fully assessing diversity beyond the D-loop and expanding interpretations based on shorter sequences (further discussion in the electronic supplementary material).
We conclude that while reintroducing primarily California sea otters to the regions analysed by Valentine et al. [39] in southern Oregon may yield better results, we contend that future Oregon reintroduction efforts should include sea otters from Washington, BC and Alaska populations, especially reintroductions occurring on the northern half of the Oregon coast. Including both northern and southern sea otter populations will reflect the hypothesized pre-fur trade hybridization zone between groups [16,42–44], and reintroducing sea otters from multiple source populations may also promote increased genetic diversity [16].
(d). Novel methodological approach
Dental calculus from historical museum specimens, in combination with archaeological dentine/bone analysis, can provide a window into past genetic diversity of extirpated populations. This study is a novel demonstration of the feasibility of successfully extracting and amplifying complete mitogenomes using dental calculus from non-human mammals. Previous analysis has been limited by the number of specimens available for sampling, and future analyses can include archaeological, historical, and modern sea otters from additional locations and in larger numbers by using dental calculus to increase the resolution of genetic patterns. Nuclear genome data (including from dental calculus) may be used to identify specific adaptations [15,78], and other applied methods such as isotopic analyses should be performed to establish past ecological contexts [79,80]. While human dental calculus has been used in genomic analyses as a source of endogenous DNA [61,62], non-human dental calculus has not been used for this purpose and our study is unique in employing this method. Given the degraded nature of sea otter DNA recovered from dental calculus from recent specimens (ca 1983) in this study (electronic supplementary material, figures S4 and S5) and others [64], we recommend using protocols designed specifically for ancient DNA when collecting calculus from skeletonized museum specimens. This approach has great potential to provide genetic data from rare museum specimens without destroying the host tissue in the service of an integrated approach to conservation challenges in the present.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
John Ossosky, Suzanne Peurach, Esther Rimer and Dave Rosenthal at the National Museum of Natural History (Washington, DC) facilitated access to teeth and sampling, as did Jon Erlandson, Pamela Endzweig and Elizabeth Kallenbach at the Museum of Natural and Cultural History (Eugene, OR), and Paul Collins at the Santa Barbara Museum of Natural History (CA). Cara Monroe and LaShanda Williams assisted with laboratory work and Tanvi Honap and Krithi Sankaranarayanan generously provided advice with data analysis. Karissa Hughes, Alexis Mychajliw, Kristen Rayfield and Robin Singleton (LMAMR) and Nelson Ting, Stephen Dueppen, Jessica Stone, Colin Brand, Diana Christie, Alexana Hickmott and Samantha Queeno (UO) provided helpful feedback. Briece Edwards shared suggestions for ethnographic data sources, and Kathy Ralls and Bob Bailey kindly provided input. We thank two anonymous, knowledgeable reviewers for their thorough and thoughtful feedback which helped to improve this manuscript. This work was supported by the University of Oklahoma, the University of Oregon Department of Anthropology Travel Funds and Cheryl Harper Fellowship, and a University of Oregon Graduate School ‘Special Opps’ grant.
Data accessibility
Raw sequence data is available through the NCBI SRA under BioProject accession PRJNA550086. Consensus sequences and the alignment file used for analysis (ModAlign.fa) are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.djh9w0vxz [75]. Specimens analysed in this study are curated at the National Museum of Natural History in Washington, DC, the Museum of Natural and Cultural History in Eugene, OR, and the Santa Barbara Museum of Natural History, CA.
Authors' contributions
H.P.W., C.A.H. and T.C.R. conceived the study. H.P.W. and C.A.H. identified sea otter samples. H.P.W., C.A.H., R.M.A. and N.D.D. conducted the ancient DNA work. M.L.M. contributed financial support and T.C.R. sampled dental calculus. H.P.W. and C.A.H. performed data analysis and wrote the first draft of the manuscript. All authors contributed substantially to the final draft.
Competing interests
We declare we have no competing interests.
References
- 1.Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci. Adv. 1, e1400253 ( 10.1126/sciadv.1400253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong D, Seddon P. 2008. Directions in reintroduction biology. Trends Ecol. Evol. 23, 20–25. ( 10.1016/j.tree.2007.10.003) [DOI] [PubMed] [Google Scholar]
- 3.Hardman B, Moro D, Calver M. 2016. Direct evidence implicates feral cat predation as the primary cause of failure of a mammal reintroduction programme. Ecol. Manag. Restor. 17, 152–158. ( 10.1111/emr.12210) [DOI] [Google Scholar]
- 4.Houde ALS, Garner SR, Neff BD. 2015. Restoring species through reintroductions: strategies for source population selection. Restor. Ecol. 23, 746–753. ( 10.1111/rec.12280) [DOI] [Google Scholar]
- 5.Mulder KP, Walde AD, Boarman WI, Woodman AP, Latch EK, Fleischer RC. 2017. No paternal genetic integration in desert tortoises (Gopherus agassizii) following translocation into an existing population. Biol. Conserv. 210, 318–324. ( 10.1016/j.biocon.2017.04.030) [DOI] [Google Scholar]
- 6.Rathbun GB, Hatfield BB, Murphey TG. 2000. Status of translocated sea otters at San Nicolas Island, California. Southwest. Nat. 45, 322 ( 10.2307/3672835) [DOI] [Google Scholar]
- 7.Bell DA, Robinson ZL, Funk WC, Fitzpatrick SW, Allendorf FW, Tallmon DA, Whiteley AR. 2019. The exciting potential and remaining uncertainties of genetic rescue. Trends Ecol. Evol. 34, 1070–1079. ( 10.1016/j.tree.2019.06.006) [DOI] [PubMed] [Google Scholar]
- 8.Flanagan SP, Forester BR, Latch EK, Aitken SN, Hoban S. 2018. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol. Appl. 11, 1035–1052. ( 10.1111/eva.12569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He X, Johansson ML, Heath DD. 2016. Role of genomics and transcriptomics in selection of reintroduction source populations. Conserv. Biol. 30, 1010–1018. ( 10.1111/cobi.12674) [DOI] [PubMed] [Google Scholar]
- 10.Frankham R, Ballou JD, Ralls K, Eldridge M, Dudash MR, Fenster CB, Lacy RC, Sunnucks P. 2017. Genetic management of fragmented animal and plant populations. Oxford, UK: Oxford University Press. [Google Scholar]
- 11.Marr MM, Brace S, Schreve DC, Barnes I. 2018. Identifying source populations for the reintroduction of the Eurasian beaver, Castor fiber L. 1758, into Britain: evidence from ancient DNA. Sci. Rep. 8, 2708 ( 10.1038/s41598-018-21173-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson S, Jameson R, Etnier M, Fleming M, Bentzen P. 2002. Loss of genetic diversity in sea otters (Enhydra lutris) associated with the fur trade of the 18th and 19th centuries. Mol. Ecol. 11, 1899–1903. ( 10.1046/j.1365-294X.2002.01599.x) [DOI] [PubMed] [Google Scholar]
- 13.Larson SE, Bodkin JL. 2015. The conservation of sea otters: a prelude. In Sea otter conservation (eds Larson S, Bodkin J, VanBlaricom G), pp. 1–17. London, UK: Elsevier Science. [Google Scholar]
- 14.Kenyon KW. 1969. The sea otter in the eastern Pacific Ocean. New York, NY: Dover Publications. [Google Scholar]
- 15.Beichman AC, et al. 2019. Aquatic adaptation and depleted diversity: a deep dive into the genomes of the sea otter and giant otter. Mol. Biol. Evol. 36, 2631–2655. ( 10.1093/molbev/msz101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larson S, Jameson R, Etnier M, Jones T, Hall R. 2012. Genetic diversity and population parameters of sea otters, Enhydra lutris, before fur trade extirpation from 1741–1911. PLoS ONE 7, e32205 ( 10.1371/journal.pone.0032205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson SE, Ralls K, Ernest H. 2015. Sea otter conservation genetics. In Sea otter conservation (eds Larson S, Bodkin J, VanBlaricom G), pp. 97–120. London, UK: Elsevier. [Google Scholar]
- 18.Ralls K, McInerney NR, Gagne RB, Ernest HB, Tinker MT, Fujii J, Maldonado J. 2017. Mitogenomes and relatedness do not predict frequency of tool-use by sea otters. Biol. Lett. 13, 20160880 ( 10.1098/rsbl.2016.0880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estes JA, Palmisano JF. 1974. Sea otters: their role in structuring nearshore communities. Science 185, 1058–1060. ( 10.1126/science.185.4156.1058) [DOI] [PubMed] [Google Scholar]
- 20.Gregr EJ, et al. 2020. Cascading social-ecological costs and benefits triggered by a recovering keystone predator. Science 368, 1243–1247. ( 10.1126/science.aay5342) [DOI] [PubMed] [Google Scholar]
- 21.Salomon AK, Wilson KBJ, White XE, Tanape N, Happynook TM. 2015. First nations perspectives on sea otter conservation in British Columbia and Alaska. In Sea otter conservation (eds Larson S, Bodkin J, VanBlaricom G), pp. 301–331. London, UK: Elsevier. [Google Scholar]
- 22.Jones RT. 2014. Empire of extinction: Russians and the North Pacific's strange beasts of the sea, 1741–1867. Oxford, UK: Oxford University Press. [Google Scholar]
- 23.Igler D. 2013. The great ocean: Pacific worlds from Captain Cook to the gold rush. Oxford, UK: Oxford University Press. [Google Scholar]
- 24.Bodkin JL. 2015. Historic and contemporary status of sea otters in the North Pacific. In Sea otter conservation (eds Larson S, Bodkin J, VanBlaricom G), pp. 43–62. London, UK: Elsevier Science. [Google Scholar]
- 25.Jameson RJ, Kenyon KW, Johnson AM, Wight HM. 1982. History and status of translocated sea otter populations in North America. Wildl. Soc. Bull. (1973–2006) 10, 100–107. [Google Scholar]
- 26.Doroff A, Burdin A. 2015. Enhydra lutris, The IUCN Red List of Threatened Species 2015 (cited 2 July 2020). See https://www.iucnredlist.org/species/7750/21939518.
- 27.Elakha Alliance. 2020. Elakha Alliance [Internet]. 2020 (cited 2 July 2020). See elakhaalliance.org.
- 28.Kone DV. 2020. An ecological assessment of a potential sea otter (Enhydra lutris) reintroduction to the Oregon coast. Masters thesis, Oregon State University, Corvallis, OR, USA. [Google Scholar]
- 29.Bailey V. 1936. Mammals and life zones of Oregon. Washington, DC: United States Department of Agriculture Bureau of Biological Survey. North American Fauna. [Google Scholar]
- 30.Hall RL, Dick R, Dave H, The Elakha Alliance. 2019. Reflections on forty years of Oregon archaeology, papers in honor of Dr Richard E (Dick) Ross, 1932–2017, vol. 10, pp. 113–129. Portland, OR: Association of Oregon Archaeologists.
- 31.Lyman RL. 1991. The prehistory of the Oregon coast: effects of excavation strategies and assemblage size on archaeological inquiry. San Diego, CA: Academic Press. [Google Scholar]
- 32.Ray VF. 1938. Lower Chinook ethnographic notes, vol. 7 Seattle, WA: University of Washington; University of Washington Publications in Anthropology. [Google Scholar]
- 33.Sauter J, Johnson B. 1974. Tillamook Indians of the Oregon coast. Portland, OR: Binford and Morts. [Google Scholar]
- 34.Burt JM, Wilson KI, Malchoff T, Mack WT, Davidson SH, Salomon AK. 2020. Enabling coexistence: navigating predator-induced regime shifts in human-ocean systems. People Nat. 2, 557–574. ( 10.1002/pan3.10090) [DOI] [Google Scholar]
- 35.Moss ML. 2020. Did Tlingit ancestors eat sea otters? Addressing intellectual property and cultural heritage through zooarchaeology. Am. Antiq. 85, 202–221. ( 10.1017/aaq.2019.101) [DOI] [Google Scholar]
- 36.Salomon AK, Burt JM, Herb I, Wilson KB, Happynook H-YT, Davidson SHA et al. 2018. Coastal voices (cited 15 May 2020). See coastalvoices.net.
- 37.Carswell LP, Speckman SG, Gill VA. 2015. Shellfish fishery conflicts and perceptions of sea otters in California and Alaska. In Sea otter conservation (eds Larson S, Bodkin J, VanBlaricom G), pp. 333–368. London, UK: Elsevier Science. [Google Scholar]
- 38.Larson SD, Hoyt ZN, Eckert GL, Gill VA. 2013. Impacts of sea otter (Enhydra lutris) predation on commercially important sea cucumbers (Parastichopus californicus) in southeast Alaska. Can. J. Fish. Aquat. Sci. 70, 1498–1507. ( 10.1139/cjfas-2013-0025) [DOI] [Google Scholar]
- 39.Valentine K, Duffield DA, Patrick LE, Hatch DR, Butler VL, Hall RL, Lehman N. 2008. Ancient DNA reveals genotypic relationships among Oregon populations of the sea otter (Enhydra lutris). Conserv. Genet. 9, 933–938. ( 10.1007/s10592-007-9422-z) [DOI] [Google Scholar]
- 40.Cronin MA, Bodkin J, Ballachey B, Estes J, Patton JC. 1996. Mitochondrial-DNA variation among subspecies and populations of sea otters (Enhydra lutris). J. Mammal. 77, 546–557. ( 10.2307/1382828) [DOI] [Google Scholar]
- 41.Larson S, Jameson R, Bodkin J, Staedler M, Bentzen P. 2002. Microsatellite DNA and mitochondrial DNA variation in remnant and translocated sea otter (Enhydra lutris) populations. J. Mammal. 83, 893–906. () [DOI] [Google Scholar]
- 42.Lyman R. 1988. Zoogeography of Oregon coast marine mammals: the last 3,000 years. Marine Mammal Sci. 4, 247–264. ( 10.1111/j.1748-7692.1988.tb00205.x) [DOI] [Google Scholar]
- 43.Wellman HP. 2018. Applied zooarchaeology and Oregon Coast sea otters (Enhydra lutris). Marine Mammal Sci. 34, 806–822. ( 10.1111/mms.12484) [DOI] [Google Scholar]
- 44.Wilson DE, Bogan MA, Brownell RL, Burdin AM, Maminov MK. 1991. Geographic variation in sea otters, Enhydra lutris. J. Mammal. 72, 22–36. ( 10.2307/1381977) [DOI] [Google Scholar]
- 45.Sanchez GM, Rick TC, Culleton BJ, Kennett DJ, Buckley M, Erlandson JM, Losey RL. 2018. Radiocarbon dating legacy collections: a Bayesian analysis of high-precision AMS 14C dates from the Par-Tee site, Oregon. J. Archaeol. Sci. Rep. 21, 833–848. ( 10.1016/j.jasrep.2018.08.033) [DOI] [Google Scholar]
- 46.Connolly TJ. 1995. Archaeological evidence for a former bay at Seaside, Oregon. Quat. Res. 43, 362–369. ( 10.1006/qres.1995.1042) [DOI] [Google Scholar]
- 47.Connolly TJ. 1992. Human responses to change in coastal geomorphology and fauna on the southern northwest coast: archaeological investigations at Seaside, Oregon. University of Oregon; University of Oregon Anthropological Papers.
- 48.Phebus GE, Drucker RM. 1979. Archaeological investigations in Seaside, Oregon: an intermediate report on the excavations of two major archaeological sites at seaside, Oregon, through September 1977. Seaside, OR: Seaside Museum and Historical Society.
- 49.Sanchez GM, Erlandson JM, Culleton BJ, Kennett DJ, Rick TC. 2016. High-resolution AMS 14C dates for the Par-Tee site (35CLT20) and prehistoric whale hunting on the Oregon Coast. Radiocarbon 58, 397–405. ( 10.1017/RDC.2016.10) [DOI] [Google Scholar]
- 50.Wellman HP, Rick TC, Rodrigues AT, Yang DY. 2017. Evaluating ancient whale exploitation on the northern Oregon coast through ancient DNA and zooarchaeological analysis. J. Island Coast. Archaeol. 12, 255–275. ( 10.1080/15564894.2016.1172382) [DOI] [Google Scholar]
- 51.Losey RJ, Yang DY. 2007. Opportunistic whale hunting on the southern northwest coast: ancient DNA, artifact, and ethnographic evidence. Am. Antiq. 72, 657–676. ( 10.2307/25470439) [DOI] [Google Scholar]
- 52.Scheffer VB. 1940. The sea otter on the Washington Coast. Pac. Northwest Quart. 31, 370–388. [Google Scholar]
- 53.Ralls K, Eagle TC, Siniff DB. 1996. Movement and spatial use patterns of California sea otters. Can. J. Zool. 74, 1841–1849. ( 10.1139/z96-207) [DOI] [Google Scholar]
- 54.Garshelis DL, Garshelis JA. 1984. Movements and management of sea otters in Alaska. J. Wildl. Manage. 48, 665–678. ( 10.2307/3801414) [DOI] [Google Scholar]
- 55.Smith EAE, Newsome SD, Estes JA, Tinker MT. 2015. The cost of reproduction: differential resource specialization in female and male California sea otters. Oecologia 178, 17–29. ( 10.1007/s00442-014-3206-1) [DOI] [PubMed] [Google Scholar]
- 56.Loughlin TR. 1980. Home range and territoriality of sea otters near Monterey, California. J. Wildl. Manage. 44, 576 ( 10.2307/3808005) [DOI] [Google Scholar]
- 57.Tarjan LM, Tinker MT. 2016. Permissible Home Range Estimation (PHRE) in restricted habitats: a new algorithm and an evaluation for sea otters. PLoS ONE 11, e0150547 ( 10.1371/journal.pone.0150547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gagne RB, Tinker MT, Gustafson KD, Ralls K, Larson S, Tarjan LM, Miller MA, Ernest HB. 2018. Measures of effective population size in sea otters reveal special considerations for wide-ranging species. Evol. Appl. 11, 1779–1790. ( 10.1111/eva.12642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang DY, Watt K. 2005. Contamination controls when preparing archaeological remains for ancient DNA analysis. J. Archaeol. Sci. 32, 331–336. ( 10.1016/j.jas.2004.09.008) [DOI] [Google Scholar]
- 60.Shapiro B, Hofreiter M (eds). 2012. Ancient DNA methods and protocols. Totowa, NJ: Humana Press; (Methods in Molecular Biology; vol. 840). [Google Scholar]
- 61.Ozga AT, Nieves-Colón MA, Honap TP, Sankaranarayanan K, Hofman CA, Milner GR, Lewis CM, Stone AC, Warinner C. 2016. Successful enrichment and recovery of whole mitochondrial genomes from ancient human dental calculus. Am. J. Phys. Anthropol. 160, 220–228. ( 10.1002/ajpa.22960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziesemer KA, et al. 2019. The efficacy of whole human genome capture on ancient dental calculus and dentin. Am. J. Phys. Anthropol. 168, 496–509. ( 10.1002/ajpa.23763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mann AE, et al. 2018. Differential preservation of endogenous human and microbial DNA in dental calculus and dentin. Sci. Rep. 8, 9822 ( 10.1038/s41598-018-28091-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Austin RM, Sholts SB, Williams L, Kistler L, Hofman CA. 2019. Opinion: to curate the molecular past, museums need a carefully considered set of best practices. Proc. Natl. Acad. Sci. USA 116, 1471–1474. ( 10.1073/pnas.1822038116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morales-Arce AY, Hofman CA, Duggan AT, Benfer AK, Katzenberg MA, McCafferty G, Warinner C. 2017. Successful reconstruction of whole mitochondrial genomes from ancient Central America and Mexico. Sci. Rep. 7, 18100 ( 10.1038/s41598-017-18356-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schubert M, Lindgreen S, Orlando L. 2016. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 1–7. ( 10.1186/s13104-016-1900-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. ( 10.1093/bioinformatics/btp698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yonezawa T, Nikaido M, Kohno N, Fukumoto Y, Okada N, Hasegawa M. 2007. Molecular phylogenetic study on the origin and evolution of Mustelidae. Gene 396, 1–12. ( 10.1016/j.gene.2006.12.040) [DOI] [PubMed] [Google Scholar]
- 69.Ginolhac A. 2011. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics 27, 2153–2155. ( 10.1093/bioinformatics/btr347) [DOI] [PubMed] [Google Scholar]
- 70.Katoh K. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. ( 10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software Version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. ( 10.1093/oxfordjournals.molbev.a026036) [DOI] [PubMed] [Google Scholar]
- 73.Leigh JW,, Bryant D. 2015. popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. ( 10.1111/2041-210X.12410) [DOI] [Google Scholar]
- 74.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. ( 10.1093/molbev/msx248) [DOI] [PubMed] [Google Scholar]
- 75.Wellman HP, Austin RM, Dagtas ND, Moss ML, Rick TC, Hofman CA. 2020. Data from: Archaeological mitogenomes illuminate the historical ecology of sea otters (Enhydra lutris) and the viability of reintroduction. Dryad Digital Repository ( 10.5061/dryad.djh9w0vxz) [DOI] [PMC free article] [PubMed]
- 76.Zobel DB. 2002. Ecosystem use by indigenous people in an Oregon coastal landscape. Northwest Sci. 76, 304–314. [Google Scholar]
- 77.Kirk R, Daugherty RD. 1974. Hunters of the whale: an adventure in northwest coast archaeology. New York: NY: William Morrow and Company. [Google Scholar]
- 78.Davis RW, Bodkin JL, Coletti HA, Monson DH, Larson SE, Carswell LP, Nichol LM. 2019. Future directions in sea otter research and management. Front Mar. Sci. 5, 510 ( 10.3389/fmars.2018.00510) [DOI] [Google Scholar]
- 79.Elliott Smith EA, et al. 2020. Reductions in the dietary niche of southern sea otters (Enhydra lutris nereis) from the Holocene to the Anthropocene. Ecol. Evol. 10, 3318–3329. ( 10.1002/ece3.6114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szpak P, Orchard TJ, McKechnie I, Gröcke DR. 2012. Historical ecology of late Holocene sea otters (Enhydra lutris) from northern British Columbia: isotopic and zooarchaeological perspectives. J. Archaeol. Sci. 39, 1553–1571. ( 10.1016/j.jas.2011.12.006) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wellman HP, Austin RM, Dagtas ND, Moss ML, Rick TC, Hofman CA. 2020. Data from: Archaeological mitogenomes illuminate the historical ecology of sea otters (Enhydra lutris) and the viability of reintroduction. Dryad Digital Repository ( 10.5061/dryad.djh9w0vxz) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Raw sequence data is available through the NCBI SRA under BioProject accession PRJNA550086. Consensus sequences and the alignment file used for analysis (ModAlign.fa) are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.djh9w0vxz [75]. Specimens analysed in this study are curated at the National Museum of Natural History in Washington, DC, the Museum of Natural and Cultural History in Eugene, OR, and the Santa Barbara Museum of Natural History, CA.