SUMMARY
Fibroblast heterogeneity has been shown within the unwounded mouse dorsal dermis, with fibroblast subpopulations being identified according to anatomical location and embryonic lineage. Using lineage tracing, we demonstrate that paired related homeobox 1 (Prrx1)-expressing fibroblasts are responsible for acute and chronic fibroses in the ventral dermis. Single-cell transcriptomics further corroborated the inherent fibrotic characteristics of Prrx1 fibroblasts during wound repair. In summary, we identify and characterize a fibroblast subpopulation in the mouse ventral dermis with intrinsic scar-forming potential.
In Brief
Fibroblasts in the mouse dermis are heterogeneous, but it is unclear which subpopulation contributes to ventral scarring. Using lineage tracing and single-cell transcriptomics, Leavitt et al. report that Prrx1-expressing fibroblasts are largely responsible for fibrosis in the ventral dermis during wound repair.
Graphical Abstract
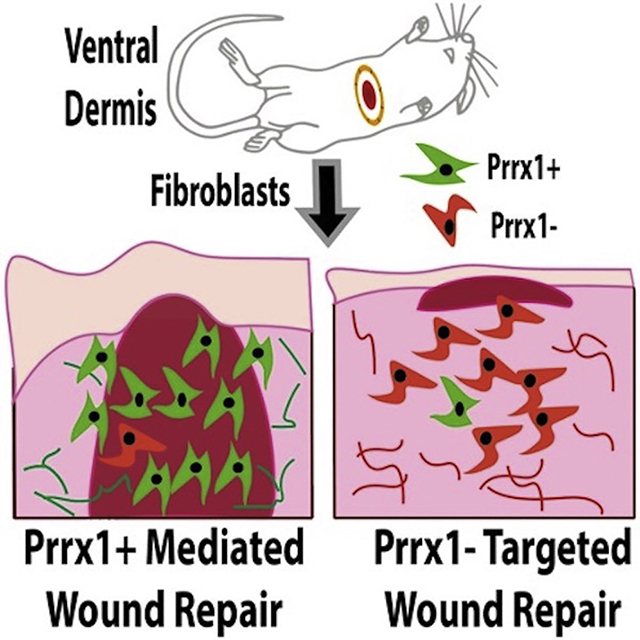
INTRODUCTION
Recent studies investigating the functional heterogeneity of dermal fibroblasts have described a number of distinct fibroblast subpopulations with differing fibrogenic potential, anatomical location, dermal niche, and cell lineage (Philippeos et al., 2018; Driskell and Watt, 2015; Driskell et al., 2013; Rinkevich et al., 2015; Tabib et al., 2018; Sorrell and Caplan, 2004; Millar, 2018; Marsh et al., 2018; Salzer et al., 2018; Shook et al., 2018; Lynch and Watt, 2018; Chang et al., 2002; Korosec et al., 2018). We previously identified a scar-forming fibroblast subpopulation in mouse dorsal dermis, marked by embryonic expression of Engrailed-1 (En1) and demonstrated that targeted depletion of these En1-positive fibroblasts (EPFs) reduces cutaneous scarring following wounding (Rinkevich et al., 2015). The adult dermis develops from anatomically distinct embryological origins, and fibroblast heterogeneity is increasingly recognized as a key driver in dermal development. However, most work to date has focused on fibroblasts in the dorsal dermis, which is derived from paraxial mesoderm (Driskell and Watt, 2015). Comparatively little is known about fibroblasts of the ventral dermis, derived from lateral plate mesoderm. Investigations into mouse dermal fibroblast heterogeneity have largely focused on unwounded skin, with limited exploration into fibroblast behavior in healing/healed wounds. Furthermore, the dermal fibroblast response throughout wound healing has not been studied extensively at a single-cell level (Guerrero-Juarez et al., 2019). Collectively, these studies have yet to fully elucidate contributions of fibroblast subpopulations during different stages of wound repair (Driskell et al., 2013).
Unlike humans, mice are quadrupedal; their dorsum is at greatest risk from trauma or attack. The human ventral dermis, by contrast, is significantly more exposed and at higher risk of injury. Nearly 2 million abdominal surgeries are performed in the US every year, each involving one or more ventral incisions (Carney et al., 2017). Furthermore, the chest has high skin tension and is among the locations most prone to hypertrophic scars and keloids (Ogawa et al., 2012). As such, elucidating fibroblast heterogeneity within the mouse ventral dermis has potentially important translational implications for humans.
Paired related homeobox 1 (Prrx1) is a transcription factor with a critical role in limb development (Martin and Olson, 2000; McKean et al., 2003). During amphibian limb regeneration, Prrx1 is specifically expressed in migrating fibroblasts on the ventral surface (McKean et al., 2003; Jones et al., 1999; Suzuki et al., 2005; Satoh et al., 2007; Jones et al., 2001). In vitro, activation of Prrx1 in skin fibroblasts is mediated through the integrin/focal adhesion kinase (FAK) signaling pathway in a manner conserved between humans and axolotls (McKean et al., 2003; Satoh et al., 2011). Recent work demonstrated the existence of Prrx1-positive fibroblasts (PPFs) within the adult mouse dermis, located in perivascular and hair follicle niches, that may expand in response to injury (Currie et al., 2019). We hypothesized that PPFs may be a profibrotic fibroblast lineage within the ventral dermis.
Until recently, limited precision of high-throughput single-cell analytical tools has challenged characterization of cell populations within heterogeneous tissue niches such as wounds. Here, we combine cell lineage tracing with high-throughput protein-barcoded single-cell RNA sequencing (scRNA-seq) to demonstrate that PPFs and Prrx1-negative fibroblasts (PNFs) represent two distinct fibroblast lineages in the mouse ventral dermis, with changes in relative abundance and fibrotic properties throughout development and during wound healing.
RESULTS
Distinct Fibroblast Lineages Are Present in the Ventral Skin
Lineage tracing of fibroblasts derived from Prrx1-expressing progenitors was performed by crossing Prrx1Cre mice with dual-fluorescent ROSA26mTmG (R26mTmG) reporter mice (Rinkevich et al., 2015). In resulting Prrx1Cre;R26mTmG offspring, PPFs express membrane-bound green fluorescent protein (GFP) expression secondary to embryonic Cre-mediated recombination. The remaining fibroblasts (PNFs) express membrane-bound tdTomato red fluorescent protein (RFP) (Figures 1A and S1A).
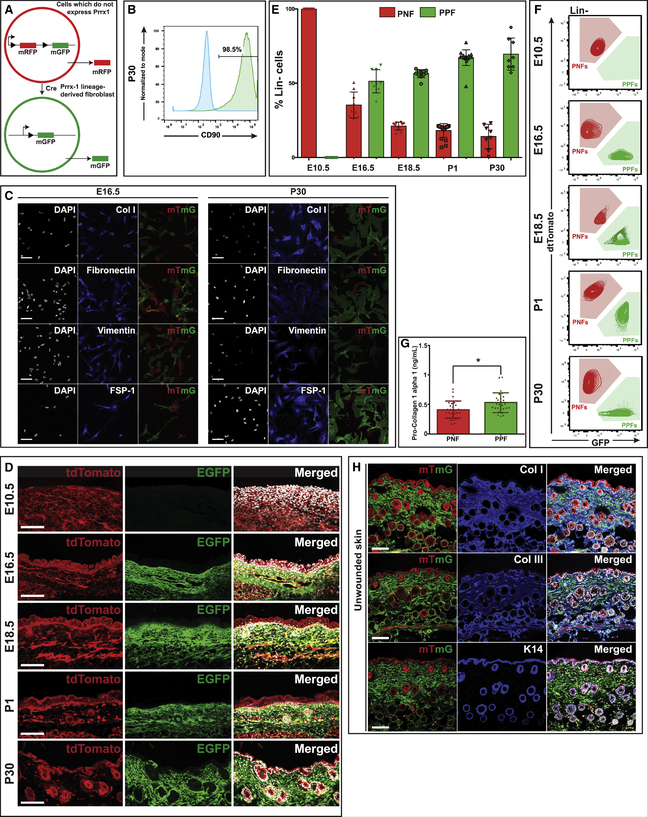
Figure 1. PPFs and PNFs Represent Two Distinct Lineages of Fibroblasts within the Ventral Dermis.
(A) Schematic demonstrating mTmG reporter system. PNFs do not express Prrx1 and are thus red due to production of the membrane monomeric red fluorescent protein (mRFP) versus PPFs (Prrx1 lineage-derived fibroblasts) that are green due to production of the membrane monomeric green fluorescent protein (mGFP).
(B) FACS analysis of PPFs harvested from ventral skin of P30 Prrx1Cre;R26mTmG mice, demonstrating expression of characteristic fibroblast marker CD90 (Thy-1) (n = 6).
(C) Immunohistochemical staining of fibroblasts (Lin− PPFs, green; Lin− PNFs, red) FACS isolated from E16.5 (left) and P30 (right) Prrx1Cre;R26mTmG mice and cultured in vitro. Immunostaining for collagen type I (first row), fibronectin (second row), vimentin (third row), and fibroblast-specific protein-1 (FSP-1) (fourth row). DAPI (white; left columns); fibroblast markers (blue; middle columns); tdTomato and EGFP merged (red and green, respectively; right columns). Scale bar, 200 μm (n = 6).
(D) Histological analysis of ventral skin harvested from Prrx1Cre;R26mTmG mice at E10.5, E16.5, E18.5, P1, and P30 demonstrating absence of PPFs at E10.5 and increasing presence of PPFs throughout the ventral dermis at later time points. tdTomato (left column), EGFP (middle column), and merged with DAPI (white) (right column). Scale bar, 100 μm (n = 9).
(E) Bar graph showing the percentage of Lin− cells that are PPFs versus PNFs as assessed by FACS in ventral skin from Prrx1Cre;R26mTmG mice at E10.5, E16.5, E18.5, P1, and P30 (n = 9).
(F) Corresponding FACS plots showing relative abundance of PPFs (Lin−GFP+) and PNFs (Lin−tdTomato+) in ventral skin from Prrx1Cre;R26mTmG mice at E10.5, E16.5, E18.5, P1, and P30 (n = 9).
(G) Intracellular pro-collagen 1-alpha 1 in freshly isolated PPFs from unwounded skin of P30 Prrx1Cre;R26mTmG mice was significantly greater than that in PNFs (*p = 0.014) (n = 6).
(H) Immunohistochemical analysis of ventral skin from P30 Prrx1Cre;R26mTmG mice demonstrating overlap of PPFs (green) with secreted collagens type I (top row) and type III (middle row). No overlap is observed with keratin 14 staining (bottom row), which localizes to the epidermis and surrounding hair follicles; tdTomato and EGFP (red and green, respectively; left column); immunostaining for ECM markers (blue; middle column); merged with DAPI (white; right column). Scale bar, 100 μm (n = 9). Data are represented as mean ± SD.
PPFs and PNFs were isolated from Prrx1Cre;R26mTmG ventral skin using fluorescence-activated cell sorting (FACS). As previously described for dorsal dermis, a negative gating strategy was used to exclude non-mesenchymal lineages (vascular, hematopoietic, epidermal, etc.) in order to prevent potential enrichment/loss of fibroblast subtypes. PPFs and PNFs are defined as Lin−GFP+RFP− and Lin−GFP−RFP+, respectively (Figure S1B). Fibroblast populations can thus be isolated without requiring cell culture (Rinkevich et al., 2015). To confirm that Prrx1 expression represents a distinct ventral fibroblast lineage, we compared Prrx1 expression to En1 using En1Cre;R26mTmG mice (En1 identifies scarring fibroblasts within the dorsal dermis; Rinkevich et al., 2015). Fluorescence imaging revealed that En1 expression is confined to dorsal skin (Figure S1C), while Prrx1 is expressed ventrally (Figure S1A).
Preliminary analysis of PPFs isolated from post-natal day 30 (P30) mouse ventral skin revealed positive expression of CD90 (thymus cell antigen 1 [Thy-1]), a well-described fibroblast marker (Saalbach et al., 1998) in 98.5% of cells (Figure 1B). In vitro characterization of PPFs and PNFs from embryonic day 16.5 (E16.5) and P30 ventral skin using immunocytochemistry revealed that both PPFs and PNFs stain positively for two extracellular matrix (ECM) components commonly synthesized by fibroblasts, collagen type I (Col I) and fibronectin, and express the fibroblast-associated markers vimentin and fibroblast-specific protein-1 (FSP-1), supporting a fibroblast identity (Figure 1C).
PPFs Produce the Majority of Collagen in Unwounded Skin
We next sought to understand how PPF and PNF populations change in relative abundance throughout dermal development, as dermal collagen content and scar-forming potential increase (Figures 1D–1G and S1B). PPFs are absent from ventral skin at E10.5, but by E16.5 they represent more than half (51.3% ± 7.8%) of ventral dermal fibroblasts (Figure 1E). PPFs continue to increase in prevalence through post-natal development (Figure 1E). Bulk RNA sequencing (RNA-seq) of PPFs and PNFs isolated from unwounded ventral skin confirmed that Prrx1 mRNA expression is higher in PPFs than in PNFs, is present at E16.5 and E18.5, and increases post-natally (Figure S1D).
Interestingly, PPFs FACS isolated from adult (P30) unwounded ventral dermis had significantly more intracellular collagen than PNFs (*p = 0.014) (Figure 1G). Immunohistochemical staining demonstrated high expression of collagen types I and III (Col I and Col III, respectively) in PPFs (Figure 1H, top and middle rows) (Weber et al., 1984). Conversely, there was no observed overlap between PPFs and keratinocyte marker keratin 14 (K14) (Figure 1H, bottom row). Together, these findings suggest that PPFs may represent the fibroblast subpopulation primarily responsible for dermal ECM production in unwounded adult ventral skin.
PPFs Contribute to Scar Formation in Acute Fibrosis
We next sought to examine PPFs’ contribution to ECM production during wound healing. Splinted full-thickness excisional wounds were created in the ventral skin of adult (P30) Prrx1Cre;R26mTmG mice (Figure 2Ai; Galiano et al., 2004). FACS analysis of wounds on postoperative day 14 (POD 14), the point of complete healing, demonstrated that PPFs represent 90.8% ± 3.5% of the lineage negative (Lin−) population in wounded skin (****p < 0.0001 versus PNFs [2.2% ± 1.3%]) (Figure 2Aii). Histology confirmed that while POD 14 scars contained some PNFs (2.2%), the vast majority of cells in scar were PPFs (90%) (Figure 2Aiii).
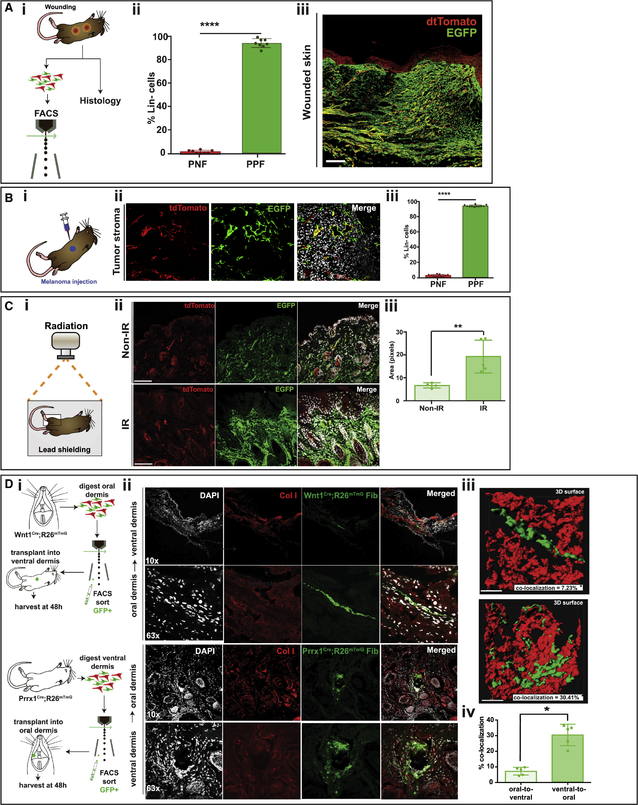
Figure 2. PPFs Are Responsible for the Majority of Connective Tissue Deposition within the Ventral Dermis and Are Key Mediators of the Cutaneous Fibrotic Response.
(A) (i) Schematic depicting the strategy for wounding and assessment of PPFs and PNFs histologically and by FACS in P30 Prrx1Cre;R26mTmG mice. (ii) Bar graph showing the abundance of PPFs versus PNFs within healed (POD 14) ventral wounds of P30 Prrx1Cre;R26mTmG mice, as assessed by FACS (n = 8) (90.83 ± 3.49 versus 2.23 ± 1.29; ****p < 0.0001). (iii) Representative histology of a fully healed (POD 14) ventral wound from P30 Prrx1Cre;R26mTmG mouse, demonstrating abundance of PPFs (EGFP+) within the scar. Scale bar, 100 μm (n = 8).
(B) (i) Schematic depicting the strategy for assessment of PPFs and PNFs histologically and by FACS within tumor stroma. Melanoma cells were injected into ventral skin of P30 Prrx1Cre;R26mTmG mice, and the injected area was harvested 10 days later. (ii) Histological analysis revealed the majority of tumor stroma was comprised of PPFs (EGFP+). tdTomato (red; left); EGFP (green; middle); EGFP merged with DAPI (DAPI in white; right). Scale bar, 100 μm (n = 10). (iii) Bar graph showing abundance of PPFs versus PNFs in tumor stroma of P30 Prrx1Cre;R26mTmG mice 10 days after melanoma cell injection (n = 11) as assessed by FACS (94.14 ± 1.21 versus 2.32 ± 1.27; ****p < 0.0001).
(C) (i) Schematic showing the overall strategy for assessing PPFs and PNFs in ventral skin of P30 Prrx1Cre;R26mTmG mice that developed chronic fibrosis following irradiation. (ii) Histological analysis revealed expansion of PPFs (EGFP+) in the irradiated (IR; right) (bottom row) compared with the non-irradiated (non-IR; left) limb (top row). tdTomato (red; left); EGFP (green; middle); tdTomato and EGFP merged with DAPI (DAPI in white; right). Scale bar, 50 μm (n = 10). (iii) Bar graph showing comparison in pixels positive for EGFP in the IR (right) (bottom row) compared with the non-IR (left) limb showing greater EGFP positivity in irradiated limbs (**p < 0.001) (n = 5).
(D) (i) Schematic depicting the strategy for determining whether PPF fibrogenic potential is cell intrinsic. Wnt1-positive fibroblasts (WPFs) were FACS isolated from oral (non-scarring) dermis of Wnt1Cre;R26mTmG mice (expressing EGFP) and transplanted into the ventral (scarring) dermis of recipient C57BL/6 wild-type (WT) mice. PPFs were FACS isolated from ventral dermis of Prrx1Cre;R26mTmG mice (expressing EGFP) and transplanted into the oral dermis of recipient WT mice. The ventral and oral dermis of recipient mice, respectively, was harvested 48 h later for histological analysis. (ii) Histological analysis of WPFs (green) transplanted in the ventral dermis of recipient WT mice at 103 (top row) and 633 (second row) magnifications, and PPFs transplanted into the oral dermis of recipient WT mice at 103 (third row) and 633 (bottom row) magnifications. DAPI (white; first column); collagen I (red; second column); WPFs (green; third column top two rows), PPFs (green; third column bottom two rows); merged (fourth column), with (iii) 3D surface-rendered images of transplanted WPFs (top) and PPFs (bottom) into the ventral and oral dermis, respectively, of recipient WT mice at 633 magnification (n = 5). (iv) Comparison of colocalization of transplanted WPFs and PPFs with collagen I fibers by Imaris revealed significantly greater colocalization with transplanted PPFs (right column) in the oral dermis compared with WPFs (left column) in the ventral dermis (*p = 0.02) (n = 5) (Videos S1 and S2). Data are represented as mean ± SD.
Furthermore, we aimed to evaluate relative contributions of PPFs and PNFs throughout wound healing and how these lineage-defined fibroblasts related to surface marker-defined subpopulations (Figure S2; Driskell et al., 2013). Contributions of papillary, lipo-, and reticular fibroblasts in unwounded skin have been described previously (Driskell et al., 2013), but their role in wounds over time remains unclear. During mouse wound repair, fibroblasts migrate into excisional wounds around POD 3–4, proliferate, deposit fibrotic matrix, and decrease in number by the time the wound is fully healed and remodeled (Landén, Li and Ståhle, 2016). We sought to examine PPFs and PNFs by FACS using surface markers previously described for candidate fibroblast subpopulations: papillary (Lin−Sca1−CD26+), reticular (Lin−Sca1−Dlk1+), and lipofibroblasts (Lin−Sca1+) (Figure S2A; Driskell et al., 2013).
Prrx1 lineage expression was present in papillary, reticular, and lipofibroblasts in unwounded and wounded skin. Relative proportions of PPF and PNF papillary fibroblasts were similar from POD 0 to POD 14 compared with unwounded skin (Figure S2Bi). By contrast, both PPF and PNF lipofibroblasts showed a significant increase at POD 4, followed by a marked decrease at POD 9 compared with unwounded skin (****p < 0.001) (Figure S2Bii). However, only the proportion of PPF reticular fibroblasts significantly increased at POD 9. By contrast, reticular PNF fibroblasts decreased over time compared with unwounded skin (****p < 0.0001) (Figure S2Biii). These findings suggest that PPF and PNF behavior in wound healing may vary in a modular fashion based on intrinsic transcriptional programming.
PPFs Contribute to Scar Formation in Chronic Fibrosis
We sought to determine whether PPFs also play a role in chronic dermal fibrosis by examining two well-studied fibrotic processes: tumor stroma formation in melanoma and cutaneous fibrosis associated with irradiation (Labrousse et al., 2004). In melanoma, local fibroblasts are recruited to support and promote tumor growth (Labrousse et al., 2004; Nakhleh et al., 1990; Garza et al., 2014). We injected cancer cells derived from malignant melanoma cell lines (B16-F10) into the ventral dermis of Prrx1Cre;R26mTmG mice and harvested the resulting tumors after 10 days (Figure 2Bi). Histological analysis demonstrated that the desmoplastic tumor stroma associated with the melanoma cells was almost entirely made up of PPFs (Figure 2Bii). FACS analysis confirmed that, similar to wounds, PPFs represented 94.1% ± 1.2% of the Lin− population in tumors (****p < 0.0001 versus PNFs [2.32% ± 1.27%]) (Figure 2Biii).
We also explored the contribution of PPFs to fibrotic tissue that develops following irradiation. The ventral right hindlimbs of adult (P30) Prrx1Cre;R26mTmG mice were irradiated and harvested after 1 month, allowing chronic effects of irradiation to develop (Figure 2Ci; Garza et al., 2014). Histological assessment of irradiated hindlimbs showed that the vast majority of fibrotic dermis was made up of PPFs (Figure 2Cii, bottom row), compared with non-irradiated hindlimb skin (Figure 2Cii, top row). Quantification of pixel density for GFP revealed significantly more EGFP+ pixels in the irradiated compared with non-irradiated hindlimbs (**p < 0.01) (Figure 2Ciii).
The Scar-Forming Potential of PPFs Is Cell Intrinsic
To determine whether the fibrotic potential of PPFs is an intrinsic property of these cells, we exploited the differing fibrogenic potential of fibroblasts from the ventral skin, which heals with scarring, and the oral dermis, which heals with little to no scarring (Wong et al., 2009). We used lineage tracing in Wnt1Cre;R26mTmG mice to label neural crest-derived oral dermal Wnt1-positive fibroblasts (WPFs) by GFP expression (Nichols and Bruce, 2006; Yoshida et al., 2008). The oral dermis of Wnt1Cre;R26mTmG mice was harvested, and WPFs were FACS isolated and transplanted into the ventral dermis (Figure 2Di). Similarly, PPFs were isolated from the ventral dermis of Prrx1Cre;R26mTmG mice and transplanted into the oral mucosa, as previously described (Figure 2Di; Rinkevich et al., 2015). Recipient sites were harvested after 48 h, and confocal imaging was used in conjunction with 3D reconstruction and quantification (Imaris v.8.1.2, Interactive Microscopy Visualization Software) to compare collagen colocalization (as a measure of fibrogenic potential) for PPFs and WPFs. We found that PPFs transplanted from ventral dermis into oral dermis led to fibrosis, while WPFs transplanted from oral dermis into ventral dermis did not (Figure 2Dii), exemplified by significantly greater colocalization with collagen I of PPFs in oral dermis compared with WPFs in ventral dermis (30.4% versus 7.2%, *p < 0.05) (Figure 2Diii and iv; Videos S1 and S2). These findings strongly suggest that the fibrogenic potential of PPFs is cell intrinsic.
PPFs Are Characterized by Globally Profibrotic Transcriptional Programming That Is Persistent throughout a Heterogeneous Wound Healing Landscape
We next sought to examine the relationship between the fibrogenic potential of PPFs and the inherent heterogeneity of fibroblasts in wound healing. Using a protein barcoding system, we isolated PPFs and PNFs via FACS for high-throughput scRNA-seq using the 10x Genomics Chromium platform (Figure 3A). Data were log normalized and evaluated using uniform manifold approximation and projection (UMAP) analysis in Seurat v.3.2 (Becht et al., 2018), which identified six transcriptionally distinct populations, agnostic to lineage (PPF versus PNF) or wound state (unwounded versus healed wound [POD 14]) within the mouse ventral dermis (Figure 3B). Automated cell annotation using SingleR v.3.11 against two murine reference sets identified each subpopulation as characteristically “fibroblast” (Figure S3Ai and ii).
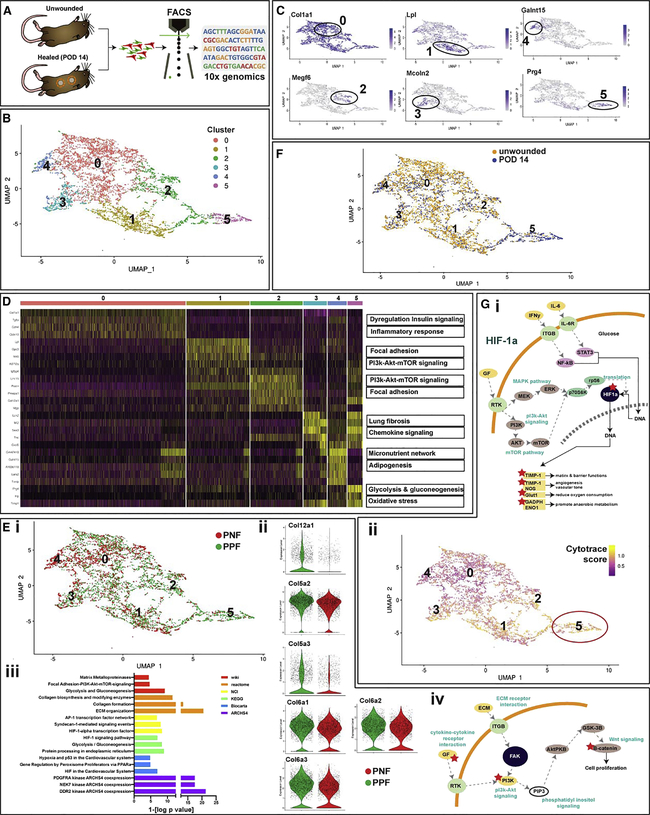
Figure 3. PPFs Are Characterized by Globally Profibrotic Transcriptional Programming Persistent throughout a Heterogeneous Wound Healing Landscape.
(A) Schematic showing strategy for isolation of PPFs and PNFs from unwounded and healed wounds (POD 14) of P30 Prrx1Cre;R26mTmG/iDTR mice by FACS for scRNA-seq analysis using 10x Chromium (data based on 20 wounds from 10 mice).
(B) (i) scRNA-seq was used to profile 7,300 cells from unwounded and scarred ventral dermis. We identified six transcriptionally distinct subpopulations, visualized using UMAP. The top 100 genes for each subgroup were used to perform GSEA against several pathway databases; top results from Wikipathway mouse 2019 are shown in the right panel.
(C) UMAP plots demonstrating highly expressed genes characteristic of each cluster.
(D) Heatmap of the most variable genes detected for each cluster. Most differences among clusters centered on collagen production at the FAK-PI3K-Akt-mTor signaling pathway.
(E) (i) UMAP plots were applied to visualize clusters by lineage agnostic to wounded state (4,792 cells were PPFs; 2,568 cells were PNFs); PPFs are shown in green, and PNFs are shown in red. Cluster 5 was comprised mostly of PPFs, while cluster 4 was primarily PNFs. (ii) Violin plots showing expression of collagen genes in PPFs (green) and PNFs (red) in cluster 5. (iii) Gene set enrichment analysis using multiple pathways revealed the top signaling pathways upregulated in PPFs versus PNFs. These were centered around the FAK-PI3K-Akt-mTor axis shown in (iv) with upregulated genes marked by asterisk (*).
(F) UMAP plots visualizing clusters by wound state in the ventral dermis, agnostic to lineage and cranial versus caudal location (3,713 cells were from unwounded dermis [blue]; 3,587 cells were from healed wound dermis [yellow]).
(G) (i) Schematic of HIF1A signaling pathway, which was significantly upregulated in PPFs of cluster 5. (ii) UMAP plots applied to visualize cells colored by CytoTRACE differentiation state, agonistic to lineage and wound state; yellow indicates high CytoTRACE score (undifferentiated state); purple indicates low CytoTRACE score (differentiated state). Cluster 5 comprised the most undifferentiated cells. Data are represented as mean ± SEM.
The top 100 genes most differentially expressed within each cluster were identified (Figure 3C; Table S1) and used to perform gene set enrichment analysis (GSEA) with EnrichR v.2.1 for additional insight into each cluster’s functional properties (Figure 3D; Chen et al., 2013). Results indicated a heterogenous tissue milieu characterized by transcriptionally distinct fibroblast subpopulations, significantly differentiated by varying collagen production and activity of the FAK-PI3K-Akt-mTor (FAK-phosphoinositide 3 kinase-protein kinase B-mammalian target of rapamycin) signaling pathway.
Our initial analysis was agnostic to phenotype; unmasking lineage, we see that PPFs and PNFs are distributed differentially in all six transcriptionally defined subpopulations (Figure 3Ei). In particular, cluster 5 comprised primarily PPFs (73.1%) versus PNFs (26.9%). We identified the top 100 differentially expressed genes significantly upregulated in PPF versus PNF cells globally (independent of cluster), which included numerous collagen genes (Figure 3Eii). These targets were then used to perform GSEA as described above. Overexpressed pathways were heavily tilted toward fibrosis, collagen production, and FAK signaling, further supporting a role for PPFs as drivers of fibrosis in wound healing (Figure 3Eiii and iv).
In our scRNA-seq pipeline, we used hashtag oligonucleotide barcoding (HTO) to precisely record the identity of pooled cells in each of our aggregated 10x samples. As such, in addition to tracking each cell’s lineage (PPF versus PNF), we traced cells according to their wound status (unwounded versus POD 14) and wound location (ventral cranial versus ventral caudal) (Figure S3B). Globally, differences between unwounded and POD 14 wound samples were associated with a combination of profibrotic and pro-regenerative programming, consistent with prior studies (Figures 3F and S3Ci and ii; Rinkevich et al., 2015). The strategic placement of two ventral dermal wounds on each mouse along the cranial-caudal axis allowed for separate barcoding and analysis of fibroblasts isolated from regions of different embryologic origins. Through this lens, we found fibroblast heterogeneity to vary with wound location (Figure S3 Di–iii). Notably, the transcriptionally defined cluster 5 comprised almost exclusively cranial region cells. Furthermore, these cells were predominantly wound-derived PPFs. Given our utilization of HTOs, with cells from each group pooled across eight mice, we had high confidence that this confluence of characteristics was not artifactual. As such, we examined cluster 5 in more detail.
As described above, we found many profibrotic, collagen production, and FAK-associated pathways differentially regulated in cluster 5 cells, although not to a greater extent than in the global PPF superset. However, unlike other subsets, this population was significantly enriched for HIF1A (hypoxia inducible factor 1 alpha)-associated signaling pathways (Figure 3Gi). This finding may suggest a potential link between hypoxia and the chronic inflammatory state underlying fibrosis. Interestingly, a computational framework to predict cell differentiation state (CytoTRACE) (Gulati et al., 2020) found PPFs in cluster 5 to be significantly less differentiated than other fibroblast populations (Figure 3Gii).
To understand whether ventral fibroblasts have similar transcriptional profiles to dorsal fibroblast subtypes, we compared gene expression for markers recently shown to be highly expressed in dorsal fibroblasts (Plikus et al., 2017). Similar to the analysis by Plikus et al. (2017), we found high expression of collagens (collagen type 1-alpha 1 [Col1a1], collagen type 3-alpha 1 [Col3a1], collagen type 5-alpha 1 [Col5a1]) and other ECM proteins (decorin [Dcn], fibulin-2 [FBln2], matrix metalloproteinase 2 [Mmp2], serpin family f member 1 [Serpinf1], and caldesmon [Cald1]) in ventral fibroblasts (Figure S4B). We also identified higher expression of bone morphogenetic protein receptor 2 (Bmpr2), apoliporotein E (Apoe), and integrin subunit-beta 1 (Itgb1) in ventral fibroblasts, paralleling our findings in the dorsal dermis (Rinkevich et al., 2015; Figure S4B). It is important to highlight, however, the incomplete overlap between transcriptional profiles of dorsal and ventral dermis fibroblasts, and thus how critical it is to consider the distinct fibroblast lineage marked by Prrx1 expression when investigating fibrosis of the ventral dermis.
PPF Ablation Reduces Connective Tissue Deposition during Scar Formation
To explore whether selective elimination of the PPF lineage could reduce scar formation, ROSA26tm1(HBEGF)Awai (R26iDTR) mice, which exhibit Cre-dependent expression of the simian diphtheria toxin (DT) receptor, were crossed with Prrx1Cre;R26mTmG mice. The resulting triple-positive offspring (Prrx1Cre;R26mTmG/iDTR) allow for both PPF identification (via GFP expression) and selective ablation (by DT administration). Splinted full-thickness excisional wounds were created in Prrx1Cre;R26mTmG/iDTR mice as described above. At PODs 0, 2, 4, and 6, wounds were treated with topical administration of either DT in phosphate-buffered saline (PBS) or PBS alone (Figure 4A). As expected, DT treatment eliminated the vast majority of PPFs in the healed wound, compared with PBS-treated wounds (Figure 4Bi). PPF ablation did not affect time to healing, marked by complete epithelialization at POD 14 in both groups (Figure S4A), but histological characteristics of the resultant scars differed dramatically. DT-treated mice had markedly reduced scarring compared with PBS-treated (control) mice (Figure 4Bii) and a more loosely organized dermis in the healed scar (Figure 4Biii).
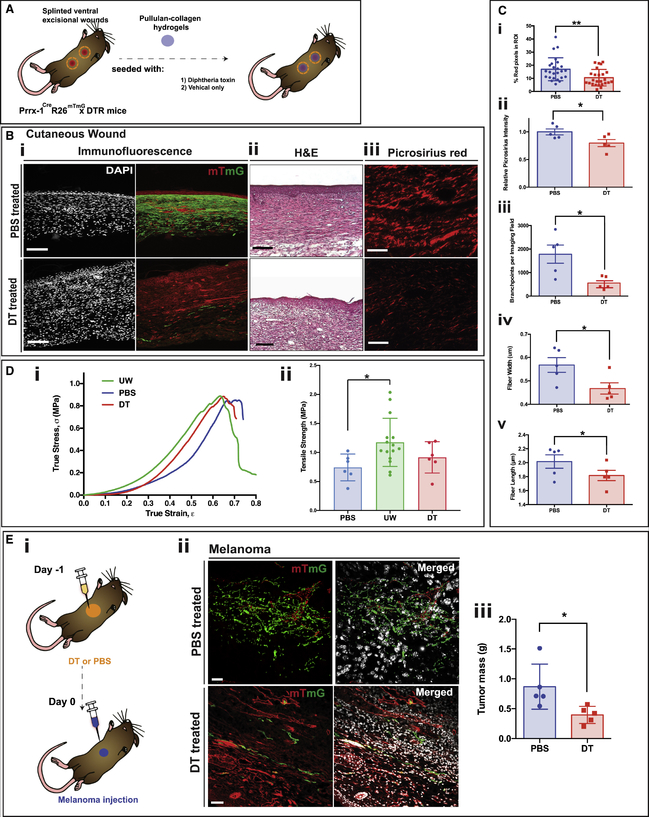
Figure 4. DTR-Based Ablation Results in Diminished Connective Tissue Deposition during Cutaneous Scar and Tumor Stroma Formation.
(A) Schematic showing strategy for DT ablation in P30 Prrx1Cre;R26mTmG/iDTR wounds using pullulan-collagen hydrogels seeded with DT (experimental group) or vehicle (PBS) only (control).
(B) (i) Fluorescent imaging of healed (POD 14) ventral excisional wounds of P30 Prrx1Cre;R26mTmG/iDTR mice, treated with PBS (control; top row) or DT (bottom row). DAPI (white; left column); tdTomato and EGFP (red and green, respectively; right column). Scale bar, 100 μm. (ii) Hematoxylin and eosin (H&E) staining of DT- and PBS-treated wounds. Scale bar, 200 μm. (iii) Picrosirius red staining of DT- (bottom) and PBS-treated (top) wounds. Scale bar, 50 μm.
(C) Quantification of connective tissue parameters in PBS- and DT-treated wounds based on collagen fiber networks stained with picrosirius red. (i) DT ablation of PPFs significantly decreased dermal collagen content, quantified using red pixel area as a surrogate for type I collagen (**p = 0.0047) (n = 25). A machine learning algorithm was used to quantify the following: (ii) overall collagen deposition (*p = 0.0428), (iii) collagen fiber branching (*p = 0.0496), (iv) fiber width (*p = 0.0338), and (v) fiber length (*p = 0.0326).
(D) (i) Representative stress-strain curves of fully healed (POD 14) wounds from Prrx1Cre;R26mTmG/iDTR mice treated with DT (red line; n = 6) or PBS (blue line; n = 6) compared with unwounded skin (UW; green line; n = 15). (ii) Bar graph showing increased mean tensile strength (MPa) in unwounded skin compared with PBS-treated wounds (*p < 0.05) of Prrx1Cre;R26mTmG/iDTR mice; DT-treated wounds (red bar), PBS-treated wounds (blue bar), and unwounded skin (green bar).
(E) (i) Schematic depicting the strategy for DT ablation and assessment on ventral melanoma tumor stroma formation; melanoma cells were injected into ventral skin of P30 Prrx1Cre;R26mTmG/iDTR mice pre-treated with either PBS or DT. Skin was harvested 10 days after injection. (ii) Representative histological images of tumor stroma harvested 10 days after injection in mice pre-treated with either PBS (top row) or DT (bottom row), revealing less PPFs in tumor stroma of DT-treated mice. tdTomato and EGFP (red and green, respectively; left column), merged with DAPI (white; right column). Scale bar, 100 μm (n = 5). (iii) Quantification of tumor mass in melanoma-cell-injected mice treated with PBS (blue bar) or DT (red bar), showing greater tumor mass in PBS-treated mice (n = 5; *p = 0.0464). Data are represented as mean ± SEM. All experiments based on six mice per control and intervention group.
Histological analysis of scars has traditionally relied on subjective visual comparisons. In order to quantitatively compare connective tissue in PBS- versus DT-treated scars, we used an image processing algorithm. Briefly, picrosirius red-stained histological sections from scars were processed to remove cells (such that only ECM was analyzed) (S.M. and M.T.L., unpublished data). These images were binarized, and various collagen fiber characteristics (e.g., average fiber length and width, branchpoint density) were quantified. DT-treated wounds, in addition to having significantly reduced total intracellular collagen (determined from positive red pixels on picrosirius red staining) compared with control wounds (**p = 0.0047) (Figure 4Ci), also exhibited significant differences in collagen fiber organization. DT-treated scars had significantly reduced overall collagen deposition (measured as relative picrosirius red intensity) (*p = 0.0428) (Figure 4Cii) and their collagen fibers had fewer branchpoints (*p = 0.0496) (Figure 4Ciii). Furthermore, individual collagen fibers in DT-treated wounds were of significantly reduced width (*p = 0.0338) (Figure 4Civ) and length (*p = 0.0326) (Figure 4Cv). Together, these data demonstrate less-fibrotic ECM architecture in scars following PPF lineage ablation.
DT- and PBS-treated Prrx1Cre;R26mTmG/iDTR wounds and unwounded skin were also tested for tensile strength (Figure 4Di and ii). Unwounded skin had significantly greater tensile strength than PBS-treated wounds at POD 14 (*p < 0.05). Interestingly, despite the reduced fibrosis associated with DT-induced PPF ablation, the overall strength of POD 14 wounds was not significantly different compared with unwounded skin (Figure 4Dii). Furthermore, stress-strain profiles of DT-treated wounds were similar to those of PBS-treated wounds (Figure 4Di and ii). This suggests that collagen organization, and not solely quantity, may be a key determinant of wound strength. Collectively, these data demonstrate that PPF ablation reduces scarring without affecting tensile strength of healed wounds, a finding with considerable translational implications.
PPF Ablation Reduces Melanoma Growth
Melanoma tumor stroma is almost entirely composed of PPFs (Figure 2Bii and iii). To explore the role of PPFs in supporting tumor growth, we assessed the effects of DT-based PPF ablation on tumor growth. Prrx1Cre;R26mTmG/iDTR mice were treated with either DT or PBS 24 h prior to transplantation of B16-F10 mouse melanoma cells into the ventral dermis (Figure 4Ei). Tumors were allowed to grow for 10 days and then harvested for analysis. Melanoma cell transplantation into PBS-treated Prrx1Cre;R26mTmG/iDTR skin produced similar histologic findings to Prrx1Cre;R26mTmG recipients (Figures 2Bii and iii and 4Eii, top row). However, DT administration successfully ablated the vast majority of PPFs within the tumor stroma and adjacent skin (Figure 4Eii, bottom row). DT ablation of PPFs was also associated with significantly decreased tumor burden as measured by tumor mass at 10 days (*p < 0.05) (Figure 4Eiii). These data suggest an important role for tumor stromal PPFs in melanoma growth.
DISCUSSION
Identification of the cells responsible for scar formation and skin fibrosis is essential for development of treatments to promote regenerative repair and decrease scarring (Guerrero-Juarez et al., 2019). In the mouse dorsal dermis, we previously identified a fibrogenic fibroblast lineage characterized by En1 expression during embryonic development (Rinkevich et al., 2015). Unlike quadrupedal mice, bipedal humans often experience ventral (e.g., abdominal, chest) wounds, through trauma or surgery. Thus, understanding the functional heterogeneity of ventral skin fibroblasts is critical to developing clinically translatable models. Here, we have identified a scar-forming Prrx1 fibroblast lineage responsible for the majority of ECM deposition in the ventral dermis following injury. Our FACS data reveal that Prrx1 expression is present in all known fibroblast subtypes including papillary, reticular, and lipofibroblasts. However, the reticular PPF subpopulation was shown to disproportionally contribute to scarring. Furthermore, we show that selective ablation of fibrogenic PPFs significantly decreased scarring without affecting wound tensile strength. These findings have important translational implications: it may be possible to minimize skin fibrosis through selective depletion of profibrotic fibroblast populations, without compromising skin’s functional integrity. Finally, we showed that PPF ablation in tumor stroma significantly reduced tumor burden. Fibroblasts are increasingly recognized as integral to tumor survival/growth (Östman and Augsten, 2009), and targeting PPFs may also provide therapeutic benefits in the setting of malignancy. In summary, our findings suggest a pivotal role for the PPF population in both acute and chronic fibroses on the mouse ventrum.
Our data show that Prrx1 expression marks an intrinsically fibrotic fibroblast lineage. We observed Prrx1 expression both in embryonic development and post-natally in unwounded skin. Future work must explore whether wound PPFs are from embryonic fibroblast progenies activated in response to wounding, or whether PPFs are present and active throughout development. Currie et al. (2019) similarly found that Prrx1 fibroblasts in the dermal perivascular and hair follicle niche increase in number in response to dermal limb injury. These data may suggest that Prrx1 expression is present throughout development and these cells are injury responsive (Currie et al., 2019). However, as limb skin has a different embryonic origin than ventrum (Tickle, 2015), the behavior of Prrx1 fibroblasts in ventrum and limb cannot be assumed to be similar.
Our scRNA-seq analyses reveal six fibroblast clusters that express Prrx1 following wounding, highlighting heterogeneity of the PPF scarring lineage. Interestingly, cluster 5 comprised primarily PPFs at POD 14, suggesting that they contributed significantly to scarring. However, as our topical ablation experiments eliminated all PPFs equally, we cannot specifically identify cluster 5 as the dominant scarring subpopulation. Future work is needed to identify whether this specific subcluster is responsible for the majority of PPF scarring.
Together with our prior study in the dorsal dermis (Rinkevich et al., 2015), we have identified at least four fibroblast lineages in mouse skin with distinct roles in skin fibrosis: PPFs and PNFs in the ventral dermis and EPFs and ENFs in the dorsal dermis. Recent advances in single-cell sequencing have permitted characterization of individual cells from heterogenous tissue such as wounds in a robust high-throughput fashion. Thus far, scRNA-seq studies in murine wound healing have focused on the dorsal dermis, either unwounded skin or a single time point during wound repair (Guerrero-Juarez et al., 2019). In this study, we integrate scRNA-seq with lineage tracing to explore transcriptional profiles of distinct subpopulations from two ventral fibroblast lineages in wound healing. This technique allowed us to overcome inherent heterogeneity in the wound milieu and to identify specific transcriptionally defined subgroups with differentially profibrotic characteristics. These findings corroborated our histologic and proteomic observations that PPFs represent an intrinsically profibrotic lineage in the ventral dermis.
In summary, we have identified and characterized a profibrotic lineage of fibroblasts in the mouse ventral dermis. This work provides extensive insight into potential therapeutic targets to modulate cutaneous fibrosis for treatment of scarring and other pathologic fibroses.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Longaker (longaker@stanford.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The RNA bulk sequencing and scRNA sequencing data generated during this study have been deposited in the NCBI’s Gene expression Omnibus and is accessible through GEO series accession number GSE159345 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159345).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All studies were conducted in accordance with Stanford University Animal Care and Use Committee guidelines. Daily care for the mice was provided by the Stanford Veterinary Service Center. Animal were housed in a controlled environment with optimal temperature and fed rodent chow ad libitum.
Mice Strains
Prrx1Cre, ROSA26mTmG (R26mTmG), and ROSA26tm1(HBEGF)Awai (R26iDTR) were obtained from The Jackson Laboratory (Bar Harbor, ME). ROSA26VT2/GK3 mice were a gift from the Chan laboratory, Stanford, CA. The R26mTmG reporter mice harbor a cell-membrane targeted double fluorescent Cre-reporter allele. Prior to Cre recombinase activity mTomato (mT) is expressed in ubiquitously. Following Cre recombinase activity, Cre expressing cells express green fluorescent protein (EGFP) (mG) in place of mTmG. Prrx1Cre mice were crossed with R26mTmG reporter mice (Rinkevich et al., 2015) to trace the lineage of a population of Prrx1-lineage–positive fibroblasts (PPFs), defined in vivo by the expression of GFP within ventral dermis of Prrx1Cre; R26mTmG offspring. To confirm that the Prrx1 expression represents a distinct fibroblast lineage, we compared Prrx1 expression to En1 using En1Cre;R26mTmG mice by crossing En1Cre mice with R26mTmG reporter mice to create En1Cre;R26mTmG mice as described by Rinkevich et al. (Rinkevich et al., 2015). To confirm the role of Prrx1 expressing fibrobalsts in fibrosis Prrx1Cre;R26mTmG mice were bred with the R26iDTR transgenic strain, to allow for the ablation of all Prrx1 lineage fibroblasts following the administration of diphtheria toxin (DT). For this study equal numbers of female and male mice ranging from the age of 8–12 weeks were allocated to experimental groups and sample size for any given experiment is detailed in the figure legend.
Mouse Melanoma Cell Line Culturing Conditions
The B16-F10 melanoma cell line (ATCC, Manassas, Virginia) was expanded in culture consisting of Dulbecco’s Modified Eagle Medium (DMEM) with 10% Fetal Bovine Serum (FBS) and 1% pen-strep for two passages prior to transplantation into Prrx1Cre; R26mTmG mice.
METHOD DETAILS
Harvesting dermal fibroblasts from ventral dermis
Adult mice were sacrificed by asphyxiation and cervical dislocation, and hair was removed using a chemical depilatory cream. Harvest from embryological samples was achieved at specific time points with the superovulation technique, as previously described (Rinkevich et al., 2015). Ventral skin was harvested immediately to preserve cell viability. Scalpel and forceps were used in the dissection, taking care to remove any adherent subcutaneous adipose tissue. The dissected skin was washed in 3x serial dilutions of betadine in phosphate buffered saline (PBS): betadine dilutions, followed by 2x rinse steps in PBS on ice to preserve cell viability. Tissue was finely minced using a scalpel and scissors until a uniform consistency was achieved. Enzymatic digest was performed by incubating tissue in 20 mL of Liberase DL (0.5 mg/ml Liberase DL in DMEM) per mouse, for 1.5 h at 37°C. Wash media (10% FBS and 1% pen-strep in DMEM) was added to quench the enzyme prior to centrifugation (300 g, 5 min, 4°C) to pellet cells. All centrifugation steps were performed under these conditions. The supernatant, including the top layer of floating adipocytes, was removed. Cells were resuspended in wash media and passed through a 100-μm filter. The cells were again centrifuged. The supernatant was removed, and the pellet was resuspended in wash media prior to passage through a 40-μm filter. The filtered suspension was again centrifuged. The entire supernatant was then carefully removed.
Fluorescence-activated cell sorting (FACS)
The mouse pelleted cells were then washed by resuspending the pellet in FACS buffer, centrifuging and removing the supernatant. The pellet was resuspended in FACS buffer and cells were stained The pellet was resuspended in FACS buffer and cells were stained with multiple antibodies in the Pacific Blue lineage channel (Lin) CD31, CD45, TIE-2, Ter119, EpCAM, and CD324 antibodies eBioscience (San Diego, California) for 30 min on ice, shielded from light as previously described (Rinkevich et al., 2015). Of note, TIE-2 and CD324 require an initial incubation with biotinylated antibodies followed by a further 20 min incubation with eFluor 450-conjugated streptavidin on ice. In addition, CD26, Sca1, and DLK1 were used to label papillary (CD26+Sca1+) reticular (Dlk1+Sca1−) and lipofibroblast (or ‘adipocyte precursors’, Sca1+) PPF and PNF fibroblast subpopulations (all 1:100). Cells were washed 3x with FACS buffer and centrifuged. DAPI was added to the final sample in FACS buffer as a viability marker. Fluorescence-activated cell sorting (FACS) (FACS Aria II instrument, BD Bioscience, san Jose, CA) was performed to isolate dermal fibroblasts, identified as DAPI-negative, CD31-negative, CD45-negative, TIE-2-negative, Ter119-negative, EpCAM-negative, and CD324-negative cells. Cells of hematopoietic, endothelial, and epithelial origins were thereby excluded. A negative gating strategy was employed to limit enrichment of unknown fibroblast subpopulations based on cell-surface marker expression. PPFs and PNFs were distinguishable based on positivity for GFP and RFP, respectively among the Lin- population.
Reciprocal transplantation
Ventral dermal fibroblasts were isolated from Prrx1Cre;R26mTmG mice, and oral dermal fibroblasts were isolated from Wnt1Cre;R26mTmG using the methods described (Rinkevich et al., 2015). The harvested cells were stained with DAPI and eFlour 450 conjugated Lin antibodies (CD31, CD45, TIE-2, Ter119, EpCAM, CD324). Viable GFP-positive CD31-negative CD45-negative Ter119-negative EpCAM-negative and CD324-negative populations were isolated from the two respective tissue types. The freshly sorted cell suspensions were centrifuged (300 g for 5 min at 4°C) and resuspended in PBS at a concentration of 100,000 cells per 10 μl. The FACS-isolated cells from the ventral dermis of Prrx1Cre;R26mTmG mice were transplanted into the buccal mucosa of recipient C57BL/6 mice using methods described previously (Rinkevich et al., 2015). The FACS-isolated cells from the oral dermis of Wnt1Cre;R26mTmG mice were transplanted into the ventral dermis of C57BL/6 mice, 100,000 cells were used per transplantation. All tissues were harvested at 48 h post transplantation for analysis after transplantation.
Immunostaining of cultured fibroblasts
FACS-isolated fibroblasts harvested from young adult (P30) Prrx1Cre;R26mTmG mice were plated into 8-well chamber slides (Nunc Lab-Tek II Chamber Slide System, Thermo Fisher, Waltham, Massachusetts) for 2 days. Cells were washed 3X in PBS, fixed in 10% formalin for 10 min at 25°C, and stained with the following anti-rabbit primary antibodies overnight at 4°C: Fibronectin, Fibroblast Specific Protein (FSP), Col I, and vimentin. Chicken anti-rabbit Alexa Fluor 647 (Thermofisher,) was utilized as a secondary antibody, incubated for 1 hour at 25°C.
Collagen production
FACS-isolated P30 Lin- PPFs and PNFs (20,000 cells each) were directly sorted assayed for intracellular collagen production using a standard ELISA (Abcam) as per the manufacturers’ instructions.
Ventral wounding
Adult age- and sex-matched male and female Prrx1Cre; R26mTmG mice were used for cutaneous wound healing experiments. Splinted, full-thickness excisional wounds were performed as previously described (Rinkevich et al., 2015). In brief, mice were induced and maintained under anesthesia using a 2% isoflurane/oxygen mixture at 3 L per minute. Ventral hair was removed with a chemical depilatory cream and skin was prepped with povidone-iodine and alcohol. Two 6-mm full-thickness circular wound were placed through the panniculus carnosus; one at the upper abdomen of each animal, directly caudal to the xyphoid process, and one immediately caudal. A circular silicone 12-mm diameter stent was secured around the perimeter of each wound with glue and 8 simple interrupted 6–0 nylon sutures (Ethicon, Somerville, New Jersey). Wounds were dressed using Tegaderm (3M, Minnesota, USA) and bolstered by Telfa non-adherent dressings (Covidien, Dublin, Republic of Ireland). Dressings were changed every other day under anesthesia until wounds had fully healed. Wounds were imaged every other day, and wound healing curves were plotted as a percentage of wound size versus days since wounding. The relative proportions of PPF and PNF subpopulations throughout wounding (POD 4, 7, 9, 14) were analyzed by FACs.
Bulk RNA-sequencing
PPF and PNF fibroblasts were collected from the ventral dermis of twenty ventral wounds from ten Prrx1Cre; R26mTmG mice using FACS (using a Lin-GFP+RFP− and Lin-GFP-RFP+ strategy as described above) at the following developmental time points for bulk RNA-sequencing in order to explore the expression of Prrx1 mRNA throughout development; E16.5, E18.5, P1 and P30. Fibroblasts were directly sorted into TRIZOL for bulk RNA sequencing by Stanford’s Functional Genomic Facility.
Single cell barcoding, library preparation, and sequencing
Single cells from twenty wounds from ten Prrx1Cre; R26mTmG mice were barcoded using the 10x Chromium Single Cell platform, and cDNA libraries were prepared according to the manufacturer’s protocol (Single Cell 3′ v3, 10x Genomics, USA). In brief, cell suspensions, reverse transcription master mix and partitioning oil were loaded on a single cell chip, then run on the Chromium Controller. Reverse Transcription was performed within the droplets at 53°C for 45min. cDNA was amplified for a 12 cycles total on a BioRad C1000 Touch thermocycler. cDNA size selection was performed using SpriSelect beads (Beckman Coulter, USA) and a ratio of SpriSelect reagent volume to sample volume of 0.6. cDNA was analyzed on an Agilent Bioanalyzer High Sensitivity DNA chip for qualitative control purposes. cDNA was fragmented using the proprietary fragmentation enzyme blend for 5min at 32°C, followed by end repair and A-tailing at 65°C for 30min. cDNA were double-sided size selected using SpriSelect beats. Sequencing adaptors were ligated to the cDNA at 20°C for 15min. cDNA was amplified using a sample-specific index oligo as primer, followed by another round of double-sided size selection using SpriSelect beads. Final libraries were analyzed on an Agilent Bioanalyzer High Sensitivity DNA chip for qualitative control purposes. cDNA libraries were sequenced on a HiSeq 4000 Illumina platform aiming for 50,000 reads per cell.
Data processing, fastq generation, and read mapping
Base calls were converted to reads with the software Cell Ranger (10x Genomics; version 3.1)’s implementation mkfastq. These were then aligned against either the GRCh38 v3.0.0 (for human) or mm10 v3.0.0 (for mouse) genomes using Cell Ranger’s count function (an implementation of STAR v2.7.0) with SC3Pv3 chemistry and 5,000 expected cells per sample (Dobin et al., 2013). Cell barcodes representative of quality cells were delineated from barcodes of apoptotic cells or background RNA based on a threshold of having at least 200 unique transcripts profiled, less than 10,000 total transcript, and less than 10% of their transcriptome of mitochondrial origin.
Data normalization, hashtag oligo demultiplexing, and cell subpopulation identification
UMIs from each cell barcode were retained for all downstream analysis. Raw UMI counts were normalized with a scale factor of 10,000 UMIs per cell and subsequently natural log transformed with a pseudocount of 1 using the R package Seurat (version 3.1.1) (Stuart et al., 2019). Highly variable genes were identified, and cells were scaled by regression to the fraction of mitochondrial transcripts. Hashtag oligos (HTOs) were demultiplexed using Seurat’s implementation HTODemux. Briefly, k-medoid clustering is performed on the normalized HTO values, after which a ‘negative’ HTO distribution is calculated. For each HTO, the cluster with the lowest average value is treated as the negative group and a negative binomial distribution is fit to this cluster. Using the 0.99 quantile of this distribution as a threshold, each cell is classified as positive or negative for each HTO. Cells that are positive for more than one HTOs are annotated as doublets and removed. Cells that are not positive for any HTO are also removed. Aggregated data was then evaluated using uniform manifold approximation and projection (UMAP) analysis over the first 15 principal components (Becht et al., 2018). Cell annotations were ascribed using SingleR (version 3.11) against the Immunological Genome Project (ImmGen) and mouse RNA-seq reference sets. The Seurat ‘table’ function was used to classify the number of PPFs and PNFs in cluster 5.
Generation of characteristic subpopulation markers and enrichment analysis
Cell-type marker lists were generated with Seurat’s native FindMarkers function with a log fold change threshold of 0.25 using the ROC test to assign predictive power to each gene. The 100 most highly ranked genes from this analysis for each cluster were used to perform gene set enrichment analysis against pathway databasess in a programmatic fashion using EnrichR (version 2.1) (Chen et al., 2013). scRNA-seq data was further analyzed using CytoTRACE (https://cytotrace.stanford.edu; Gulati et al., 2020) an algorithm enabling robust reconstruction of cellular differentiation trajectories, to trace dynamic chromatin changes using the mouse lung fibroblast dataset.
Groin irradiation
The right hind limbs of female Prrx1Cre; R26mTmG mice were irradiated using a protocol previously described (Garza et al., 2014; Luan et al., 2016). In brief, 30 Gy external beam radiation was administered to the right hind limb in six 5 Gy doses delivered every two days for 12 days total. Lead shielding was used to ensure only the right hind limb was irradiated. The non-irradiated left hind limb served as an internal control. A one-month recovery period followed irradiation to allow for the development of contracture and chronic limb changes.
Analysis of tumor stroma formation and fibrotic response
The B16-F10 melanoma cell line (ATCC, Manassas, Virginia) was expanded in culture for two passages. The melanoma cells were then prepared for injection by mixing 5.0 × 105 cells in 40 μL of a 1:1 mixture of Matrigel (Corning, New York, USA) and PBS was performed. Adult male and female Prrx1Cre;R26mTmG mice were anesthetized and hair was removed via shaving and application of the depilatory agent Nair®. The cells were transplanted into the ventral skin via intradermal injection. After 10 days, a palpable tumor had formed, and the injected area was harvested for histological and FACS analysis.
Histological sample preparation
Immediately following tissue harvest, samples were placed in 10% formalin for 12 to 16 h at 4°C. Samples were then washed with PBS and soaked in 30% sucrose in PBS for 3–5 days in preparation for embedding. Tissue blocks were prepared by embedding in Tissue-Tek® O.C.T. (Sakura Finetek, Torrance, California) frozen in a dry ice/ethanol bath. Frozen blocks were sectioned at a thickness of 8-μm and then transferred to Superfrost Plus microscope slides (Fisherbrand).
Immunohistochemistry
Standardized protocols were utilized for both H&E as well as Picrosirius Red staining, without any modifications. For immunostaining of frozen sections, slides were fixed in 10% formalin for 10 min, blocked in 1X Power Block for 1 hour, and then incubated with primary antibody for 12–16 h. Primary antibodies used for staining of frozen sections included: type I collagen, type III collagen, keratin-14. Slides were then incubated with Alexa Fluor 647-conjugated anti-rabbit antibodies (Thermofisher).
Analysis of collagen fiber characteristics
Automated quantification of collagen fiber characteristics was performed using MATLAB with Image Processing Toolbox installed. Images of Picrosirius Red-stained histological specimens (63x magnification) were first segmented into red (mature fibers) and green (immature fibers) channels using color deconvolution. Next, images were de-noised using adaptive filtering (wiener2, 3-by-3 neighborhood), binarized (im2bw), eroded (diamond structuring element), and dilated (line structuring element). Finally, the bwmorph function was used to skeletonize binary images, thereby tracing collagen fibers. Individual fiber properties (length, width, orientation, persistence, branch points) were gathered using the regionprops command. Differences between means were compared by two-tailed Student’s t test.
Microscopy
Fluorescent images were captured with laser scanning confocal microscopy using a Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany). The frame size of the image was 1600 × 900. Confocal images were taken with a 0.3 μm z-step size. Bright-field and polarized light images were taken with Leica DM4000B or DM5000B microscopes and Leica DFC550 camera.
Image processing
The confocal z-stack images were analyzed using extensive three-dimensional reconstructing software IMARIS 8.1.2 software (Interactive Microscopy Visualization Software) (Bitplane). The surfaces of collagen I surface and of the transplanted Wnt1Cre;R26mTmG and Prr1Cre;R26mTmG fibroblasts were reconstructed. The percent of surface contact between collagen I and the transplanted fibroblasts was determined by the colocalization module.
DTR-based ablation of PPFs during wound healing
Functional assays to confirm the role of PPFs in fibrosis were performed by breeding Prrx1Cre;R26mTmG mice with the R26iDTR transgenic strain, which will allow for ablation of all Prrx1 lineage fibroblasts with the administration of DT. These Prrx1Cre; R26mTmG/iDTR mice were subjected to wound healing experiments using a splinted, full-thickness excisional wound model, as described in the section “Ventral wounding.” Mice were either treated with topical application of 20 ng DT in 1 μL PBS (n = 6) or 1 μL PBS alone (n = 6) at post-operative days 0, 2, 4, and 6 during every other day dressing changes, which continued until wounds were fully healed.
Tensile strength testing
Fully healed wounds from Prrx1Cre; R26mTmG/iDTR mice treated with either DT or PBS as described above, was performed usinga microtester (model 5848, Instron, Norwood, Massachusetts) equipped with a 100 N load cell as previously described (Rinkevich et al., 2015). Briefly, the tissue was attached to custom grips with double-sided tape, providing a gauge length of 1 cm. The tissue specimen was stretched until a break in the skin was detected, observed as a decrease in stress despite increasing strain. Change in length divided by gauge length was used to calculate true strain. True stress was determined by dividing force by the original tissue cross-sectional area. Ultimate tensile strength corresponds to the greatest true stress achieved prior to breakage.
DTR-based ablation of PPFs prior to tumor injection
The ventral skin of adult (8-to-10-week old) age- and sex-matched Prrx1Cre; R26mTmG/iDTR mice was treated with a chemical depilatory cream prior to pre-treatment with either DT or PBS. Intradermal injection of 80 ng DT in 20 μL PBS or 20 μL of PBS alone was performed at four evenly spaced locations around the circumference of a circle 6 mm in diameter (n = 5, total volume of 80 μL administered in both groups). After 24 h, 5.0 × 105 B16-F10 melanoma cells were transplanted intradermally into the center of the marked circle on the ventral skin, as described above. Tumors were again harvested for analysis at 10 days.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 8.0.2 (GraphPad Software, La Jolla, California). All values are expressed as mean ± standard deviation (SD). Statistical significance between groups was determined using an unpaired Student’s t test assuming two-tailed distribution and unequal variances if not stated in the figure legends. The value of n refers to the number of mice used in the mice study part. A p value < 0.05 was considered statistically significant. For all figures, asterisks denote statistical significance at the following levels: * p < 0.05, ** p < 0.001, *** p < 0.001, **** p < 0.0001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse Brilliant Violet 605, Sca-1 | Biolegend | Cat# 108133;RRID:AB_2562275 |
| Anti-Mouse Monoclonal eFluor 450, CD45 | Thermo Fisher Scientific | Cat# 48-0451-82; RRID:AB_1518806 |
| Anti-Mouse Monoclonal APC, DLK1 | R&D Systems | Cat# FAB8634A |
| Anti-Mouse Monoclonal PerCP-Cyanine5.5, CD26 | Thermo Fisher Scientific | Cat# 45-0261-82; RRID:AB_1548738 |
| Anti-Mouse Monoclonal eFluor 450, TER-119 | Thermo Fisher Scientific | Cat# 48-5921-82; RRID:AB_1518808 |
| Anti-Mouse Monoclonal eFluor 450, CD324 | Thermo Fisher Scientific | Cat# 13-3249-82; RRID:AB_1659688 |
| Anti-Mouse Monoclonal eFluor 450, TIE-2 | Thermo Fisher Scientific | Cat# 13-5987-82; RRID:AB_466848 |
| Anti-Mouse Monoclonal eFluor 450, CD326 | Thermo Fisher Scientific | Cat# 48-5791-82; RRID:AB_10717090 |
| Anti-Mouse Monoclonal eFluor 450, CD31 | Biolegend | Cat# 303114; RRID:AB_2114316 |
| Anti-Mouse Monoclonal eFluor 450, CD45 | Thermo Fisher Scientific | Cat# 48-0451-82; RRID:AB_1518806 |
| Rabbit Polyclonal, Fibronectin | Abcam | Cat# ab2413; RRID:AB_2262874 |
| Rabbit Polyclonal, FSP | Abcam | Cat# ab27957; RRID:AB_2183775 |
| Rabbit Polyclonal, Col1 | Abcam | Cat# ab34710; RRID:AB_731684 |
| Rabbit Polyclonal, Vimentin | Abcam | Cat#ab137321 |
| Rabbit Polyclonal, Col3 | Abcam | Cat# ab59436; RRID:AB_941099 |
| Rabbit Monoclonal, Cytokeratin 14 | Abcam | Cat# ab181595; RRID:AB_2811031 |
| Anti-rabbit Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21443; RRID:AB_2535861 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fluromont-G | Southern Biotech | Cat# 0100-01 |
| Ethanol | GoldShield | Cat# 64175 |
| Permount | Fisher Chemicals | Cat# SP15 |
| Triton x-100 | Sigma | Cat# X100 |
| DAPI-(4,6-Diamido-2-Phenylindole, dihydrochloride) | Thermo Fisher Scientific | Cat# D1306; RRID:AB_2629482 |
| Hematoxylin | Sigma | Cat# H3136 |
| Eosin | Sigma | Cat# HT1101128 |
| Liberase | Sigma | Cat# 5401119001 |
| Dulbecco Modified Eagle Medium | Sigma | Cat# D5796 |
| Phosphate Buffer Saline | Sigma | Cat# P5368 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10428026 |
| Tween-20 | Sigma | Cat# 113322465001 |
| 1x Power Block | Roche | Cat# Hk085-5k |
| Trypsin Antigen Retrieval Kit | Abcam | Cat# 970 |
| Matrigel | Corning | Cat# 356234 |
| Tissue Tek Optimal Cutting Temperature (OCT) | Sakura | Cat# 4583 |
| 1,4-Dithiothreitol | Sigma | Cat# 10197777001 |
| Critical Commercial Assays | ||
| Collagen ELISA | Abcam | Cat# Ab210966 |
| Chromium Next GEM Chip H Single Cell Kit | 10x Genomics | Cat# 1000161 |
| PicroSirius Red Staining Kit | Abcam | Cat# Ab150681 |
| Deposited Data | ||
| RNA Bulk Sequencing Data | This data has been deposited in NCBI’s Gene expression Omnibus. | GSE159345 |
| scRNA-Sequencing Data | This data has been deposited in NCBI’s Gene expression Omnibus. | GSE159345 |
| Experimental Models: Cell Lines | ||
| Mouse B16F-10 Melanoma Cells | ATCC | Cat# CRL-6475 |
| Experimental Models: Organisms/Strains | ||
| Prrx1Cre | The Jackson Laboratory | JAX stock # 005584 |
| R26Mtmg | The Jackson Laboratory | JAX stock # 007676 |
| C57/BL/6J | The Jackson Laboratory | JAX stock # 000664 |
| Wnt1Cre | The Jackson Laboratory | JAX stock #022137 |
| R26iDTR | The Jackson Laboratory | JAX stock # 007900 |
| En1Cre | The Jackson Laboratory | JAX stock #007916 |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | RRID:SCR_003070 |
| Adobe Photoshop CC | Adobe | RRID:SCR_014199 |
| Adobe Illustrator CC | Adobe | RRID:SCR_010279 |
| Prism 5 | Graph Pad | RRID:SCR_002798 |
| Imaris | Oxford Instruments | RRID:SCR_007370 |
| MATLAB | MATLAB | RRID:SCR_001622 |
| Other | ||
| 6-0 nylon sutures | Ethicon | Cat# 1856G |
| Tegaderm Dressings | 3M | Cat# 1626w |
| Telfa Dressings | Covidien | Cat# 2132 |
Highlights.
Fibroblast subpopulations exist in the mouse dermis
Prrx1-expressing fibroblasts contribute to ventral skin scarring
Prrx1 defines fibroblasts in the mouse dermis with scar-forming potential
ACKNOWLEDGMENTS
We would like to acknowledge Patty Lovelace and Catherine Crumpton in the Shared FACS Facility in the Lokey Stem Cell Institute, the Dauskardt lab at Stanford University for use of microtester, and Vida Shokoohi at the Stanford Functional Genomics Facility for technical help with sequencing library preparation. This work was supported by the Hagey Laboratory for Pediatric Regenerative Medicine; the Gunn/Olivier Research Fund; the Stinehart-Reed Fund (to M.T.L.); NIH grants R01 GM116892 (to H.P.L. and M.T.L.), P50-HG007735 (to H.Y.C.), R0I DE 027346 (to D.C.W.), and R01 GM136659 (to M.T.L.); the Scleroderma Research Foundation (to H.Y.C.); the Plastic Surgery Research Foundation (to M.R.B.); the U.K. Fullbright Commission (to M.G.); the Sarnoff Cardiovascular Research Foundation (to A.H.S.); and F.P. Johnson, Jr. (to H.Y.C. and M.T.L.). H.Y.C. is an investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
H.Y.C. is a co-founder of Accent Therapeutics and Boundless Bio and an advisor to 10x Genomics, Arsenal Bioscience, and Spring Discovery.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108356.
REFERENCES
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38. [DOI] [PubMed] [Google Scholar]
- Carney MJ, Weissler JM, Fox JP, Tecce MG, Hsu JY, and Fischer JP (2017). Trends in open abdominal surgery in the United States-Observations from 9,950,759 discharges using the 2009–2013 National Inpatient Sample (NIS) datasets. Am. J. Surg 214, 287–292. [DOI] [PubMed] [Google Scholar]
- Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, and Brown PO (2002). Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. USA 99, 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie JD, Grosser L, Murawala P, Schuez M, Michel M, Tanaka EM, and Sandoval-Guzman T (2019). The Prrx1 limb enhancer marks an adult population of injury-responsive, multipotent dermal fibroblasts. BioRxiv. 10.1101/524124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, and Watt FM (2015). Understanding fibroblast heterogeneity in the skin. Trends Cell Biol. 25, 92–99. [DOI] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, and Watt FM (2013). Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano RD, Michaels J 5th, Dobryansky M, Levine JP, and Gurtner GC (2004). Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen. 12, 485–492. [DOI] [PubMed] [Google Scholar]
- Garza RM, Paik KJ, Chung MT, Duscher D, Gurtner GC, Longaker MT, and Wan DC (2014). Studies in fat grafting: Part III. Fat grafting irradiated tissue–improved skin quality and decreased fat graft retention. Plast. Reconstr. Surg. 134, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 10, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJ, Ilagan F, Kuo AH, Hsieh RW, and Cai S (2020). Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Jones FS, Zhou B, and Rabinovitch M (1999). Induction of vascular smooth muscle cell tenascin-C gene expression by denatured type I collagen is dependent upon a beta3 integrin-mediated mitogen-activated protein kinase pathway and a 122-base pair promoter element. J. Cell Sci. 112, 435–445. [DOI] [PubMed] [Google Scholar]
- Jones FS, Meech R, Edelman DB, Oakey RJ, and Jones PL (2001). Prx1 controls vascular smooth muscle cell proliferation and tenascin-C expression and is upregulated with Prx2 in pulmonary vascular disease. Circ. Res. 89, 131–138. [DOI] [PubMed] [Google Scholar]
- Korosec A, Frech S, Gesslbauer B, Vierhapper M, Radtke C, Petzelbauer P, and Lichtenberger BM (2018). Lineage Identity and Location within the Dermis Determine the Function of Papillary and Reticular Fibroblasts in Human Skin. J. Invest. Dermatol. 139, 342–351. [DOI] [PubMed] [Google Scholar]
- Labrousse AL, Ntayi C, Hornebeck W, and Bernard P (2004). Stromal reaction in cutaneous melanoma. Crit. Rev. Oncol. Hematol. 49, 269–275. [DOI] [PubMed] [Google Scholar]
- Landén NX, Li D, and Ståhle M (2016). Transition from inflammation to proliferation: a critical step during wound healing. Cell. Mol. Life Sci. 73, 3861–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan A, Duscher D, Whittam AJ, Paik KJ, Zielins ER, Brett EA, Atashroo DA, Hu MS, Lee GK, Gurtner GC, et al. (2016). Cell-assisted lipotransfer improves volume retention in irradiated recipient sites and rescues radiation-induced skin changes. Stem Cells 34, 668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, and Watt FM (2018). Fibroblast heterogeneity: implications for human disease. J. Clin. Invest. 128, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh E, Gonzalez DG, Lathrop EA, Boucher J, and Greco V (2018). Positional stability and membrane occupancy define skin fibroblast homeostasis in vivo. Cell 175, 1620–1633.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JF, and Olson EN (2000). Identification of a prx1 limb enhancer. Genesis 26, 225–229. [PubMed] [Google Scholar]
- McKean DM, Sisbarro L, Ilic D, Kaplan-Alburquerque N, Nemenoff R, Weiser-Evans M, Kern MJ, and Jones PL (2003). FAK induces expression of Prx1 to promote tenascin-C-dependent fibroblast migration. J. Cell Biol. 161, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SE (2018). Revitalizing Aging Skin through Diet. Cell 175, 1461–1463. [DOI] [PubMed] [Google Scholar]
- Nakhleh RE, Wick MR, Rocamora A, Swanson PE, and Dehner LP (1990). Morphologic diversity in malignant melanomas. Am. J. Clin. Pathol. 93, 731–740. [DOI] [PubMed] [Google Scholar]
- Nichols DH, and Bruce LL (2006). Migratory routes and fates of cells transcribing the Wnt-1 gene in the murine hindbrain. Dev. Dyn. 235, 285–300. [DOI] [PubMed] [Google Scholar]
- Ogawa R, Okai K, Tokumura F, Mori K, Ohmori Y, Huang C, Hyakusoku H, and Akaishi S (2012). The relationship between skin stretching/contraction and pathologic scarring: the important role of mechanical forces in keloid generation. Wound Repair Regen. 20, 149–157. [DOI] [PubMed] [Google Scholar]
- Östman A, and Augsten M (2009). Cancer-associated fibroblasts and tumor growth–bystanders turning into key players. Curr. Opin. Genet. Dev. 19, 67–73. [DOI] [PubMed] [Google Scholar]
- Philippeos C, Telerman SB, Oulès B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, and Watt FM (2018). Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J. Invest. Dermatol. 138, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Guerrero-Juarez CF, Ito M, Li YR, Dedhia PH, Zheng Y, Shao M, Gay DL, Ramos R, Hsi TC, et al. (2017). Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, et al. (2015). Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalbach A, Kraft R, Herrmann K, Haustein U-F, and Anderegg U (1998). The monoclonal antibody AS02 recognizes a protein on human fibroblasts being highly homologous to Thy-1. Arch. Dermatol. Res. 290, 360–366. [DOI] [PubMed] [Google Scholar]
- Salzer MC, Lafzi A, Berenguer-Llergo A, Youssif C, Castellanos A, Solanas G, Peixoto FO, Attolini CS-O, Prats N, and Aguilera M (2018). Identity Noise and Adipogenic Traits Characterize Dermal Fibroblast Aging. Cell 175, 1575–1590.e22. [DOI] [PubMed] [Google Scholar]
- Satoh A, Gardiner DM, Bryant SV, and Endo T (2007). Nerve-induced ectopic limb blastemas in the Axolotl are equivalent to amputation-induced blastemas. Dev. Biol. 312, 231–244. [DOI] [PubMed] [Google Scholar]
- Satoh A, Makanae A, Hirata A, and Satou Y (2011). Blastema induction in aneurogenic state and Prrx-1 regulation by MMPs and FGFs in Ambystoma mexicanum limb regeneration. Dev. Biol. 355, 263–274. [DOI] [PubMed] [Google Scholar]
- Shook BA, Wasko RR, Rivera-Gonzalez GC, Salazar-Gatzimas E, López-Giráldez F, Dash BC, Muñ oz-Rojas AR, Aultman KD, Zwick RK, and Lei V (2018). Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362, eaar2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell JM, and Caplan AI (2004). Fibroblast heterogeneity: more than skin deep. J. Cell Sci. 117, 667–675. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Satoh A, Ide H, and Tamura K (2005). Nerve-dependent and -independent events in blastema formation during Xenopus froglet limb regeneration. Dev. Biol. 286, 361–375. [DOI] [PubMed] [Google Scholar]
- Tabib T, Morse C, Wang T, Chen W, and Lafyatis R (2018). SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin. J. Invest. Dermatol. 138, 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickle C (2015). How the embryo makes a limb: determination, polarity and identity. J. Anat. 227, 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L, Kirsch E, Müller P, and Krieg T (1984). Collagen type distribution and macromolecular organization of connective tissue in different layers of human skin. J. Invest. Dermatol. 82, 156–160. [DOI] [PubMed] [Google Scholar]
- Wong JW, Gallant-Behm C, Wiebe C, Mak K, Hart DA, Larjava H, and Häkkinen L (2009). Wound healing in oral mucosa results in reduced scar formation as compared with skin: evidence from the red Duroc pig model and humans. Wound Repair Regen. 17, 717–729. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, and Iseki S (2008). Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev. 125, 797–808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA bulk sequencing and scRNA sequencing data generated during this study have been deposited in the NCBI’s Gene expression Omnibus and is accessible through GEO series accession number GSE159345 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159345).