Abstract
Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), found in cooked meat, is a known food carcinogen that causes several types of cancer, including breast cancer, as PhIP metabolites produce DNA adduct and DNA strand breaks. Curcumin, obtained from the rhizome of Curcuma longa, has potent anticancer activity. To date, no study has examined the interaction of PhIP with curcumin in breast epithelial cells. The present study demonstrates the mechanisms by which curcumin inhibits PhIP-induced cytotoxicity in normal breast epithelial cells (MCF-10A). Curcumin significantly inhibited PhIP-induced DNA adduct formation and DNA double stand breaks with a concomitant decrease in reactive oxygen species (ROS) production. The expression of Nrf2, FOXO targets; DNA repair genes BRCA-1, H2AFX and PARP-1; and tumor suppressor P16 was studied to evaluate the influence on these core signaling pathways. PhIP induced the expression of various antioxidant and DNA repair genes. However, co-treatment with curcumin inhibited this expression. PhIP suppressed the expression of the tumor suppressor P16 gene, whereas curcumin co-treatment increased its expression. Caspase-3 and -9 were slightly suppressed by curcumin with a consequent inhibition of cell death. These results suggest that curcumin appears to be an effective anti-PhIP food additive likely acting through multiple molecular targets.
Keywords: Phytochemicals, Cancer prevention, Reactive oxygen species, DNA damage, DNA adduct, Heterocyclic amines
Introduction
The prevention of cancer through diet is categorized as one of the most effective ways to reduce cancer incidence [1,2]. A few studies have demonstrated an association between an elevated risk of breast cancer and high consumption of well-done meat [3]. This correlation between increased cancer risk and meat preparation is most likely due to the production of high levels of heterocyclic amines [4]. In 2006, the nonprofit health organization ‘Physicians Committee for Responsible Medicine’ tested samples of grilled chicken from national fast food giants (McDonald’s, Burger King, Outback, Chick-fil-A, Applebee’s, Chili’s and TGI Friday’s) and found it to contain various carcinogenic compounds classified as heterocyclic amines (HCAs) [5]. Therefore, people who primarily consume fast food might be at greater risk of developing cancer because of HCAs but such a diet can also lead to obesity, which increases cancer risk as well. Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most abundantly found HCA in the human diet [3]. Several studies have shown that PhIP can induce tumors in breast, prostate and colon tissue [6–8] and rodent models [9–12]. N-Hydroxy derivatives are formed by the oxidation of PhIP by cytochrome P-450 1A2 (CYP1A2) [13]. Acetylation or sulfation of these derivatives results in the formation of a free radical that covalently binds to the guanine residues of DNA at the C8 position. These changes lead to the formation of adducts [14] which transverse GC → TA and cause G-rich repetitive sequences to undergo frameshift mutations [15]. Due to these mutations, PhIP exhibits genotoxicity, which in turn leads to DNA damage, chromosome aberrations, micronuclei formation, and sister chromatid exchange [16–18]. Breast epithelial cells contain all the machinery to metabolize HCA and the genotoxic effects of these metabolites may lead to breast cancer [6].
Curcumin (diferuloylmethane) is a polyphenol and major component of the spice turmeric. Turmeric is derived from the rhizome of the Indian plant Curcuma longa, which is a member of the Zingiberacae (ginger) family and used in various food preparations. Curcumin inhibits cell proliferation and has anticancer effects [19]. Recently, several researchers have demonstrated the anticancer effect of curcumin in prostate [20,21], breast [22–25], colon [26–28], and liver cancer [29]. Thus, curcumin has gained interest as a dietary supplement because there is substantial evidence in pre-clinical models that curcumin is a potent chemopreventive dietary agent [30–32]. Cole and colleagues demonstrated the inhibition of PhIP-induced DNA strand breaks by the antioxidant diallyl sulfide (found in garlic) in MCF-10A cells [33]. However, none of the studies have investigated the effect of curcumin on PhIP-induced carcinogenicity. Previous studies have shown that PhIP induces the production of reactive oxygen species (ROS) and DNA adduct formation [6,7,15,16]. Phytochemicals like curcumin are able to inhibit DNA adduct formation [34]. We hypothesized that curcumin may be a potential food additive that may be inhibitory to PhIP-induced carcinogenicity by inhibiting ROS production, DNA adduct formation and DNA strand breaks. In the present study, we have explored the molecular mechanisms by which curcumin inhibits PhIP-induced ROS production, DNA adduct formation and DNA damage using MCF-10A normal breast epithelial cells as a model system.
Materials and methods
Chemicals and reagents
RPMI 1640, 1× Hank’s Balanced Salt Solution (HBSS) and horse serum were obtained from Cellgro (Manassas, VA). Trypsin-EDTA was obtained from Gibco (Invitrogen), penicillin/streptomycin was obtained from MB Chemicals. Ultrapure normal melting point agarose was obtained from Invitrogen (Carlsbad, CA). Epidermal growth factorwas obtained from BD Biosciences (San Jose, CA). Dimethyl sulfoxide (DMSO), insulin, hydrocortisone, Triton X-100, PBS, NaOH, Trizma base, NaCl, diethylpyrocarbonate (DEPC), ethanol and curcumin were all purchased from Sigma (St. Louis, MO). PhIP was purchased from Toronto Research Chemicals (Toronto, Ontario). PhIP was dissolved in a minimum volume of DMSO (0.1 volume) and a 10 mM stock solution was made by adding the remaining volume (0.9 volume) of culture media. Acridine orange/propidium iodide (AOPI) and Annexin-V-PI solution were purchased from Nexelom Biosciences (Lawrence, MA). 2’,7’-Dichlorodihydrofluorescein diacetate (DCFDA) was purchased from Molecular Probes, Inc. (Eugene, OR). Comet microscope slides were purchased from Trevigen, Inc. (Gaithersburg, MD).
Cell culture
MCF-10A human breast epithelial cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured in a humidified incubator at 37 °C under 5% CO2 atmospheric conditions in RPMI media supplemented with 10 μg/ml insulin, 20 ng/ml epidermal growth factor, 10 mg/ml hydrocortisone, 5% horse serum and 1% penicillin-streptomycin (10,000 U/ml). Experiments were conducted within 25 passages.
Dose curve
MCF-10A cells were treated with PhIP at various concentrations ranging from 0.001 μM (1 nM) to 500 μM for 24 h and 48 h to study PhIP cytotoxicity. Five thousand cells were plated per well in 96-well plates and treated after one day with varying concentrations of PhIP. Cell death was analyzed using a Cell counting kit-8 (Dojindo Laboratories, CA, USA). After treatment, a 10 μl aliquot of WTS Solution (Dojindo Laboratories, Santa Clara, CA, USA) was added to each well and incubated at 37 °C for 2 h to allow color development. The plates were analyzed on a Bio-Tek Synergy HT microplate reader using Gen5 2.00 software and the absorbance at 480 nm was determined for each sample. In this colorimetric assay, viable cells convert the WTS tetrazolium compound to a formazan product soluble in culture media. The amount of formazan product formed is directly proportional to the number of living cells in the culture.
PhIP and curcumin treatment and cell survival assays
In separate experiments, MCF-10A cells were treated with or without PhIP (50 and 250 μM) in the presence or absence of curcumin (25–200 μM) and cell viability determined. Cell viability was assessed using the cell counting kit-8 (Dojindo Laboratories) as described above. MCF-10A cells treated with 0.01% DMSO served as negative controls. Cell viability was expressed as percent survival which is calculated based on the formula 100 × OD of test/OD of control. The control OD value was normalized to 100% viability. In a parallel experiment, treated and control cells were stained with AOPI and counted using a Cellometer Vision CBA instrument; cell viability was calculated and results were compared to verify the results using cell counting kit-8.
For all other experiments PhIP (at 50 μM or 250 μM) and curcumin (at 150 μM) were used for 24 h. These PhIP concentrations were chosen based on a PhIP dose curve showing significant cell death; curcumin at 100 and 150 μM inhibited PhIP-induced cell death. Therefore, curcumin at 150 μM concentration was used to determine the mechanism. Cells were pretreated with curcumin 15 minutes before dosing with PhIP (50 or 250 μM).
ROS assay
MCF-10A cells were grown in 96 well plates and treated with or without PhIP (50 and 250 μM) in the presence and absence of curcumin (150 μM). After 24 hours, the cells were rinsed 3 times with 1× HBSS to remove curcumin or PhIP. The cells were incubated with 5 μM dichlorodihydrofluorescein diacetate (DCF) (Molecular Probes, Inc., Eugene, OR) for 45 minutes. The cells were again washed with 1× HBSS twice; 100 μL of 1XHBSS was added to each well and the fluorescence was measured using a Biotek, Synergy HT instrument with an excitation of 475–495 and emission of 518–528. Similarly, microscopic images were taken for control and treated cells to record the comparative fluorescence immediately using an Olympus DP 71 microscope.
Anti DNA adduct analysis
The effect of curcumin on PhIP-induced DNA adduct formation was determined using an immunofluorescence assay. MCF-10A cells were grown on coverslips and treated with or without PhIP in the presence and absence of curcumin for 24 h. Following treatment, cells were fixed with 3.7% PFA (paraformaldehyde) and permeabilized (0.2% Triton-X-100) at room temperature. After blocking with 5% BSA (bovine serum albumin), DNA adducts were detected with an anti-DNA adduct primary antibody (1:50 dilution) [35] and visualized using an Alexa Fluor 455 anti-rabbit secondary antibody (Invitrogen). Slides were counterstained with 4’,6-diamidino-2-phenylindole (DAPI nuclear stain) from Calbiochem (San Diego, CA).
Alkaline comet assay (single cell-gel electrophoresis)
The alkaline comet assay was performed as described by Singh et al. [36] with minor modifications. Briefly, three hundred thousand cells were treated as indicated in six-well culture plates for 24 h at 37 °C. Cells treated with vehicle (0.01% DMSO) served as the negative vehicle control and PhIP-treated cells as the positive control. Following treatments, cells were washed with ice-cold PBS, trypsinized and centrifuged at 1000 rpm for 5 min. Subsequently, 100 μl of the cell suspension containing 2 × 104 cells was mixed with 900 μl of 0.75% low-melting point agarose and immediately spread on comet microscope slides. The cell-gel sandwich was incubated to allow gel solidification and stored at 4 °C. The slides were immersed in ice-cold lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% Triton X-100, pH 10) for 1 h at 4 °C to remove cell proteins and break down cell membranes. After lysis, slides were placed in freshly prepared electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13) for 30 min to allow DNA unwinding before electrophoresis. Electrophoresis was performed for 30 min at 25 V (300 mA). All of the above steps were conducted under low light in a refrigerated chamber to prevent additional DNA damage. After electrophoresis, the slides were neutralized (0.4 M Tris, pH 7.5), washed and stained with propidium iodide (2.5 μg/ml). After drying overnight at room temperature, slides were viewed on an inverted fluorescence microscope (Olympus DP 71) and images were transferred to a computer with a digital camera.
Comet analysis
A total of 75 cells were scored (25 cells per individual experiment and a total of 3 experiments) to determine the olive tail moment for each treatment. Imaging was performed with the Comet analysis system (Loats Associate System, Westminster, MD). This software defines head and tail regions and evaluates a range of derived parameters including tail moment, an index of DNA damage that considers both the tail length (comet length), and the fraction of DNA in the comet tail (TM = % DNA in tail × tail length)/100) to evaluate the length of DNA migration as an indicator of DNA damage [37].
Annexin-V–FITC staining
Cell death evaluation was performed using annexin V–PI assay for apoptosis. After a 24-hour treatment of MCF-10A cells with or without PhIP in the presence and absence of curcumin cells were harvested, washed twice with cold PBS and re-suspended in 1× binding buffer (1.0 mmol/l HEPES [4-(2-hydroxyethl)-1-piperazineethanesulfonic acid], pH = 7.4, 140 mmol/l NaOH, 2.5 mmol/l CaCl2) (Bio Legend, San Diego, CA) and stained with Annexin V–FITC and propidium iodide (PI); assays were then evaluated for apoptosis using Cellometer Vision Image cytometry according to the manufacturer’s protocol (Nexcelom Bioscience LLC, Lawrence, MA). Annexin V–FITC and PI-positive cells were either in the end stages of apoptosis or undergoing necrosis and were considered dead. PI-positive cells were considered necrotic.
RT-PCR analysis
RT-PCR was performed as per standard protocols. Briefly, total RNA was isolated from treated and untreated cells using Trizol (Invitrogen, San Diego, CA) according to the manufacturer’s instructions and used for first strand cDNA synthesis using MMLV reverse transcriptase (Promega, Madison, WI). The cDNA was then used as a template for PCR amplification. The PCR primers for specific genes were designed based on the nucleotide sequences available in GenBank. Primers for different genes were designed using NCBI pick primer software. Reverse transcription-PCR (RT-PCR) was repeated several times (at least twice) with each RNA sample. Hypoxanthine phosphoribosyltransferase (HPRT) was used as the internal standard. The sequences of primers are listed in Supplementary Table S1.
Western blot analysis
MCF-10A cells were treated with or without PhIP in the presence and absence ofcurcumin for 24 h. Following treatment, cells were lysed by incubation with ice-cold lysis buffer (50 mM HEPES, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.25% deoxycholate) supplemented with protease inhibitor cocktail (Calbiochem, San Diego, CA). Lysates were clarified by centrifugation, boiled in Laemmli buffer, and separated on 10% mini-gels followed by electrophoretic transfer to nitrocellulose. The blots were blocked with 5% bovine serum albumin in PBS containing Tween 20 and incubated with primary antibodies overnight at 4 °C. The following primary antibodies were tested: monoclonal anti-caspase-3, polyclonal anti-caspase-9, polyclonal anti-PARP-1, anti GPX-1, anti-catalase (Cell Signaling Technologies), anti-cleaved caspase-3 and anti-β-actin antibodies (Sigma Aldrich). All the primary antibodies were used at a 1:1000 dilution except for that recognizing β-actin (1:2000). After overnight incubation the appropriate HRP-conjugated secondary antibody or antimouse for monoclonal antibody (Bio-Rad Laboratories, Hercules, CA) was used, and the bands were visualized using chemiluminescence according to the manufacturer’s instructions (Pierce, Rockford, Ill). The band intensity of immunoblots were quantified using ImageJ software (http://imagej.nih.gov/ij/) for different proteins and treatments. The absolute band intensity was converted to relative expression as compared to control values.
Statistical analysis
Results are expressed as means ± SEM from a minimum of three independent experiments. Data were analyzed by Student t-test using Graphpad Prism or ANOVA with a Student–Newman–Keuls post-hoc test using Graphpad Instat software.
Results
Dose response of PhIP on the viability of MCF-10A cells
The effect of PhIP on the viability of MCF 10A breast epithelial cells was determined. Lower doses of PhIP (1 nM–10 μM) had no significant effect on MCF-10A breast epithelial cell viability. However, at higher doses PhIP decreased cell viability in a dose-dependent manner. Cells treated with 50 μM PhIP showed 80% (±6.09, P < 0.05) viability, whereas those treated with 250 μM showed 33.6% (±1.90, p < 0.05) viability as compared to the control (100% viability) (Fig. 1a). Above 300 μM, PhIP did not induce any further significant decrease in cell viability. Therefore, 50 μM and 250 μM concentrations of PhIP were selected for subsequent co-treatment experiments with curcumin. A dose response of curcumin (25–200 μM) on 50 μM and 250 μM PhIP-treated MCF-10A cells was also determined. Our results show that curcumin at a concentration of 150 μM significantly inhibited PhIP-induced reductions in viability at 24 h, with cells treated with 50 μM PhIP plus 150 μM curcumin, and 250 μM PhIP plus 150 μM curcumin showing 101% (±3.95) and 96.86% (±8.17) viability, respectively. These results suggest that in breast epithelial cells curcumin can inhibit PhIP-induced cytotoxicity in a dose-dependent manner at 24 hours (Fig. 1b). Forty-eight hours of PhIP treatment did not induce additional pronounced cytotoxicity. Therefore, in all experiments we have used 50 μM and 250 μM of PhIP and co-treated with 150 μM curcumin for 24 hr to determine the anticancer effects.
Fig. 1.
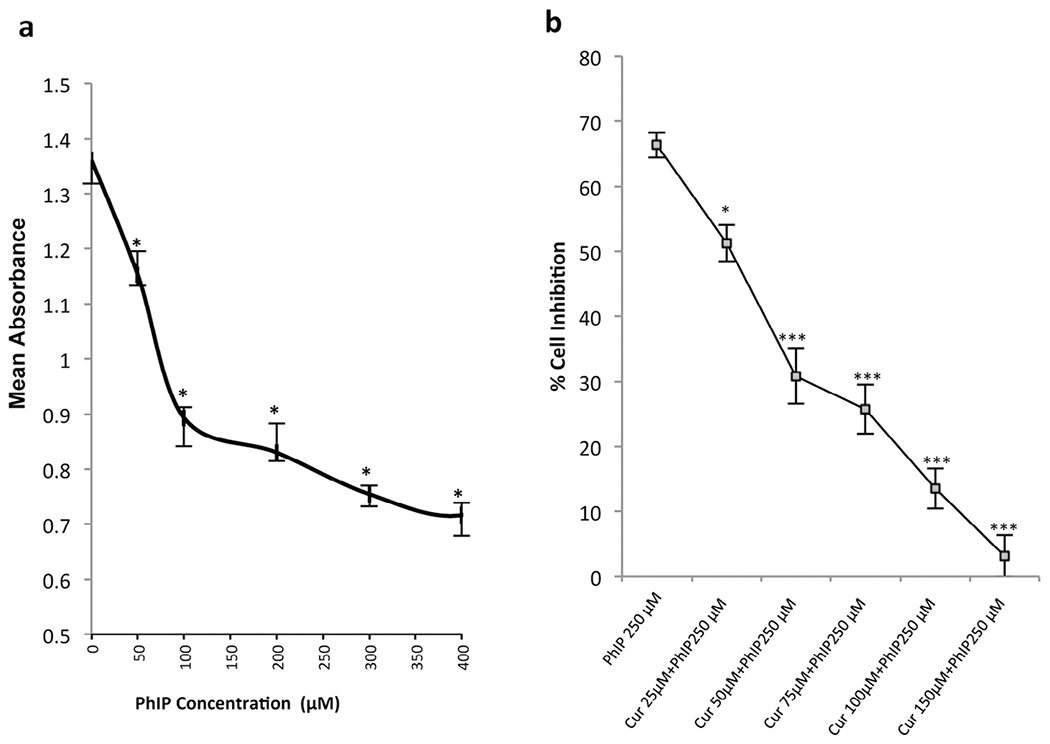
(a) The effect of PhIP at various concentrations on cell number in MCF-10A cells. PhIP concentrations ranging from 50 μM to 400 μM were used to treat MCF-10A cells for 24 hours. Values represent the means ± SEM of 3 separate experiments; *P < 0.05 by an ANOVA followed by a Student–Newman–Keuls post-hoc test. PhIP shows cytotoxicity of 20% at a 50 μM concentration in MCF-10A cells, but at a 250 μM concentration PhIP imparted cytotoxicity of almost 60%. (b) Effect of various doses of curcumin on 250 μM PhIP induced toxicity in MCF-10A cells. 250 μM PhIP was used to treat MCF-10A cells and the inhibition of its cytotoxic effect was studied upon cotreatment with various concentrations of curcumin. All doses of curcumin produced reversal of PhIP-induced cytotoxicity but curcumin at 150 μM exhibited the greatest degree of reversal of cytotoxicity (p < 0.05). Values are mean ± SEM from three independent experiments performed in triplicate. The statistical significance is expressed as *indicates p < 0.05, **indicates p < 0.01, and *** indicates a p < 0.001.
Curcumin inhibits PhIP-induced ROS production
The antioxidant capacity of curcumin was analyzed in the absence and in the presence of PhIP, a well-known peroxidant agent. Its efficiency was evaluated in terms of inhibition of intracellular reactive oxygen species (ROS) production induced spontaneously or in the presence of PhIP.
Intracellular free radical species were detected by measuring the fluorescence intensity values due to the oxidation of DCF and expressed as relative fluorescence units (RFU) with 1.0 and 10.0 μM H202 used as a positive control. In the absence of PhIP, ROS production was limited (2.66 ± 0.33) but increased significantly in the presence of PhIP in a dose-dependent manner (50 μM: 6.33 ± 0.33) and (250 μM: 8.33 ± 0.33). Co-treatment of MCF-10A cells with curcumin resulted in a significant decrease in PhIP-induced ROS production (3.14 ± 0.33 and 3.63 ± 0.33 with 50 and 250 μM PhIP respectively) such that ROS were reduced almost to control values (Fig. 2a, b).
Fig. 2.
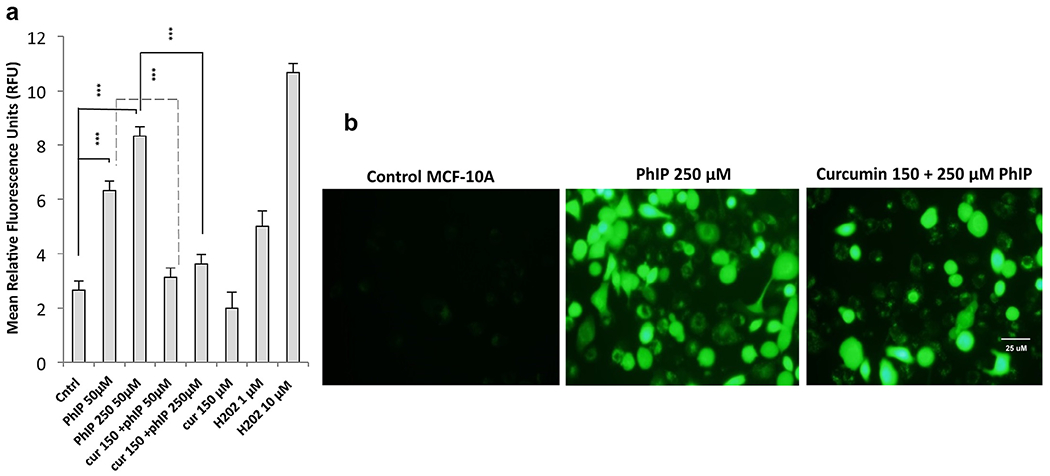
(a) Quantitative fluorescence intensity analysis of ROS production in PhIP-treated MCF-10A cells upon co-treatment with curcumin. MCF-10A cells were treated with or without PhIP in the presence and absence of curcumin for 24 hours and then fluorescence intensity was measured with DCF-DA (5 μg) within 45 minutes. Co-treatment with curcumin reduced the PhIP-stimulated ROS production in MCF-10A cells. Values are mean ± SEM from three independent experiments performed in triplicate. The statistical significance is expressed as ***p < 0.001. (b) Relative DCF-DA fluorescence emission from cells treated with PhIP ± curcumin. MCF-10A cells were observed with an inverted fluorescence microscope equipped with a FITC filter. Shown are cells expressing fluorescence which represents ROS.
PhIP-treated MCF-10A cells exhibit DNA adduct formation which is reduced in the presence of curcumin
DNA adduct formation was determined using an immunofluorescence method with an anti-DNA adduct primary antibody [35]. DNA adducts accumulated in a dose-dependent manner in both 50 and 250 μM PhIP-treated breast epithelial MCF 10A cells. When MCF 10A cells were co-treated with curcumin (150 μM), PhIP-induced DNA adduct formation was noticeably reduced (Fig. 3). These results clearly demonstrate that PhIP causes formation of DNA adducts within 24 h in MCF-10A cells, whereas curcumin inhibits the PhIP-induced DNA adduct formation.
Fig. 3.
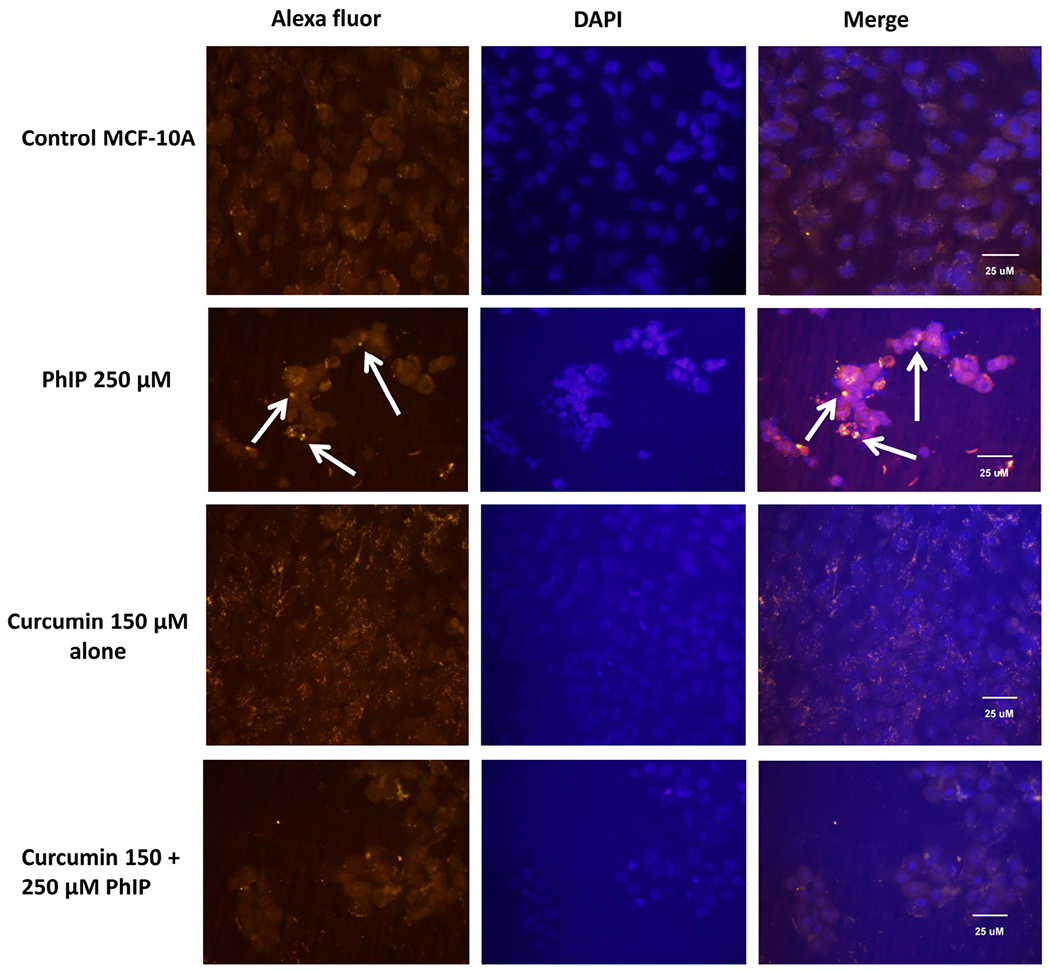
Immunofluorescence assay of DNA adduct formation using an anti-DNA adduct antibody. MCF-10A ceils (untreated, treated with PhIP, co-treated with curcumin and PhIP or treated with curcumin alone) were incubated with anti-DNA adduct primary antibodies and an Alexa fluor-conjugated secondary antibody. Nuclei were stained with DAPI. Cells showing DNA adduct formation are indicated by arrows.
Curcumin inhibits double-strand DNA damage caused by PhIP
The ability of curcumin to inhibit PhIP-induced DNA double strand breaks in MCF-10A human breast epithelial cells was then determined using single-cell gel electrophoresis (the comet assay) using the mean olive tail moment (OTM). Tail moment is defined as the product of the tail length and the fraction of total DNA in the tail. Tail moment incorporates a measure of the smallest detectable size of migrating DNA (reflected in the comet tail length) and the number of relaxed/broken pieces represented by the intensity of the DNA found in the tail [34]. OTM in control cell populations ranged from 0.0 to 0.92 with an average of 0.155 ± 0.022 (SEM). PhIP-treated cells displayed a significant increase in DNA strand breaks as the mean OTM for PhIP was 1.02 ± 0.173 and 1.50 ± 0.173 for 50 μM or 250 μM PhIP, respectively (Fig. 4a). Previous studies showed that the DNA strand breaks that were produced after 24 h were repaired after 48 and 72 h [23]; therefore, we conducted studies only for 24 h. Treatment with curcumin (150 μM) had no effect on the production of DNA strand breaks compared to the controls (mean OTM 0.17 ± 0.026). However, pretreatment with curcumin inhibited DNA double strand breaks induced by PhIP after 24 h. Thus, the mean olive tail moments in cells co-treated with PhIP and curcumin were reduced to 0.26 ± 0.034 and 0.53 ± 0.055 for 50 and 250 μM PhIP, respectively. Fig. 4a illustrates the DNA strand breaks induced by PhIP and the inhibition of these strand breaks by curcumin. Individual cells with no DNA strand breaks appear as spheres with no tail. However, the cells that contain DNA damage appear as spheres with tails that resemble comets. This result suggests that curcumin has an inhibitory effect on PhIP-induced DNA strand breakage in human breast epithelial cells. These results also support the DNA adduct data since the reduction in DNA adduct formation in the presence of curcumin represents decreased DNA damage.
Fig. 4.
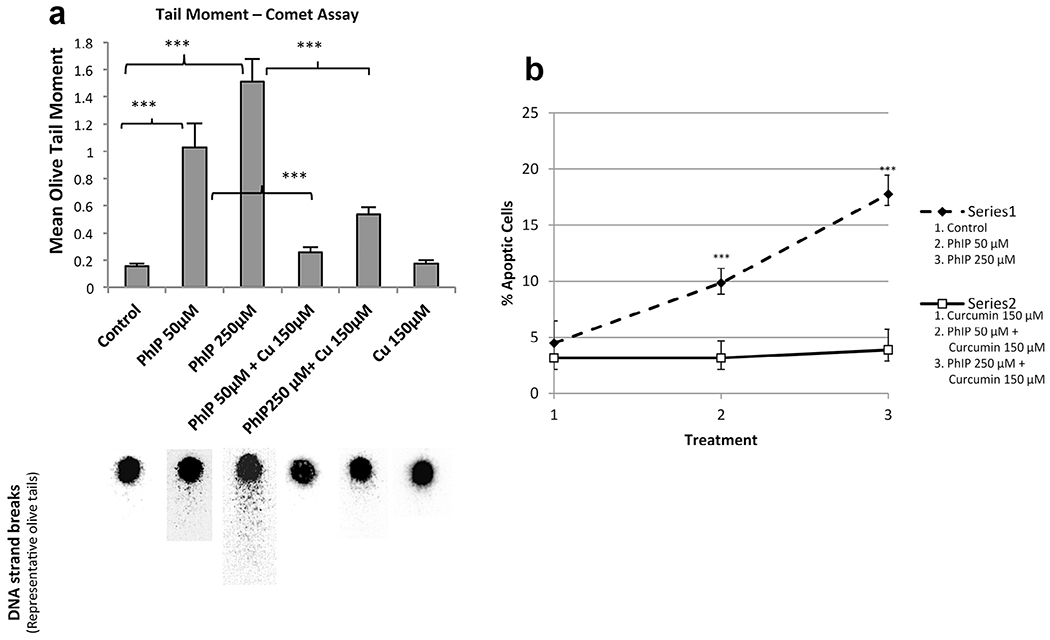
(a) Upper panel – Inhibition of PhIP-induced DNA strand breaks by curcumin at 24 h. MCF 10A cells were treated as indicated and analyzed for comet tail moment. Each bar on the graph represents the mean olive tail moment ± SEM of three independent experiments in which at least 25 individual cells per treatment per experiment were scored; ***P < 0.001. Lower panel – Representative pictures for each condition showing the head and comet tail, which represent DNA double strand damage. (b) Annexin-V–FITC analyses. Apoptotic cells obtained upon treatment with or without PhIP in the presence or absence of curcumin for 24 h. PhIP treatment of MCF-10A cells increased the number of apoptotic cells but upon co-treatment with curcumin (150 μM) the number of apoptotic cells was reduced almost to control values; ***p < 0.001.
PhIP-induced apoptosis is inhibited by curcumin
Annexin-V–FITC and PI staining were performed to analyze ceil death induced by PhIP as well as the effect of curcumin on this response. The results show that the percentage of apoptotic cells increased in a dose-dependent manner upon PhIP treatment (p < 0.05). Similarly, the number of necrotic cells was significantly increased by 250 μM PhIP. Curcumin inhibited the PhIP-induced increase in apoptosis (Fig. 4b).
PhIP induced the expression of NRF2, FOXO, and DNA repair genes whereas it suppressed the expression of P-16, and this altered expression is reverted in the presence of curcumin
To understand the interaction of PhIP and curcumin at the molecular level reverse transcriptase PCR (RT-PCR) was performed. Oxidative stress signals through NRF-2 [Nuclear factor (erythroid-derived 2)-like 2] and its targets such as NQO-1 [NAD(P)H quionine oxidoreductase-1], GPX-1 [glutathione peroxidase] and GSR [glutathione reductase], as well as FOXO [forkhead box protein] targets such as CAT [catalase], GADD-45 [growth arrest and DNA damage-inducible 45] and PRDX-3 [Thioredoxin-dependent peroxide reductase], and the expression of these genes were monitored. H2AX [histone H2A], BRCA-1 [breast cancer 1, early onset] and P-16 (cyclin-dependent kinase inhibitor 2A) were also evaluated. The results of RT-PCR for these gene targets are shown in Fig.5a.
Fig. 5.
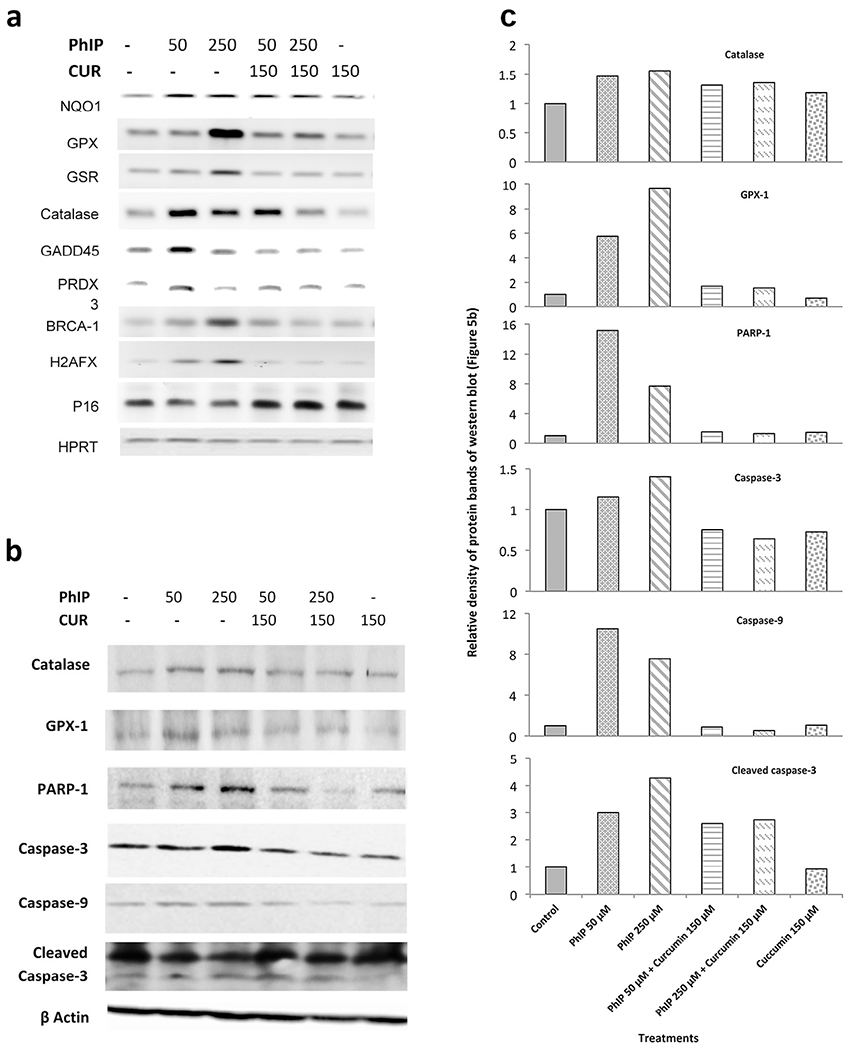
(a) Effect of PhIP and curcumin alone and in combination on Nrf2, FOXO, BRCA-1, H2AFX and P16 signaling pathways, with HPRT used as a normalization control. MCF 10A cells were treated for 24 h, total RNA was isolated and RT-PCR was applied to amplify specific gene products. The sequences of forward and reverse primers are given in Supplementary Table S1. Comparative band intensity was used to determine the induction or suppression of each transcript. All results were repeated at least twice with similar expression. (b) Western blot analysis results of PhIP-treated MCF-10A cells with and without co-treatment with curcumin. The antibodies used were: a) anti-caspase-3, b) anti-caspase-9, c) anti-GPX-1, d) anti-catalase, e) anti-PARP-1, and f) anti-β-actin as a loading control. All results were repeated at least once with similar results. (c) Relative band intensity of western blot, expressed as compared to control.
The NQO-1 gene is associated with prevention of reduction of quinones by an electron, which further activates free radical generation. PhIP treatment induced the expression of NQO-1 inMCF-10A breast epithelial cells, and curcumin consistently inhibited the PhIP-induced expression of NQO-1. These data are in agreement with the observed ROS production results. GPX-1 is an important antioxidant gene in humans. This gene has a major role in detoxification of hydrogen peroxide by scavenging ROS. The results show that PhIP induced the expression of GPX-1 transcripts; however, curcumin maintained GPX-1 expression near basal levels, presumably by suppressing PhIP-induced changes in oxidative stress. The expression of GADD-45 and PRDX-3 was also induced by PhIP treatment, especially at the lower dose of PhIP (50 μM). Co-treatment with curcumin maintained the mRNA levels of both GADD-45 and PRDX-3 near basal values, indicating either less PhIP-induced ROS production in the presence of curcumin or an ability of curcumin to act as an efficient antioxidant; in this case induction of antioxidant enzymes such as GPX, catalase, GADD-45 or PRDX-3 is not required.
BRCA-1 is found in all humans and is considered a caretaker gene involved in repairing DNA damage or destroying the damaged cells if the DNA cannot be repaired. PhIP treatment of MCF-10A cells induced the expression of BRCA-1 in a dose-dependent manner, indicating that PhIP causes DNA damage, as was also observed with the comet assay. However, curcumin inhibited the PhIP-elevated expression of BRCA-1 in MCF-10A cells, suggesting that curcumin plays a key role in protecting cells from PhIP-induced double-strand DNA breaks (consistent with the comet assay data) and consequently the induction of BRCA-1 expression is prevented. This clearly indicates that curcumin counteracts PhIP toxicity and therefore excess BRCA-1 is not required.
H2AFX (H2A histone family, member X) is one of several genes coding for histone H2A. PhIP treatment induces the expression of H2AFX indicating DNA damage and co-treatment with curcumin maintained basal levels of H2AFX, again indicating that curcumin had an inhibitory effect on PhIP-induced DNA damage.
Cyclin-dependent kinase inhibitor 2A (P16), which is also known as multiple tumor suppressor protein, plays an important role in regulating the cell cycle and reducing the risk of developing cancers, notably melanoma. P16 expression is suppressed by PhIP as compared to control; however, P16 expression was maintained in breast epithelial cells co-treated with curcumin. Since MCF-10A cells are P53 deficient, the expression of the P16 tumor suppressor is important to reduce PhIP carcinogenicity. Thus, down-regulation of the P16 transcript by PhIP could result in a carcinogenic effect, such that maintenance of P16 expression by curcumin suggests that this agent should reduce PhIP-induced carcinogenicity. The house-keeping gene hypoxanthine phosphoribosyl transferase (HPRT) was expressed uniformly in all groups (Fig. 5a).
Curcumin regulates DNA repair, apoptotic and antioxidant proteins
We analyzed the expression of antioxidant proteins to confirm the RT-PCR results. Our results have shown that PhIP treatment increases GPX-1 protein levels, and that co-treatment with curcumin blocks this effect in MCF-10A cells. Another antioxidant protein catalase also shows changes in protein expression in the presence of PhIP and curcumin similar to those observed with GPX-1 (Fig. 5b). Thus levels of GPX-1 and catalase protein correlate with expression of their transcripts. The band intensity for each treatment was calculated using ImageJ software and relative expression was calculated compared to control to determine the treatment effect on protein expression (Fig. 5c).
We then analyzed the expression of apoptotic proteins. Results revealed that PhIP treatment increased the expression of both caspase-3 and caspase-9; but co-treatment with curcumin reduced this PhIP-induced expression, although curcumin had no effect itself (Fig. 5b, c). The increased levels of cleaved caspase-3 also revealed that caspase-3 activation is increased in PhIP-treated samples as compared to all other groups (Fig. 5b, c).
We next analyzed the effects of PhIP and curcumin on the DNA repair protein PARP-1 in MCF-10A cells. PhIP increased the expression of total PARP-1; but co-treatment with curcumin of MCF-10A cells reduced the PhIP-elicited expression of PARP-1 to almost control levels. Treatment with curcumin alone showed similar levels to control (Fig. 5b, c).
Discussion
Breast cancer is one of the leading causes of death in women [38,39], and diet plays a major role in the development of the disease [1,2]. A direct relationship between red meat consumption and the PhIP associated development of breast cancer has been demonstrated [3]. In breast epithelial cells, PhIP induces mutagenesis by forming DNA adducts [40]. Since previous studies have demonstrated that phytochemicals can inhibit DNA adduct formation [41], inhibition of breast cancer formation using phytochemicals is a major area of research [42–44]. This is the first study demonstrating the mechanism by which curcumin can inhibit the carcinogenic effect of PhIP in MCF-10A breast epithelial cells. Normal breast epithelial cells have an inherent capacity to bioactivate PhIP which then causes DNA damage [33]. Our cytotoxicity data reveal that curcumin can inhibit PhIP-induced cell death. Previous studies with diallyl sulfide (at a 100 μM concentration) have also shown the ability of this agent to inhibit PhIP-induced cytotoxicity in normal breast epithelial cells [33]. We have observed that normal breast epithelial MCF-10A cells exhibit DNA adduct formation after 24 hrs of PhIP treatment at 50 and 250 μM concentrations. Bio-activated PhIP also causes the production of ROS; the effect of PhIP on both these processes has been established previously [45]. PhIP-induced DNA adducts and ROS lead to DNA double strand breaks, as observed in the present study. The cumulative effect of these factors affects cells’ normal behavior and is responsible for the decrease in cell viability. Indeed, annexin-V–FITC and PI analysis showed that cell death observed at the 50 μM PhIP concentration was due to apoptosis. However, the higher concentration of PhIP (250 μM) led to a significant number of both apoptotic and necrotic cells. Breast epithelial cells that evade cell death have the capability of transforming into breast cancer [46]. Many studies demonstrate that an imbalance in the production and detoxification of ROS may lead to various cancers [47,48]. Our results show that curcumin reduced DNA adduct formation, decreased DNA double strand breaks and reduced ROS production to basal levels to result in an inhibition of PhIP-induced cell death.
NAD(P)H quinone oxidoreductase 1 (NQO-1) is a Nrf2 target gene, which catalyzes the reduction and detoxification of highly reactive quinones that can cause redox cycling and finally lead to oxidative stress. The Nrf2 antioxidant response pathway is considered to be the primary cellular defense against the cytotoxic effects of oxidative stress [49]. PhIP treatment of MCF-10A cells induced NQO-1 expression; however, curcumin co-treatment returns the expression of NQO-1 to a value similar to the control. In this study GSR expression was also induced by PhIP and curcumin treatment maintains the GSR expression at control levels even in the presence of PhIP. Studies have shown that GSTs are induced by Nrf2 activation and represent an important route of detoxification [50]. The GADD-45 gene plays a major role in preventing transformation of normal cells into a malignant phenotype [43]. The regulation of this gene is mediated by DNA damage induced by alkylating agents, serum depletion or UV radiation. Our results show that PhIP treatment induced GADD-45 expression with higher levels at 50 μM PhIP as compared to 250 μM PhIP. Co-treatment of curcumin reduced GADD-45 expression. Hence GADD-45 may have a role in the inhibition of PhIP-induced carcinogenesis in normal breast epithelial cells.
The peroxiredoxin-3 (PRDX-3) gene was also induced by PhIP at the lower concentration; however, curcumin co-treatment down-regulated PRDX-3 expression. Cao and colleagues demonstrated that the PRDX gene product is responsible for inhibition of ROS, and that mice that lack this gene die prematurely of cancer due to excessive ROS production [51]. Our finding shows that PhIP induces ROS production, suggesting that PRDX-3 expression is induced to inhibit ROS. However, as curcumin itself can inhibit ROS production, induction of PRDX-3 is no longer necessary in its presence. Similarly, the antioxidant catalase gene was also induced by PhIP treatment but curcumin treatment inhibited this PhIP-induced catalase expression. Catalase is very important for protecting cells from oxidative damage and ROS, again suggesting an ability of curcumin to block ROS production thereby precluding the necessity of anti-oxidant protection.
Our results clearly show that many antioxidant genes are induced in the presence of PhIP. Both the Nrf2 and FOXO pathways are up-regulated by PhIP to scavenge the elevated ROS and protect cells from DNA adduct formation and the resulting DNA damage. Increased expression of H2AFX and BRCA-1 in the PhIP-treated group as well as data from comet assays, ROS monitoring and immunofluorescence with anti-DNA adduct antibodies supports this idea. Previous studies have shown that BRCA-1, P-53 and other tumor suppressor genes are able to increase GADD-45 expression [52–54]. Catalase and GADD-45 are associated with the FOXO pathway; however, these two genes perform different functions of detoxification and DNA repair, respectively.
Recent studies have shown that BRCA-1 regulates oxidative stress, which has a major role in cancer development. In our study, PhIP treatment increased the expression of BRCA-1 indicating that oxidative stress was induced by PhIP (also confirmed by the DCF assay); curcumin co-treatment reduced the oxidative stress allowing the expression of BRCA-1 to return toward basal levels. Studies have shown that BRCA-1 regulates Nrf2-dependent antioxidant signaling by physically interacting with Nrf2 and promoting its stability and activation [55].
In addition to increased BRCA-1, PhIP-treated cells also showed elevated levels of H2AFX. The capacity of BRCA-1 to bind directly to DNA contributes to its ability to inhibit nuclease activity. BRCA-1 also co-localizes with γ-H2AX in DNA double-strand break repair foci. This association plays a role in recruiting repair factors [56]. Increased expression of H2AFX in PhIP-treated MCF-10A cells may be due to its association with BRCA-1, which together repair the DNA damage induced by PhIP. Nevertheless it appears that the upregulation of these genes is unable to repair the DSBs to return the cell environment to normal. Indeed, the results of ROS measurements, DNA adduct formation and DNA strand breaks (comet assays) demonstrate a high degree of genetic disruption by PhIP. These findings prove that PhIP has carcinogenic effects and ultimately causes breast cancer as reported previously [6]. Thus, although PhIP induces antioxidant and DNA repair mechanisms through the Nrf2 and FOXO pathways, this response does not completely inhibit ROS or DNA adduct formation. Curcumin, however, inhibits both ROS and DNA adduct generation to rescue DNA damage (Fig. 6).
Fig. 6.
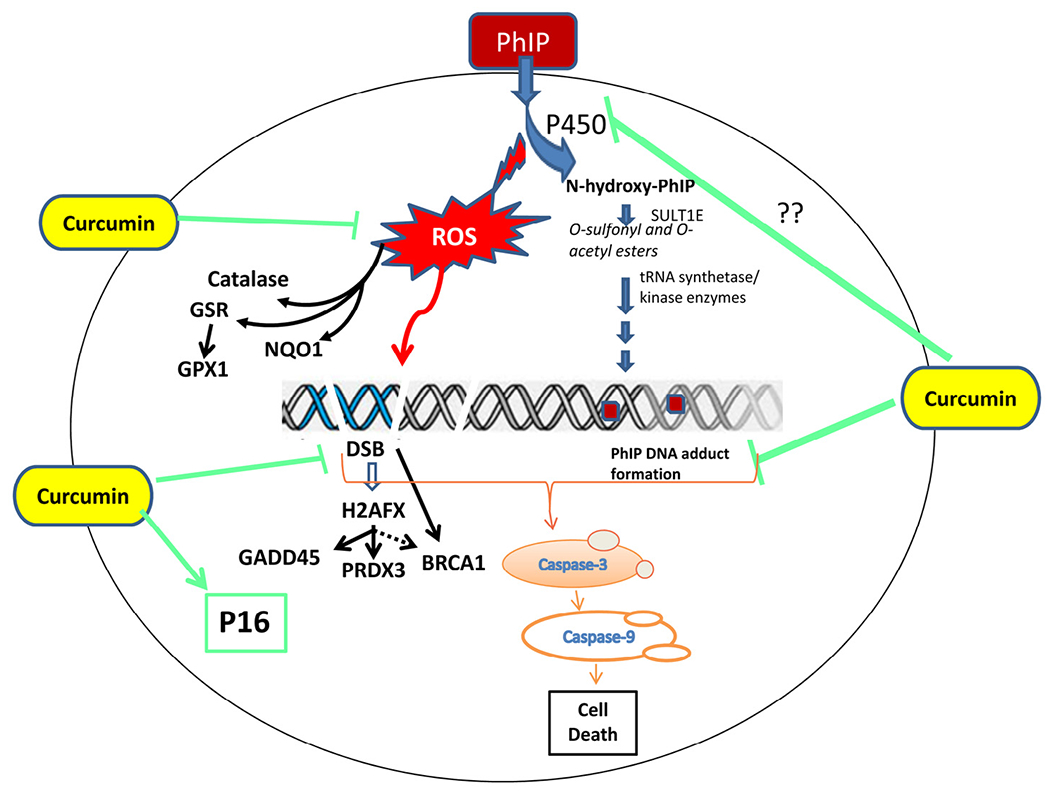
Concept map showing PhIP-induced carcinogenicity and protection by curcumin.
Our results show that curcumin modulate PhIP induced cytotoxic effects by inducing P16. Several studies have shown that this kinase (P16) is involved in cellular senescence [57,58]. Cellular senescence is a normal biological process, which is initiated in response to a range of intrinsic and extrinsic factors that function to remove irreparable damage and potentially harmful/damaged or aging cells [59]. Consequently, cellular senescence prevents cancer by permanently arresting cell cycle, preventing transmission of damage in daughter cells and by reducing the proliferative pool of damage cells [58–60]. Several studies have shown that curcumin is involved in cell cycle control via upregulation of P16 [61,62]. P16 induced cellular senescence by curcumin has been reported in breast cancer associated fibroblasts [63,64]. In the present study, increased levels of P16 were observed up to 24 hours in MCF 10A cells co-treated with PhIP and curcumin as compared to cells that were treated only with PhIP. However, further studies are required to demonstrate whether P16 levels are maintained in co-treated cells leading to cellular senescence.
We also analyzed the proteins involved in apoptotic cell death by western blot. Activation of caspase-3 has a significant role in the execution phase of apoptosis [65]. Results also show that cleavage of capase-3 was reduced in PhIP-treated MCF-10A cells. Curcumin significantly increased the cleavage of caspase-3; but cell death was not observed in these curcumin co-treated groups. Studies have previously shown that activation of caspases is associated not only with cell death but also with cell differentiation [66].
We conclude that in addition to DNA adduct formation, oxidative DNA damage is crucial to PhIP-induced carcinogenicity. Previously, Sato et al. [67] have shown that ROS production during the metabolism of heterocyclic amines including PhIP occurs through NADPH/P450, suggesting that production of ROS in PHIP-treated MCF-10A cells is due to the metabolism of PhIP to N-hydroxy-PhIP through p450 detoxification. Therefore, DNA double strand breaks in PhIP-treated cells are likely to arise through two mechanisms: (i) the ROS generated directly contribute to DSB; and (ii) PhIP-DNA adduct formation leads to DNA strand breaks (Fig. 6). Since curcumin inhibits both PhIP-induced ROS production and DNA adduct formation, this agent ultimately reduces DNA-DSB. In addition, curcumin might improve DNA repair mechanisms, and together these responses reduce the possibility of DNA mutations. Such multiple mechanisms of action of curcumin in cancer cells have been documented previously [68]. Our results also indicate that curcumin modulates PhIP-induced effects through the regulation of multiple cell signaling pathways including antioxidant, DNA repair, apoptotic (caspase activation pathway including caspase-3 and -9) and tumor suppressor pathways (p16) to minimize the damage caused by the food carcinogen PhIP (Fig. 6).
Supplementary Material
Acknowledgements
This work was supported by the Department of Defense, U.S. Army Medical Research and Material Command (W81XWH-10-1-1042), and National Institutes of Health, Research Infrastructure in Minority Institution Grant (2P20MD001085-08). Dr. Bollag is supported by a VA Research Career Scientist Award. The contents of this article do not represent the views of the Department of Veterans Affairs or the United States Government.
Abbreviations:
- AOPI
acridine orange/propidium iodide
- BRCA-1
breast cancer 1 early onset
- CAT
catalase
- DCFDA
2’,7’ dichlorodihydrofluorescein diacetate
- DEPC
diethylpyrocarbonate
- DMSO
dimethyl sulfoxide
- FOXO
forkhead box protein
- GADD-45
growth arrest and DNA damage-inducible 45
- GPX-1
glutathione peroxidase]
- GSR
glutathione reductase
- H2AX
histone H2A, family member X
- HBSS
Hank balanced salt solution
- HCA
heterocyclic amines
- HPRT
hypoxanthine phosphoribosyl transferase
- NQO-1
[NAD(P)H quionine oxidoreductase-1]
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- OTM
olive tail moment
- PhIP
amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- P-16
cyclin-dependent kinase inhibitor 2A
- PARP-1
poly[ADP-ribose] polymerase 1
- PRDX-3
thioredoxin-dependent peroxide reductase
- RFU
relative fluorescence units
- ROS
reactive oxygen species
Footnotes
Conflict of interest
The authors have no conflict of interests.
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.canlet.2015.05.017.
References
- [1].Pal D, et al. , Dietary-induced cancer prevention: an expanding research arena of emerging diet related to healthcare system, J. Adv. Pharm. Technol. Res. 3 (2012) 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].He K, et al. , Types of fish consumed and fish preparation methods in relation to pancreatic cancer incidence: the VITAL Cohort Study, Am. J. Epidemiol. 177 (2013) 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Butler LM, et al. , Heterocyclic amines, meat intake, and association with colon cancer in a population-based study, Am. J. Epidemiol. 157 (2003) 434–445. [DOI] [PubMed] [Google Scholar]
- [4].Smith JS, et al. , Effect of marinades on the formation of heterocyclic amines in grilled beef steaks,J. Food Sci. 73 (2008) 100–105. [DOI] [PubMed] [Google Scholar]
- [5].Kinburn D, Fast-food grilled chicken contains dangerous carcinogen, laboratory tests reveal. Physicians Committee for Responsible Medicine. Published Sept. 28, 2006.<http://www.pcrm.org/media/news/fast-food-grilled-chicken-contains-dangerous>, 2006. (accessed 24.06.14).
- [6].Lauber SN, Gooderham NJ, The cooked meat derived genotoxic carcinogen 2-amino-3-methylimidazo[4,5-b]pyridine has potent hormone-like activity: mechanistic support for a role in breast cancer, Cancer Res. 67 (2007) 9597–9602. [DOI] [PubMed] [Google Scholar]
- [7].Tang D, et al. , Grilled meat consumption and PhIP-DNA adducts in prostate carcinogenesis, Cancer Epidemiol. Biomarkers Prev. 16 (2007) 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pool-Zobel BL, et al. , Induction of DNA damage by risk factors of colon cancer in human colon cells derived from biopsies, Mutat. Res. 375 (1997) 105–115. [DOI] [PubMed] [Google Scholar]
- [9].lto N, Hasegawa R, Sano M, et al. , A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), Carcinogenesis 12(1991) 1503–1506. [DOI] [PubMed] [Google Scholar]
- [10].Kato T, Ohgaki H, Hasegawa H, et al. , Carcinogenicity in rats of a mutagenic compound, 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline, Carcinogenesis 9 (1988) 71–73. [DOI] [PubMed] [Google Scholar]
- [11].Kato T, Migita H, Ohgaki H, et al. , Induction of tumors in the Zymbal gland, oral cavity, colon, skin and mammary gland of F344 rats by a mutagenic compound, 2-amino-3,4-dimethylimidazo[4,5-f]quinoline, Carcinogenesis 10 (1989) 601–603. [DOI] [PubMed] [Google Scholar]
- [12].Shirai T, Sano M, Tamano S, et al. , The prostate: a target for carcinogenicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) derived from cooked foods, Cancer Res. 57 (1997) 195–198. [PubMed] [Google Scholar]
- [13].Turesky RJ, Le Marchand L, Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines, Chem. Res. Toxicol. 24 (2011) 1169–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Turteland KW, et al. , Macromolecular adduct and metabolism of heterocyclic amines in humans and rodents at low doses, Cancer Lett. 143(1999) 149–155. [DOI] [PubMed] [Google Scholar]
- [15].Morgenthaler PM, et al. , Analysis of mutation induced by 2-amino-1methyl-6-phenylimidazo [4,5-b]pyridine (PhIP) in human lymphob-lastodial cells, Carcinogenesis 16(1995) 713–718. [DOI] [PubMed] [Google Scholar]
- [16].Gooderham N, et al. ,Molecular and genetic toxicology of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), Mutat. Res. 506 (2002) 91–99. [DOI] [PubMed] [Google Scholar]
- [17].Otsuka C, et al. , Cytogenic effects of a food mutagen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and its metabolite 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (N-OH PhIP) on human and Chinese hamster cells in vitro, Mutat. Res. 367 (1996) 115–121. [DOI] [PubMed] [Google Scholar]
- [18].Wu R, et al. , Differential effect of acetyl transferase expression on the genotoxicity of heterocyclic amines in CHO cells, Mutat. Res. 390 (1997) 93–103. [DOI] [PubMed] [Google Scholar]
- [19].Reason W, et al. , Curcumin: a review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma, Mol. Cancer 10 (2011) 12, doi: 10.1186/1476-4598-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wei X, et al. , Effects of pyridine analogs of curcumin on growth, apoptosis and NF-κB activity in prostate cancer PC-3 cells, Anticancer Res. 33 (2013) 1343–1350. [PubMed] [Google Scholar]
- [21].Cheng TS, et al. , Curcumin-targeting pericellular serine protease matriptase role in suppression of prostate cancer cell invasion, tumor growth, and metastasis, Cancer Prev. Res. (Phila.) 6 (2013) 495–505. [DOI] [PubMed] [Google Scholar]
- [22].Sun XD, et al. , Curcumin induces apoptosis of triple negative breast cancer cells by inhibition of EGFR expression, Mol. Med. Rep. 6 (2012) 1267–1270. [DOI] [PubMed] [Google Scholar]
- [23].Liu D, et al. , The effect of curcumin on breast cancer cells, J. Breast Cancer 16 (2013)133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Masuelli L, et al. , Curcumin induces apoptosis in breast cancer cell lines and delays the growth of mammary tumors in neu transgenic mice, J. Biol. Regul. Homeost.Agents 27 (2013) 105–119. [PubMed] [Google Scholar]
- [25].Sinha D, et al. , Chemopreventive and chemotherapeutic potential of curcumin in breast cancer, Curr. Drug Targets 13 (2012) 1799–1819. [DOI] [PubMed] [Google Scholar]
- [26].Lu WD, et al. , Effect of curcumin on human colon cancer multidrug resistance in vitro and in vivo, Clinics 86 (2013) 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shehzad A, et al. , Curcumin induces apoptosis in human colorectal carcinoma (HCT-15) cells by regulating expression of Prp4 and p53, Mol. Cells 35 (2013) 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Collett GP, et al. , Curcumin modifies Apcmin apoptosis resistance and inhibits 2-amino 1-methyl-6-phenylimidazol[4,5-b]pyridine (PhIP) induced tumor formation in Apcmin mice, Carcinogenesis 22 (2001)821–825. [DOI] [PubMed] [Google Scholar]
- [29].Fan H, et al. , Curcumin induces apoptosis of HepG2 cells via inhibiting fatty acid synthase, Target. Oncol 9(2014)279–286, doi: 10.1007/S11523-013-0286-5 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- [30].Bachmeier B, et al. , The chemopreventive polyphenol curcumin prevents hematogenous breast cancer metastases in immunodeficient mice, Cell. Physiol. Biochem. 19 (2007) 137–152. [DOI] [PubMed] [Google Scholar]
- [31].Bachmeier BE, et al. , Curcumin downregulates the inflammatory cytokines CXCL1 and -2 in breast cancer cells via NFkappaB, Carcinogenesis 29 (2008) 779–789. [DOI] [PubMed] [Google Scholar]
- [32].Chuang SE, et al. , Curcumin-containing diet inhibits diethylnitrosamine-induced murine hepatocarcinogenesis, Carcinogenesis 21 (2000) 331–335. [DOI] [PubMed] [Google Scholar]
- [33].Aboyade-Cole A, et al. , Diallyl sulfide inhibits PhIP-induced cell death via the inhibition of DNA strand breaks in normal human breast epithelial cells, Oncol. Rep. 20 (2008) 319–323. [PMC free article] [PubMed] [Google Scholar]
- [34].Metry KJ, et al. , Effect of N-acetyltransferase 2 polymorphism on tumor target tissue DNA adduct levels in rapid and slow acetylator congenic rats administered 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine or 2-amino-3,8-dimethylimidazo-[4,5-f]quinoxaline, Drug Metab. Dispos. 37 (2009) 2123–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Takahashi S, et al. , Immunohistochemical demonstration of carcinogen-DNA adducts in tissues of rats given 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP): detection in Parafilm-embedded sections and tissue distribution, Cancer Res. 58 (1998) 4307–4313. [PubMed] [Google Scholar]
- [36].Singh NP, et al. , A simple technique for quantitation of low levels of DNA damage in individual cells, Exp. Cell Res. 175 (1988) 184–191. [DOI] [PubMed] [Google Scholar]
- [37].Duez P, et al. , Validation of raw data measurements in the comet assay, Talanta 63 (2004) 879–886. [DOI] [PubMed] [Google Scholar]
- [38].Jain A, Targeting breast cancer, in: Singh SR, Mishra MK (Eds.), Signaling Gene Regulation and Cancer, Nova Science Publishers, Inc., 2013, pp. 199–222. [Google Scholar]
- [39].Redig AJ, et al. , Breast cancer as a systemic disease: a view of metastasis, J. Intern. Med. 274 (2013) 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhu J, et al. , Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine DNA adducts in normal breast tissues and risk of breast cancer, Cancer Epidemiol. Biomarkers Prev. 12(2003) 830–837. [PubMed] [Google Scholar]
- [41].Yu Z, et al. , Indole-3-carbinol in the maternal diet provides chemoprotection for the fetus against transplacental carcinogenesis by the polycyclic aromatic hydrocarbon dibenzo[a,l]pyrene, Carcinogenesis 27 (2006) 2116–2123. [DOI] [PubMed] [Google Scholar]
- [42].McCann SE, et al. , Dietary lignan intakes in relation to survival among women with breast cancer: the Western New York Exposures and Breast Cancer (WEB) Study, Breast Cancer Res. Treat. 122 (2010) 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Guha N, et al. , Soy isoflavones and risk of cancer recurrence in a cohort of breast cancer survivors: the Life After Cancer Epidemiology study, Breast Cancer Res. Treat. 118 (2009) 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lynn SA, et al. , Whole blueberry powder modulates the growth and metastasis of MDA-MB-231 triple negative breast tumors in nude mice,J. Nutr. 141(2011) 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bendaly J, et al. , Role of human CYP1A1 and NAT2 in 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-induced mutagenicity and DNA adducts, Xenobiotica 39 (2009) 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Glaab W, et al. , Specificity of mutations induced by the food-associated heterocyclic amine 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in colon cancer cell lines defective in mismatch repair, Cancer Res. 60 (2001)4921–4925. [PubMed] [Google Scholar]
- [47].Nogueira V, et al. , Molecular pathways: reactive oxygen species homeostasis in cancer cells and implications for cancer therapy, Clin. Cancer Res. 19 (2013) 4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Waris G, Ashan H, Reactive oxygen species: role in the development of cancer and various chronic conditions, J. Carcinog 5 (2006) 14–19, doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hayes JD, et al. , The Nrf2 transcription factor contributes both to the basal expression of glutathione S transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin, Biochem. Soc. Trans. 28 (2000) 33–41. [DOI] [PubMed] [Google Scholar]
- [50].Fornace AJ Jr., et al. , Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents, Mol. Cell. Biol. 9(1989) 4196–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cao J, et al. , Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity, EMBO J. 28 (2009) 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhan Q, GADD-45a, a p53-and BRCA1-regulated stress protein, in cellular response to DNA damage, Mutat. Res. 569 (2005) 133–143. [DOI] [PubMed] [Google Scholar]
- [53].Salvador JM, et al. , GADD-45 in stress signaling, cell cycle control, and apoptosis, Adv. Exp. Med. Biol. 793 (2013) 1–19. [DOI] [PubMed] [Google Scholar]
- [54].Starita LM, et al. , The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair, Curr. Opin. Cell Biol. 15 (2003) 345–350. [DOI] [PubMed] [Google Scholar]
- [55].Gorrini C, et al. , BRCA-1 interects with Nrf2 to regulate antioxidant signaling and cell survival, J. Exp. Med. 210(2013) 1529–1544, doi: 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Caestecker KW, et al. , The role of BRCA1 in DNA double-strand repair: past and present, Exp. Cell Res. 319 (2013) 575–587. [DOI] [PubMed] [Google Scholar]
- [57].Lynch MD, How does cellular senescence prevent cancer?, DNA Cell Biol. 25 (2006) 69–78. [DOI] [PubMed] [Google Scholar]
- [58].Campisi J, Fagagna F, Cellular senescence: when bad things happen to good cells, Nat. Rev. Mol. Cell Biol. 8 (2007) 729–740. [DOI] [PubMed] [Google Scholar]
- [59].Sabin RJ, Cellular senescence – its role in cancer and the response to ionizing radiation, Genome Integr. 2 (2011) 7–17, doi: 10.1186/2041-9414-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wright WE,Shay JW, Cellular senescence as a tumor-protection mechanism: the essential role of counting, Curr. Opin. Genet. Dev. 11 (2001)98–103. [DOI] [PubMed] [Google Scholar]
- [61].Wilken R, et al. , Curcumin: a review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma, Mol. Cancer 10 (2011) 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Singh M, Singh N, Curcumin counteracts the proliferative effect of estradiol and induces apoptosis in cervical cancer cells, Mol. Cell. Biochem. 347 (2011) 1–11, doi: 10.1007/s11010-010-0606-3. [DOI] [PubMed] [Google Scholar]
- [63].Hendrayani SF, et al. , Curcumin triggers p16-dependent senescence in active breast cancer-associated fibroblasts and suppresses their paracrine procarcinogenic effects, Neoplasia 15 (2013) 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Capparelli C, et al. , CDK inhibitors (p16/p19/p21) induce senescence and autophagy in cancer-associated fibroblasts, “fueling” tumor growth via paracrine interactions, without an increase in neo-angiogenesis, Cell Cycle 11 (2012) 3599–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Simon DJ, et al. , A caspase cascade regulating developmental axon degeneration,J. Neurosci. 32 (2012) 17540–17553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Allen R, et al. , Oxidative stress and gene regulation, Free Radic. Biol. Med. 28 (2000) 463–499. [DOI] [PubMed] [Google Scholar]
- [67].Sato K, et al. , Evidence of direct generation of oxygen free radicals from heterocyclic amines by NADPH/cytochrome P-450 reductase in vitro, Jpn J. Cancer Res. 83 (1992) 1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ravindran J, et al. , Curcumin and cancer cells: how many ways can curry kill tumor cells selectively, AAPS J. 11 (2009) 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


