Abstract
The classical definition posits hybrid sterility as a phenomenon when two parental taxa each of which is fertile produce a hybrid that is sterile. The first hybrid sterility gene in vertebrates, Prdm9, coding for a histone methyltransferase, was identified in crosses between two laboratory mouse strains derived from Mus mus musculus and M. m. domesticus subspecies. The unique function of PRDM9 protein in the initiation of meiotic recombination led to the discovery of the basic molecular mechanism of hybrid sterility in laboratory crosses. However, the role of this protein as a component of reproductive barrier outside the laboratory model remained unclear. Here, we show that the Prdm9 allelic incompatibilities represent the primary cause of reduced fertility in intersubspecific hybrids between M. m. musculus and M. m. domesticus including 16 musculus and domesticus wild-derived strains. Disruption of fertility phenotypes correlated with the rate of failure of synapsis between homologous chromosomes in meiosis I and with early meiotic arrest. All phenotypes were restored to normal when the domesticus Prdm9dom2 allele was substituted with the Prdm9dom2H humanized variant. To conclude, our data show for the first time the male infertility of wild-derived musculus and domesticus subspecies F1 hybrids controlled by Prdm9 as the major hybrid sterility gene. The impairment of fertility surrogates, testes weight and sperm count, correlated with increasing difficulties of meiotic synapsis of homologous chromosomes and with meiotic arrest, which we suppose reflect the increasing asymmetry of PRDM9-dependent DNA double-strand breaks.
Keywords: reproductive isolation, Prdm9 polymorphism, meiotic chromosome synapsis, HORMAD2, synaptonemal complex
Introduction
The biological species concept defines species as groups of interbreeding natural populations reproductively isolated from other such groups (Mayr 1963). One postzygotic reproductive isolation mechanism, the sterility of interspecific hybrids, has attracted human curiosity since Aristotle discussed the infertility of mules. Charles Darwin foresaw hybrid sterility as a by-product of evolution, rather than a naturally selected trait (Darwin 1859). To reconcile Darwinian evolution with genetics, Dobzhansky and Muller (Dobzhansky 1936, 1951; Muller 1942; Muller and Pontecorvo 1942) proposed a genetic model explaining hybrid sterility as an incompatibility between independently diverged genes. During the last 80 years, most of our genetic knowledge on hybrid sterility in animals came from the studies of Drosophila species (Naveira and Maside 1998; Coyne and Orr 2004; Presgraves 2008; Maheshwari and Barbash 2011), nonetheless, the main principles of hybrid sterility proved valid for many other animal and plant species hybrids (Schilthuizen et al. 2011). These include Haldane’s rule stating that the heterogametic (XY or ZW) sex is preferentially affected in hybrids (Haldane 1922), or the large X-effect (Coyne’s rule) referring to the predominant role of the X chromosome in contrast to autosomes (Dobzhansky 1951; Forejt 1996; Coyne and Orr 2004; Good et al. 2008; Presgraves 2018), Perhaps surprisingly, given the extensive genetic studies in species such as fruit flies, yeasts, and house mice, the genetic architecture of hybrid sterility as well as the molecular mechanisms behind these rules remain mostly unclear (reviewed in Maheshwari and Barbash [2011], Phifer-Rixey and Nachman [2015], Dion-Cote and Barbash [2017], Mack and Nachman [2017], Payseur et al. [2018]).
Although hybrid sterility usually behaves as a complex polygenic trait, a handful of discrete hybrid sterility and hybrid inviability genes such as OdsH, JYAlpha, Hmr, Nup96, and Ovd have been described mostly in Drosophila (Ting et al. 1998; Barbash et al. 2003; Presgraves et al. 2003; Masly et al. 2006; Phadnis and Orr 2009). Forejt and Ivanyi (1974) mapped the first hybrid sterility genetic locus in vertebrates (Hybrid sterility 1, Hst1) in crosses of wild male mice of the Mus musculus musculus (hereafter referred to as musculus) subspecies with females of laboratory inbred strains of the Mus musculus domesticus (hereafter domesticus) origin. Later, by positional cloning, they identified the Hst1 locus with the Prdm9 gene coding for PR/SET domain-containing 9 protein (Mihola et al. 2009). The current model of Prdm9-controlled hybrid sterility refers to male progeny of an intersubspecific cross of musculus wild-derived PWD strain females and C57BL/6J (hereafter B6) domesticus laboratory strain males. Hybrid descendants of the cross are viable and healthy individuals except that all males are completely sterile. The phenotype of hybrid sterility includes small testes, absence of sperm, meiotic arrest at the mid-to-late pachytene stage (Forejt 1996; Forejt et al. 2012), impairment of transcriptional inactivation of the sex chromosomes (MSCI), and incomplete meiotic chromosome synapsis (Forejt 1984; Bhattacharyya et al. 2013; Gregorova et al. 2018). Obeying Haldane’s rule, the female hybrids are fertile, though with partially defective oogenesis (Bhattacharyya et al. 2014). In contrast to other mouse studies, which observed a complex polygenic nature often of dozens of hybrid incompatibilities (Tucker et al. 1992; Payseur et al. 2004; Macholan et al. 2007, 2011; Duvaux et al. 2011; Janoušek et al. 2012; Turner et al. 2012; Morgan et al. 2020), the genomic architecture of the F1 hybrid sterility of (PWD×B6)F1 males appears relatively straightforward. The three main components consist of the intersubspecific allelic combination Prdm9PWD/B6, the PWD allelic form of the X-linked Hstx2 locus (Storchova et al. 2004; Bhattacharyya et al. 2014; Lustyk et al. 2019), and the PWD/B6 heterozygosity of homeologous autosomes (homeologs are homologous autosomes from related (sub)species) (Gregorova et al. 2018).
PRDM9 is a histone methyltransferase enzyme that initiates meiotic recombination (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010) by directly binding DNA via its C-terminal zinc finger (ZnF) domain. The ZnF domain is highly polymorphic within and between mouse subspecies and determines the allelic specificity of PRDM9 DNA-binding sites. After binding to DNA, PRDM9 defines specific sites (hotspots) where meiotic recombination may occur (Hayashi and Matsui 2006; Smagulova et al. 2011; Wu et al. 2013; Baker et al. 2014; Eram et al. 2014; Powers et al. 2016) by inserting trimethylation marks on the lysine 4 and lysine 36 residues of histone 3 (H3K4me3 and H3K36me3). The PRDM9-marked hotspots guide SPO11 protein to induce programed DNA double-strand breaks (DSBs) along the chromosomes, which are repaired by homologous recombination either as crossovers or noncrossovers (gene conversions) (for review, see Hunter [2015] and Smith and Nambiar [2020]). The lack of PRDM9 leads to redistribution of recombination events toward the functional elements such as promoters that are also marked by H3K4me3 (Smagulova et al. 2016) resulting in sterility in both sexes (Hayashi and Matsui 2006) (but see Mihola et al. [2019]).
The molecular mechanism by which Prdm9 acts as a mouse hybrid sterility gene is less clear. The “asymmetry-due-to erosion” hypothesis posits the differential erosion of PRDM9 binding sites in musculus and domesticus subspecies as the initial event (Davies et al. 2016). The evolutionary erosion is a well-documented feature of recombination as well as Prdm9 hotspots in several species (Boulton et al. 1997; Myers et al. 2010; Baker et al. 2015). The erosion at the evolutionary scale is explained by biased gene conversion that favors repair using the template with lower PRDM9 affinity, because initiation of DNA DSBs is preferred at the PRDM9 binding sites with higher affinity. In this way, a subset of PRDM9 recognition sequences is gradually blurred in one subspecies but remains intact in the other. In (PWD×B6)F1 intersubspecific hybrids, the asymmetric hotspots occur predominantly on a “nonself chromosome” where the recognition motifs are intact; the PRDM9B6 hotspots appear mostly on PWD chromosomes and PRDM9PWD hotspot on B6 chromosomes. It is presumed that the SPO11-induced DSBs at such asymmetric hotspots are difficult to repair from the homologous chromosome or are repaired using the sister chromatid as a template (Hinch et al. 2019), resulting in asynapsis and apoptosis. Many aspects of this “asymmetry-due-to erosion” hypothesis remain unsolved, such as the role of the second major hybrid sterility factor located within the X-linked Hstx2 locus, the significance of “default,” Prdm9-independent hotspots (Hayashi and Matsui 2006; Smagulova et al. 2016), or the effect of PRDM9 binding sites within repetitive sequences (Yamada et al. 2017). However, several pieces of evidence supporting this idea have been gathered (Gregorova et al. 2018; Wang et al. 2018; Gergelits et al. 2019).
To sum up, the Prdm9-dependent model of hybrid sterility has all principal attributes of classical F1 hybrid sterility (Coyne and Orr 2004) including underdominance (Prdm9 heterozygosity-dependent sterility), male-limited infertility following Haldanés rule, and the large X-effect reflected by the interacting X-linked Hstx2 locus (Lustyk et al. 2019). However, virtually nothing is known about Prdm9 allelic incompatibilities in the hybrids of wild mice, known to display a high level of intrasubspecific Prdm9 polymorphism (Buard et al. 2014; Kono et al. 2014; Vara et al. 2019), an unorthodox feature for a hybrid sterility gene. To investigate the role of Prdm9-dependent reproductive isolation in natural mouse populations, we tested ten different Prdm9 alleles from 16 European populations of musculus and domesticus subspecies for their effect on fertility of intersubspecific F1 hybrids. We found that aberrant synapsis of meiotic chromosomes is indeed a Prdm9-dependent trait negatively correlated with surrogate fertility phenotypes—number of produced sperm cells and testes weight. Moreover, despite a continuum of variation in fertility-related phenotypes, full hybrid sterility occurred when the asynapsis rate exceeded a certain threshold level. Our results provide the first direct evidence that incompatibilities between Prdm9 alleles segregating in natural populations can result in F1 hybrid sterility.
Results
Identification of Intersubspecific Introgressions in Genomes of Wild-Derived Strains
All 16 wild-derived mouse strains originated from founders trapped in Central and Western Europe (Gregorova and Forejt 2000; Pialek et al. 2007; Martincová et al. 2019) (see also Materials and Methods and https://housemice.cz/en; last accessed July 11, 2020). The sampling sites were located on both sides of the European natural hybrid zone separating musculus and domesticus subspecies (Macholán et al. 2007; Geraldes et al. 2008; Duvaux et al. 2011; Baird and Macholan 2012), whose genomes diverged ∼0.5 Ma (She et al. 1990; Boursot et al. 1996; Salcedo et al. 2007; Geraldes et al. 2008) (fig. 1A). Since genomic introgression occurs in both directions across the hybrid zone (Payseur et al. 2004; Teeter et al. 2007; Dufkova et al. 2011; Macholan et al. 2011; Janoušek et al. 2012), we first searched for possible genomic admixtures from the other subspecies. Such introgressions could influence local meiotic pairing between musculus and domesticus homologs, as modeled by hybrids of the intersubspecific chromosome substitution strains (Gregorova et al. 2018). To identify the introgressed genomic segments, we used a high-density SNP genotyping array (Morgan et al. 2015) in eight musculus and seven domesticus wild-derived inbred strains. The wild-derived PWD/Ph (hereafter PWD) and classical laboratory strain C57BL/6J (hereafter B6) inbred strains were included as a reference. The highest admixture of musculus in domesticus strains was 7.01% in the B6 reference strain; it is well known that most classical inbred strains, although primarily of domesticus ancestry, have significant musculus component (Yang et al. 2011). The remaining seven domesticus strains (SPLY, SIN, SIT, SCHUNT, STRB, STRA, STAIL) all showed <1% of the genome (autosomes + X chromosome) of musculus ancestry (0.07–0.97%). The SPLY strain differs from other domesticus strains by the presence of a musculus Y chromosome, and C57BL/6J is confirmed to carry a different musculus Y chromosome (Nagamine et al. 1992; Morgan and Pardo-Manuel de Villena 2017) (supplementary table S1 and fig. S1, Supplementary Material online). The SCHEST strain became extinct before genotyping could be performed. Remarkably, many introgressed segments were found recurrently; seven domesticus strains (excluding B6 reference) displayed in total 73 musculus segments, of which 35 occurred only once and the remaining 38 consisted of 12 distinct segments shared by 2–5 strains (supplementary fig. S1 and table S2, Supplementary Material online). Eight musculus strains displayed 91 islands of the domesticus sequence. Eighty-one occurred only once, whereas ten consisted of five distinct islands shared by two strains. These regions could represent introgression events that have risen to high frequency in source populations, or (less likely) artifacts in ancestry assignment. All musculus strains except PWD showed <1% (0.02–0.6%) of the domesticus ancestry. The PWD genome includes 4.89% (133.5 Mb) of domesticus segments, of which 26.52 and 44.2 Mb are situated on the chromosomes 9 and 14, respectively (supplementary table S3, Supplementary Material online, see also Yang et al. [2011]). Thus, besides these two PWD chromosomes, no chromosome in any other studied strain displayed >27-Mb introgression, a threshold previously shown to be sufficient for rescuing meiotic synapsis of intersubspecific homologs (Gregorova et al. 2018).
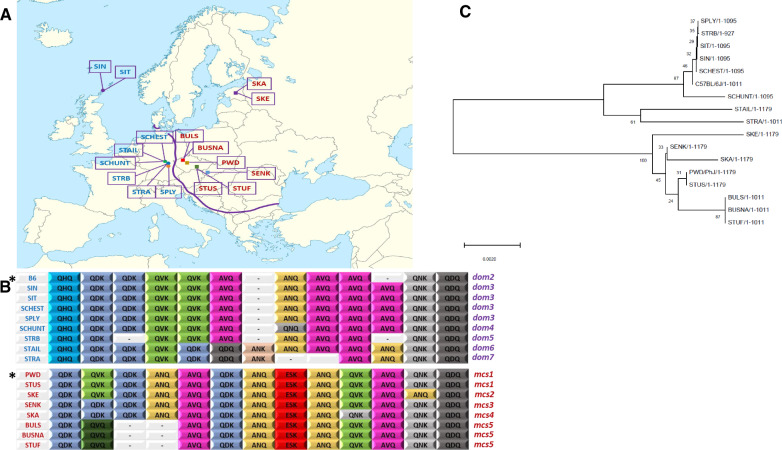
Fig. 1.
Geographic distribution of founders of wild-derived strains and their Prdm9 ZnF arrays. (A) Localities of origin of eight musculus (squares) and eight domesticus (circles) strains along the musculus/domesticus hybrid zone (Baird and Macholan 2012; Ďureje et al. 2012; Macholan et al. 2019). Musculus strains: SKA, Kaerepere, Estonia; SKE, Keava, Estonia; BULS and BUSNA, Buškovice, Czech Republic; PWD, Kunratice (Prague), Czech Republic; STUF and STUS, Studenec, Czech Republic; SENK, Šenkvice, Slovak Republic. Domesticus strains: STRA and STRB, Straas, Bavaria, Germany; STAIL and SCHUNT, Schweben, Hessen, Germany; SPLY, Plössen, Bavaria, Germany; SIN and SIT, Scar, Sanday Island, Orkneys, Scotland. The blank map of Europe by courtesy of Wikimedia Commons (https://en.wikipedia.org/wiki/File:Europe_blank_laea_location_map.svg#filehistory). (B) PRDM9 zinc finger arrays of all strains. The first, nonvariant ZnF separated from ZnF domain is not shown. Three DNA-binding amino acids represent each ZnF. Included are allelic variants of reference strains B6 and PWD. (C) Phylogenetic relationships between Prdm9 alleles estimated using the neighbor-joining method in MEGA X (Kumar et al. 2018). The numbers next to each node represent bootstrapping values.
ZnF Sequence, Nomenclature, Geography, and Phylogeny of the Studied Prdm9 Alleles
The PRDM9 ZnF haplotypes of individual wild-derived strains were identified after amplification and sequencing of PRDM9 exon12, which contains the entire ZnF array except for the first, invariant ZnF repeat. Sequences of individual ZnFs were translated, and three highly variable DNA-binding amino acids at positions −1, +3, and +6 (Oliver et al. 2009) were used to identify each ZnF type. Five distinct haplotypes were present in eight musculus strains, and five haplotypes in eight domesticus strains (fig. 1B). The PRDM9 ZnF sequences were deposited to GenBank under accession numbers MT252946–MT252960. To follow the official nomenclature (International Committee on Standardized Genetic Nomenclature for Mice; http://www.informatics.jax.org/mgihome/nomen/index.shtml; last accessed July 11, 2020), we applied serial numbers to the newly identified Prdm9msc# musculus and Prdm9dom# domesticus alleles. The only so far registered musculus allele Prdm9msc (PWD/Ph inbred strain) was given number 1, Prdm9msc1.
Only the PWD/Ph and STUS wild-derived musculus strains, from the localities Kunratice and Studenec, Czech Republic, carried the Prdm9msc1 allele (fig 1B). Three other strains displayed variants of PRDM9msc1 protein, which we designated PRDM9msc2 and PRDM9msc3 and which differ only in a single ZnF (SKE strain from Estonia, substitution of QNK by ANQ in the 13th ZnF, and SENK strain from Slovakia, substitution of QVK by QDK in the third ZnF). The PRDM9msc4 differs by two ZnFs (SKA strain from Estonia—substitution of QVK by QDK in the third ZnF and QVK by QNK in the 11th ZnF). The remaining three musculus strains (BULS and BUSNA) carry the same Prdm9msc5 allele, which differs from Prdm9msc1 by deletion of sequence for the fourth and fifth ZnFs and by substitution of QVK by QVQ in the third ZnF (fig. 1B). Notably, all five substitutions but one occurred at the QVK ZnF, and always at amino acid positions responsible for DNA binding, namely positions +3 or +6 of the alpha-helix of the C2H2 ZnF.
The Prdm9dom2domesticus allele present in the reference B6 strain and in many other laboratory inbred strains was absent among the eight domesticus wild-derived strains. The allele most similar to Prdm9dom2 was Prdm9dom3 found in four wild-derived domesticus strains. The same allele has already been described in several laboratory inbred strains (C3H/HeJ, CBA/CaJ, NOD/LtJ, PERA/EiJ, and WSB/EiJ). The founders of strains carrying Prdm9dom3 came from Orkney Islands, UK (SIN and SIT); Bavaria, Germany (SPLY); and Hesse, Germany (SCHEST). SCHUNT, also from Hesse carries the Prdm9dom4, which differs from Prdm9dom3 only at ZnF8 (substitution of ANQ by QNQ) (fig. 1B). The Prdm9dom5 allele (strain STRB) from Bavaria differs from Prdm9dom2 only by a deletion of the fourth ZnF. The remaining two alleles Prdm9dom6 (STAIL, Hessen) and Prdm9dom7 (STRA, Bavaria) appear the most distant from the Prdm9dom2 reference.
The Prdm9dom3 allele was the most frequent domesticus allele identified in two large-scale studies of wild mice, occurring in 23.1% (9/39) and 35% (7/20) cases in widely separated sites along the European hybrid zone (Buard et al. 2014; Kono et al. 2014). Admittedly, our choice of strains was not completely random, since SIN and SCHEST strains, which both turned out to be Prdm9dom3, were sequenced based on our previous knowledge of male sterility of their hybrids (see below). The Prdm9msc1 allele of the PWD reference and STUS strains was the most common allele among the musculus samples (30%, 6/20) (Buard et al. 2014), though only in 7.4% (2/27) of mice in Eastern Europe and Asia screened in a previous (Kono et al. 2014).
The evolutionary relationships between the zinc finger arrays of Prdm9 alleles of individual wild-derived strains were estimated using MEGA X (Kumar et al. 2018) (fig. 1C). The musculus alleles can be divided into two subgroups—the group of BULS, BUSNA, and STUF and the group of “PWD-like” alleles SKA, SKE, and SENK. In the domesticus group, STRA and STAIL alleles seem evolutionarily distant forming a separate group from the remaining alleles. The distribution of musculus and domesticus alleles respected the hybrid zone between both subspecies, but within each subspecies territory showed limited local clustering.
Prdm9 Controls Fertility Phenotypes of Intersubspecific Hybrids
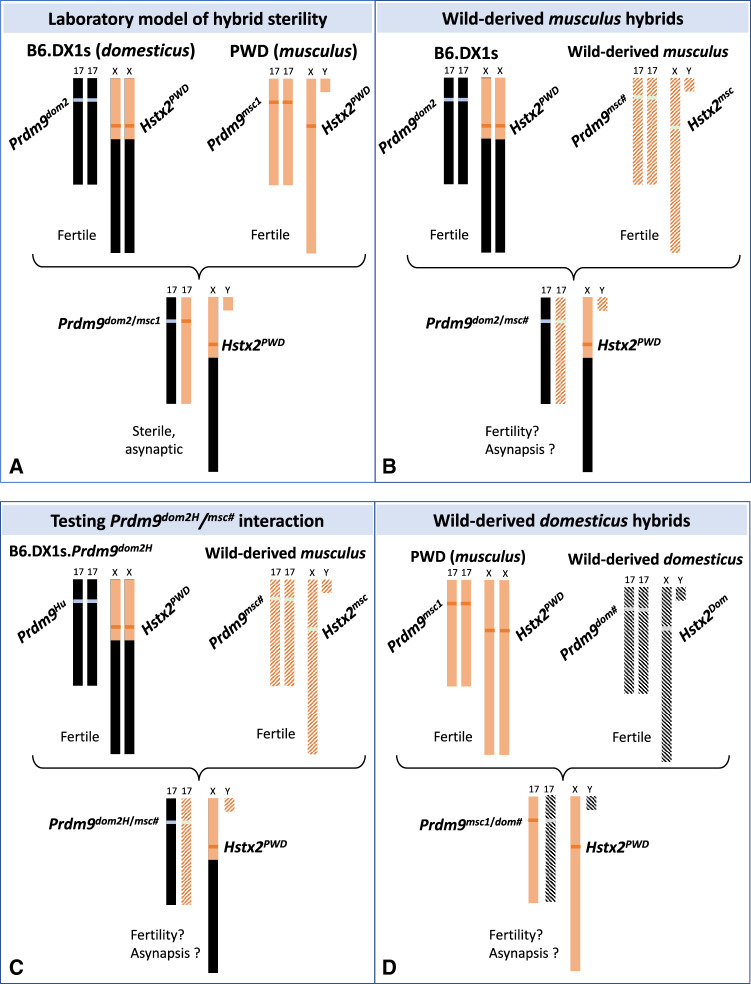
Three main components, intersubspecific allelic heterozygosity Prdm9msc1/dom2, Hstx2PWD locus on Chromosome X, and F1 hybrid genetic background, define the genomic architecture of the (PWD×B6)F1 laboratory model of hybrid sterility (Bhattacharyya et al. 2013, 2014; Gregorova et al. 2018; Lustyk et al. 2019) (fig. 2A). To evaluate the general validity of this model for other wild-derived musculus Prdm9msc# alleles, we designed crosses where Hstx2PWD and Prdm9dom2 alleles remained constant, and the only variables in the F1 hybrid genome were the wild-derived Prdm9msc# allele and the musculus portion of the genomic background (fig. 2B). The effect of a “humanized” Prdm9dom2H allele (Davies et al. 2016) known to rescue sterility of (PWD×B6)F1 hybrids was assessed in hybrids with wild-derived Prdm9msc# variants (fig. 2C). Similarly, to test domesticus Prdm9 alleles, PWD females were crossed with the domesticus wild-derived males. In this cross, both Hstx2PWD and Prdm9msc1 were controlled, whereas the wild-derived Prdm9dom# and the domesticus part of genetic background varied (fig. 2D). The synapsis of meiotic chromosomes was visualized by immunostaining of axial elements of synaptonemal complexes by antibody against SYCP3 protein on spermatocyte nuclei spreads. Furthermore, the HORMA domain-containing protein-2 (HORMAD2), which is a meiosis-specific protein known to accumulate on unsynapsed chromosome axes (Wojtasz et al. 2009) was used to visualize asynapsed homologs (Bhattacharyya et al. 2013, 2014; Gregorova et al. 2018). To score the synapsis failure, one asynapsis event was defined as one HORMAD2 positive structure, mostly an unsynapsed univalent. Normal asynapsis events, namely HORMAD2 positive nonhomologous parts of X and Y sex chromosomes as well as the unpaired axial elements of cells at earlier stages (leptotene, zygotene) were not scored as asynapsis events. To differentiate between zygotene and pachytene stages of the first meiotic prophase, we utilized phosphorylated histone H2AX at Ser 139 (γH2AX) immunostaining (fig. 3). Phosphorylation of H2AX is an early mark of DNA DSBs. In wild-type spermatocytes, it decorates unsynapsed autosomal stretches at the zygotene stage and transcriptionally inactive X and Y chromosomes engulfed within the XY (sex) body in the pachytene spermatocytes (Wojtasz et al. 2012). Altogether 10,333 immunostained pachynemas were analyzed, and relation between meiotic chromosome asynapsis rate and fertility phenotypes was studied.
Fig. 2.
Scheme of F1 hybrid genotypes to test the role of PRDM9 alleles in F1 hybrid fertility. (A) The laboratory model of (PWD×B6)F1 hybrid sterility. The first parent is female. Hybrid sterility depends on Prdm9 msc1/dom2 heterozygosity, presence of PWD allele of Hybrid sterility X2 (Hstx2PWD) on chromosome X and on interaction of homeologous (domesticus/musculus) autosomes. (B) The Hstx2PWD and Prdm9dom2 alleles interact with the Prdm9 allele from a wild-derived musculus In B6.DX1s×wild-derived musculus F1 hybrid. (C) The same cross as in (B) but Prdm9dom2 is replaced by humanized Prdm9dom2H, known to restore Prdm9-dependent sterility. (D) The same Prdm9 and Hstx2 genotype as in (A) but Prdm9dom2 is replaced by a wild-derived domesticus allele.
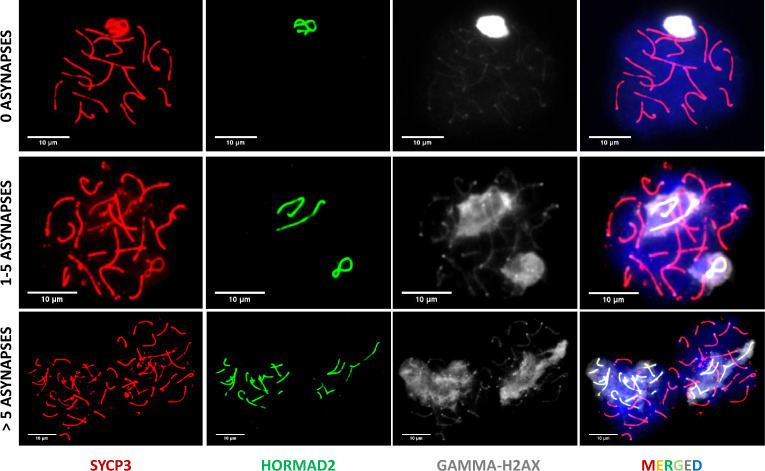
Fig. 3.
Representative immunomicroscopy images of chromosome synapsis in spreads of pachynema nuclei. Synaptonemal complexes are visualized by immunostaining SYCP3 protein (red), asynapsed parts of the XY sex chromosomes and unsynapsed autosomes are decorated by anti HORMAD2 antibody (green) and DNA is counterstained with DAPI (blue). The images illustrate a fully synapsed cell (top panel), where only the nonhomologous parts of sex chromosomes are decorated by HORMAD2 and the cells of two categories, 1–5 asynapses (middle panel) and >5 asynapses (bottom panel). One asynapsis is defined as one HORMAD2-stained structure, most often representing a univalent, an unpaired homolog.
The Wild-Derived musculus Prdm9msc# Alleles Control Fertility Phenotypes of Hybrids
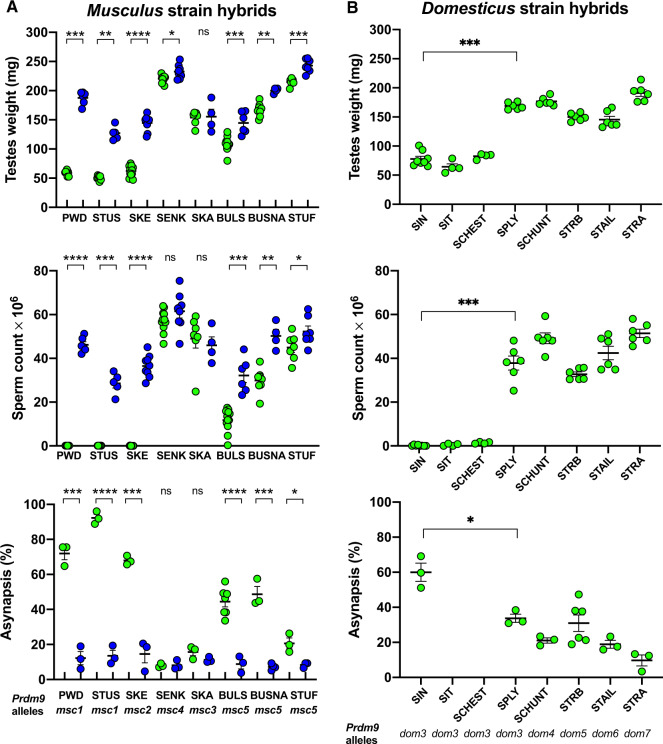
Females of congenic domesticus strain B6.PWD-Chr X.1s (hereafter B6.DX1s) carrying the hybrid sterility-augmenting Hstx2PWD locus (Bhattacharyya et al. 2014; Lustyk et al. 2019) were crossed with males of the eight musculus wild-derived inbred strains to ensure the same Hstx2 allele in hybrids. Their F1 hybrid male progeny was phenotyped at 60-day postpartum (dpp) by determining the absolute testes weight, the number of sperm cells in both epididymides, and asynapsis rate (see Materials and Methods for using absolute testes weight values). Prdm9 alleles were determined after fertility data were obtained. Phenotyping of (B6.DX1s×musculus)F1 hybrids revealed a considerable variation between both fertility phenotypes and asynapsis rates, dependent on the wild-derived Prdm9 allele. Besides the PWD reference strain, only STUS and SKE produced fully sterile hybrids (no sperm and >90% of asynapsed pachynemas). STUS was the only strain that shared the Prdm9msc1 allele with PWD (fig. 4A). The most related to Prdm9 msc1 were the Prdm9msc2 and Prdm9msc3 alleles of SKE and SENK strains, which each showed a single ZnF substitution in their ZnF arrays. Following the predicted importance of ZnF3 to ZnF6 for specificity of the DNA-binding motif (Paigen and Petkov 2018), a single substitution of ZnF at position 3 (QVK to QNK amino acid triplets) in Prdm9msc3 of the SENK strain was associated with restoration of full male fertility and only 8% of asynaptic pachynemas. In contrast, the SKE males with the substituted ZnF13 (QNK to ANQ) produced sterile hybrids with 0–0.17×106 sperm cells, small testes, and 68.2% of pachynemas showing asynapsis (fig. 4A).
Fig. 4.
Fertility phenotypes and chromosome pairing of intersubspecific mouse hybrids. (A) Hybrids from crosses of musculus wild-derived inbred strain males and B6.DX1s (green circles) or B6.DX1s. Prdm9dom2H “humanized” domesticus females (blue circles). Only Prdm9msc1 and related Prdm9msc2 alleles support high asynapsis and full meiotic arrest or quasi sterility. The Prdm9 control of fertility phenotypes is shown by improved meiotic chromosome pairing and fertility parameters in hybrids carrying humanized alleles compared with the Prdm9dom2 allele. (B) Hybrids of PWD musculus females and wild-derived domesticus males. All wild-derived strains that carry the Prdm9dom3 allele except for SPLY, produced sterile hybrids. Asynapsis rate was significantly higher in sterile SIN hybrids than in fertile SPLY carrying the same Prdm9dom3 allele. Significance of fertility improvement caused by the humanized Prdm9 allele was evaluated by Mann–Whitney U test for testes weight and sperm count and by unpaired two-tailed t-test for asynapsis. P values <0.0001****, 0.0001–0.001***, 0.001–0.01**, 0.01–0.05*, >0.05 ns.
To further investigate the contribution of Prdm9 to these fertility phenotype variations, we employed B6.DX1s mice with ´humanized´ Prdm9dom2H gene. This engineered allele is identical with Prdm9dom2 except for the substituted sequence of the ZnF array coming from the human PRDM9b ortholog. In (PWD×B6.Prdm9dom2H)F1 hybrid males, humanization of Prdm9 improved symmetric binding of PRDM9 equally on paternal and maternal chromosomes and reversed their sterility (Davies et al. 2016). We used the B6.DX1s.Prdm9dom2H mice with humanized Prdm9dom2H and Hstx2PWD locus to probe the involvement of Prdm9 in fertility impairment of hybrids with wild-derived musculus strains. Provided that the low testes weight and sperm count were caused by Prdm9-unrelated hybrid sterility gene(s), the expected increase of symmetric PRDM9 binding caused by humanized allele should not improve the fertility scores. On the contrary, if the decreased fertility of hybrids is (predominantly) under the Prdm9 control, the humanized allele should restore fertility to normal values. Indeed, in crosses of B6.DX1s.Prdm9dom2H females with males of wild-derived musculus strains, we observed complete reversal of male sterility of PWD, STUS, and SKE hybrids and recovery of physiologically normal synapsis of homologous chromosomes (fig. 4A). The fertility phenotypes of hybrids of the remaining five strains were improved proportionally to the degree of their spermatogenic damage. In all cases of lower sperm counts and testes weights in the (B6.DX1s×musculus) hybrids, we saw an increase in sperm production after the Prdm9dom2 allele was replaced by the humanized one. The alleles of BULS and BUSNA showed a mean sperm count increase from 11.8 to 32.2 million and from 29.9 to 50.2 million, respectively. The asynapsis rates were lowered accordingly. In the case of SKE allele, asynapsis decreased from 68.0% down to 14.6%. The Prdm9dom2H allele also reduced the mean asynapsis rate of pachynemas in the hybrids of SKA (15.7–11.2%), BULS (from 44.4% to 8.8%), BUSNA (from 48.7% to 7.3%), and STUF (from 20.6% to 8.4%) strains. The reduction of the asynapsis rate coincided with a complete lack of cells with more than five asynapses along with a decrease of the cell population with one to six asynapses (fig 4A and supplementary fig. S2, Supplementary Material online).
We can conclude that the sterility of the studied F1 hybrids was Prdm9-dependent since the humanized Prdm9 allele completely rescued meiotic pairing and spermatogenic arrest. The persisting differences in the testes weight and sperm count between hybrids with humanized Prdm9 allele can indicate either the presence of minor Prdm9-independent hybrid incompatibilities or genetically controlled physiological variations between these quantitative traits (Le Roy et al. 2001). These findings provide the first evidence for the Prdm9 behaving as a hybrid sterility gene outside the (PWD×B6)F1 laboratory model.
Fertility Phenotypes Are Controlled by the Wild-Derived domesticus Prdm9dom# Alleles
The (PWD×domesticus)F1 hybrids revealed varying degrees of fertility impairment estimated by the testes weight, sperm count, and meiotic asynapsis rate. The PRMD9dom3 ZnF array is identical with the reference PRMD9dom2 of the B6 laboratory strain except for an extra copy of AVQ ZnF at position 11 (fig. 1B). After crosses with PWD females, the SIN, SIT, and SCHEST males carrying the Prdm9dom3 allele produced sterile hybrids (Prdm9dom3/msc1, Hstx2PWD) with sperm count close to zero and mean testes weight <80 mg. They significantly differed from other Prdm9dom3 strains, the wild-derived SPLY and laboratory strain C3H/Di (not shown), which produced fertile hybrids (see fig. 4B for statistics). Both C3H/Di and SPLY are almost entirely domesticus on the autosomes and X chromosome, but carry a musculus Y chromosome. It is unclear which modifiers in the SPLY genome ensure fertility restoration. The involvement of the Y chromosome is unlikely (see Discussion). The asynapsis estimates were unavailable for SIT and SCHEST hybrids, but the asynapsis rate of SIN and SPLY hybrids correlated with the weight of testes and the number of produced sperm (see below).
The STRB Prdm9dom5 allele also differs from the Prdm9dom2 allele only at a single ZnF repeat, in this case by deletion of the fourth ZnF at the N-terminal part of ZnF array, which is essential for the DNA-binding pattern. In accordance with the importance of the N-terminus of the array for the DNA-binding pattern, the (PWD×STRB)F1 hybrids showed physiologically normal values of sperm count (mean 32.7 million) and 27.7% asynapsis rate. The SCHUNT strain also differs by a single ZnF substitution (at position 8), in this case from Prdm9dom3. Still, the (PWD×SCHUNT)F1 hybrids displayed the highest sperm count of 49.2 million and asynapsis rate of 21%. The most phylogenetically distant from Prdm9dom2 were Prdm9dom6 and Prdm9dom7 alleles of STAIL and STRA strains, which produced fertile hybrids with sperm counts of 42.4 and 51.4 million, respectively. They also exhibited the lowest asynapsis rates among the studied domesticus alleles—18.9% and 9.7%, respectively (fig. 4B).
To conclude, out of the five wild-derived domesticus alleles only the Prdm9dom3 allele generated sterile hybrids. The interference of genetic background was apparent from the fertility of SPLY hybrids, which carry the Prdm9dom3 allele. The SPLY differs from other domesticus strains (fig. 4B) by the presence of the Y chromosome of musculus origin, but other factor(s) in the SPLY genome can be responsible for the fertility rescue. The latter seems to be the more likely explanation as many classical laboratory strains including B6 strain also carry the musculus type of Y chromosome (Bishop et al. 1985; Yang et al. 2011) but produce sterile males in crosses with PWD mice.
Meiotic Asynapsis Varies within and between Musculus and Domesticus Subspecies
The most likely, though not exclusive explanation for the deficiency of proper synapsis of subspecific homeologs seems to be the asymmetry of PRDM9 binding due to differential erasure and resulting heterozygosity of PRDM9 binding motifs (Davies et al. 2016; Hinch et al. 2019). In principle, such asymmetry can vary in hybrids depending on differential erasure of the parental recombination hotspots but it drops to zero in inbred strains because of the sequence identity of homologs. To analyze Prdm9-related variation in meiotic chromosome synapsis, we compared the overall asynapsis rates of intersubspecific hybrids (musculus×domesticus) to intrasubspecific hybrids (domesticus×domesticus and musculus×musculus) and control inbred strains (fig. 5A and B; supplementary fig. 2, Supplementary Material online) and their relation to fertility phenotypes. In intersubspecific musculus crosses, the asynapsis rate was reduced from median 45.7% to 9.4% by the humanized Prdm9dom2H allele (P < 0.0001, Mann–Whitney test). The musculus intrasubspecific crosses showed median 11.3% asynapsis rate, which was significantly lower than interspecific average (P < 0.0001, Mann–Whitney test), but significantly higher than the asynapsis rate in the humanized crosses (P = 0.046, Mann–Whitney test). As predicted from the absence of hotspot asymmetry, the lowest value of asynapsis, 3.5%, was observed in the PWD strain (compared with humanized crosses P = 0.0144, Mann–Whitney test, fig. 5A). Similarly, in domesticus hybrids, the asynapsis rate was significantly higher in intersubspecific (median 23.4%) compared with the intrasubspecific hybrids (9.9%, P < 0.0001, Mann–Whitney test, fig. 5B), which was significantly higher than in the B6 inbred males (2.2%, P = 0.0484, unpaired t-test). The unexpectedly increased asynapsis rate observed in B6.DX1s compared with B6 (12.6%, P = 0.0032, unpaired t-test) cannot be explained by hotspot asymmetry and will be analyzed separately.
Fig. 5.
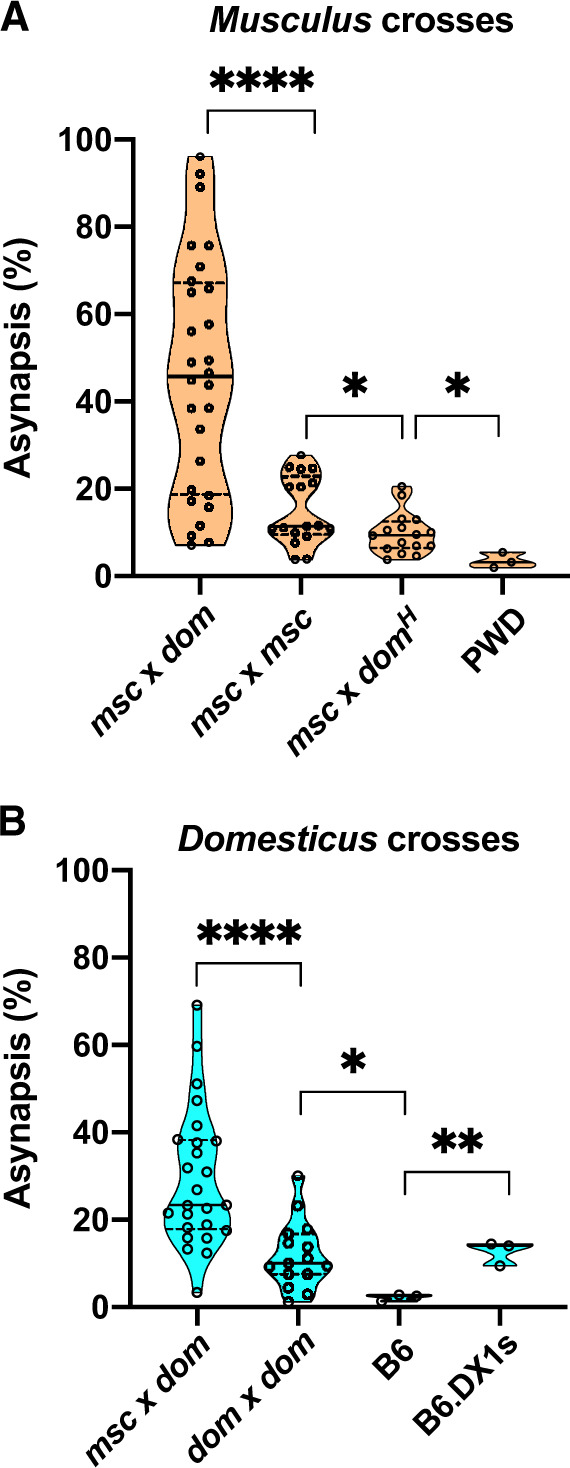
Asynapsis rate in inter- and intrasubspecific crosses. (A) The overall asynapsis rate in intrasubspecific crosses of wild-derived musculus strains does not exceed 30%. The role of Prdm9 in intrasubspecific crosses follows from the significant difference between musculus intrasubspecific and “humanized” hybrids. The lowest asynapsis rate was observed in PWD and B6 inbred strains. (B) The significant difference between inter and intrasubspecific crosses and between intrasubspecific hybrid and inbred B6 mice is apparent in domesticus crosses as well. Note the significant elevation of asynapsis on B6 background caused by introgression of 67-Mb od PWD sequence at the X chromosome centromeric end. Dashed lines in the violin plots denote quartiles, full line medians. Msc, dom, and domH denote musculus, domesticus, and humanized domesticus (Prdm9dom2H). P values <0.0001****, 0.001–0.01**, 0.01–0.05*.
The sperm count and testes weight showed a positive correlation in all hybrids (fig. 6A). Testes <80 mg produced no sperm or extremely low number of sperm cells in the epididymides and occurred exclusively in intersubspecific hybrids. Sterility was consistently associated with high asynapsis rate (range 46–96%) (fig. 7A and B; supplementary fig. S3, Supplementary Material online). In fertile intersubspecific hybrids, the asynapsis rate showed continuous variation inversely proportional to the testes weight and sperm count. Asynapsis and fertility phenotypes were not significantly correlated in intraspecific or humanized interspecific hybrids.
Fig. 6.
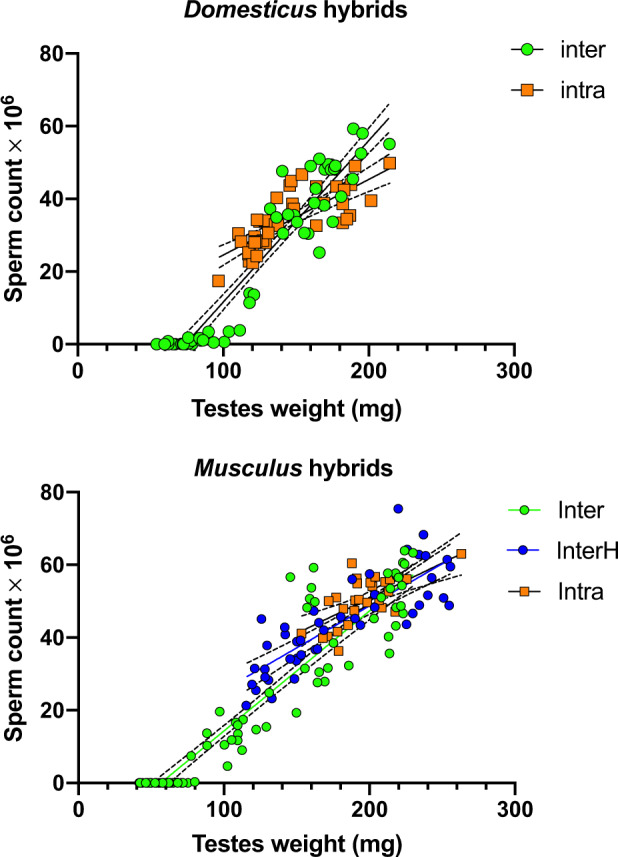
Correlation between fertility phenotypes in hybrid males. Testes weight and sperm count are positively correlated in both types of hybrid males in interspecific (Pearson’s r = 0.95, P < 0.0001 for domesticus and musculus), musculus “humanized” interspecific hybrids (r = 0.83, P < 0.0001), and intraspecific domesticus and musculus hybrids (r = 0.78 and 0.66, P < 0.0001 for domesticus and musculus). The F1 hybrids with testes weight <80 mg are fully sterile, producing virtually no sperm. Dashed lines denote 95% CI profile likelihood.
Fig. 7.
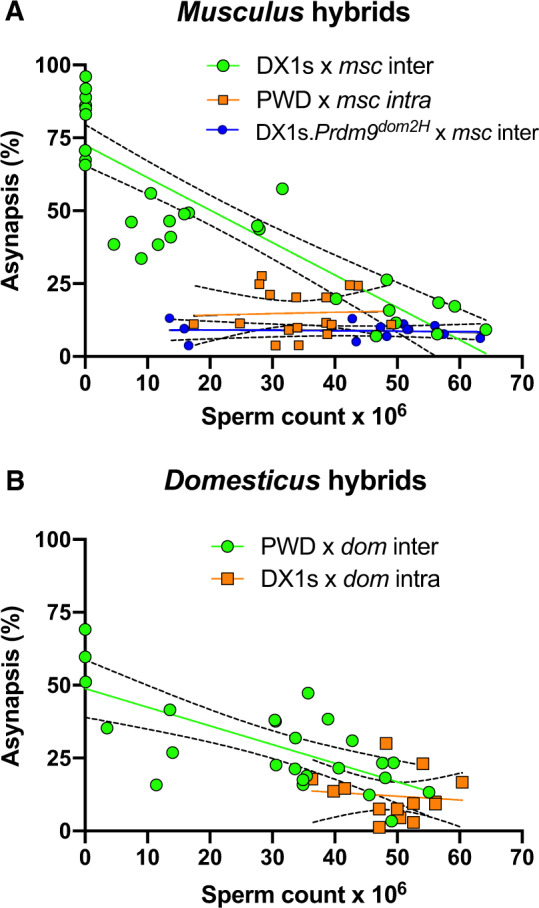
Correlation between asynapsis and sperm count in hybrid males. (A) Sperm count was positively correlated only in musculus intersubspecific hybrids (green circles, r = 0.87, P < 0.0001) but did not show correlation in intrasubspecific (orange squares, r = 0.05, P = 0.86) and “humanized” hybrids (blue circles, r = 0.08, P = 0.78). (B) In addition, in domesticus crosses, only intersubspecific hybrids showed a high positive correlation between sperm count and asynapsis (green circles, r = 0.70, P < 0.0001). Dashed lines denote the first and the third quartile of the data sets.
Discussion
The Prdm9 gene is the first hybrid sterility gene identified in vertebrates. In this study, we investigated whether it contributes to hybrid sterility outside the (PWD×B6)F1 laboratory hybrids where it was first described and where it has been studied until now (Forejt and Ivanyi 1974; Forejt 1996; Gregorová et al. 1996; Trachtulec et al. 1997; Mihola et al. 2009; Forejt et al. 2012; Bhattacharyya et al. 2013; Davies et al. 2016; Smagulova et al. 2016; Gregorova et al. 2018).
Variations in the Fertility of Wild-Derived Intersubspecific Hybrids Is Prdm9 Dependent
To verify the role of various Prdm9 alleles in hybrid sterility of mice from natural populations, we substituted one of the parents in the laboratory model of hybrid sterility with a wild-derived inbred strain from the same subspecies and compared the fertility and meiotic phenotypes of the newly derived hybrid males to that of the laboratory hybrids. Since in both experimental layouts, the genotype at a second major hybrid sterility locus was always the same (Hstx2PWD), the differences in fertility and meiotic phenotypes between laboratory model and a newly designed hybrid could only be attributed to the introduced wild-derived Prdm9 allele and/or additional genetic factor(s) in the wild-derived part of the genetic background (fig. 2).
In this experimental setup, the wild-derived domesticus males from SIN, SIT, and SCHEST strains produced infertile hybrids with PWD musculus females. They all shared the domesticus Prdm9dom3 allele, whose amino acid sequence is most similar to the B6 Prdm9dom2 reference “sterility” allele. The Prdm9dom3 allele was also identified in the WSB/EiJ strain (Parvanov et al. 2010) derived from North American wild domesticus mice (Centreville, Maryland, Harr 2006, that produced quasi-sterile hybrids with PWD females, White et al. 2011). Moreover, in the same study, the QTL analysis of a (WSB×PWD)F2 population revealed an underdominant QTL locus for sperm count on proximal chromosome 17, suggesting involvement of Prdm9dom3. Thus, altogether four wild-derived domesticus strains with unrelated genetic backgrounds from Germany, Scotland, and the United States carry Prdm9dom3 associated with hybrid sterility.
Admittedly, Prdm9dom3 was also found in the SPLY and C3H/Di strains, which produce fertile hybrids. Three explanations of fertility of these hybrids can be considered. First, the association of Prdm9msc1/dom3 and sterility of hybrids could be coincidental. We consider this unlikely based on the concordance of PRDM9dom3 ZnF variant with sterility of hybrids of four unrelated domesticus strains and association of PRDM9msc1 and PRDM9msc2 ZnF variants with sterility in musculus hybrids (fig. 4A). Second, autosomal and/or Y chromosome introgression of musculus genome in SPLY and C3H/Di could be responsible for restored fertility. This too seems unlikely, because SPLY and C3H were shown to carry <1% and 5% of the musculus autosomal genome, respectively (supplementary table S1, Supplementary Material online), an admixture too low to attenuate hybrid sterility (Gregorova et al. 2018). Likewise, the idea that presence of a musculus-type Y chromosome in SPLY and C3H (in contrast to other tested domesticus strains carrying Prdm9dom3) (Abe et al. 2004; Morgan and Pardo-Manuel de Villena 2017) could mitigate Prdm9-related sterility is not supported by available evidence. In our study (supplementary table S4, Supplementary Material online) as well as in various previous crosses, hybrid sterility clearly segregated with the Prdm9 alleles and the X but not Y chromosome (White et al. 2011; Campbell et al. 2012; Dzur-Gejdosova et al. 2012; Bhattacharyya et al. 2013; Gregorova et al. 2018). The X–Y intragenomic conflict caused by the copy-number imbalance of the Slx/Slxl1 and Sly gene families in hybrids does lead to a hybrid sterility-like incompatibility (Cocquet et al. 2009, 2012; Campbell et al. 2012). However, Prdm9-related infertility operates at the first meiotic prophase, upstream of the postmeiotic defects associated with X–Y intragenomic conflict. Notwithstanding, it is highly likely that the X–Y conflict can operate as a hybrid sterility barrier independently of Prdm9, and, in the case of partial Prdm9-controlled hybrid fertility with attenuated early meiotic arrest, the X–Y interactions could contribute to the strength of the overall reproductive barrier between subspecies (Martincová et al. 2019). The third explanation seems most likely so far. The renewed fertility of Prdm9dom3 hybrids could be either a consequence of presumed low level of erosion of PRDM9dom3 binding sites in SPLY and C3H strains, or due to action of polymorphic accessory factors promoting PRDM9 binding (Mahgoub et al. 2020; Spruce et al. 2020).
Hybrid male sterility was also observed in musculus wild-derived hybrids, where it was restricted to the Prdm9msc1 allele (STUS and PWD strains) and closely similar Prdm9msc2 (SKE) allele. To distinguish the effect of Prdm9 gene from unrelated hybrid sterility factor(s) in the genetic background, we substituted the domesticus Prdm9dom2 of the B6.DX1s parent with humanized Prdm9dom2H carrying the ZnF array from the human PRDM9B variant. Because the human PRDM9 binding sites have no history of evolutionary erasure in the naïve mouse genome, Prdm9dom2H is thought to reverse sterility of (PWD×B6. Prdm9dom2H)F1 hybrids by reducing PRDM9 hotspot asymmetry (Davies et al. 2016). Consequently, if the Prdm9-dependent erasure of DSB hotspots is the main cause of fertility impairment of B6.DX1s×musculus hybrids, we can expect the humanized allele to improve their fertility phenotypes. Indeed, as expected, the humanized Prdm9dom2H gene significantly increased the testes weight, number of sperm, and normalized meiotic chromosome synapsis in intersubspecific hybrids, thus confirming the absence of a major Prdm9-unrelated mechanism of male sterility.
Variation of Meiotic Synapsis and Fertility Parameters within Subspecies
We found that meiotic asynapsis rate >60% is diagnostic for full hybrid sterility in musculus and domesticus intersubspecific F1 hybrids. Remarkably, variation of asynapsis was also found in intrasubspecific crosses within the range of 1.2–30% in intra-domesticus hybrids and 3.8–27.6% in intra-musculus hybrids. It is tempting to speculate that the increased levels of asynapsis in intrasubspecific crosses represent a cytological counterpart of hotspot asymmetry between populations within the same subspecies. To verify the idea, direct estimation of the DSB hotspot asymmetry in intraspecific F1 hybrids and their parental strains are necessary.
Prdm9-Dependent Hybrid Sterility in Musculus/Domesticus Hybrid Zone?
Our relatively simple model of hybrid sterility composed of Prdm9, Hstx2, and genomic asymmetry of PRDM9 binding sites contrasts with the complex polygenic control found in wild mice from the European hybrid zone or from the laboratory intercrosses between wild-derived inbred strains. We believe that the difference can be explained by multiplicity of unrelated reproductive isolation mechanisms differentially revealed by different experimental approaches. For instance, the studies of gene flow across the hybrid zone (Tucker et al. 1992; Macholan et al. 2007; Teeter et al. 2007, 2010; Janoušek et al. 2012) can reveal regions in the genome that are resistant to introgression and potentially carry genes affecting any form of reproductive fitness at prezygotic and/or postzygotic level. In addition, the genome-wide association studies of male fertility in two feline interspecies models (Davis et al. 2015) and in mice from the hybrid zone (Turner and Harr 2014) disclosed complex genetic networks potentially affecting unrelated phenotypes of reproductive isolation. The mouse study included strong GWAS interactions between loci on Chromosomes X and 17, which, however, clearly mapped outside the Prdm9 and Hstx2 loci. The Prdm9-dependent hybrid sterility requires F1 hybrid genetic background since even small stretches of consubspecific sequence present in multiple chromosomes can rescue meiotic synapsis and fertility of hybrids (Gregorova et al. 2018).
The laboratory crosses of wild-derived mouse strains designed to map fertility phenotypes (Good et al. 2007; Vyskocilova et al. 2009; Oka et al. 2010; White et al. 2011, 2012; Wang et al. 2015; Larson et al. 2018; Schwahn et al. 2018) or gene misexpression in hybrid testis (Mack et al. 2016; Mack and Nachman 2017; Morgan et al. 2020) are much closer to our F1 hybrid sterility model. However, although the classical F1 hybrid sterility is governed by underdominance, the fertility of intersubspecific backcrosses and F2 crosses is affected by more frequent autosomal recessive incompatibilities (Coyne and Orr 2004; White et al. 2011). Indeed, in a study of (PWD×WSB)F2 hybrid population (White et al. 2011), out of 19 autosomal QTLs only three were underdominant, two of them for sperm density and testes size on chromosome 17. Admittedly, there must be some additional genetic factors in Prdm9-controlled hybrid sterility, besides Prdm9, Hstx2, and background heterozygosity, that modify fertility of F1 hybrids. The hybrid sterility factors on chromosomes 3, 9, and 13 polymorphic between PWD and STUS musculus strains could represent possible examples (Bhattacharyya et al. 2014).
Previously, it was shown that the intersubspecific F1 hybrids are virtually missing in the central parts of the European zone so that a pure form of F1 hybrid sterility, as seen in (PWD×B6) laboratory crosses, could hardly function as a major reproductive barrier (Macholan et al. 2007; Teeter et al. 2010; Albrechtova et al. 2012; Turner et al. 2012). On the other hand, we know that the Prdm9-dependent meiotic arrest and associated asynapsis between homeologous autosomes gradually weakens, but does not disappear in intersubspecific laboratory backcrosses (Dzur-Gejdosova et al. 2012; Gregorova et al. 2018). We can speculate that similar intersubspecific and consubspecific genomic mixture reduces the Prdm9 hotspot asymmetry and results in the prevalence of recessive incompatibilities within the hybrid zone. Future studies will be necessary to elucidate the role of the Prdm9-controlled hybrid sterility in maintenance of the current hybrid zone between the musculus and domesticus subspecies of house mouse.
To conclude, our data show for the first time that the male infertility of wild-derived musculus and domesticus subspecies F1 hybrids is controlled by Prdm9 as the major hybrid sterility gene. The impaired fertility surrogates, testes weight and sperm count, negatively correlated with increasing difficulties of meiotic synapsis of homologous chromosomes and meiotic arrest, which might reflect the increasing asymmetry of PRDM9-dependent DNA DSBs.
Materials and Methods
Mice
Except for the PWD/Ph strain (Gregorova and Forejt 2000), all wild-derived inbred strains (F13–F61) in this study have been developed and maintained in the breeding facility of the Institute of Vertebrate Biology in Studenec (Pialek et al. 2007; Baird and Macholan 2012; Albrechtová et al. 2014; Martincová et al. 2019), with licenses for maintaining the mice and experimental work (61,974/2017-MZE-17214 and 62,065/2017-MZE-17214, respectively). The localities of origin of the musculus strains are: SKA/Jpia—Kaerepere, Estonia, [N: 58° 57′, E: 24° 50′], SKE/Jpia—Keava, Estonia [N: 58° 56′ 27″, E: 24° 54′], BULS/Jpia and BUSNA/Jpia—Buškovice, Czech Republic [N: 50° 13′, E: 13° 23′], PWD/Ph—Kunratice (Prague), Czech Republic, STUF/Jpia and STUS/Jpia—Studenec, Czech Republic [N: 49° 12′, E: 16° 04′], SENK/Jpia—Šenkvice, Slovak Republic [N: 48° 18′, E: 17° 21′. The domesticus strains: STRA/Jpia and STRB/Jpia—Straas, Bavaria, Germany [N: 50° 11′, E: 11° 46′], STAIL/Jpia, SCHEST/Jpia and SCHUNT/Jpia—Schweben, Hessen, Germany [N: 50° 26′, E: 9° 35′], SPLY/Jpia—Plössen, Bavaria, Germany [N: 49° 51′ 18″, E: 11° 47′], SIN/Jpia and SIT/Jpia—Scar, Whitemill Bay, Sanday Island, Orkneys, Scotland [N: 59° 18′, E: −2° 33′].
The C57BL/6J-Chr X.1sPWD/Ph/ForeJ (in short, B6.DX1s) is a consomic strain carrying 69.6 Mb of the proximal PWD sequence on the genetic background of the B6/J strain (Bhattacharyya et al. 2014). Coisogenic strain C57BL/6-Prdm9TmHu (abbreviated B6.Prdm9dom2H) (Davies et al. 2016) was kindly provided by Dr Simon Myers, Oxford University, UK. All mice were maintained in the Specific Pathogen-Free Facilities, in accordance to animal care protocols approved by the Committee on the Ethics of Animal Experiments of the Institute (No. 141/2012). The animal care obeyed the Czech Republic Act for Experimental Work with Animals (Decree No. 207/2004 Sb and Acts Nos 246/92 Sb and 77/2004 Sb), fully compatible with the corresponding regulations and standards of the European Union (Council Directive 86/609/EEC and Appendix A of the Council of Europe Convention ETS123).
Genotyping of Mouse Strains and Quality Control
One male and one female from each of 17 strains of interest (including PWK/Ph and C57BL/6J) were genotyped at 143,259 markers with the GigaMUGA array (Neogen Europe, Ayr, Scotland). An updated annotation for the GigaMUGA array (https://kbroman.org/MUGAarrays/new_annotations.html; last accessed July 11, 2020) was used for all subsequent analyses. All samples had <10% missing calls, a useful cutoff for this platform (Morgan et al. 2015), and <5% heterozygous calls, as expected for inbred strains. The sex of each sample was confirmed by comparing hybridization intensities for X- and Y-linked markers.
Ancestry Inference
To define ancestry-informative markers, we used published genotypes on the GigaMUGA array from 15 wild-caught M. m. domesticus and 5 wild-caught M. m. musculus individuals (Morgan 2015). The domesticus set includes mice trapped in Greece, Spain, Italy, Switzerland, and the eastern United States. The musculus set includes mice trapped in China, Poland, Germany, and Russia. All trapping locations were far from the musculus–domesticus hybrid zone, so these individuals are presumed to be pure representatives of their subspecies.
Autosomes and X Chromosome
Markers with absolute allele frequency difference >0.75 between the musculus and domesticus reference groups and <25% missing genotypes within each group were retained as putative ancestry-informative sites. A total of 28,146 unique autosomal and 1,303 X-linked sites were retained. Genotypes for inbred strains of interest were recoded as musculus, domesticus, or heterozygous at each marker according to the consensus allele in each reference group. A hidden Markov model (HMM) was then used to decode ancestry along the genome of each inbred strain. The HMM has two hidden states, musculus and domesticus. (Given the low observed residual heterozygosity in these strains, a musculus/domesticus heterozygous state was not modeled.) The probability of transition between states between consecutive sites is 1e-5; the probability of observing the opposite subspecies’ allele is 5e-2; and probability of observing a heterozygous genotype is 1e-3. Although these parameters were set arbitrarily, we confirmed that the output of the procedure is relatively insensitive to their values. The HMM was applied separately to observed genotypes of each sample, and the posterior decoding was obtained by the Viterbi algorithm. Per-site ancestry was aggregated into blocks by simple run-length encoding. For strains previously analyzed using the Mouse Diversity Array (Yang et al. 2011), we confirmed by manual inspection that introgressed blocks >5 Mb in size were recovered in both platforms. Analyses were performed in “R” v3.3.2 using packages “argyle” (https://github.com/andrewparkermorgan/argyle; last accessed July 11, 2020) and “HMM” (https://cran.r-project.org/package=HMM; last accessed July 11, 2020). The ancestry origin of C3H/Di strain was extrapolated from Mouse Phylogeny Viewer (http://msub.csbio.unc.edu/; last accessed July 11, 2020).
Sequencing of Mouse Prdm9 Alleles and Phylogeny
Partially purified DNA from mouse spleens was isolated by Puregene Core Kit A (QIAGEN 158267) according to the manufactureŕs instructions. DNA template of 50–100 ng was used for amplification of Prdm9 Exon12 in a reaction mixture containing primers for Prdm9 Exon12: Exon12-L1—TGAGATCTGAGGAAAGTAAGAG and Exon12-R—TGCTGTTGGCTTTCTCATTC with a concentration of 0.4 µM of each primer, 0.2 mM dNTP, 2 mM MgCl2, and Taq DNA Polymerase, recombinant (1 U/ml) (Thermofisher Scientific EP0404) by 0.15 U per reaction. Samples were amplified by BIOER XP Cycler under the PCR program: 94 °C for 5 min and 40 cycles at 94 °C for 30 s, 61 °C for 1 min, 68 °C for 2 min, and a single step after the cycles of 72 °C for 7 min. The PCR products were resolved in 2% agarose gel allowing visual estimation of the quality and homogeneity of amplified DNA. The samples were prepared for sequencing as ExoSAP-treated samples. The obtained sequencing data were visualized and extracted by the Chromas Lite 2.1 application.
In crosses between heterozygous Prdm9dom2H/+ females versus wild males, the type of Prdm9 allele in the hybrid offspring was checked by forward primer (5′-TTCTGCCATCACTTCCTTCGGTGA-3′) and reverse primer (5′-TCTGAAGCCCAACTATTTCATTAATACCCC-3′). A 677-bp amplicon was obtained from the humanized allele and a 491-bp amplicon was obtained from the wild-type allele (Davies et al. 2016). The reaction mixture was the same as for the sequencing of Prdm9 Exon12 by PCR program: 95 °C for 2 min and 40 cycles at 95 °C for 30 s, 55 °C for 40 s, 72 °C for 40 s, and a single step after the cycles of 72 °C for 3 min.
The evolutionary relationships of among Prdm9 alleles were inferred using the neighbor-joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length = 0.03369633 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. This analysis involved 17 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd+Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 1,263 positions in the final data set. Evolutionary analyses were conducted in MEGA X (Kumar et al. 2018).
Fertility Phenotyping
Males were euthanized by cervical dislocation at 60 dpp and their body weight, the weight of paired testes in milligrams (TW), and sperm count in million was determined. Since the correlation between the body weight and testes weight was not significant in hybrid males (supplementary fig. S4, Supplementary Material online), we used the absolute weight of paired testes as one of the fertility phenotypes. Spermatozoa were released from the whole epididymides, and the number of sperm heads was counted in 25 squares of a Bürker chamber using an Olympus CX41 microscope under 200× magnification (for details, see Vyskočilová et al. [2005]). For each F1 cross, four to eight hybrid males were examined for testes weight and sperm count.
Immunostaining and Asynapsis Rate Determination
For immunocytochemistry, the spread spermatocyte nuclei were prepared as described (Anderson et al. 1999) with modifications. Briefly, a single-cell suspension of spermatogenic cells in 0.1 M sucrose with protease inhibitors (Roche) was dropped on 1% paraformaldehyde-treated slides and allowed to settle for 3 h in a humidified box at 4 °C. After brief washing in distilled water and PBS and blocking with 5% goat sera in PBS (vol/vol), the cells were immunolabeled using a standard protocol with the following antibodies: anti-HORMAD2 (1:700, rabbit polyclonal antibody, a gift from Attila Toth) and SYCP3 (1:100, mouse monoclonal antibody, Santa Cruz, #74569). Centromere painting was done by human antibodies from autoimmune serum, AB-Incorporated, 15-235. Rabbit α-γ-H2AX (1:1,000, Rabbit polyclonal anti gamma H2A.X, ABCAM, ab2893) identified early DSBs and unsynapsed parts of autosomes and sex chromosomes. Secondary antibodies were used at 1:300 dilutions and incubated at 4 °C for 60 min: goat anti-Mouse IgG-AlexaFluor568 (MolecularProbes, A-11031), goat anti-Rabbit IgG-AlexaFluor647 (MolecularProbes, A-21245), goat anti-Human IgG-AlexaFluor647 (MolecularProbes, A-21445), goat anti-Rabbit IgG-AlexaFluor488 (MolecularProbes, A-11034).
The images were acquired and examined using a Nikon Eclipse 400 microscope with a motorized stage control using a Plan Fluor objective, 60× (MRH00601; Nikon) and captured using a DS-QiMc monochrome CCD camera (Nikon) and the NIS-Elements program (Nikon). The images were processed using the Image J software (Schneider et al. 2012). For each sample, we analyzed between 68 and 121 pachynemas. The number of asynapses per nucleus was scored in each pachynema. One asynapsis was equal to one HORMAD2 stained element, excluding XY chromosomes. The asynapsis rate was represented as the percentage of pachynemas with asynapses out of the total number of the checked pachynemas of each male. A minimum of three males per F1 cross was used for asynapsis rate estimation.
Statistics
The statistical significance for the testes weight and sperm count was assessed by two-tailed Mann–Whitney test. Asynapsis rate was evaluated by unpaired t-test in the GraphPad Prism version 8.4.0. for MacOS.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Simon Myers for the B6.Prdm9dom2H mice, Attila Toth for HORMAD2 antibody, and Sarka Takacova for comments. This work was funded by LQ1604 Project of the National Sustainability Program II from the Ministry of Education, Youth and Sports of the Czech Republic, and by Czech Science Foundation (Grant Nos. GA CR No. 17-04364S to E.P., 20-04075S to J.F., and 19-12774S to J.P.).
References
- Abe K, Noguchi H, Tagawa K, Yuzuriha M, Toyoda A, Kojima T, Ezawa K, Saitou N, Hattori M, Sakaki Y, et al. 2004. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 14(12):2439–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtova J, Albrecht T, Baird SJ, Macholan M, Rudolfsen G, Munclinger P, Tucker PK, Pialek J.. 2012. Sperm-related phenotypes implicated in both maintenance and breakdown of a natural species barrier in the house mouse. Proc R Soc B. 279(1748):4803–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtová J, Albrecht T, Ďureje L, Pallazola VA, Piálek J.. 2014. Sperm morphology in two house mouse subspecies: do wild-derived strains and wild mice tell the same story? PLoS One 9(12):e115669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LK,ReevesAWebbLMAshleyT.. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SJE, Macholan M.. 2012. What can the Mus musculus musculus/M. m. domesticus hybrid zone tell us about speciation? In: Macholan M, Baird SJ, Muclinger P, Pialek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press; p. 334–372. [Google Scholar]
- Baker CL, Kajita S, Walker M, Saxl RL, Raghupathy N, Choi K, Petkov PM, Paigen K.. 2015. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 11(1):e1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Walker M, Kajita S, Petkov PM, Paigen K.. 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 24(5):724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash DA, Siino DF, Tarone AM, Roote J.. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci U S A. 100(9):5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B.. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327(5967):836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P, Simecek P, Forejt J.. 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci U S A. 110(6):E468–E477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Reifova R, Gregorova S, Simecek P, Gergelits V, Mistrik M, Martincova I, Pialek J, Forejt J.. 2014. X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS Genet. 10(2):e1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CE, Boursot P, Baron B, Bonhomme F, Hatat D.. 1985. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature 315(6014):70–72. [DOI] [PubMed] [Google Scholar]
- Boulton A, Myers RS, Redfield RJ.. 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci U S A. 94(15):8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot P, Din W, Anand R, Darviche D, Dod B, Von Deimling F, Talwar GP, Bonhomme F.. 1996. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J Evol Biol. 9(4):391–415. [Google Scholar]
- Buard J, Rivals E, Dunoyer de Segonzac D, Garres C, Caminade P, de Massy B, Boursot P.. 2014. Diversity of Prdm9zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One 9(1):e85021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Good JM, Dean MD, Tucker PK, Nachman MW.. 2012. The contribution of the Y chromosome to hybrid male sterility in house mice. Genetics 191(4):1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS.. 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 8(9):e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS.. 2009. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 7(11):e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection or the preservation of favored races in the struggle for life. London: Murray. [PMC free article] [PubMed] [Google Scholar]
- Davies B, Hatton E, Altemose N, Hussin JG, Pratto F, Zhang G, Hinch AG, Moralli D, Biggs D, Diaz R, et al. 2016. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530(7589):171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BW, Seabury CM, Brashear WA, Li G, Roelke-Parker M, Murphy WJ.. 2015. Mechanisms underlying mammalian hybrid sterility in two feline interspecies models. Mol Biol Evol . 32(10):2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion-Cote AM, Barbash DA.. 2017. Beyond speciation genes: an overview of genome stability in evolution and speciation. Curr Opin Genet Dev. 47:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. 1951. Genetics and the origin of species. New York: Columbia University. [Google Scholar]
- Dobzhansky T. 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21(2):113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufkova P, Macholan M, Pialek J.. 2011. Inference of selection and stochastic effects in the house mouse hybrid zone. Evolution 65:993–1010. [DOI] [PubMed] [Google Scholar]
- Ďureje Ľ, Macholán M, Baird SJE, Piálek J.. 2012. The mouse hybrid zone in Central Europe: from morphology to molecules. Folia Zool. 61(3–4):308–318. [Google Scholar]
- Duvaux L, Belkhir K, Boulesteix M, Boursot P.. 2011. Isolation and gene flow: inferring the speciation history of European house mice. Mol Ecol. 20(24):5248–5264. [DOI] [PubMed] [Google Scholar]
- Dzur-Gejdosova M, Simecek P, Gregorova S, Bhattacharyya T, Forejt J.. 2012. Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution 66(11):3321–3335. [DOI] [PubMed] [Google Scholar]
- Eram MS, Bustos SP, Lima-Fernandes E, Siarheyeva A, Senisterra G, Hajian T, Chau I, Duan S, Wu H, Dombrovski L, et al. 2014. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J Biol Chem. 289(17):12177–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791. [DOI] [PubMed] [Google Scholar]
- Forejt J. 1996. Hybrid sterility in the mouse. Trends Genet. 12(10):412–417. [DOI] [PubMed] [Google Scholar]
- Forejt J. 1984. X-inactivation and its role in male sterility In: Bennett M, Gropp A, Wolf U, editors. Chromosomes today. London: Geroge Allen and Unwin; p. 117–127. [Google Scholar]
- Forejt J, Ivanyi P.. 1974. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet Res. 24(2):189–206. [DOI] [PubMed] [Google Scholar]
- Forejt J, Pialek J, Trachtulec Z.. 2012. Hybrid male sterility genes in the mouse subspecific crosses In: Macholan M, Baird SJE, Muclinger P, Pialek J, editors. Evolution of the house mouse. Cambridge: Cambridge University Press; p. 482–503. [Google Scholar]
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachman MW.. 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 17(24):5349–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergelits V, Parvanov E, Simecek P, Forejt J.. 2019. Chromosome-wide distribution and characterization of intersubspecific meiotic noncrossovers in mice. bioRxiv. 792226.
- Good JM, Dean MD, Nachman MW.. 2008. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179(4):2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW.. 2007. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evol Int J Org Evol. 62:50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorova S, Forejt J.. 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies – a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol (Praha). 46:31–41. [DOI] [PubMed] [Google Scholar]
- Gregorova S, Gergelits V, Chvatalova I, Bhattacharyya T, Valiskova B, Fotopulosova V, Jansa P, Wiatrowska D, Forejt J.. 2018. Modulation of Prdm9-controlled meiotic chromosome asynapsis overrides hybrid sterility in mice. Elife 7:pii e34282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorová S, Mňuková-Fajdelová M, Trachtulec Z, Čapková J, Loudová M, Hoglund M, Hamvas R, Lehrach H, Vincek V, Klein J, et al. 1996. Sub-milliMorgan map of the proximal part of mouse chromosome 17 including the hybrid sterility 1 gene. Mamm Genome. 7(2):107–113. [DOI] [PubMed] [Google Scholar]
- Haldane J. 1922. Sex ration and unisexual sterility in animal hybrids. J Gen. 12(2):101–109. [Google Scholar]
- Harr B. 2006. Genomic islands of differentiation between house mouse subspecies. Genome Res. 16(6):730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Matsui Y.. 2006. Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell Cycle 5(6):615–620. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Zhang G, Becker PW, Moralli D, Hinch R, Davies B, Bowden R, Donnelly P.. 2019. Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm. Science 363(6433):eaau8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 7:a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoušek V, Wang L, Luzynski KEN, Dufková P, Vyskočilová MM, Nachman MW, Munclinger P, Macholán M, Piálek J, Tucker PK.. 2012. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol. 21(12):3032–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono H, Tamura M, Osada N, Suzuki H, Abe K, Moriwaki K, Ohta K, Shiroishi T.. 2014. Prdm9 polymorphism unveils mouse evolutionary tracks. DNA Res. 21(3):315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EL, Vanderpool D, Sarver BAJ, Callahan C, Keeble S, Provencio LL, Kessler MD, Stewart V, Nordquist E, Dean MD, et al. 2018. The evolution of polymorphic hybrid incompatibilities in house mice. Genetics 209(3):845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy I, Tordjman S, Migliore-Samour D, Degrelle H, Roubertoux PL.. 2001. Genetic architecture of testis and seminal vesicle weights in mice. Genetics 158(1):333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustyk D, Kinsky S, Ullrich KK, Yancoskie M, Kasikova L, Gergelits V, Sedlacek R, Chan YF, Odenthal-Hesse L, Forejt J, et al. 2019. Genomic structure of Hstx2 modifier of Prdm9-dependent hybrid male sterility in mice. Genetics 213(3):1047–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack KL, Campbell P, Nachman MW.. 2016. Gene regulation and speciation in house mice. Genome Res. 26(4):451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack KL, Nachman MW.. 2017. Gene regulation and speciation. Trends Genet. 33(1):68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA.. 2011. The genetics of hybrid incompatibilities. Annu Rev Genet. 45(1):331–355. [DOI] [PubMed] [Google Scholar]
- Mahgoub M, Paiano J, Bruno M, Wu W, Pathuri S, Zhang X, Ralls S, Cheng X, Nussenzweig A, Macfarlan TS.. 2020. Dual histone methyl reader ZCWPW1 facilitates repair of meiotic double strand breaks in male mice. Elife 9:e53360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholan M, Baird SJ, Dufkova P, Munclinger P, Bimova BV, Pialek J.. 2011. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution 65(5):1428–1446. [DOI] [PubMed] [Google Scholar]
- Macholan M, Baird SJE, Fornuskova A, Martincova I, Rubík P, Ďureje Ľ, Heitlinger E, Piálek J.. 2019. Widespread introgression of the Mus musculus musculus Y chromosome in Central Europe. bioRxiv 12.23.887471.
- Macholan M, Munclinger P, Sugerkova M, Dufkova P, Bimova B, Bozikova E, Zima J, Pialek J.. 2007. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61(4):746–771. [DOI] [PubMed] [Google Scholar]
- Macholán M, Vyskočilová M, Bonhomme F, Kryštufek B, Orth A, Vohralík V.. 2007. Genetic variation and phylogeography of free-living mouse species (genus Mus) in the Balkans and the Middle East. Mol Ecol. 16(22):4774–4788. [DOI] [PubMed] [Google Scholar]
- Martincová I, Ďureje Ľ, Kreisinger J, Macholán M, Piálek J.. 2019. Phenotypic effects of the Y chromosome are variable and structured in hybrids among house mouse recombinant lines. Ecol Evol. 9(10):6124–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Jones CD, Noor MA, Locke J, Orr HA.. 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313(5792):1448–1450. [DOI] [PubMed] [Google Scholar]
- Mayr E. 1963. Animal species and evolution. Cambridge: Harvard University Press. [Google Scholar]
- Mihola O, Pratto F, Brick K, Linhartova E, Kobets T, Flachs P, Baker CL, Sedlacek R, Paigen K, Petkov PM, et al. 2019. Histone methyltransferase PRDM9 is not essential for meiosis in male mice. Genome Res. 29(7):1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J.. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323(5912):373–375. [DOI] [PubMed] [Google Scholar]
- Morgan AP. 2015. argyle: an R package for analysis of Illumina genotyping arrays. G3 (Bethesda) 6:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AP, Fu CP, Kao CY, Welsh CE, Didion JP, Yadgary L, Hyacinth L, Ferris MT, Bell TA, Miller DR.. 2015. The Mouse Universal Genotyping Array: from substrains to subspecies. G3 (Bethesda) 6:263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AP, Pardo-Manuel de Villena F.. 2017. Sequence and structural diversity of mouse Y chromosomes. Mol Biol Evol. 34(12):3186–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan K, Harr B, White MA, Payseur BA, Turner LM.. 2020. Disrupted gene networks in subfertile hybrid house mice. Mol Biol Evol. 37(6):1547–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Pontecorvo G.. 1942. Recessive genes causing interspecific sterility and other disharmonies between Drosophila melanogaster and simulans. Genetics. 27:157. [Google Scholar]
- Muller HJ. 1942. Isolation mechanisms, evolution and temperature. Biol Symp. 6:71–125. [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P.. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327(5967):876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine CM, Nishioka Y, Moriwaki K, Boursot P, Bonhomme F, Lau YF.. 1992. The musculus-type Y chromosome of the laboratory mouse is of Asian origin. Mamm Genome. 3(2):84–91. [DOI] [PubMed] [Google Scholar]
- Naveira H, Maside X.. 1998. The genetics of hybrid male sterility in Drosophila In: Howard D, Berlocher S, editors. Endless forms. Oxford: Oxford University. p. 330–338. [Google Scholar]
- Oka A, Mita A, Takada Y, Koseki H, Shiroishi T.. 2010. Reproductive isolation in hybrid mice due to spermatogenesis defects at three meiotic stages. Genetics 186(1):339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP.. 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5(12):e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Petkov PM.. 2018. PRDM9 and its role in genetic recombination. Trends Genet. 34(4):291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K.. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327(5967):835–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW.. 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58(9):2064–2078. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Presgraves DC, Filatov DA.. 2018. Introduction: sex chromosomes and speciation. Mol Ecol. 27(19):3745–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Orr HA.. 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323(5912):376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, Nachman MW.. 2015. Insights into mammalian biology from the wild house mouse Mus musculus. Elife 4:e05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pialek J, Vyskocilova M, Bimova B, Havelkova D, Pialkova J, Dufkova P, Bencova V, Dureje L, Albrecht T, Hauffe HC, et al. 2007. Development of unique house mouse resources suitable for evolutionary studies of speciation. J Hered. 99(1):34–44. [DOI] [PubMed] [Google Scholar]
- Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, Paigen K.. 2016. The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet. 12(6):e1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2018. Evaluating genomic signatures of “the large X-effect” during complex speciation. Mol Ecol. 27(19):3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24(7):336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA.. 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423(6941):715–719. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M.. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol . 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- Salcedo T, Geraldes A, Nachman MW.. 2007. Nucleotide variation in wild and inbred mice. Genetics 177(4):2277–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J, Bonhomme F, Boursot P, Thaler L, Catzeflis F.. 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol J Linn Soc. 41(1–3):83–103. [Google Scholar]
- Schilthuizen M, Giesbers MC, Beukeboom LW.. 2011. Haldane’s rule in the 21st century. Heredity 107(2):95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwahn DJ, Wang RJ, White MA, Payseur BA.. 2018. Genetic dissection of hybrid male sterility across stages of spermatogenesis. Genetics 210(4):1453–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV.. 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30(3):266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV.. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472(7343):375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR, Nambiar M.. 2020. New solutions to old problems: molecular mechanisms of meiotic crossover control. Trends Genet. 36(5):337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce C, Dlamini S, Ananda G, Bronkema N, Tian H, Paigen K, Carter GW, Baker CL.. 2020. HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev. 34(5–6):398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova R, Gregorova S, Buckiova D, Kyselova V, Divina P, Forejt J.. 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome. 15(7):515–524. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S.. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 101(30):11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, Bakewell MA, Thibodeau LM, O’Brien JE, Krenz JG, Sans-Fuentes MA, Nachman MW, Tucker PK.. 2007. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18(1):67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Thibodeau LM, Gompert Z, Buerkle CA, Nachman MW, Tucker PK.. 2010. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution 64(2):472–485. [DOI] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI.. 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282(5393):1501–1504. [DOI] [PubMed] [Google Scholar]
- Trachtulec Z, Mňuková-Fajdelová M, Hamvas RMJ, Gregorová S, Mayer WE, Lehrach HR, Vincek V, Forejt J, Klein J.. 1997. Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm Genome. 8(5):312–316. [DOI] [PubMed] [Google Scholar]
- Tucker P, Sage R, Warner J, Wilson A, Eicher E.. 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46(4):1146–1163. [DOI] [PubMed] [Google Scholar]
- Turner LM, Harr B.. 2014. Genome-wide mapping in a house mouse hybrid zone reveals hybrid sterility loci and Dobzhansky-Muller interactions. Elife 3:e02504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Schwahn DJ, Harr B.. 2012. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution 66(2):443–458. [DOI] [PubMed] [Google Scholar]
- Vara C, Capilla L, Ferretti L, Ledda A, Sánchez-Guillén RA, Gabriel SI, Albert-Lizandra G, Florit-Sabater B, Bello-Rodríguez J, Ventura J, et al. 2019. PRDM9 diversity at fine geographical scale reveals contrasting evolutionary patterns and functional constraints in natural populations of house mice. Mol Biol Evol. 36(8):1686–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyskocilova M, Prazanova G, Pialek J.. 2009. Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mamm Genome. 20:83–91. [DOI] [PubMed] [Google Scholar]
- Vyskočilová M, Trachtulec Z, Forejt J, Piálek J.. 2005. Does geography matter in hybrid sterility in house mouse? Biol J Linn Soc. 84(3):663–674. [Google Scholar]
- Wang L, Valiskova B, Forejt J.. 2018. Cisplatin-induced DNA double-strand breaks promote meiotic chromosome synapsis in PRDM9-controlled mouse hybrid sterility. Elife 7:e42511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, White MA, Payseur BA.. 2015. The pace of hybrid incompatibility evolution in house mice. Genetics 201(1):229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Steffy B, Wiltshire T, Payseur BA.. 2011. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics 189(1):289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Stubbings M, Dumont BL, Payseur BA.. 2012. Genetics and evolution of hybrid male sterility in house mice. Genetics 191(3):917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Remenyi A, Turner JMA, et al. 2012. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev. 26(9):958–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5(10):e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mathioudakis N, Diagouraga B, Dong A, Dombrovski L, Baudat F, Cusack S, de Massy B, Kadlec J.. 2013. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep. 5(1):13–20. [DOI] [PubMed] [Google Scholar]
- Yamada S, Kim S, Tischfield SE, Jasin M, Lange J, Keeney S.. 2017. Genomic and chromatin features shaping meiotic double-strand break formation and repair in mice. Cell Cycle 16(20):1870–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, et al. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 43(7):648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.