Abstract
Gene gain and loss are crucial factors that shape the evolutionary success of diverse organisms. In the past two decades, more attention has been paid to the significance of gene gain through gene duplication or de novo genes. However, gene loss through natural loss-of-function (LoF) mutations, which is prevalent in the genomes of diverse organisms, has been largely ignored. With the development of sequencing techniques, many genomes have been sequenced across diverse species and can be used to study the evolutionary patterns of gene loss. In this review, we summarize recent advances in research on various aspects of LoF mutations, including their identification, evolutionary dynamics in natural populations, and functional effects. In particular, we discuss how LoF mutations can provide insights into the minimum gene set (or the essential gene set) of an organism. Furthermore, we emphasize their potential impact on adaptation. At the genome level, although most LoF mutations are neutral or deleterious, at least some of them are under positive selection and may contribute to biodiversity and adaptation. Overall, we highlight the importance of natural LoF mutations as a robust framework for understanding biological questions in general.
Key words: adaptive evolution, biodiversity, essential genes, loss-of-function, natural variation
Loss-of-function mutations are prevalent in the genomes of diverse organisms. In natural populations, loss-of-function mutations are natural gene knockouts that can provide insights into gene function. Furthermore, loss-of-function mutation is a strategy for adaptation.
Introduction
Gene gain and loss are prevalent in the genomes of diverse organisms and contribute to genetic variation (Albalat and Canestro, 2016; Cheetham et al., 2020). Since Susumu Ohno proposed the importance of gene duplication in evolution (Ohno, 1970), much attention has been paid to whole-genome duplication, gene duplication, and the origin of new genes (Chen et al., 2010; Guo, 2013; Palmieri et al., 2014; Zhao et al., 2014; Li et al., 2016). However, the importance of gene loss (or pseudogene formation) has been almost entirely ignored for a long time, mainly because pseudogenes may not produce full-length proteins and are regarded as putatively non-functional (Li et al., 1981; Vanin, 1985; Harrison et al., 2003; Torrents et al., 2003). Nevertheless, the “less is more” hypothesis proposes that gene loss may be an adaptive evolutionary process that is beneficial to organisms (Olson, 1999).
Two major mechanisms can cause gene loss: physical removal events (recombination or the mobilization of transposable or viral elements) that lead to the fragment deletion of one or more genes, and deleterious mutations at gene coding regions that cause loss-of-function (LoF) mutations (Albalat and Canestro, 2016). In this review, we focus on LoF mutations, which contribute to a large proportion of gene loss and have rarely been systematically explored in plants.
An LoF mutation was first observed in the 5S DNA gene of Xenopus laevis. This gene could not encode a functional 5S rRNA and was named a pseudogene (Jacq et al., 1977). Subsequently, many pseudogenes caused by LoF mutations were reported in Escherichia coli (Inokuchi et al., 1979), yeast (Fink, 1987), mammals (Lee et al., 1983; Torrents et al., 2003; Zheng et al., 2018), and plants (Zou et al., 2009; Yang et al., 2011). Since the development of whole-genome sequencing and methods of homolog detection, there have been many studies at the whole-genome level on pseudogenes derived from duplicated or retrotransposed genes through LoF mutations (Harrison et al., 2003; Torrents et al., 2003; Xie et al., 2019). At the population level within species, there have been few studies of LoF mutations, and these have focused mainly on humans (Balasubramanian et al., 2011; MacArthur et al., 2012; Narasimhan et al., 2016; Saleheen et al., 2017); however, we recently carried out a study in the model plant Arabidopsis thaliana (Xu et al., 2019). In this review, we emphasize the importance of LoF mutations for adaptive evolution in natural populations.
Genetic Variations that Cause LoF Mutations
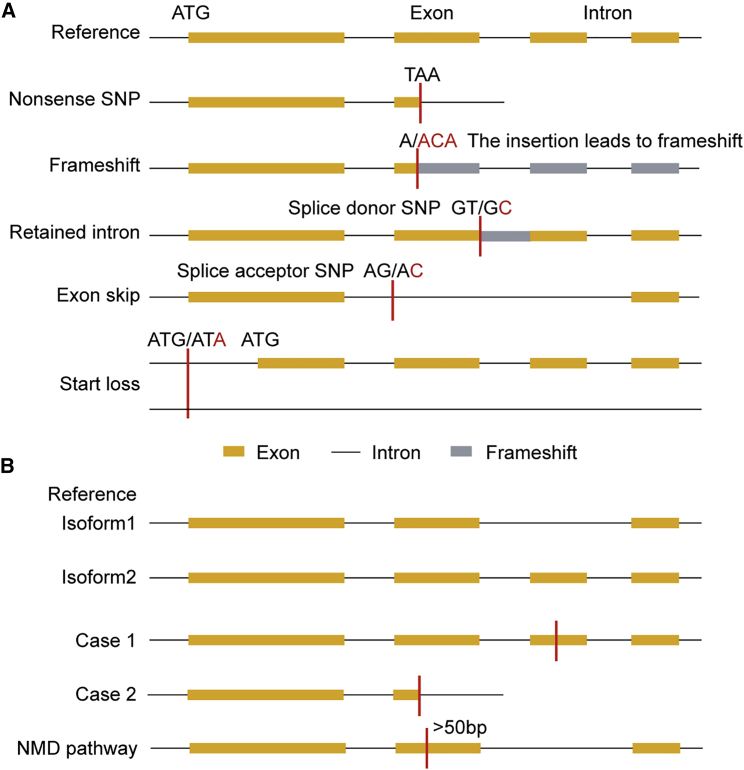
There are four major genetic variations that can lead to LoF mutations (Figure 1A). First, a nonsense SNP may lead to a premature stop codon, producing a truncated protein sequence. For example, in Arabidopsis, FRIGIDA (FRI) alleles with premature stop codons explain a large fraction of flowering-time variation (Le Corre et al., 2002; Lempe et al., 2005; Shindo et al., 2005; Werner et al., 2005; Atwell et al., 2010). Second, a SNP that occurs at a canonical splice site may affect splicing. Specifically, a SNP that occurs in a splice donor site causes intron retention in the mRNA, whereas a SNP that occurs in a splice acceptor site removes an exon from the original mRNA. Splice site variations may eventually lead to frameshifts or premature stop codons (Balasubramanian et al., 2011). In the Arabidopsis relative Capsella rubella, splice site mutations in the FLOWERING LOCUS C gene cause a premature stop codon that promotes flowering (Guo et al., 2012). Third, insertion or deletion variants with non-integral multiples of 3 located in the gene coding region can lead to frameshifts by disrupting the full-length transcript (MacArthur et al., 2012; Lim et al., 2013). Fourth, the loss of an initiation codon can lead to LoF mutations. The loss of transcription start codon (ATG) variations prevents gene transcription if there is no alternative start codon near the mutation. For example, in humans, the loss of the start codon in the FRMD7 gene leads to idiopathic infantile nystagmus disease (Choi et al., 2015).
Figure 1.
Evolutionary Events Lead to LoF Mutations.
(A) Different kinds of LoF mutations.
(B) LoF mutations affect different isoforms and the mechanism of the NMD pathway. The LoF mutation only affects a part of the gene transcript in case 1 and affects all transcripts in case 2. Orange rectangles indicate exons, gray rectangles indicate frameshift sequences of exons or retained intron sequences, black lines between them indicate introns, and red lines indicate LoF mutations caused by loss of a start codon or gain of a premature stop codon.
Given that most genes in a genome can be transcribed into multiple transcripts, only mutations that affect all transcripts are regarded as LoF mutations in many studies (Figure 1B) (Balasubramanian et al., 2011; MacArthur et al., 2012; Kaiser et al., 2015).
LoF Mutations Are Abundant in Diverse Species
With the rapid development of sequencing technologies, LoF mutations have been studied in diverse species at the population level. Almost every genome contains many LoF mutations in either heterozygous or homozygous states. Most LoF mutations in a genome tend to be present at low allele frequencies and have a heterozygous status (Lek et al., 2016; Minikel et al., 2020). In selfing or consanguineous outcrossing species, there is a higher ratio of homozygous LoF mutations (Saleheen et al., 2017; Minikel et al., 2020). In humans, an analysis of 7597 genomes identified 17 764 stop-gain variants and 13 915 frameshift variants within 11 369 protein-coding genes (Rausell et al., 2014). Based on 1432 whole exome sequences from five isolated European populations, 173 homozygous LoF mutations were identified within 167 genes (Kaiser et al., 2015). Analysis of whole genomes sequencing from 2636 Icelanders and chip-genotype data from 101 584 additional individuals identified 6795 autosomal LoF variants within 4924 genes (Sulem et al., 2015). Our recent study of 1071 A. thaliana genomes from worldwide accessions revealed 60 819 LoF variants within 12 907 genes (Xu et al., 2019).
The evolutionary pattern of LoF variants in natural populations can be influenced by various factors. Redundant genes within larger gene families are more likely to gain LoF mutations due to the presence of paralogs that buffer their functional effects (MacArthur et al., 2012; Xu et al., 2019). Furthermore, our recent study revealed that the level of nucleotide diversity and the density of transposable elements are positively correlated with the presence of LoF mutations (Xu et al., 2019). Given that these three factors are roughly associated with sequence evolution rate, rapid sequence evolution may be positively correlated with the frequency of LoF mutations (Xu et al., 2019).
Genes with LoF mutations may be regulated at the mRNA or protein level. If the LoF truncation is located further than 50 base pairs upstream of the last exon, the truncated transcript may be eliminated by the nonsense-mediated decay (NMD) pathway (Figure 1B) (Balasubramanian et al., 2011). In humans, based on 1151 individual genomes from 56 worldwide populations, 55% of genes are predicted to undergo transcript degradation through the NMD pathway (Yngvadottir et al., 2009). Nevertheless, only a small fraction of transcripts predicted to be regulated by the NMD pathway are eventually degraded in this manner. For example, MacArthur et al. (2012) found that transcription is decreased in only 25% of the genes predicted to be regulated by the NMD pathway. By contrast, at the protein level, a truncated protein may be degraded due to an unstable protein-folding state (Williams et al., 2003).
Functional Effects of LoF Mutations
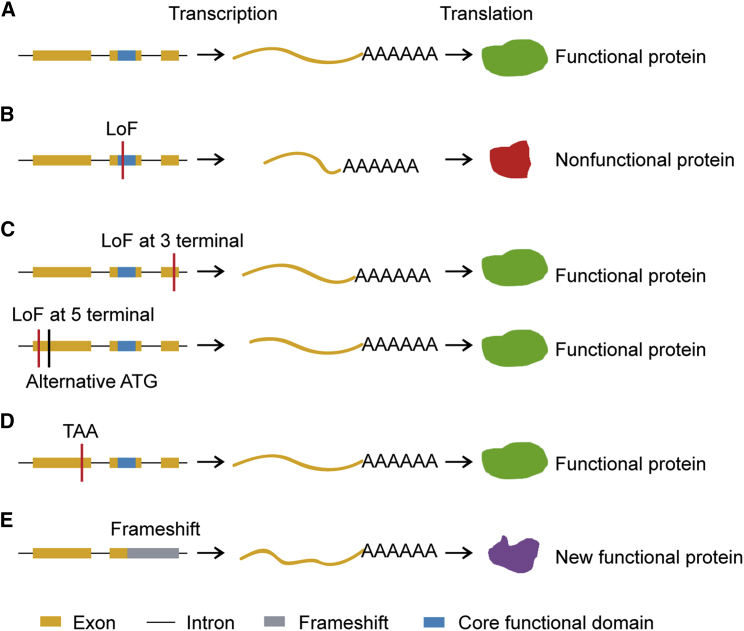
Compared with functional protein-coding genes (Figure 2A), LoF mutations are natural gene knockouts that can provide insight into gene function in diverse organisms (Cheetham et al., 2020). For example, natural variants at the FRI locus from more than 1000 Arabidopsis genomes have helped to reveal the function of the FRI central domain (Zhang and Jimenez-Gomez, 2020). In A. thaliana, natural brx LoF alleles confer root adaptation to acidic soil, and a natural knockout allele of ARMADILLO REPEAT-CONTAINING KINESIN1 causes root hair branching (Gujas et al., 2012; Rishmawi et al., 2014). In addition, different LoF mutations of duplicated genes in diverse A. thaliana populations can lead to hybrid incompatibility (Blevins et al., 2017). In rice, a natural LoF mutation in GSE5, which encodes a plasma membrane-associated protein, contributes to grain size diversity (Duan et al., 2017).
Figure 2.
Functional Effects of LoF Mutations.
(A) Functional genes without LoF mutation.
(B) Gene knockout with LoF mutations.
(C) Gene translated into functional protein if LoF mutations are located at 3′ or 5′ termini.
(D) Transcriptional readthrough of the premature stop codon.
(E) Gene translated to a new functional protein with the LoF mutation. Orange rectangles indicate exons, black lines between them indicate introns, red lines indicate LoF mutations, blue rectangles indicate the core functional domain of the gene, and gray rectangle indicates frameshift. Orange lines with polyadenylic acid indicate mRNA.
In the human fungal pathogen Cryptococcus deuterogattii, an LoF mutation in the mismatch repair component MSH2 can affect phenotypic diversity and drug resistance (Billmyre et al., 2017). In budding yeast, loss-of-start and nonsense mutations in AMN1 (antagonist of mitotic exit network) cause a transition from multicellularity to unicellularity (Kuzdzal-Fick et al., 2019). In humans, much more attention has been paid to the effects of LoF mutations that are correlated with Mendelian diseases. The precise annotation of LoF mutations has played an important role in the diagnosis of rare diseases (Cummings et al., 2020). LoF mutations have been found in defensive genes of many individuals and may lead to diseases (Saleheen et al., 2017). For example, in 9% of the European population, two LoF variants occurred independently in the filaggrin (FLG) gene and were associated with an increased risk of atopic dermatitis (Palmer et al., 2006). Consanguineous unions can confer more homozygous LoF variants, making them good systems for complex disease study (Saleheen et al., 2017). Based on a population with a high rate of consanguinity, association studies between 49 138 LoF mutations and more than 200 biochemical and disease traits revealed many LoF mutations in diverse disease genes (Saleheen et al., 2017).
However, not all LoF mutations lead to complete functional knockouts (Figure 2B). First, the effect of an LoF mutation on a gene depends on its location. Many LoF mutations at the 5′ or 3′ ends of affected genes do not completely destroy their functions (Figure 2C) (de Valles-Ibanez et al., 2016; MacArthur et al., 2012), given that an alternative start codon at the 5′ end may rescue the transcript, and an LoF mutation at the 3′ end will probably not remove the core functional domain. Second, the truncated proteins may act as dominant-negative mutations. For example, the nonsense allele of sex-determining region Y-box transcription factor 9 (MiniSOX9) acts in a dominant-negative manner to buffer the effect of the wild-type allele (Abdel-Samad et al., 2011). Third, a functional pseudo-pseudogene may exist. In Drosophila sechellia, the transcriptional readthrough of the premature stop codon in the chemosensory variant ionotropic glutamate receptor repertoire (Ir75a) can encode a functional receptor (Figure 2D) (Prieto-Godino et al., 2016). Fourth, LoF can be an effective evolutionary mechanism for generating new functional genes with physicochemical properties similar to those of the genes from which they originated, as recently demonstrated (Figure 2E) (Bartonek et al., 2020).
Essential Genes in Natural Populations
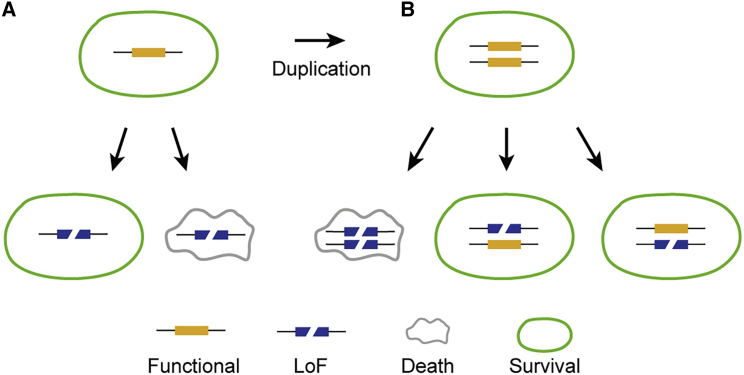
Essential genes are those genes that are necessary for survival (Meinke et al., 2008). An individual cannot survive if LoF mutations have occurred in essential genes; otherwise, these genes would be considered non-essential (Figure 3A). Essential genes in one species may be functionally redundant in another related species if they have undergone duplication (Figure 3B). The number of essential genes, namely the minimal gene set required for survival, remains a fundamental biological question (Koonin, 2003; Meinke et al., 2008). Beyond its theoretical importance, the study of essential genes in diverse organisms is crucial for synthetic biology, which requires essential gene lists to synthesize new cell lines or new organisms. Despite knowing the number of essential gene families in a lineage such as the green plants, which are estimated to contain approximately 2745–2928 core gene families (Van Bel et al., 2012; Guo, 2013), estimating the number of genes that are essential for an organism is more challenging (Meinke, 2020).
Figure 3.
The Identification of Essential Genes through LoF Mutations.
(A) The identification of single-copy essential genes. The individuals could not survive when LoF mutations occurred in essential genes.
(B) The identification of multiple copies of essential genes. When duplication events occurred in essential genes, either one of the copies would be redundant. The individual could survive if LoF mutations occurred in only one of the copies.
In the laboratory, the essential genes of an organism are usually assessed based on the fitness of knockout mutants of each gene in the genome. Prokaryotic organisms are frequently used to validate essential genes, largely because of their small gene number and ease of culture and phenotyping. Here, we have summarized essential gene studies across diverse species (Table 1). Based on mutant studies, there are 4.7%–38% essential genes across different human pathogens (Table 1) (Ji et al., 2001; Akerley et al., 2002; Kobayashi et al., 2003; Roemer et al., 2003; Poulsen et al., 2019). In the yeast Saccharomyces cerevisiae, 1105 (18.7%) of 5916 studied genes were found to be essential for growth on rich glucose medium (Giaever et al., 2002). In E. coli, 303 (7.1%) of 4288 studied genes were essential (Baba et al., 2006). In mice, 38 (42%) of 90 studied genes were shown to be essential for mouse viability, and essential genes were more likely to be part of a protein complex (White et al., 2013). Based on genetic studies of A. thaliana mutants, 358 genes (1.3% of all genes in the genome) were demonstrated to be essential (Meinke et al., 2008).
Table 1.
Essential Gene Numbers in Diverse Species.
| Species | Method | Gene number (percentage) | Reference |
|---|---|---|---|
| Bacillus subtilis | Mutants | 192 (4.7%a) | Kobayashi et al. (2003) |
| Candida albicans | Mutants | 567 (9%a) | Roemer et al. (2003) |
| Haemophilus influenza | Mutants | 259 (38%a) | Akerley et al. (2002) |
| Pseudomonas aeruginosa | Mutants | 321 (6.6%a) | Poulsen et al. (2019) |
| Staphylococcus aureus | Antisense RNA | 150 (5.6%a) | Ji et al. (2001) |
| Escherichia coli | Mutants | 303 (7.1%b) | Baba et al. (2006) |
| Saccharomyces cerevisiae | Mutants | 1105 (18.7%b) | Giaever et al. (2002) |
| Arabidopsis thaliana | Mutants | 358 (1.3%a) | Meinke et al. (2008) |
| Arabidopsis thaliana | Machine learning prediction | 2675 (9.8%a) | Lloyd et al. (2015) |
| Arabidopsis thaliana | LoF at population level | 9249 (34%a) | Xu et al. (2019) |
| Mice | Mutants | 38 (42%b) | White et al. (2013) |
| Human | LoF at population level | 194 (3%b) | Narasimhan et al. (2016) |
| Human | LoF at population level | 3230 (17.7%a) | Lek et al. (2016) |
Indicates the percentage of all genes in the genome.
Indicates the percentage of studied genes.
LoF mutations in the genomes of natural populations provide direct evidence to delimit essential genes (Albalat and Canestro, 2016). Specifically, genes without any LoF mutations in natural populations must be crucial for survival and can be regarded as essential gene candidates. Recent LoF studies in humans and A. thaliana have indicated that essential genes (lethal genes) are depleted in genes with LoF mutations (Xu et al., 2019; Karczewski et al., 2020). In an LoF mutation study of 3222 closely related adults of British Pakistani heritage, there were 194 recessive lethal equivalent LoF mutations in the heterozygous state (3% of studied genes) (Narasimhan et al., 2016). In an exome sequencing study of 60 706 humans, 3230 genes were LoF intolerant (17.7% of all genes in the genome), and LoF in these genes may cause survival or reproductive disadvantage (Lek et al., 2016). In A. thaliana, our recent study revealed that at least 34% of protein-coding genes do not have any LoF variants based on 1071 genomes from worldwide accessions. These genes are probably essential for the survival of A. thaliana in its natural environment (Xu et al., 2019). In contrast to the 358 genes identified as essential in a genetic study of mutants (Meinke et al., 2008) or the 2675 essential genes predicted by machine-learning methods (Lloyd et al., 2015), our essential gene number is much larger but understandable. Because growth conditions in laboratories are much better than in natural habitats, plants probably tolerate more LoF mutations when grown in laboratories. Therefore, the definition of essential genes is context dependent, reflecting the niche that the organisms inhabit.
LoF Mutations Are Crucial for Adaptation and Diversification
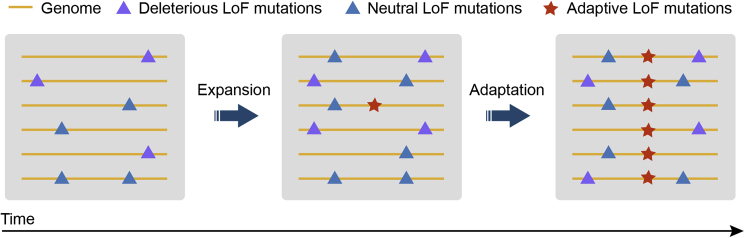
LoF mutations in the genome may be neutral, deleterious, or advantageous. Neutral or less-deleterious LoF mutations can be tolerated and may even accumulate during range expansion (Figure 4). However, compared with SNPs at the whole-genome level, LoF mutations are biased toward low allele frequencies, indicating that they are mostly deleterious and under purifying selection (MacArthur et al., 2012; The 1000 Genomes Project Consortium, 2010; Xu et al., 2019).
Figure 4.
Adaptive Evolution of LoF Mutations.
LoF mutations can be neutral, deleterious, or advantageous. Deleterious LoF mutations are usually present at low allele frequencies in natural populations due to purifying selection. Neutral or beneficial LoF mutations can be tolerated or may accumulate during range expansion, and beneficial LoF mutations may be fixed by positive selection.
Nevertheless, the “less is more” hypothesis proposes that gene loss may be beneficial to organisms (Olson, 1999). Adaptive LoF mutations have been observed frequently in bacteria and yeast. An analysis of bacterial fitness in more than 100 different conditions revealed that LoF mutations can provide fitness benefits (Hottes et al., 2013). In the yeast S. cerevisiae, aquaporin genes are critical for freeze-thaw tolerance (Tanghe et al., 2004). However, sensitive strains have lost the function of aquaporin genes independently at least six times to adapt to high-sugar substrates (Will et al., 2010). Adaptive LoF mutations have also been found in plants. In A. thaliana, about 1% of LoF mutations are under positive selection, and the LoF allele of the KUK gene is correlated with longer roots (Xu et al., 2019). Overall, gene loss may be an adaptive process; adaptive LoF mutations may be quickly fixed in specific scenarios such as range expansion to new niches (Figure 4).
LoF mutations play important roles in the evolution and diversification of diverse organisms. In plants, a premature stop codon in GL4 caused smaller grain size and loss of seed shattering during African rice domestication (Wu et al., 2017). Furthermore, LoF mutations have been found to act as evolutionary hotspots for changing plant-pollinator communication and speciation. The floral scent is an important chemical signal between plants and pollinators (Klahre et al., 2011). In the genus Petunia (Solanaceae), a premature stop codon in the gene encoding cinnamate-CoA ligase 1 (CNL1), which produces phenylalanine-derived volatiles, eliminates the scent of Petunia exserta. As a result, P. exserta cannot attract hawkmoths and shifts its pollinator from hawkmoths to hummingbirds (Segatto et al., 2014; Amrad et al., 2016). A similar phenomenon was observed in Capsella (Brassicaceae), which independently lost CNL1 and scent twice, contributing to the transition from the outcrossing species Capsella grandiflora to the selfing species Capsella rubella (Sas et al., 2016). Similarly, flower color plays an important role in pollinator attraction (Bradshaw and Schemske, 2003). In Petunia axillaris, an LoF mutation in ANTHOCYANIN2 (AN2) occurred independently at least five times, changing the flower color from violet-red to white compared with Petunia integrifolia and influencing the shift in pollinator attraction from bees to hawkmoths (Hoballah et al., 2007).
In the Drosophila relative Scaptomyza flava, LoF mutations in odorant receptor genes resulted in a transition to herbivory from its yeast-feeding relatives (Goldman-Huertas et al., 2015). In mammals, LoF mutations in proto-Xist and its four flanking protein genes are associated with the emergence of X-chromosome inactivation and played an important role in the divergence of eutherians and marsupials (Duret et al., 2006). LoF mutations have also been found to be beneficial in human evolution. A CASP12 LoF allele is known to promote resistance to severe sepsis (Saleh et al., 2004), and rare LoF mutations in SLC30A8 can protect against type 2 diabetes (Flannick et al., 2014).
Future Directions
LoF mutations are prevalent in natural populations of diverse species. Species-wide studies could be performed to understand the genome-wide distribution patterns, functional effects, and evolutionary importance of LoF mutations. Several studies of LoF mutations at the genome level have been performed in natural populations (MacArthur et al., 2012; Xu et al., 2019). In particular, our recent study revealed that the level of nucleotide diversity, the density of transposable elements, and gene family size are positively correlated with the presence of LoF mutations (Xu et al., 2019). However, LoF studies are mostly limited to humans, chimpanzees, and Arabidopsis. More species should be studied to understand the evolutionary patterns and importance of LoF mutations. Many crucial questions about LoF mutations need to be addressed in the future.
First, the identification of LoF mutations is usually reference biased, based on comparison with one reference genome. For example, compared with the LoF mutations caused by stop-gain mutations, stop-loss mutations identified in other accessions may be premature stop codons in the reference genome instead. Pan-genome studies in diverse species would compensate for this reference bias (Zhao et al., 2018; Golicz et al., 2020; Liu et al., 2020).
Second, the evolutionary effects of natural LoF mutations on other genes in the same pathway may be complicated. When genes become non-functional, other genes in the same pathway may accumulate LoF mutations as well (Zufall and Rausher, 2004). Such an evolutionary tendency could explain Dollo's law, which posits the irreversibility of character elimination (Gould, 1970). More cases in diverse species are needed to understand the evolutionary consequences of LoF mutations.
Third, the functional effects of LoF mutations are interesting to study. Genome-wide study of LoF variants can provide information on gene lethality based on the frequency of LoF alleles for a given gene in natural environments (population gene lethality) (Albalat and Canestro, 2016). Our recent study revealed that 34% of the genes in A. thaliana are probably essential in the natural environment (Xu et al., 2019). However, essential genes validated by functional analysis may have LoF mutations as well. In our recent study of A. thaliana, 11.7% of the essential genes validated by functional analysis had LoF mutations (Xu et al., 2019). In yeast S. cerevisiae, a study of 1106 essential gene knockouts found that 88 (9%) of them could survive through adaptive evolution, and these were defined as evolvable essential genes (Liu et al., 2015). In addition, an essential gene mutation study of E. coli and S. cerevisiae revealed that revertant mutants could occur in essential gene mutations when cells were grown under stress conditions, causing the mutant to regain its gene function to some extent (Kheir Gouda et al., 2019; Rodrigues and Shakhnovich, 2019). The study of revertant LoF mutations provides insight into the drug resistance of microorganisms, which is useful in synthetic biology in medicine. How individuals with natural mutations in essential genes can survive in natural populations is an intriguing question for in-depth study.
Fourth, most studies of LoF mutations have focused on variations in coding regions; however, a mutation in the promoter or UTR could also affect gene expression (Yang et al., 2018; Niu et al., 2019; Xu et al., 2020). Synthetic biology can offer interesting insights into the beneficial effects of regulator loss. For example, in Chinese hamster ovary cells, repressor loss in the promoter region of PuroR leads to high gene expression and drug resistance (Farquhar et al., 2019).
Finally, LoF mutations that are abundant in natural populations can provide valuable genetic resources for the functional study of genes and are particularly useful for crop breeding. New genes (gene duplication and de novo genes) or new functional alleles can make fruitful contributions to crop breeding. For instance, the COLD1 allele from the wild rice Oryza rufipogon confers chilling tolerance (Ma et al., 2015), and the 27-kDa γ-zein gene duplication contributes to maize protein quality through endosperm modification (Liu et al., 2016). However, the agricultural importance of LoF mutations is largely unknown. In plants, several studies have reported that LoF mutations can also increase crop yield. For example, LoF of GW2, a gene that encodes a RING-type E3 ubiquitin ligase, can increase grain width and weight (Song et al., 2007). Similarly, an LoF mutation of MEI2-LIKE PROTEIN4 (OML4) leads to large and heavy grains in rice (Oryza sativa) (Lyu et al., 2020). Nevertheless, there have been no studies of LoF mutations at the genome level in crops to date.
Overall, there are abundant natural LoF mutations in the genomes of diverse organisms, and this important genetic variation is correlated with biodiversity and adaptation. In particular, LoF mutations in natural populations provide an invaluable resource and a robust framework for gaining theoretical biological insight while simultaneously improving crop breeding in the context of climate change.
Funding
This work was supported by the National Natural Science Foundation of China (31925004), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27010305), and the Innovative Academy of Seed Design, Chinese Academy of Sciences.
Authors Contributions
Y.L.G. conceived the study. Y.C.X. and Y.L.G. wrote the manuscript.
Acknowledgments
We thank members of the Guo lab for suggestions and comments about this work, and especially the anonymous reviewers for their help improving the manuscript. We apologize for our inability to cite many relevant publications due to space limitations. No conflict of interest declared.
Published: August 13, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
References
- Abdel-Samad R., Zalzali H., Rammah C., Giraud J., Naudin C., Dupasquier S., Poulat F., Boizet-Bonhoure B., Lumbroso S., Mouzat K. MiniSOX9, a dominant-negative variant in colon cancer cells. Oncogene. 2011;30:2493–2503. doi: 10.1038/onc.2010.621. [DOI] [PubMed] [Google Scholar]
- Akerley B.J., Rubin E.J., Novick V.L., Amaya K., Judson N., Mekalanos J.J. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U S A. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R., Canestro C. Evolution by gene loss. Nat. Rev. Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- Amrad A., Moser M., Mandel T., de Vries M., Schuurink R.C., Freitas L., Kuhlemeier C. Gain and loss of floral scent production through changes in structural genes during pollinator-mediated speciation. Curr. Biol. 2016;26:3303–3312. doi: 10.1016/j.cub.2016.10.023. [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y.S., Vilhjalmsson B.J., Willems G., Horton M., Li Y., Meng D., Platt A., Tarone A.M., Hu T.T. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Habegger L., Frankish A., MacArthur D.G., Harte R., Tyler-Smith C., Harrow J., Gerstein M. Gene inactivation and its implications for annotation in the era of personal genomics. Genes Dev. 2011;25:1–10. doi: 10.1101/gad.1968411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartonek L., Braun D., Zagrovic B. Frameshifting preserves key physicochemical properties of proteins. Proc. Natl. Acad. Sci. U S A. 2020;117:5907–5912. doi: 10.1073/pnas.1911203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billmyre R.B., Clancey S.A., Heitman J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. eLife. 2017;6:e28802. doi: 10.7554/eLife.28802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T., Wang J., Pflieger D., Pontvianne F., Pikaard C.S. Hybrid incompatibility caused by an epiallele. Proc. Natl. Acad. Sci. U S A. 2017;114:3702–3707. doi: 10.1073/pnas.1700368114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H.D., Schemske D.W. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- Cheetham S.W., Faulkner G.J., Dinger M.E. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 2020;21:191–201. doi: 10.1038/s41576-019-0196-1. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang Y.E., Long M. New genes in Drosophila quickly become essential. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.H., Shin J.H., Seo J.H., Jung J.H., Choi K.D. A start codon mutation of the FRMD7 gene in two Korean families with idiopathic infantile nystagmus. Sci. Rep. 2015;5:13003. doi: 10.1038/srep13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings B.B., Karczewski K.J., Kosmicki J.A., Seaby E.G., Watts N.A., Singer-Berk M., Mudge J.M., Karjalainen J., Satterstrom F.K., O'Donnell-Luria A.H. Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. doi: 10.1038/s41586-020-2329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Valles-Ibanez G., Hernandez-Rodriguez J., Prado-Martinez J., Luisi P., Marques-Bonet T., Casals F. Genetic load of loss-of-function polymorphic variants in great apes. Genome Biol. Evol. 2016;8:871–877. doi: 10.1093/gbe/evw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P., Xu J., Zeng D., Zhang B., Geng M., Zhang G., Huang K., Huang L., Xu R., Ge S. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant. 2017;10:685–694. doi: 10.1016/j.molp.2017.03.009. [DOI] [PubMed] [Google Scholar]
- Duret L., Chureau C., Samain S., Weissenbach J., Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Farquhar K.S., Charlebois D.A., Szenk M., Cohen J., Nevozhay D., Balazsi G. Role of network-mediated stochasticity in mammalian drug resistance. Nat. Commun. 2019;10:2766. doi: 10.1038/s41467-019-10330-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G.R. Pseudogenes in yeast. Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B., Grarup N., Burtt N.P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Goldman-Huertas B., Mitchell R.F., Lapoint R.T., Faucher C.P., Hildebrand J.G., Whiteman N.K. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc. Natl. Acad. Sci. U S A. 2015;112:3026–3031. doi: 10.1073/pnas.1424656112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz A.A., Bayer P.E., Bhalla P.L., Batley J., Edwards D. Pangenomics comes of age: from bacteria to plant and animal applications. Trends Genet. 2020;36:132–145. doi: 10.1016/j.tig.2019.11.006. [DOI] [PubMed] [Google Scholar]
- Gould S.J. Dollo on Dollo's law: irreversibility and the status of evolutionary laws. J. Hist. Biol. 1970;3:189–212. doi: 10.1007/BF00137351. [DOI] [PubMed] [Google Scholar]
- Gujas B., Alonso-Blanco C., Hardtke C.S. Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr. Biol. 2012;22:1962–1968. doi: 10.1016/j.cub.2012.08.026. [DOI] [PubMed] [Google Scholar]
- Guo Y.L. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 2013;73:941–951. doi: 10.1111/tpj.12089. [DOI] [PubMed] [Google Scholar]
- Guo Y.L., Todesco M., Hagmann J., Das S., Weigel D. Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics. 2012;192:729–739. doi: 10.1534/genetics.112.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P.M., Milburn D., Zhang Z., Bertone P., Gerstein M. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res. 2003;31:1033–1037. doi: 10.1093/nar/gkg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah M.E., Gubitz T., Stuurman J., Broger L., Barone M., Mandel T., Dell'Olivo A., Arnold M., Kuhlemeier C. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell. 2007;19:779–790. doi: 10.1105/tpc.106.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes A.K., Freddolino P.L., Khare A., Donnell Z.N., Liu J.C., Tavazoie S. Bacterial adaptation through loss of function. PLoS Genet. 2013;9:e1003617. doi: 10.1371/journal.pgen.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi H., Kodaira M., Yamamoto K., Ozeki H. Variability of 3'-terminal region of transfer rna2gln gene in E. coli - unequal crossover with presumptive pseudogenes. Jpn. J. Genet. 1979;54:437. [Google Scholar]
- Jacq C., Miller J.R., Brownlee G.G. A pseudogene in 5S DNA of Xenopus laevis. Cell. 1977;12:109–120. doi: 10.1016/0092-8674(77)90189-1. [DOI] [PubMed] [Google Scholar]
- Ji Y.D., Zhang B., Van Horn S.F., Warren P., Woodnutt G., Burnham M.K.R., Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- Kaiser V.B., Svinti V., Prendergast J.G., Chau Y.Y., Campbell A., Patarcic I., Barroso I., Joshi P.K., Hastie N.D., Miljkovic A. Homozygous loss-of-function variants in European cosmopolitan and isolate populations. Hum. Mol. Genet. 2015;24:5464–5474. doi: 10.1093/hmg/ddv272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheir Gouda M., Manhart M., Balazsi G. Evolutionary regain of lost gene circuit function. Proc. Natl. Acad. Sci. U S A. 2019;116:25162–25171. doi: 10.1073/pnas.1912257116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Gurba A., Hermann K., Saxenhofer M., Bossolini E., Guerin P.M., Kuhlemeier C. Pollinator choice in Petunia depends on two major genetic Loci for floral scent production. Curr. Biol. 2011;21:730–739. doi: 10.1016/j.cub.2011.03.059. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Ehrlich S.D., Albertini A., Amati G., Andersen K.K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- Kuzdzal-Fick J.J., Chen L., Balazsi G. Disadvantages and benefits of evolved unicellularity versus multicellularity in budding yeast. Ecol. Evol. 2019;9:8509–8523. doi: 10.1002/ece3.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre V., Roux F., Reboud X. DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol. Biol. Evol. 2002;19:1261–1271. doi: 10.1093/oxfordjournals.molbev.a004187. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Lewis S.A., Wilde C.D., Cowan N.J. Evolutionary history of a multigene family—an expressed human beta-tubulin gene and 3 processed pseudogenes. Cell. 1983;33:477–487. doi: 10.1016/0092-8674(83)90429-4. [DOI] [PubMed] [Google Scholar]
- Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1:109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Gojobori T., Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Li Z.W., Chen X., Wu Q., Hagmann J., Han T.S., Zou Y.P., Ge S., Guo Y.L. On the origin of de novo genes in Arabidopsis thaliana populations. Genome Biol. Evol. 2016;8:2190–2202. doi: 10.1093/gbe/evw164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.T., Raychaudhuri S., Sanders S.J., Stevens C., Sabo A., MacArthur D.G., Neale B.M., Kirby A., Ruderfer D.M., Fromer M. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yong M.Y., Yurieva M., Srinivasan K.G., Liu J., Lim J.S., Poidinger M., Wright G.D., Zolezzi F., Choi H. Gene essentiality is a quantitative property linked to cellular evolvability. Cell. 2015;163:1388–1399. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- Liu H., Shi J., Sun C., Gong H., Fan X., Qiu F., Huang X., Feng Q., Zheng X., Yuan N. Gene duplication confers enhanced expression of 27-kDa gamma-zein for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. U S A. 2016;113:4964–4969. doi: 10.1073/pnas.1601352113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Du H., Li P., Shen Y., Peng H., Liu S., Zhou G.A., Zhang H., Liu Z., Shi M. Pan-genome of wild and cultivated soybeans. Cell. 2020 doi: 10.1016/j.cell.2020.1005.1023. [DOI] [PubMed] [Google Scholar]
- Lloyd J.P., Seddon A.E., Moghe G.D., Simenc M.C., Shiu S.-H. Characteristics of plant essential genes allow for within- and between-species prediction of lethal mutant phenotypes. Plant Cell. 2015;27:2133–2147. doi: 10.1105/tpc.15.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J., Wang D., Duan P., Liu Y., Huang K., Zeng D., Zhang L., Dong G., Li Y., Xu R. Control of grain size and weight by the GSK2-LARGE1/OML4 pathway in rice. Plant Cell. 2020;32:1905–1918. doi: 10.1105/tpc.19.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Dai X., Xu Y., Luo W., Zheng X., Zeng D., Pan Y., Lin X., Liu H., Zhang D. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D., Muralla R., Sweeney C., Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 2008;13:483–491. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Meinke D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2020;226:306–325. doi: 10.1111/nph.16071. [DOI] [PubMed] [Google Scholar]
- Minikel E.V., Karczewski K.J., Martin H.C., Cummings B.B., Whiffin N., Rhodes D., Alfoldi J., Trembath R.C., van Heel D.A., Daly M.J. Evaluating drug targets through human loss-of-function genetic variation. Nature. 2020;581:459–464. doi: 10.1038/s41586-020-2267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V.M., Hunt K.A., Mason D., Baker C.L., Karczewski K.J., Barnes M.R., Barnett A.H., Bates C., Bellary S., Bockett N.A. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X.M., Xu Y.C., Li Z.W., Bian Y.T., Hou X.H., Chen J.F., Zou Y.P., Jiang J., Wu Q., Ge S. Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl. Acad. Sci. U S A. 2019;116:6908–6913. doi: 10.1073/pnas.1811498116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Allen & Unwin; Springer-Verlag; London, New York: 1970. Evolution by Gene Duplication. [Google Scholar]
- Olson M.V. When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.N., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P., Goudie D.R., Sandilands A., Campbell L.E., Smith F.J. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- Palmieri N., Kosiol C., Schlötterer C. The life cycle of Drosophila orphan genes. eLife. 2014;3:e01311. doi: 10.7554/eLife.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen B.E., Yang R., Clatworthy A.E., White T., Osmulski S.J., Li L., Penaranda C., Lander E.S., Shoresh N., Hung D.T. Defining the core essential genome of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 2019;116:10072–10080. doi: 10.1073/pnas.1900570116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Godino L.L., Rytz R., Bargeton B., Abuin L., Arguello J.R., Peraro M.D., Benton R. Olfactory receptor pseudo-pseudogenes. Nature. 2016;539:93–97. doi: 10.1038/nature19824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausell A., Mohammadi P., McLaren P.J., Bartha I., Xenarios I., Fellay J., Telenti A. Analysis of stop-gain and frameshift variants in human innate immunity genes. PLoS Comput. Biol. 2014;10:e1003757. doi: 10.1371/journal.pcbi.1003757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishmawi L., Sun H., Schneeberger K., Hulskamp M., Schrader A. Rapid identification of a natural knockout allele of ARMADILLO REPEAT-CONTAINING KINESIN1 that causes root hair branching by mapping-by-sequencing. Plant Physiol. 2014;166:1280–1287. doi: 10.1104/pp.114.244046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J.V., Shakhnovich E.I. Adaptation to mutational inactivation of an essential gene converges to an accessible suboptimal fitness peak. eLife. 2019;8:e50509. doi: 10.7554/eLife.50509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T., Jiang B., Davison J., Ketela T., Veillette K., Breton A., Tandia F., Linteau A., Sillaots S., Marta C. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003;50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- Saleh M., Vaillancourt J.P., Graham R.K., Huyck M., Srinivasula S.M., Alnemri E.S., Steinberg M.H., Nolan V., Baldwin C.T., Hotchkiss R.S. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- Saleheen D., Natarajan P., Armean I.M., Zhao W., Rasheed A., Khetarpal S.A., Won H.H., Karczewski K.J., O'Donnell-Luria A.H., Samocha K.E. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas C., Muller F., Kappel C., Kent T.V., Wright S.I., Hilker M., Lenhard M. Repeated inactivation of the first committed enzyme underlies the loss of benzaldehyde emission after the selfing transition in Capsella. Curr. Biol. 2016;26:3313–3319. doi: 10.1016/j.cub.2016.10.026. [DOI] [PubMed] [Google Scholar]
- Segatto A.L., Caze A.L., Turchetto C., Klahre U., Kuhlemeier C., Bonatto S.L., Freitas L.B. Nuclear and plastid markers reveal the persistence of genetic identity: a new perspective on the evolutionary history of Petunia exserta. Mol. Phylogenet. Evol. 2014;70:504–512. doi: 10.1016/j.ympev.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Shindo C., Aranzana M.J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005;138:1163–1173. doi: 10.1104/pp.105.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Sulem P., Helgason H., Oddson A., Stefansson H., Gudjonsson S.A., Zink F., Hjartarson E., Sigurdsson G.T., Jonasdottir A., Jonasdottir A. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- Tanghe A., Van Dijck P., Colavizza D., Thevelein J.M. Aquaporin-mediated improvement of freeze tolerance of Saccharomyces cerevisiae is restricted to rapid freezing conditions. Appl. Environ. Microb. 2004;70:3377–3382. doi: 10.1128/AEM.70.6.3377-3382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents D., Suyama M., Zdobnov E., Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158:590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin E.F. Processed pseudogenes—characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- Werner J.D., Borevitz J.O., Uhlenhaut N.H., Ecker J.R., Chory J., Weigel D. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics. 2005;170:1197–1207. doi: 10.1534/genetics.104.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.K., Gerdin A.K., Karp N.A., Ryder E., Buljan M., Bussell J.N., Salisbury J., Clare S., Ingham N.J., Podrini C. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will J.L., Kim H.S., Clarke J., Painter J.C., Fay J.C., Gasch A.P. Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 2010;6:e1000893. doi: 10.1371/journal.pgen.1000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S., Chasman D.I., Hau D.D., Hui B., Lau A.Y., Glover J.N. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J. Biol. Chem. 2003;278:53007–53016. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- Wu W., Liu X., Wang M., Meyer R.S., Luo X., Ndjiondjop M.N., Tan L., Zhang J., Wu J., Cai H. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants. 2017;3:17064. doi: 10.1038/nplants.2017.64. [DOI] [PubMed] [Google Scholar]
- Xie J.B., Li Y., Liu X.M., Zhao Y.Y., Li B.L., Ingvarsson P.K., Zhang D.Q. Evolutionary origins of pseudogenes and their association with regulatory sequences in plants. Plant Cell. 2019;31:563–578. doi: 10.1105/tpc.18.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Gokcumen O., Khurana E. Loss-of-function tolerance of enhancers in the human genome. PLoS Genet. 2020;16:e1008663. doi: 10.1371/journal.pgen.1008663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.C., Niu X.M., Li X.X., He W., Chen J.F., Zou Y.P., Wu Q., Zhang Y.E., Busch W., Guo Y.L. Adaptation and phenotypic diversification in Arabidopsis through loss-of-function mutations in protein-coding genes. Plant Cell. 2019;31:1012–1025. doi: 10.1105/tpc.18.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Takuno S., Waters E.R., Gaut B.S. Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol. Biol. Evol. 2011;28:1193–1203. doi: 10.1093/molbev/msq298. [DOI] [PubMed] [Google Scholar]
- Yang L., Wang H.N., Hou X.H., Zou Y.P., Han T.S., Niu X.M., Zhang J., Zhao Z., Todesco M., Balasubramanian S. Parallel evolution of common allelic variants confers flowering diversity in Capsella rubella. Plant Cell. 2018;30:1322–1336. doi: 10.1105/tpc.18.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yngvadottir B., Xue Y., Searle S., Hunt S., Delgado M., Morrison J., Whittaker P., Deloukas P., Tyler-Smith C. A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am. J. Hum. Genet. 2009;84:224–234. doi: 10.1016/j.ajhg.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jimenez-Gomez J.M. Functional analysis of FRIGIDA using naturally occurring variation in Arabidopsis thaliana. Plant J. 2020;103:154–165. doi: 10.1111/tpj.14716. [DOI] [PubMed] [Google Scholar]
- Zhao L., Saelao P., Jones C.D., Begun D.J. Origin and spread of de novo genes in Drosophila melanogaster populations. Science. 2014;343:769–772. doi: 10.1126/science.1248286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Feng Q., Lu H., Li Y., Wang A., Tian Q., Zhan Q., Lu Y., Zhang L., Huang T. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018;50:278–284. doi: 10.1038/s41588-018-0041-z. [DOI] [PubMed] [Google Scholar]
- Zheng L.L., Zhou K.R., Liu S., Zhang D.Y., Wang Z.L., Chen Z.R., Yang J.H., Qu L.H. dreamBase: DNA modification, RNA regulation and protein binding of expressed pseudogenes in human health and disease. Nucleic Acids Res. 2018;46:D85–D91. doi: 10.1093/nar/gkx972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Lehti-Shiu M.D., Thibaud-Nissen F., Prakash T., Buell C.R., Shiu S.H. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 2009;151:3–15. doi: 10.1104/pp.109.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall R.A., Rausher M.D. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature. 2004;428:847–850. doi: 10.1038/nature02489. [DOI] [PubMed] [Google Scholar]