Abstract
Structural variants (SVs) are a largely unstudied feature of plant genome evolution, despite the fact that SVs contribute substantially to phenotypes. In this study, we discovered SVs across a population sample of 347 high-coverage, resequenced genomes of Asian rice (Oryza sativa) and its wild ancestor (O. rufipogon). In addition to this short-read data set, we also inferred SVs from whole-genome assemblies and long-read data. Comparisons among data sets revealed different features of genome variability. For example, genome alignment identified a large (∼4.3 Mb) inversion in indica rice varieties relative to japonica varieties, and long-read analyses suggest that ∼9% of genes from the outgroup (O. longistaminata) are hemizygous. We focused, however, on the resequencing sample to investigate the population genomics of SVs. Clustering analyses with SVs recapitulated the rice cultivar groups that were also inferred from SNPs. However, the site-frequency spectrum of each SV type—which included inversions, duplications, deletions, translocations, and mobile element insertions—was skewed toward lower frequency variants than synonymous SNPs, suggesting that SVs may be predominantly deleterious. Among transposable elements, SINE and mariner insertions were found at especially low frequency. We also used SVs to study domestication by contrasting between rice and O. rufipogon. Cultivated genomes contained ∼25% more derived SVs and mobile element insertions than O. rufipogon, indicating that SVs contribute to the cost of domestication in rice. Peaks of SV divergence were enriched for known domestication genes, but we also detected hundreds of genes gained and lost during domestication, some of which were enriched for traits of agronomic interest.
Keywords: structural variation, rice, domestication, transposable element insertions, gene gain and loss
Introduction
Structural variants (SVs) are commonly defined as differences between individuals in genome order or DNA content that span >50 bases in length (Alkan et al. 2011; Tattini et al. 2015; Gaut et al. 2018). They remain a relatively mysterious feature of plant genomes, for at least three reasons. The first is their contribution to phenotypes. Numerous examples indicate that they can affect phenotypes (Żmieńko et al. 2014; Gaut et al. 2018), such as the transposable element (TE) that inserted near a grapevine myb gene and caused a shift in berry color from red to green (Kobayashi et al. 2004). More generally, however, it is not clear how often and how many SVs contribute to phenotypic traits. The second is their prevalence. The fact that plant genomes vary substantially in size among individuals (Díez et al. 2013; Gordon et al. 2017; Roessler et al. 2019) suggests that SVs are common, and we now know that they contribute to remarkable fluidity in genic content. For example, ∼15% of genes within grapevine cultivars are hemizyogous due to SVs that cause allelic loss (Zhou et al. 2019) and 10% of genes are nonsyntenic between maize inbred lines (Sun et al. 2018). Yet, for most plant species, we have little idea of the full extent of SVs, how they vary across different SV types—such as insertions, deletions, duplications, inversions, and translocations—or the rate at which they originate. A third reason is that little is known about the population frequencies of individual SV events. This frequency information is necessary to assess the forces that shape the evolutionary fate of SVs, to evaluate their associations with phenotypes and to use them as a tool for understanding important processes like adaptation, speciation, and domestication.
The first step toward addressing all of these questions is to identify SVs in population samples. Substantial progress has been made on SV detection over the last decade using paired-end, short-read resequencing data (for a review, see Alkan et al. 2011). These approaches typically map resequencing data to a single reference genome and then use coverage statistics and/or information from errant read orientations to detect an SV event. Most studies suggest that SV inference can be reasonably accurate if coverage is sufficient—that is, usually well above 10× coverage (Layer et al. 2014). However, the analysis of short-read data clearly underestimates some SV events, such as large chromosomal inversions (Mahmoud et al. 2019). For that reason, it is crucial to begin to use other types of data, including long-read sequencing data and whole-genome comparisons, to complement and confirm SVs inferred from short-read data (Zhou et al. 2019).
Here, we use a variety of data sources to detect SVs in Asian rice (Oryza sativa) and its wild relative O. rufipogon, with our primary goal to evaluate SVs as a tool to study domestication. The domestication history of the two main varieties of Asian rice (O. sativa ssp. japonica and ssp. indica; hereafter, japonica and indica) had been enigmatic until the emergence of a recent consensus. Under this consensus, japonica was domesticated from O. rufipogon (hereafter, rufipogon) in Southern China, ∼10,000 years ago (ya) or perhaps earlier (Gross and Zhao 2014; Choi et al. 2017). This primary event was facilitated by selection for agronomic phenotypes that shifted allele frequencies at domestication genes like sh4, which contributes to a nonshattering phenotype (Li et al. 2006), and qSW5, which affects grain width (Shomura et al. 2008). The uncertainty centered on the origin of indica. The recent consensus posits that indica domestication was both geographically and temporally separate from japonica domestication, having occurred on the Asian subcontinent as recently as ∼4,500 ya (Fuller et al. 2010), but indica domestication was aided by the introgression of beneficial domestication alleles from japonica (Huang and Han 2016; Choi and Purugganan 2018a, 2018b).
This consensus has been achieved primarily through the study of population genetics on large samples of resequenced rice genomes. These studies have not only substantiated that japonica and indica are genetically distinct, they have also supported other discernible groups within domesticated rice, such as the aus and aromatic varieties and temperate versus tropical japonica (Garris et al. 2005; Huang et al. 2012; Wang et al. 2018; Zhao et al. 2018). They have also documented complex relationships between cultivated rice and wild O. rufipogon, because the latter commonly—and perhaps usually—bears the signature of introgression from cultivated accessions (Wang et al. 2017). They have also been an important for identifying putative domestication genes—such as Sh4, qSW5, qSH1, prog1, sd1, Wx, Badh2, Rc, and others—via the identification of selective sweep regions (Huang et al. 2012; Zhao et al. 2018). Finally, they have helped establish that the domestication of Asian rice was associated with a genetic cost, as measured by higher frequencies and numbers of putatively deleterious SNPs in domesticates compared with wild isolates (Lu et al. 2006; Günther and Schmid 2010; Liu et al. 2017).
Large population samples have also been the basis to study SVs in Asian rice. For example, Fuentes et al. (2019) provided a catalog of SVs from 3,000 cultivated rice accessions. This study included accessions with an average coverage of 14×, so that some accessions may have had coverage too low for reliable SV detection. However, the study also employed unusual rigor by applying a suite of ten different SV detection programs. Once detected, the study looked at several SV features, such as their distribution across chromosomes and their concordance with SNPs. More recently, Carpentier et al. (2019) employed the same data set to identify SV events caused by retrotransposon insertions. This study ultimately established that mobile element insertions (MEIs) tend to be found in only a handful of 3,000 individuals, suggesting either recent TE activity in the domesticate or selection against MEIs reaching high frequency (Wright et al. 2001; Lockton et al. 2008; Zhou et al. 2019). Both studies relied on the comparison of two individuals to verify their methods and SV calls (Carpentier et al. 2019; Fuentes et al. 2019), meaning that the confirmation of SVs has been limited across these large samples.
Although SVs have been studied in Asian rice, these previous studies either did not investigate SVs in wild germplasm (Carpentier et al. 2019; Fuentes et al. 2019) or were based on small (n = 13) samples of wild accessions (Zhao et al. 2018). As a result, comparisons of SV diversity between wild and cultivated rice have been limited, and SVs have not been used to gain insights into rice domestication. Thus far, SV population frequencies have been compared only between wild and cultivated grapevines (Zhou et al. 2019). In this case, SVs provided unique insights into regions of the genome that have been under artificial selection, and they also reflected an increased genetic burden associated with clonal propagation. It remains an open question, however, whether other domesticated plants have increased SV burdens relative to their wild relatives, whether SVs will commonly provide useful insights into genomic regions subjected to positive selection and whether domestication commonly includes shifts in genic content. The identification of genes gained and lost during domestication is an area of growing emphasis, because such genes may contribute to agronomic phenotypes (Gaut et al. 2018).
Here, we take advantage of the remarkable array of publicly available rice data, including high-coverage resequencing data, long-read data, and genome assemblies to study SVs in cultivated and wild rice accessions. After inferring SVs from these different data sets, we assess the reliability of SV calls across data sets, including a comparison to a previous SV study (Fuentes et al. 2019). We then focus on a highly curated set of biallelic SVs: (1) to compare SV population frequencies between wild and cultivated taxa, (2) to investigate MEI frequencies across different TE families, and (3) to evaluate SVs with respect to common features associated with domestication, such as its genetic cost and signatures of positive selection.
Results
Comparing SVs among Short-Read, Long-Read, and Whole-Genome Data
We gathered three data sets to identify, contrast, and confirm SV calls: (1) 14 whole-genome assemblies, (2) Pacific Biosciences (PacBio) SMRT long reads from a subset of ten accessions included in whole-genome assembly, and (3) paired-end resequencing data from 347 wild and cultivated accessions that met our filtering criteria (see Materials and Methods), especially high enough mapped coverage (>15×) to aid accurate SV detection.
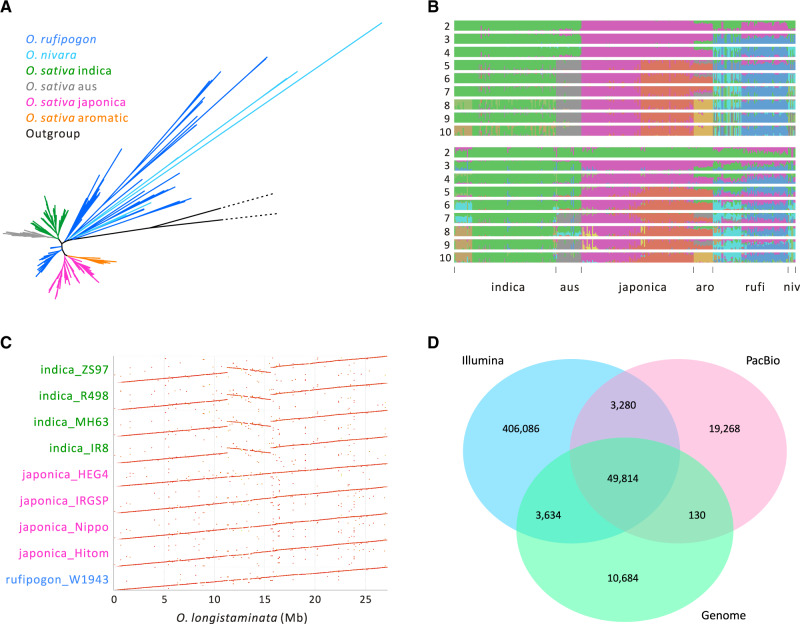
Focusing first on the resequencing data, we identified 38,717,560 SNPs and small variants (i.e., indels <50 bp). As expected (Garris et al. 2005; Huang et al. 2012; Wang et al. 2018; Zhao et al. 2018), phylogenetic analyses based on SNPs revealed clear group-specific clades, with the aus-indica clade separated from the japonica-aromatic clade (fig. 1A). The exception to clear separation was O. nivara, which nested within rufipogon clades, but this result was consistent with previous analyses based on wide sampling of O. nivara and rufipogon (Cai et al. 2019). In the phylogeny, rufipogon was represented by two major clades: one that branched next to the outgroups (O. meridionalis and O. longistaminata) and derived primarily from India and Southeast Asia, and a second that rooted between indica and japonica and included accessions primarily from China. We include this information to emphasize population structure among wild rice accessions, which prompted us to include only one clade for domestication analyses (see below).
Fig. 1.
Features of the data and SV data sets (A) A phylogeny based on SNPs of n = 347 accessions of Asian rice with outgroups Oryza meridionalis and O. longistaminata. (B) Population structure inference based on SNPs (top) and SVs (below) for the short-read data set of 347 individuals. The accessions are arranged in the same order for the SNP and SV plots, the x-axis labels denote the different groups, with “aro,” “rufi,” and “niv” referring to aromatic, rufipogon, and O. nivara. (C) A dotplot of chromosome 6 showing the large (∼4.3 Mb) inversion in indica accessions relative to the O. longistaminata outgroup. The inversion is not shared with the japonica accessions in our sample. (D) A Venn diagram based on the combined results from three SV types (DEL, DUP, and INV) that compares SVs among three data sets based on short reads (Illumina, n = 347), long reads (SMRT, n = 10), and genome alignments (n = 14). Results for each SV type separately are available in supplementary figure S3, Supplementary Material online.
We called SVs from the set of 347 individuals using two approaches, following Zhou et al. (2019). First, to identify deletion (DEL), duplication (DUP), inversion (INV), and translocation (TRA) events, we combined population calls from Delly (Rausch et al. 2012) and Lumpy (Layer et al. 2014). The combined SVs were then filtered based on minimal quality scores and also on the requirement that a single SV had exact breakpoints shared among accessions (see Materials and Methods). Although this pipeline also calls insertion (INS) events, previous studies (Mahmoud et al. 2019; Zhou et al. 2019) suggest that INS are not accurately detected from short reads. INS may also overlap with MEIs, which we called using separate methods (see below). For those reasons, we excluded INS inferences from further analyses. Altogether, the approach led to a highly curated set of 824,390 biallelic SVs across the entire data set of 347 accessions (table 1 and supplementary table S1, Supplementary Material online).
Table 1.
The Number of Variants Detected by Illumina Short Reads, PacBio Long Reads, and Genome Alignment.
| Variant Type | Illumina | PacBio | Genome Alignment |
|
|---|---|---|---|---|
| LASTZ | MUMmer | |||
| Number of samples | 347 | 10 | 14 | 14 |
| TRA | 76,835 | 85,279 | 1,332 | |
| DUP | 48,132 | 213 | 3,700 | 2 |
| DEL | 72,930 | 60,747 | 62,953 | 48,153 |
| INV | 341,752 | 11,532 | 159 | 343 |
| INS | 103,447 | 70,065 | 113 | |
| MEI | 284,741 | 270,708 | 246,340 | |
| Total | 824,390 | 531,926 | 390,823 | |
We used multiple methods to assess the accuracy of the SV calls. First, as in previous papers (Gordon et al. 2017; Fuentes et al. 2019; Zhou et al. 2019), we assessed the congruence of population structure between SNP and SV calls from the same data—that is, the 347 resequenced individuals—to see if they provided similar insights into population structure and history. To do so, we applied NGSadmix to the SNP data, allowing the number of groups (K) to range from 2 to 10. At K = 10, the SNP data recapitulated the expected population groupings—that is, indica and japonica were separate as were two distinct clades of rufipogon (fig. 1B). We then applied ADMIXTURE to the full data set of SVs, which identified the same major groups (fig. 1B). Per-individual assignments were strongly correlated between SV and SNP results (Pearson r = 0.853, P < 2.2 × 10−16), suggesting the suitability of our SV calls for population genetic analyses.
Second, we compared our SV inferences with those from Fuentes et al. (2019), whose SVs were based on ten detection methods and 3,000 cultivars (Wang et al. 2018). For this comparison, we included only DEL, DUP, and INV events, because the size and location of TRA events were not always clear, thus leading to the potential for improper comparison. Our DEL, DUP, and INV SV calls overlapped with 90.14% of those identified by Fuentes et al. (2019). We did have an additional 9.64% of events, however, due to the added diversity within the rufipogon and outgroup samples. Nonetheless, the vast majority of SV inferences within cultivated rice agreed with previous work, suggesting that our combination of high-coverage data with two SV-callers performed well.
Third, we compared SVs based on short-read data with SVs inferred from two other types of sequence data: whole-genome assemblies and SMRT reads (supplementary table S2, Supplementary Material online). SVs were inferred from genome assemblies based on pairwise genome alignments using MUMmer (Marçais et al. 2018) and LASTZ (Harris 2007) (see Materials and Methods). MUMmer detects DEL, DUP, INV, TRA, and INS events, and we further delineated INS events into MEI and non-MEI events. Across the pairwise comparison of 14 genomes, including an O. longistaminata (Reuscher et al. 2018) outgroup, we detected a total of 390,823 SVs (table 1 and supplementary table S1, Supplementary Material online). Notably, the SVs included a ∼4.3-Mb (coordinates: 11,296,942–15,576,712 bp in the O. longistaminata reference) homozygous inversion that spanned the centromere of chromosome 6 and differed between the four indica assemblies and assemblies from other Oryza species (fig. 1C and supplementary figs. S1 and S2, Supplementary Material online).
In addition to whole-genome assemblies, we collected raw SMRT reads from the subset of six indica, three japonica, and one O. longistaminata accession that were used in genome alignment (table 1 and supplementary table S2, Supplementary Material online) and then mapped SMRT reads to the Nipponbare genome (Zhang et al. 2018) using Minimap2 (Li 2018). SVs were called across all ten samples using the Sniffles pipeline (Sedlazeck et al. 2018), identifying 531,926 SVs (table 1 and supplementary table S1, Supplementary Material online), again detecting DEL, DUP, INV, TRA, and INS events and further separating INS events into MEI and non-MEI events. SMRT reads also provide the unique opportunity to investigate within-genome hemizygosity, based on the presence of alternative reads that do and do not span a genomic region (Vondras et al. 2019; Zhou et al. 2019). Applying this approach to a japonica individual (Nipponbare), an indica individual (93-11), and a wild outcrossing relative O. longistaminata, we detected similar numbers of presence–absence variants (PAVs) in Nipponbare (257 DELs and 31 DUPs) and 93-11 (306 DELs and 18 DUPs). For these two accessions, only 0.73% (385) and 0.35% (171) of genes were hemizygous, whereas 56-fold more PAVs were found in the outcrossing O. longistaminata accession, resulting in estimated hemizygosity for 8.89% (or 3,895) genes.
Finally, we compared the three data sets for three SV categories that were most comparable across methods: DEL, DUP, and INV events. We first contrasted the genome and SMRT-read data sets. For the three categories, more SVs were detected with SMRT-reads (72,492) than with genome assemblies (64,262), despite the fact that data for the former (n = 10) were a subset of the latter (n = 14; supplementary table S2, Supplementary Material online). Notably, the pipeline for genome alignments detected 10-fold fewer INV events, whereas SMRT reads yielded 10-fold fewer DUP events (supplementary fig. S3, Supplementary Material online), suggesting systematic biases in detection methods. Altogether, however, the two data sets had 57.53% of DEL, DUP, and INV events in common (fig. 1D and supplementary fig. S3, Supplementary Material online).
We then compared short-read SVs to the two other data sets, but this was an inherently biased process because the sample size was much larger for short-read data, yielding more SVs despite extensive filtering. Given this bias, we examined the overlap among data sets in a directional manner, asking: How often do Illumina SV calls identify SVs found within the other two data sets? The Illumina data set included 70.07% of DEL events identified within the whole-genome and SMRT-read data sets, 75.24% of DUP events, and a lower proportion (43.22%) of INV events (supplementary fig. S3, Supplementary Material online). Summing across the three SV types, the short-read data identified 65.35% of SVs found in the two other data sets (fig. 1D). Notably, this level of correspondence (65.35%) exceeded that between the genome and SMRT-read data sets (57.53%), again suggesting the short-read population calls were reasonable.
Population Properties of SVs
Given concordant information with SNPs and SVs and notable overlaps among long-read and short-read SVs, we focused on short-read SVs to investigate population dynamics. For simplicity, we narrowed our focus to the three groups with the largest population samples: rufipogon, indica (n = 96), and japonica (n = 106). For rufipogon, we examined only the clade of n = 40 accessions that appeared to be truly wild, based on their phylogenetic position (fig. 1A), recognizing that combining the two distinct clades would produce skewed population statistics.
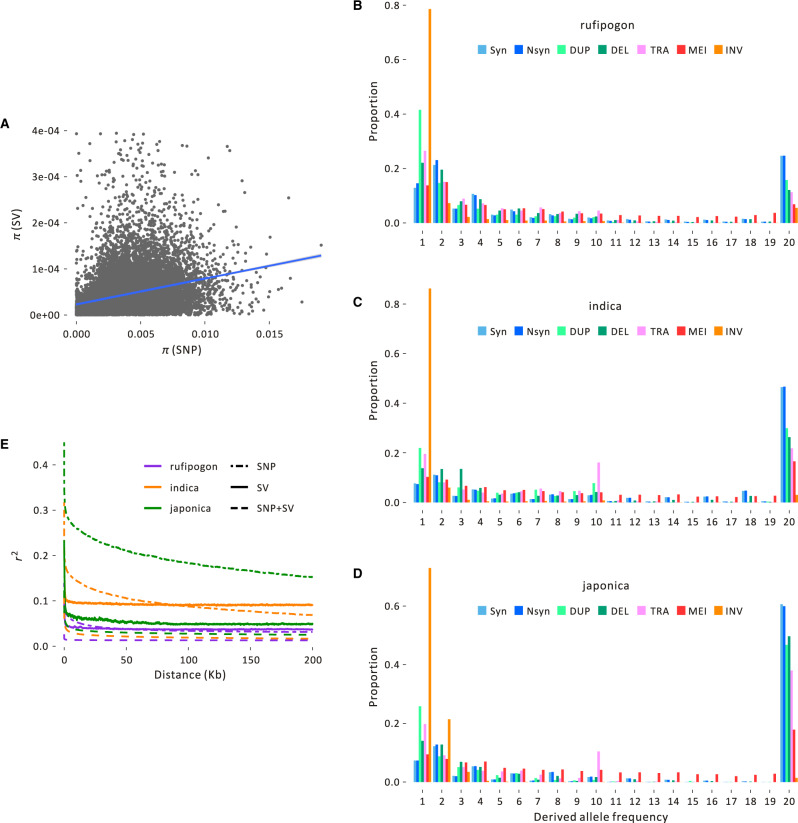
We first characterized chromosomal positions of SVs, plotting SV diversity using sliding window analyses for each taxon (e.g., supplementary figs. S4–S6, Supplementary Material online). Visually, there were no compelling patterns that suggested particular regions were more prone to specific SV events. However, SV and SNP diversity were slightly but significantly correlated across chromosomal windows in all three population groups (Pearson r = 0.0332, P = 1.07 × 10−5 for rufipogon; Pearson r = 0.0637, P = 2.2 × 10−16 for indica; Pearson r = 0.0494, P = 1.07 × 10−10 for japonica; fig. 2A and supplementary fig. S7, Supplementary Material online).
Fig. 2.
Population information about SVs. (A) The plot graphs SNP and SV average pairwise diversity (π) for the rufipogon sample across 20-kb windows of the genome, with the line indicating the correlation, which is weakly positive but significant (Pearson r = 0.0332, P = 1.07 × 10−5). Similar graphs for the japonica and indica samples are in supplementary figure S7, Supplementary Material online. Plots (B), (C), and (D) show the unfolded SFS of different types of SVs in (B) rufipogon, (C) indica, and (D) japonica. Each SFS contains synonymous SNPs (Syn), nonsynonymous SNPs (Nsyn), and SV events that fall into the duplication (DUP), deletion (DEL) translocation (TRA), mobile element insertion (MEI), and inversion (INV) categories. (E) The decay of linkage disequilibrium (LD) of SNPs and SVs measured by r2 for the three population groups based on SNPs, SVs, and SNPs+SVs.
We also calculated the unfolded site-frequency spectra (SFS) of the three taxa for a sample of ten individuals with high-coverage and little missing data (fig. 2B–D). Each SFS included four SV types (DUP, DEL, TRA, and INV), along with sSNPs and nSNPs and 284,741 MEIs, which we called separately using two separate pipelines (see Materials and Methods). The SFSs revealed three salient features of SV polymorphism. First, there were demonstrable differences among taxa, because there was a higher proportion of fixed variants (and fewer intermediate variants) in cultivated rice compared with rufipogon. The U-shape of the SFS from cultivated rice had been noted previously and is consistent with both enhanced genetic drift during a domestication bottleneck (Caicedo et al. 2007) and a shift in mating system. Second, in all three taxa, there was a lower proportion of fixed SVs than fixed sSNPs and nSNPs. The distributions for each SV type were significantly different from the sSNP distribution in all three taxa (P < 0.01, Kolmogorov–Smirnov test). Assuming sSNPs provide a reasonable “neutral” control, the leftward shift in the SFS suggests either that SV variants were deleterious, on an average, or that they have higher mutation rates than SNPs such that many new events have not had the opportunity to rise in frequency. Finally, the SFS varied among SV types. In all three taxa, INV events had the most extreme SFS; in each group, >90% of INV events were identified in three or fewer individuals, suggesting either strong selection or perhaps detection biases (see Discussion).
The SFSs suggest that MEIs and all SV types have lower population frequencies, on an average, than sSNPs in all three taxa. As a consequence, SVs may have generally lower linkage disequilibrium (LD) values than SNPs, with the potential for a faster decay of LD over physical distance. We calculated LD in each of the three taxa based on SNPs, SVs, and both SNPs+SVs, using the squared correlation coefficients (r2). The SNP data confirmed previous observations that LD decays more slowly in japonica than either indica or rufipogon (Mather et al. 2007) (fig. 2E). For example, r2 for SNPs remained ∼0.2 over a distance of ∼100 kb for japonica, but was ∼0.1 for indica and <0.05 for rufipogon over the same physical distance. Note, however, that SVs had lower r2 values than SNPs for all taxa, with values that exceeded 0.1 only over very short (<15 kb) distances. The r2 values were even lower when based on both SNP+SV data (fig. 2E). These results may have important implications for detecting the effect of SVs on phenotypes.
SVs and Domestication
It is an open question in other domesticated taxa whether SV burden increases as a consequence of domestication, whether SVs provide useful insights into genomic regions potentially subjected to positive selection during domestication and whether domestication shifts genic content. We investigated these features with our set of SVs, using the previously described population samples of indica (n = 96) and japonica (n = 106) and rufipogon (n = 40) for all analyses.
SV Burden
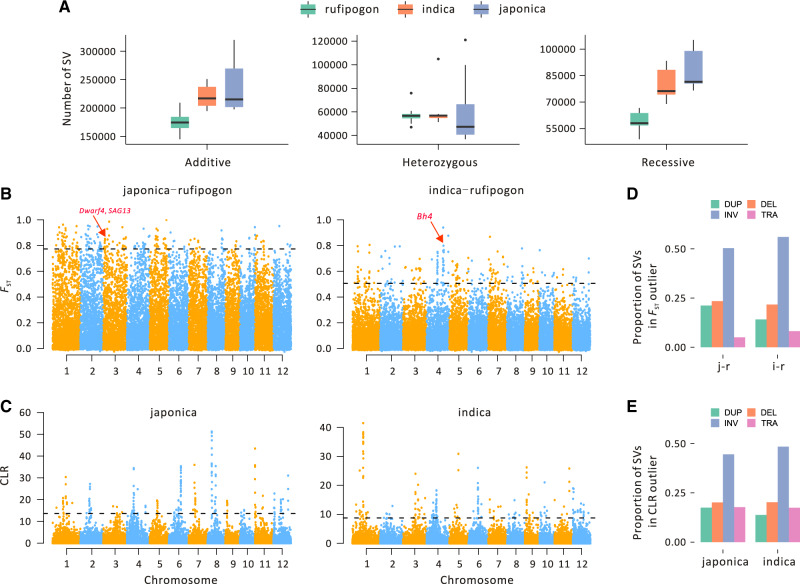
Because the SFS of SVs and MEIs imply that they may be deleterious, we predicted that they contribute to the deleterious load, reflecting the cost of domestication in rice (Lu et al. 2006; Günther and Schmid 2010; Liu et al. 2017). We evaluated cost by calculating the additive SV+MEI burden per individual, which is the number of derived heterozygous sites (the heterozygote burden) plus two times the number of homozygous variants (the recessive burden) (Henn et al. 2016). Comparing the additive SV+MEI burden across the three taxa, it was 35% and 25% higher on an average for japonica and indica relative to rufipogon (P < 0.005 for both contrasts, t-test; fig. 3A). Given the differences in mating system between cultivated and wild rice, we also expected the recessive burden to be the primary contributor to differences in the additive burden. This expectation held, because the recessive burden was >72% of the additive burden for both cultivars, but the proportion was lower (67%) for rufipogon (fig. 3A). These patterns—that is, higher additive and especially recessive burdens—held across variant types, with the apparent exception of DEL events (supplementary fig. S8, Supplementary Material online).
Fig. 3.
Feature of SVs associated with domestication. (A) The genetic load of SVs for rufipogon, japonica, and indica. The selfing cultivars have a higher recessive (homozygous) load and correspondingly larger total SV burden, suggesting a cost of domestication. (B) Manhattan plots of FST values between rufipogon, based on SVs within 20-kb windows, with japonica on the left and indica on the right. The corresponding Manhattan plots for SNPs are provided in supplementary figure S10, Supplementary Material online. (C) Manhattan plots of CLR values for japonica (left) and indica (right), based on SVs within 20-kb windows. The corresponding Manhattan plots for SNPs are provided in supplementary figure S12, Supplementary Material online. The proportion of different types of SVs under the selective sweeps detected by FST (D) and SweeD (E) analyses.
Divergence between Domesticated Taxa and Rufipogon
In theory, the genes that contribute to domestication can be identified as regions of marked chromosomal divergence between wild and cultivated samples. We compared rufipogon to both indica and japonica by estimating SNP and SV divergence in fixed 20-kb windows across the genome. We calculated divergence with two measures (FST and Dxy; supplementary figs. S9 and S10, Supplementary Material online) but focus on FST results here for simplicity. Across the entire genome, mean FST estimates were substantially higher for SNPs (indica–rufipogon 0.293 ± 0.134; japonica–rufipogon 0.485 ± 0.181) than for SVs (indica–rufipogon 0.122 ± 0.079; japonica–rufipogon 0.259 ± 0.141), reflecting the fact that SVs were typically at lower population frequencies (fig. 2A).
We contrasted the two cultivars to rufipogon and ranked the top 1% FST windows (or 187 of 18,654 windows throughout the genome) for both SNPs and SVs (fig. 3B and supplementary fig. S10, Supplementary Material online). Only a small number of FST windows were within the top 1% for both SNPs and SVs (supplementary fig. S11, Supplementary Material online); for example, we detected 26 such windows for the indica–rufipogon comparison. Although a small number, 26 is far more than the ∼2 windows expected at random (P < 10−3, permutation test), suggesting that the SNP and SV data do capture some common signatures (as is expected, given that they are in the same genomic window). Similarly, we detected 12 such windows in the japonica–rufipogon comparison, again representing an enrichment over a random draw (P < 10−3, permutation test). Of these, only one window overlapped between japonica and indica; it contained a gene (LOC_Os02g43800) that was annotated as a retrotransposon protein. Thus, arguably the strongest signal of positive selection based on FST—that is, in a window identified from both SNPs and SVs across both cultivated taxa—contained a gene without obvious agronomic implications.
We examined the shared SNP–SV windows for potential candidate genes (supplementary table S3, Supplementary Material online). For example, of the 82 genes contained in the 26 windows for the indica–rufipogon comparison, 31 were annotated as expressed or hypothetical proteins, and 14 were annotated as TE-related; neither category were obvious candidates to contribute to agronomic phenotypes. Most of the remaining 37 genes were assigned putative functions, including a ribosomal protein, a male sterility protein, small auxin up-regulated genes, receptor like-kinase genes, and genes with other functions. GO analyses indicated that the 82 genes were enriched for a variety of putative functions, including cellular components extrinsic to membranes and biological processes related to superoxides (supplementary table S4, Supplementary Material online). Similarly, the shared SNP–SV peaks between japonica and rufipogon contained 36 genes in 12 windows, with 15 genes assigned putative functions and GO enrichment in DNA replication and other functions. For completeness, we also analyzed the set of genes identified in SNP-only (489) or SV-only (374) peaks for each taxon (supplementary table S3, Supplementary Material online). Not surprisingly, the genes were enriched for a variety of GO-based functions (supplementary table S4, Supplementary Material online).
Because we were interested in the utility of SVs to detect selection events, we also took a candidate gene approach to assess whether SVs enhanced their identification. To do this, we focused on a set of 15 known domestication and improvement genes (Huang et al. 2012; Wang et al. 2018) to ascertain whether they were identified in FST scans more often than expected at random (table 2). Among the 15, six were within the top 1% of FST windows between rufipogon and either indica or japonica (table 2); three of these were found with SNPs alone (including TAC1, a gene implicated in tillering, and shattering genes Sh1 and Sh4) and three more with SVs alone (Bh4, Dwarf4, and SAG13) (table 2). Overall, the set of 15 genes was highly enriched to be within FST peaks at the 1% and 10% levels for both SNPs (P < 1.19 × 10−15 for 1% peaks, P < 6.67 × 10−8 for 10% peaks, Wilcoxson–Mann–Whitney test) and SVs (P < 2.20 × 10−16 for 1% peaks, P < 2.54 × 10−14 for 10% peaks, Wilcoxson–Mann–Whitney test), suggesting that SVs do have some utility for identifying domestication genes. We note, however, that candidate gene enrichment using SVs was not as evident based on Dxy (supplementary table S5, Supplementary Material online).
Table 2.
Previously Identified Putative Domestication or Improvement Genes with Their ID, Function, and % Ranking in FST Windows.
| Locus | Gene ID (MSU7/IRGSP1.0) | Function |
F
ST (i–r) |
F
ST (j–r) |
||
|---|---|---|---|---|---|---|
| SNPs | SVs | SNPs | SVs | |||
| Sh4 | LOC_Os04g57530/Os04g0670900 | Shattering | 0.18 | 17.05 | 5.08 | 81.02 |
| qSW5 | LOC_Os05g09520/Os05g0187500 | Grain width | 97.09 | 1.44 | 84.78 | 68 |
| qSH1 | LOC_Os01g62920/Os01g0848400 | Shattering | 58.21 | 94.51 | 5.06 | 96.65 |
| Sd1 | LOC_Os01g66100/Os01g0883800 | Semidwarfing | 31.78 | 51.96 | 5.64 | 5.99 |
| Wx | LOC_Os06g04200/Os06g0133000 | Grain quality | 32.07 | 42.17 | 27.02 | 58.23 |
| Badh2.1 | LOC_Os04g39020/Os04g0464200 | Flavor or fragrance | 84.57 | 70.14 | 7.46 | 64.06 |
| Rc | LOC_Os07g11020/Os07g0211500 | Red pericarp | 47.21 | 52.79 | 7.01 | 55.86 |
| Bh4 | LOC_Os04g38660/Os04g0460200, LOC_Os04g38670/Os04g0460200, LOC_Os04g38680/Os04g0460300 | Hull color | 3.18 | 0.95 | 6.51 | 4.67 |
| PROG1 | LOC_Os07g05900/Os07g0153600 | Tiller angle | 1.79 | 2.16 | 14.18 | 6.17 |
| OsCl | LOC_Os06g10350/Os06g0205100 | Leaf sheath color | 26.58 | 75.83 | 14.01 | 73.61 |
| TAC1 | LOC_Os09g35980/Os09g0529300 | Tiller angle | 77.05 | 31.3 | 0.85 | 54.12 |
| Dwarf4 | LOC_Os03g12660/Os03g0227700 | Leaf architecture | 9.2 | 80.98 | 1.53 | 0.35 |
| SAG13 | LOC_Os03g16230/Os03g0269100 | Senescence | 48.3 | 47.08 | 30.39 | 0.56 |
| TB1 | LOC_Os03g49880/Os03g0706500 | Tillering | 2.22 | 81.45 | 5.23 | 78.2 |
| Sh1 | LOC_Os03g44710/Os03g0650000 | Shattering | 0.8 | 4.46 | 23.75 | 18.3 |
Finally, given the consensus that domestication genes were introgressed into indica from japonica (Choi and Purugganan 2018a), a previous study remarked that domestication genes should be enriched in regions of low divergence between the two cultivars (Huang et al. 2012). We tested this notion by examining the candidate set and their corresponding window rankings in FST windows calculated between indica and japonica. None of the 15 genes was located in an FST trough, as defined by windows ranking in the lowest 99% percentile, but three of the genes (Wx1, Bh4, and PROG1) had either SNP or SV values >90% (supplementary table S6, Supplementary Material online), which is a significant enrichment (P < 5.88 × 10−3, Wilcoxson–Mann–Whitney test). In contrast to genes within FST troughs, several of the 15 genes were located in FST peaks between indica and japonica, including Dwarf4, Sd1, and TB1 (supplementary table S6, Supplementary Material online). Altogether, these analyses reinforce the complex history of Asian rice domestication by suggesting that some, but not all, known domestication genes may have a history of introgression between japonica and indica.
Selective Sweeps within Domesticates
We also searched for taxon-specific signals of selective sweeps using the composite likelihood method (Pavlidis et al. 2013). We again investigated the top 1% of 20-kb windows for both SVs and SNPs (fig. 3C and supplementary fig. S12, Supplementary Material online). Qualitatively the results exhibited fewer but clearer peaks compared with FST (fig. 3B) or Dxy analyses (supplementary fig. S9, Supplementary Material online). The selection signals detected in SVs are dominated by INVs (∼50%) in both FST (fig. 3D) and SweeD (fig. 3E) analyses. However, there was little correspondence between the top 1% windows based on SVs and SNPs (supplementary fig. S13, Supplementary Material online); the two data types shared two windows in common for rufipogon, only one for indica, and ten for japonica, which is the only taxon that had shared windows more often than expected by chance (P < 10−3, permutation test). None of these windows contained any obvious candidate genes (supplementary table S7, Supplementary Material online) or the previously identified set of 15 domestication/improvement genes. For completeness, we have listed all of the genes within CLR peaks (supplementary table S8, Supplementary Material online) with their GO enrichment categories (supplementary table S9, Supplementary Material online).
Gene Gain and Loss
We focused on the subset of SVs that included genes and determined whether they were private (i.e., variable within only one taxon) or fixed (i.e., in alternative states) between taxa. For example, between rufipogon and japonica, we detected 114,840 SVs that were private in rufipogon; 144,600 that were private in japonica; 180,798 that were shared SVs between taxa; and only one fixed SV, corresponding to one gene that was annotated as related to retrotransposition. Focusing on the subset of private SVs that include genic DUP and DEL events, we found 148 genes gained and 138 lost in our japonica sample relative to the rufipogon sample. Similarly, we detected 3,410 genes gained and 181 lost in indica relative to rufipogon (supplementary table S10, Supplementary Material online). Some of the genes lost during domestication had validated functions related to physiological and morphological traits, such as sterility (LOC_Os01g11054/Os01g0208700), culm leaf (LOC_Os04g39780/Os04g0473900), flowering (LOC_Os03g05680/Os03g0151300), and stress tolerance (LOC_Os01g64970/Os01g0869900). In addition to these functions, genes inferred to be gained during domestication involved functions that contribute to eating quality, including starch storage and biosynthesis (e.g., LOC_Os01g65670/Os01g0878700, LOC_Os02g32350/Os02g0523500, LOC_Os03g09250/Os03g0192700, LOC_Os03g49350/Os03g0700400, LOC_Os03g52340/Os03g0733800, and LOC_Os09g26880/Os09g0440300) (Wang et al. 2018). A complete list of private genes and their GO enrichments are listed in supplementary tables S10 and S11, Supplementary Material online; the important point is that SV analyses between Asian rice and rufipogon yield reasonable candidate genes for traits involved in domestication or improvement.
MEIs for Specific TE Families
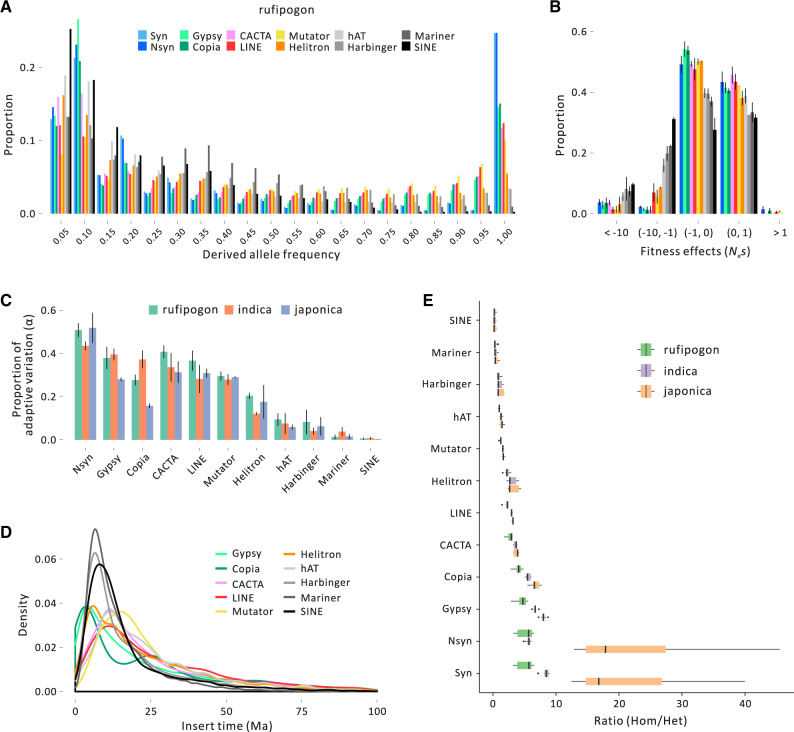
We called MEIs separately and assigned them to specific families, providing an opportunity to compare population dynamics among TE families and types. We separated MEIs into ten distinct TE families—Gypsy, Copia, LINE, SINE, CACTA, hAT, Mutator, Harbinger, Mariner, and Helitron elements—and calculated their SFSs based on calls from two methods, PoPoolationTE2 and TE-locate. Focusing on the PoPoolationTE2 results from rufipogon for simplicity (fig. 4A), but with similar results from two methods used to call MEIs (supplementary figs. S14 and S15, Supplementary Material online), all TE families had fixation frequencies lower than sSNPs, with each distribution significantly different from sSNPs (P < 0.01, Kolmogorov–Smirnov test). However, there were also marked differences among TE families (fig. 4A and supplementary figs. S14 and S15, Supplementary Material online). The most obvious deviation was for SINE and Mariner elements, for which only a small proportion of MEIs were fixed; hAT and Harbinger elements also demonstrated a substantial leftward trend relative to Gypsy, Copia, and other element types. Consistent with this observation, estimation of the distribution of fitness effects (DFEs) suggests that selection was more severe against these four TE families (fig. 4B), with the lowest proportion of putatively adaptive variants (α) for SINEs (α = 0.12%) followed by Mariner (α = 1.52%), Harbinger (α = 6.32%), and hAT (α = 5.91%) (fig. 4C).
Fig. 4.
Features of TE diversity in rice. Plot (A) provides the SFS for ten element types along with synonymous SNPs (Syn) and nonsynonymous SNPs (Nsyn). This plot is for rufipogon; analogous plots for japonica and indica are provided in supplementary figures S14 and S15, Supplementary Material online. (B) The inferred distribution of fitness effects (DFE) in rufipogon relative to nSNPs. The y-axis provides the proportion of TE insertions, and the x-axis reports Nes. The color scheme for TE families is the same as (A). (C) The estimated proportion of adaptive variation (α) for each TE family and each of the three taxonomic groups. (D) Distributions of inferred insertion times for TE families in the Nipponbare reference. (E) The ratio of homozygous to heterozygous MEI variants in the three taxa for each TE family, which shows that the families under strong selection have relatively fewer homozygous variants.
What might cause apparent differences in population dynamics among TE families? One explanation concerns the timing of TE insertion; if SINEs have been active more recently, it is possible that the SFS reflects a lack of sufficient time for insertions to reach fixation. To test this idea, we estimated the insertion time of individual elements within the japonica reference (see Methods), producing a distribution of insertion times for all ten families (fig. 4D). The distribution of insertion times was similar among TE families (P > 0.05, t-test) with Gypsy and Copia elements (but not SINEs) biased toward slightly more recent insertions. Hence, more recent activity does not seem to be an adequate explanation for the SFS of SINE and Mariner elements. Another explanation is stronger purifying selection against some element families. To assess this idea, we examined the distribution of MEIs relative to genes in the japonica reference. We found that a lower proportion of SINE, Mariner, Harbinger, and hAT MEIs were inserted within exons relative to other TE families (supplementary fig. S14, Supplementary Material online). This observation could be fueled by insertion biases, but they may also point to stronger selection against these four families. Consistent with the latter interpretation, the ratio of homozygous to heterozygous variants was lower for these four families than for the other families (fig. 4E), suggesting stronger selection when these elements are uncovered from a heterozygous state and experience recessive selection.
Discussion
SVs remain unexplored for most crops, and this is particularly true with respect to comparing population frequencies between crops and their wild relatives. Here, we have performed a genome-wide analysis of SVs in Asian rice and its wild progenitor O. rufipogon, with the goal of understanding more about the evolutionary processes that act on them and their fate during domestication. Most of our inferences have been based on SVs called from a large (347 accession) data set consisting of high-coverage (average 50×, median 28×), short-read data. Given these calls, it is important to note two important caveats. First, we have focused on biallelic SVs that were useful for population genetic inference, meaning that the SVs were filtered both for quality and to avoid complex events, such as overlapping SVs. Given this curation, it is important to convey that we did not expect, nor intend, our set of SVs to represent a comprehensive catalog, as was the intent for a previous study that lacked rufipogon samples (Fuentes et al. 2019). Second, like previous studies (Carpentier et al. 2019; Fuentes et al. 2019), all of our inferences rely on mapping to the Nipponbare genome, which may introduce a reference bias that makes it more difficult to identify novel SVs from non-japonica samples. Nonetheless, three features of our SV calls suggest they are reasonable: (1) they provide population structure information that is highly concordant to (and strongly correlated with) information from SNPs (fig. 1B), (2) they overlap substantially (>90%) with SVs reported previously, based on different data sets and analytical methods (Fuentes et al. 2019), and (3) there is some agreement across data types, where overlap is less impressive but still substantial (fig. 1D).
SV Comparisons among Data Sets
Our results are consistent with previous work claiming that different data types provide different information. For example, genome alignments can be poor for detecting heterozygous SVs, because primary assemblies ignore alternative haplotypes (Mahmoud et al. 2019). Instead, they tend to be best at identifying large SV events, such as large insertions (Nattestad and Schatz 2016). In contrast, SMRT-read mapping should be efficient for most SV detection, outperforming short-read data (Sedlazeck et al. 2018; Chaisson et al. 2019). However, the development of SMRT-read methods is still nascent (Mahmoud et al. 2019) and by no means perfected, as perhaps evidenced by the very low number of DUP events that were detected with SMRT reads (table 1 and supplementary table S1, Supplementary Material online). Finally, paired-end short reads are probably accurate for SV calls, given sufficient coverage (Layer et al. 2014), but they often miss large and complex SV events (Sedlazeck et al. 2018).
Given the strength and limitations of different approaches, our comparative data sets provide different insights into rice SVs. For example, whole-genome alignments reveal the presence of a large centromeric inversion on chromosome 6 that differentiates indica from japonica and the outgroups in our sample (fig. 1C). This inversion has been reported previously to contain 404 genes (Du et al. 2017), and it was also identified in O. brachyantha compared with indica assemblies (Liao et al. 2018). Another SV feature is genic hemizygosity, which can be estimated reasonably from SMRT reads (Zhou et al. 2019). Based on remapping SMRT reads to the Nipponbare reference, we estimate that <1% of genes are hemizygous in a japonica and an indica accession, which is consistent with the expectation of high homozygosity for predominantly selfed lineages. These estimates are low enough that they may reflect the false-positive rate of the method. In contrast, ∼9% of genes are hemizygous for the O. longistaminata individual. Superficially 9% seems high, but it is similar to the ∼10% PAV differences between inbred lines of maize (Sun et al. 2018) and lower than the ∼10–14% genic hemizygosity of grapes (Vondras et al. 2019; Zhou et al. 2019). This observation adds to a growing appreciation that outcrossed plants harbor a substantial fraction of SVs that lead to genic hemizygosity.
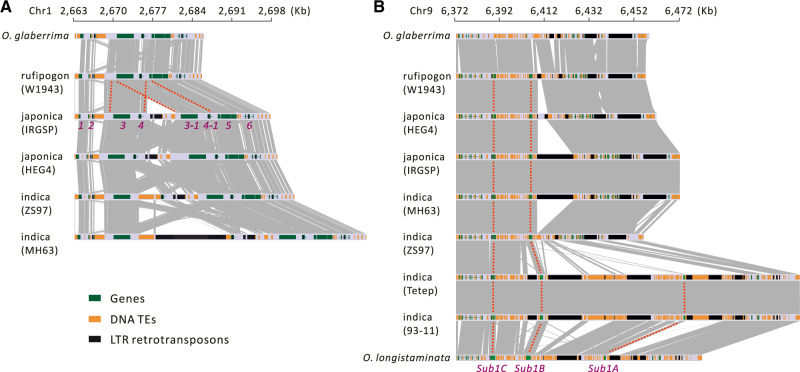
Our filtering of short-read SVs was purposefully biased against the detection of complex, overlapping SV regions, because we sought to identify discrete biallelic loci for population genetic analysis. It is worth emphasizing, however, that complex SV regions do exist. For example, we used genome alignments to investigate SVs in the region surrounding an NBS-LRR gene (LOC_Os01g05600/Os01g0149350) that was detected as gained during domestication, based on short-read SVs. In this region the cultivated individuals in our sample have an additional NBS-LRR gene relative to the wild individuals; among cultivars, the region is marked by the movement of both DNA and RNA transposons that alter distances among genes (fig. 5A). Similarly, we examined the Submergence1 (SUB1) region (fig. 5B), which contains CNVs that affect flooding tolerance (Xu et al. 2006; Mickelbart et al. 2015). This locus contains a cluster of three ethylene response factor (ERF) genes, Sub1A, Sub1B, and Sub1C. Among them, only the Sub1A-1 (an allele of the Sub1A gene) confers flooding tolerance and it was present in only a few indica cultivars (fig. 5B). Among the cultivars investigated, the region has expanded nearly 2-fold in 93-11 and Tetep due to transposon element insertions and a genic copy number variant (Sub1A-2). Altogether, these analyses accentuate the prevalence of SVs in rice (Wang et al. 2018; Fuentes et al. 2019) and the fluidity of Oryza genomes (Stein et al. 2018; Zhao et al. 2018).
Fig. 5.
Two example regions of complex SVs across Oryza taxa. (A) A segmental tandem duplication of an NBS-LRR-encoding gene (LOC_Os01g05600/Os01g0149350) that was found to be gained in indica and japonica relative to rufipogon. The synteny map is shown for a region corresponding to a 35-kb region in japonica (Nipponbare). (B) Gene and TE copy number variation in a 100-kb region of chromosome 9 that includes the Sub1A gene, which confers flooding tolerance. Both an indica accession and O. longistaminata contain three copies of genes. For both (A) and (B), gene copies are tracked by dotted red lines.
SVs Are Typically at Lower Population Frequencies than SNPs
In each of the three taxa, the SFS of each SV type differs significantly from the SFS of “neutral” (4-fold synonymous) sSNPs. Previous work has shown that plant SVs tend to be deleterious in plant populations (Flagel et al. 2014; Gaut et al. 2018; Zhou et al. 2019). Our results are consistent with this view, but such differences could also be caused by different mutational mechanisms and rates. Nonetheless, the SFSs also suggests heterogeneity among SVs, because they follow an apparent hierarchy in which INV events have the most left-leaning SFS, followed by MEI, TRA, DEL, and DUP events. One pertinent question is whether detection biases somehow fuel this heterogeneity because, for example, INV events have a lower percentage of overlap among data sets than DEL and DUP events (fig. 1D and supplementary fig. S3, Supplementary Material online). There are systematic biases for all SV types—for example, ascertainment biases—that may tend to skew the SFS leftward (Emerson et al. 2008). The question here is whether INVs are especially prone to these biases. We do not believe that this is the case for three reasons: (1) the SFS are based on the individuals with highest coverage, which limits false-negative results (Cridland and Thornton 2010); (2) some studies suggest similar false-discovery rate across SV types (Layer et al. 2014); and (3) SMRT-read analyses also indicate that INV events are found at low frequency, because 86% of INVs are found in only one indica individual, which is significantly lower than other SV types (P < 0.05, t-test). Overall, our results suggest that polymorphic INV events are at especially low frequencies in population samples, suggesting the possibility that they are particularly deleterious.
SVs are typically found at lower population frequencies than SNPs, which affects LD. We have shown that LD between SNPs and SVs declines far more rapidly than that of SNPs alone and slightly more rapidly that of SVs alone (fig. 2E). This relationship may provide a challenge for association analyses, because it implies that causative SVs in rice will not be easily tagged by anchoring SNPs. There is remarkably little information about LD based on SVs, but thus far it seems as if SVs are usually not in high LD with SNPs across plants. For example, 20% of maize copy number variants (Chia et al. 2012), 27% of maize SVs (Sudmant et al. 2015), and ∼70% of arabidopsis MEIs cannot be anchored by nearby SNPs (Stuart et al. 2016). SVs appear to have substantial explanatory power when they are included in GWA analyses (Chia et al. 2012; Yao et al. 2015; Fuentes et al. 2019), but they may require methods that incorporate the possibility of detecting associations with SVs (Voichek and Weigel 2020).
We also examined the SFS for potential differences in the population dynamics of MEIs from different TE families, given that previous work focused only on the frequency of retrotransposons and only in cultivated accessions (Carpentier et al. 2019). We found that several element families, but particularly SINE elements, have dramatically different population frequencies than other element families. Relatively few SINEs were fixed, and most were found at low frequency. This finding does not appear to be due to more recent activity (fig. 4D) but rather to stronger selection, as implied by the low frequency of SINEs in and near genes (supplementary figs. S14 and S15, Supplementary Material online) and the relative dearth of homozygous variants (fig. 4E). This last observation supports the growing consensus that deleterious variants can accrue as heterozygotes because they are typically under recessive selection (Zhou et al. 2017; Huber et al. 2018; Zhou et al. 2019), but the mating system of rice ensures that new, heterozygous TE insertions do not remain heterozygous for long. We note that in our study retrotransposon insertions (Gypsy and Copia elements) were not found at particularly low frequencies based on two detection methods. That is, unlike Carpentier et al. (2019), we did not find that ∼50% of retrotransposon insertions are at low (<5%) population frequencies (fig. 4A and supplementary figs. S14 and S15, Supplementary Material online). We suspect that the principal difference between their study and ours is that they focused on full-length—and therefore presumably functional and recently active—elements.
SVs and Domestication
To date, the fate of SVs during domestication has been investigated in only one domesticated taxon, grapevines (Zhou et al. 2019), where SVs provided insights into regions of the genome that may have been under artificial selection and into the SV burden associated with cultivation. Grapevines are, however, distinct from rice and other annuals in that they are clonally propagated and lack evidence of a domestication bottleneck (Myles et al. 2011), a history that leads to the accumulation of recessive deleterious mutations that do not affect load (Zhou et al. 2017) and leads to increased SV numbers in the domesticate (Zhou et al. 2019). Here, we have characterized SVs in both cultivated and wild rice to begin to extend the many previous studies of rice domestication to include this novel genomic component.
Domestication bottlenecks accelerate genetic drift, which can contribute to a cost of domestication. An appropriate measure of cost is the average number of deleterious variants per genome (dg) (Moyers et al. 2018). Interestingly, dg is not expected to vary substantially before and after a demographic shift under some conditions, such as a strict genetic bottleneck with outcrossing and additive (h = 0.5) variants (Simons et al. 2014). However, it can vary substantially with deviations from these conditions (Henn et al. 2016). For example, clonal variants tend to accrue recessive deleterious variants over time, because these variants are under recessive selection and permanently held in a heterozygous state (Zhou et al. 2017). Similarly, forward simulations have shown that moderately and weakly deleterious variants accumulate under various demographic regimes (Robinson et al. 2018). Here, we have shown that the SV burden is elevated by 25–35% in our japonica and indica samples relative to rufipogon (fig. 3A). Although the estimated increase of dg undoubtedly depends on the composition of the samples under comparison, this observation is consistent with previous studies suggesting a cost of domestication in Asian rice (Lu et al. 2006; Günther and Schmid 2010; Liu et al. 2017). Nonetheless, the potential for an SV-associated cost in rice has not been demonstrated previously.
Positive selection can also be pervasive during domestication (Wright et al. 2005; Doebley et al. 2006; Hufford et al. 2012; Zhou et al. 2017). We examined the SV and SNP data for signals of positive selection throughout the genome, relying either on divergence between rufipogon or signals of selective sweeps within cultivars. For divergence, as measured by FST, only a small number of windows fell within the top 1% of peaks for both SVs and SNPs, with 12 windows for japonica–rufipogon and 26 for indica–rufipogon. Although small, the numbers are enriched relative to random expectation. None of these windows contains obvious candidate genes, at least under our examination, but the lists of genes within FST peaks may prove useful for researchers studying domestication and improvement traits in indica and japonica (supplementary tables S3 and S4, Supplementary Material online). The shared windows between SNPs and SVs also do not include any of the set of 15 domestication genes (table 2), but these genes are enriched significantly in high-ranking FST peaks when SNPs and SVs are considered separately, lending some credibility to the basic approach. Importantly, FST analyses suggest that SV calls aid the detection of divergent genomic regions, because some of the 15 genes were detected with SVs only (table 2). We thus believe that some of the regions identified by SV divergence between rufipogon and cultivated rice represent bona fide selection events.
We also searched for signals of positive selection using population-specific signals of selective sweeps. One expects a priori that SVs have lower power to detect selection, given that there are fewer of them and that they tend to be at lower standing population frequencies than SNPs (so that the relative effect of sweeps is less readily detectable relative to background frequencies). Consistent with this premise, few windows of putative selection overlapped between SNPs and SVs, although there was a slight enrichment for overlapping windows in japonica. These regions again yielded no obvious candidate genes for contributing to domestication traits, but again the list of genes may prove useful for functional analyses in rice (supplementary tables S8 and S9, Supplementary Material online).
Finally, our analysis of private SVs in cultivars versus rufipogon yielded a set of genes enriched for agronomic traits. We found hundreds of genes that differed between rufipogon and either indica or japonica (supplementary tables S10 and S11, Supplementary Material online). Many of them are implicated to contribute to abiotic stresses, such as salt tolerance, and eating quality, including starch storage and biosynthesis. Clearly, our work represents only a preliminary resolution of this question in rice; further work is necessary both to confirm the private status of genes across additional samples and to substantiate their function and potential phenotypic impacts. With few exceptions (Zhao et al. 2018; Hübner et al. 2019), the topic of gene gain and loss during domestication has been systematically understudied (Gaut et al. 2018). Our results nonetheless suggest that the genomic effects of domestication include substantial genic variation.
Materials and Methods
Data Samples and Preprocessing
We collected three kinds of data to detect SVs: whole-genome assemblies, PacBio SMRT reads, and paired-end short-read (Illumina) data. For whole-genomes, we downloaded 14 assemblies from previous publications (supplementary table S2, Supplementary Material online), including an O. longistaminata outgroup (Reuscher et al. 2018) that was used to infer the ancestral state of SVs. For the PacBio data, we gathered SMRT reads for ten accessions that were a subset of the whole-genome data set (supplementary table S2, Supplementary Material online) and included data from six indica, three japonica, and one O. longistaminata accession. For the third data set, we compiled paired-end, short-read resequencing data, requiring a minimum of 15× coverage per genome. We downloaded an initial set of genome sequences representing 393 individuals. The data for this article are all publicly available, with their sources listed in supplementary tables S12 and S13, Supplementary Material online.
Both SMRT and Illumina reads were preprocessed. SMRT reads were extracted and filtered from h5 files using Dextractor v1.0 (https://github.com/thegenemyers/DEXTRACTOR) with a minimum length 1,000 and a minimum quality score 750. Paired-end Illumina reads were trimmed to remove adapters and low-quality bases (<20) and filtered for reads <40 bp using Trimmomatic 0.36 (Bolger et al. 2014). The quality of raw and filtered reads was computed using FastQC 1.0.0 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Filtered short reads were then mapped to the reference genome japonica Nipponbare (Zhang et al. 2018) using BWA-MEM (Li and Durbin 2010). Reads with mapping qualities <10 were filtered to remove nonuniquely mapped reads using SAMtools 1.9 (Li 2011). To account for the occurrence of PCR duplicates introduced during library construction, we used MarkDuplicates in the picard-tools v1.119 (https://github.com/broadinstitute/picard) to remove reads with identical external coordinates and insert lengths. The bam files were then sorted and indexed using SAMtools for downstream analyses.
Joint Variant Calls across Population Samples
SNP Calling with Short-Read Data
We used an initial data set of 393 individuals with >15× mapped coverage that represented the five major rice subpopulations (Garris et al. 2005) (temperate japonica, tropical japonica, indica, aus, and aromatic), two wild relatives (O. rufipogon and O. nivara, which is often considered an annual form of rufipogon), and two outgroup species (O. meridionalis and O. longistaminata). We mapped these resequencing data to an updated version of the Nipponbare genome (Zhang et al. 2018), called SNPs, and then subjected the sample to clustering analyses. SNPs and short (<50 bp) indels were called for the entire data set of 393 individuals using the HaplotypeCaller in GATK v4.1.2.0. SNPs were filtered using the VariationFiltration in GATK v4.1.2.0, according to the following criteria: variant quality (QD) >2.0, quality score (QUAL) >40.0, mapping quality (MQ) >30.0, and <80% missing genotypes across all samples. More than 90% of the reads uniquely mapped to the reference genome after filtering (90.22% ± 3.79% for outgroup samples and 93.79% ± 5.02% for other samples). SNP variants were then annotated to be synonymous or nonsynonymous according to the gene annotation from MSU7 Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) (Kawahara et al. 2013) using the SnpEff v4.0 (Cingolani et al. 2012) with the structural annotation of the reference based on Maker version 2.31.8 (Holt and Yandell 2011). These SNP variants were used for population assignment and inference but not for estimates of nucleotide diversity (see below).
Based on population structure and phylogenetic analyses of SNPs from the 393 accessions, we detected 46 individuals (11.39%) that did not cluster with their reported group of origin, suggesting they were misidentified in public databases (supplementary table S12, Supplementary Material online). After their removal, the curated data set consisted of 347 accessions: 244 cultivated rice representing indica (n = 96), japonica (n = 106), aus (n = 24), and aromatic (n = 18) varieties; 97 wild rice, including O. rufipogon (n = 90) and O. nivara (n = 7) accessions; and outgroup accessions from O. meridionalis (n = 3) and O. longistaminata (n = 3) (supplementary table S13, Supplementary Material online). The mean coverage among the 347 accessions was 50×, with a range of 15× to 333× (supplementary table S13, Supplementary Material online), providing sufficient coverage for high-sensitivity SV calls (Layer et al. 2014).
SV Discovery
We used separate methods to infer SVs in the three different data sets. For genome alignment, we performed comparisons between pairs of assemblies with MUMmer v4 (Marçais et al. 2018). The minimum length of a single exact match (-l 1000) and a cluster of matches (-c 1000), and the maximum diagonal difference between two adjacent anchors in a cluster (-D 5) were set using the nucmer program (nucmer -maxmatch -noextend). Dot plots were generated to visualize chromosomal collinearity and large SVs, using mummerplot. SVs between pairs of assemblies were discovered using the show-diff program in MUMmer v4; collinear regions and SV breakpoints were shown using the show-coords program. The chromosome 6 inversion and other major SVs were verified in the IGV browser (Robinson et al. 2017) from bam files that mapped the SMRT and/or Illumina reads to both the O. longistaminata (Reuscher et al. 2018) and the Nipponbare (Zhang et al. 2018) assemblies. Genome assembly-based SV discovery was also performed using the pipeline previously described (Liao et al. 2020). Briefly, soft masked target and query genomes were first aligned using LASTZ (Harris 2007), and then processed with CHAIN/NET/NETSYNTENY tools (Kent et al. 2003) to construct the syntenic blocks. SV calling and further filtering were processed using custom perl scripts which are available at https://github.com/yiliao1022/LASTZ_SV_pipeline.
To infer SVs from SMRT reads, we mapped SMRT reads from each accession to the Nipponbare reference genome (Zhang et al. 2018) using minimap2 v2.15 (Li 2018). The population calling model of the Sniffles pipeline (Sedlazeck et al. 2018) was used to genotype SV across all ten accessions (supplementary table S2, Supplementary Material online). The SVs calls were then filtered following (Zhou et al. 2019) by removing SVs with: (1) flags “IMPRECISE” and “UNRESOLVED,” (2) length <50 bp, and (3) support by fewer than four SMRT reads.
SV calls from paired-end Illumina reads were based on two methods: DELLY2 (Rausch et al. 2012) and LUMPY 0.2.13 (Layer et al. 2014). Both programs were used to call and genotype SVs across the 347 accessions as a single sample. Technically, this means that SVs were called with ∼18,000× coverage of the reference genome. For DELLY, SV calling was performed with the recommended workflow (Rausch et al. 2012). For LUMPY, read lengths and insert sizes were extracted from bam files for each sample using SAMtools 1.9 (Li 2011), and the SVs were genotyped using SVTyper (Layer et al. 2014). Both DELLY and LUMPY SV calls were filtered following Zhou et al. (2019). SV calls from DELLY and LUMPY were merged using SURVIVOR v1.0.3 (Jeffares et al. 2017). We excluded SVs that overlapped existing TE annotations, based on the RepeatMasker version 1.332 (http://www.repeatmasker.org) output using a curated rice TE library (Ou et al. 2019). The final SV calls were filtered with the additional criteria, including length >50 bp, missing genotype <80%, and identical breakpoints across all 347 individuals.
Mobile Element Insertions
We used separate approaches to examine MEIs, because these are often large enough that they are called incorrectly by short-read SV callers. Insertion frequencies of mobile elements in population samples were detected using PoPoolationTE2 (Kofler et al. 2011) using Illumina PE reads across the 347 accessions using four steps. First, the sequences of all TEs with length >50 bp were extracted from an existing TE annotation across all major TE families of reference genome japonica Nipponbare (Zhang et al. 2018). These TE regions were then masked in the reference. The TE-merged reference was generated by merging TE sequences and the masked reference genome (Kofler et al. 2016). Second, Illumina PE reads were mapped to the TE-merged reference using BWA-MEM (Li and Durbin 2010). Third, the insertion frequencies of TEs across population samples were identified, using the recommended workflow of PoPoolationTE2 with the joint algorithm and default parameters. Finally, the MEIs were genotyped for each individual based on the number of supporting reads; an MEI or non-MEI allele was genotyped as missing when there were <4 reads at the breakpoints that support either allele. An MEI supported by <4 reads was genotyped as 0/0 (homozygous non-MEI); an MEI supported by most of the reads (<4 reads support non-MEI) was genotyped as 1/1 (homozygous MEI); and the remaining cases were genotyped as 0/1 (heterozygous MEI).
To assess the results of MEIs detected by PoPoolationTE2, we also identified MEIs across all 347 accessions using TE-locate (Platzer et al. 2012), which is reported to have high sensitivity and precision in the detection of reference TE insertions (Vendrell-Mir et al. 2019). We again based our inferences on the TE reference in gff3 format. We included all major TE families with length >50 bp, which were extracted from the TE annotation of reference genome japonica Nipponbare (Zhang et al. 2018). Using the TE reference as the input file, the insertion events of TEs were then identified using the program TE_loacate.pl. Insertion events with <4 supporting reads were marked as missing, and >5 reads were considered evidence of presence of the TE in an individual. For this analysis, insertion events were further filtered if supported by less than five individuals (1% minimum frequency) across population samples.
Population Genetic Analyses
Our variant calls resulted in filtered bam files, a vcf file for SNPs, a vcf for SVs, and MEI genotypes based on population samples that were used in downstream evolutionary genomic analyses. Only biallelic variants were used. The annotation files used in this article, along with the unfiltered vcfs are available at https://zenodo.org/deposit/3758509.
Population Structure
We used the SNP vcfs to examine population structure. These analyses were performed in ANGSD v0.929 (Korneliussen et al. 2014) using genotype likelihoods in the beagle file as an input to NGSadmix. Population structure inference was based on SNP variants with a minimal quality score of 20 and a minimal mapping quality of 30. The number of genetic clusters (K) ranged from 2 to 10, and the maximum iteration of the EM algorithm was set to 2,000. The filtered SNPs across all samples were used to construct a phylogenetic tree in the FastTree v2.1.11 program with GTR+CAT model (Price et al. 2009) with O. longistaminata and O. meridionalis used as outgroups. The assignment tables from SNP and SVs were compared by flattening the matrix and calculating the Pearson correlation coefficient.
The population structure inference for SV variations were conducted using ADMIXTURE 1.3.0 with a block relaxation algorithm (Alexander et al. 2009). The termination criterion for the algorithm was when the log-likelihood increased by <0.0001 between iterations. The binary fileset (.bed) as ADMIXTURE’s input was created from the SV vcf by PLINK 1.9.0 (Purcell et al. 2007). For downstream population genetic analyses, we only chose samples with clear classifications supported by population structure analyses.
Population Genetic Statistics
Given SNP calls on the filtered set of 347 accessions, we used ANGSD v0.929 (Korneliussen et al. 2014) to estimate genome-wide nucleotide diversity, primarily to assess whether our samples reflected the well-substantiated hierarchy of diversity within Oryza and were therefore reasonable representatives of genetic diversity. The data did indeed recapitulate the known hierarchy—that is, rufipogon had higher diversity (θw = 0.0213 ± 0.0022, π = 0.0129 ± 0.0015, n = 90) than indica (θw = 0.0100 ± 0.0017, π = 0.0094 ± 0.0018, n = 96), which was more diverse than japonica (θw = 0.0057 ± 0.0012, π = 0.0039 ± 0.0012, n = 106). Because these diversity estimates were based on genomic likelihoods, they were higher and likely more accurate (Kim et al. 2011) than previous genome-wide reports based on filtered SNPs (Huang et al. 2012).
Linkage disequilibrium decay along physical distance was measured by the squared correlation coefficients (r2) between all pairs of SNPs, SVs, and all variants (SNPs + SVs) within a physical distance of 300 kb using PopLDdecay (Zhang et al. 2019). Genome-wide genetic diversity was assessed from genotype likelihood in the ANGSD v0.929 (Korneliussen et al. 2014), based on SNP variants. The −doSaf option was used to calculate the site allele frequency likelihood at all sites, and then the −realSFS was used to obtain a maximum likelihood estimate of the unfolded SFS using the EM algorithm (Kim et al. 2011). Population genetic statistics, including Watterson’s θw and pairwise differences π, were calculated for each population group using the thetaStat program (Korneliussen et al. 2014). Genetic diversity (π) for SNPs and SVs in each group was compared using VCFtools v0.1.15 (Danecek et al. 2011).
The unfolded SFS was calculated from the allele counts for each position using three O. longistaminata and three O. meridionalis accessions as outgroups. For japonica, indica, and rufipogon, we downsampled to ten samples with the highest coverage and the least missing data to calculate the SFS for each variant type, including sSNPs that were outside outlier windows based on SweeD analyses (see below), nSNPs (Nsyn), and SVs (DEL, DUP, TRA, INV, MEI). For MEIs, we also classified them into ten families, including four retrotransposon families (Gypsy, Copia, LINE, SINE) and six DNA transposon families (CACTA, Mutator, Helitron, hAT, Harbinger, Mariner), and calculated the SFS for each family. The number of derived alleles were calculated for each type of variant using O. longistaminata and O. meridionalis as outgroups. We excluded sites with missing data at all six outgroup samples in the SFS estimation. The genetic burden was calculated under additive (2 × homozygous variants + number of heterozygous variants) (Henn et al. 2016; Zhou et al. 2017).
Selection on SVs and SNPs
SweeD v3.3.2 (Pavlidis et al. 2013) was used to detect genomic signatures of selection in the indica, japonica, and rufipogon samples, based on a sliding window size of 20 kb. The genes underlying the outlier windows were then annotated based on the MSU7 annotation (Kawahara et al. 2013). Gene Ontology (GO) analyses for these genes were run in agriGO v2.0 (Tian et al. 2017).
We used the SweeD results to define neutral sSNPs, because we assumed that sSNPs outside putative selective sweeps were neutral. The neutral sSNPs were used for calculating the SFS; the sSNP SFS was compared with the SFS of other variant types using the Kolmogorov–Smirnov test in R v3.5.1. For the SFS of individual TEs, we used the unfolded SFS to estimate DFE and α, using polyDFE v.2.0 (Tataru and Bataillon 2019). The results were presented with 95% confidence intervals obtained from the inferred discretized DFEs from 20 bootstrap data sets.
Pairwise genetic differentiation (FST) and genetic divergence (Dxy) for SNPs and SVs along chromosome between each pair of three groups, indica, japonica, and rufipogon, were estimated using VCFtools v0.1.15 (Danecek et al. 2011) and PBScan v1.0 (Hämälä and Savolainen 2019) with 20-kb fixed windows. Genes underlying the FST and Dxy outlier windows were annotated based on the MSU7 annotation (Kawahara et al. 2013). GO analyses were conducted in agriGO v2.0 (Tian et al. 2017).
To examine gene gain and loss during domestication, we identified private sites in each species, and also identified shared and fixed sites between each pair of the three groups (japonica, indica, and rufipogon). The corresponding genes were inferred based on the MSU7 annotation (Kawahara et al. 2013). GO analyses were conducted in agriGO v2.0 (Tian et al. 2017) for each category.
TE Analyses on the Nipponbare Reference
To calculate parameters such as TE insertion time and distance from gene, we focused on the Nipponbare reference and relied on its TE annotation (Zhang et al. 2018) and the gene annotation from MSU7 annotation (Kawahara et al. 2013). Given the TE annotations, a multiple alignment file was generated for each TE family using MAFFT v7.305b with FFT-NS-2 method (Katoh et al. 2002; Katoh and Standley 2013). A consensus sequence of each TE family was extracted from the multiple alignment, and the sequence divergence between each TE copy and the consensus sequence was calculated using EMBOSS 6.5.7.0 (Rice et al. 2000). The TE insert time for each TE copy was estimated based on the sequence divergence (dk) and a substitution rate 6.5 × 10−9 substitutions per site per year (Gaut et al. 1996).
Data Availability
The gene and TE annotations of the reference genome and raw SV calls are available at https://zenodo.org/deposit/3758509. The LASTZ Pipeline for SV detection is available at https://github.com/yiliao1022/lastz_sv_pipeline.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Note.—FST (i–r) refers to FST between indica and rufipogon, with additional columns for japonica–rufipogon (j–r). Cells are colored as to whether the gene is in a 1% peak (dark gray), a 10% peak (medium gray), or an extreme valley (≥90%) (light gray) based on either SNPs or SVs.
Supplementary Material
Acknowledgments
B.S.G. is supported by National Science Foundation Grant Nos 1741627 and 1655808. J.J.E. is supported by National Institutes of Health Grant No. R01GM123303-1. Y.K. was supported by the China Scholarship Council (201808360265) and the National Natural Science Foundation of China (31901222). This work was made possible, in part, through access to the Genomics High-Throughput Facility Shared Resource of the Cancer Center Support Grant CA62203 at the University of California, Irvine, and NIH shared-instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01.
References
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Coe BP, Eichler EE.. 2011. Genome structural variation discovery and genotyping. Nat Rev Genet. 12(5):363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Zhou L, Ren N-N, Xu X, Liu R, Huang L, Zheng X-M, Meng Q-L, Du Y-S, Wang M-X, et al. 2019. Parallel speciation of wild rice associated with habitat shifts. Mol Biol Evol. 36(5):875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, et al. 2007. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3(9):e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier M-C, Manfroi E, Wei F-J, Wu H-P, Lasserre E, Llauro C, Debladis E, Akakpo R, Hsing Y-I, Panaud O.. 2019. Retrotranspositional landscape of Asian rice revealed by 3000 genomes. Nat Commun. 10(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, Gardner EJ, Rodriguez OL, Guo L, Collins RL, et al. 2019. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 10(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, Doebley J, Elshire RJ, Gaut B, Geller L, Glaubitz JC, et al. 2012. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 44(7):803–807. [DOI] [PubMed] [Google Scholar]
- Choi JY, Platts AE, Fuller DQ, Hsing Y-I, Wing RA, Purugganan MD.. 2017. The rice paradox: multiple origins but single domestication in Asian rice. Mol Biol Evol. 34(4):969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Purugganan MD.. 2018. a. Multiple origin but single domestication led to Oryza sativa. G3 ( Bethesda) 8(3):797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Purugganan MD.. 2018. b. Evolutionary epigenomics of retrotransposon-mediated methylation spreading in rice. Mol Biol Evol. 35(2):365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin )6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Thornton KR.. 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol. 2:83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez CM, Gaut BS, Meca E, Scheinvar E, Montes-Hernandez S, Eguiarte LE, Tenaillon MI.. 2013. Genome size variation in wild and cultivated maize along altitudinal gradients. New Phytol. 199(1):264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD.. 2006. The molecular genetics of crop domestication. Cell 127(7):1309–1321. [DOI] [PubMed] [Google Scholar]
- Du H, Yu Y, Ma Y, Gao Q, Cao Y, Chen Z, Ma B, Qi M, Li Y, Zhao X, et al. 2017. Sequencing and de novo assembly of a near complete indica rice genome. Nat Commun. 8(1):15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M.. 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320(5883):1629–1631. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Willis JH, Vision TJ.. 2014. The standing pool of genomic structural variation in a natural population of Mimulus guttatus. Genome Biol Evol. 6(1):53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes RR, Chebotarov D, Duitama J, Smith S, De la Hoz JF, Mohiyuddin M, Wing RA, McNally KL, Tatarinova T, Grigoriev A, et al. 2019. Structural variants in 3000 rice genomes. Genome Res. 29(5):870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DQ, Sato Y-I, Castillo C, Qin L, Weisskopf AR, Kingwell-Banham EJ, Song J, Ahn S-M, van Etten J.. 2010. Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol Anthropol Sci. 2(2):115–131. [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S.. 2005. Genetic structure and diversity in Oryza sativa L. Genetics 169(3):1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT.. 1996. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci U S A. 93(19):10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Seymour DK, Liu Q, Zhou Y.. 2018. Demography and its effects on genomic variation in crop domestication. Nat Plants. 4(8):512–520. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, Stritt C, Roulin AC, Schackwitz W, Tyler L, et al. 2017. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun. 8(1):2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Zhao Z.. 2014. Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad Sci U S A. 111(17):6190–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther T, Schmid KJ.. 2010. Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor Appl Genet. 121(1):157–168. [DOI] [PubMed] [Google Scholar]
- Hämälä T, Savolainen O.. 2019. Genomic patterns of local adaptation under gene flow in Arabidopsis lyrata. Mol Biol Evol. 36(11):2557–2571. [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignmnet of genomic DNA [PhD thesis]. University Park (PA): Penn State Univ. [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci U S A. 113(4):E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Han B.. 2016. Rice domestication occurred through single origin and multiple introgressions. Nat Plants. 2:15207. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490(7421):497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CD, Durvasula A, Hancock AM, Lohmueller KE.. 2018. Gene expression drives the evolution of dominance. Nat Commun. 9(1):2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner S, Bercovich N, Todesco M, Mandel JR, Odenheimer J, Ziegler E, Lee JS, Baute GJ, Owens GL, Grassa CJ, et al. 2019. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants. 5(1):54–62. [DOI] [PubMed] [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia J-M, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, et al. 2012. Comparative population genomics of maize domestication and improvement. Nat Genet. 44(7):808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ.. 2017. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 8(1):14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D.. 2003. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 100(20):11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Lohmueller KE, Albrechtsen A, Li Y, Korneliussen T, Tian G, Grarup N, Jiang T, Andersen G, Witte D, et al. 2011. Estimation of allele frequency and association mapping using next-generation sequencing data. BMC Bioinformatics 12(1):231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H.. 2004. Retrotransposon-induced mutations in grape skin color. Science 304(5673):982–982. [DOI] [PubMed] [Google Scholar]
- Kofler R, Langmüller AM, Nouhaud P, Otte KA, Schlötterer C.. 2016. Suitability of different mapping algorithms for genome-wide polymorphism scans with Pool-Seq data. G3 ( Bethesda) 6(11):3507–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Pandey RV, Schlötterer C.. 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27(24):3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15(1):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM.. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15(6):R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T.. 2006. Rice domestication by reducing shattering. Science 311(5769):1936–1939. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Zhang X, Chakraborty M, Emerson JJ.. 2020. Topologically associating domains and their role in the evolution of genome structure and function in Drosophila. bioRxiv Available from: https://www.biorxiv.org/content/10.1101/2020.05.13.094516v1. [DOI] [PMC free article] [PubMed]
- Liao Y, Zhang X, Li B, Liu T, Chen J, Bai Z, Wang M, Shi J, Walling JG, Wing RA, et al. 2018. Comparison of Oryza sativa and Oryza brachyantha genomes reveals selection-driven gene escape from the centromeric regions. Plant Cell 30(8):1729–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhou Y, Morrell PL, Gaut BS.. 2017. Deleterious variants in Asian rice and the potential cost of domestication. Mol Biol Evol. 34(4):908–924. [DOI] [PubMed] [Google Scholar]
- Lockton S, Ross-Ibarra J, Gaut BS.. 2008. Demography and weak selection drive patterns of transposable element diversity in natural populations of Arabidopsis lyrata. Proc Natl Acad Sci U S A. 105(37):13965–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tang T, Tang H, Huang J, Shi S, Wu C-I.. 2006. The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 22(3):126–131. [DOI] [PubMed] [Google Scholar]
- Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ.. 2019. Structural variant calling: the long and the short of it. Genome Biol. 20(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A.. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 14(1):e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD.. 2007. The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 177(4):2223–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J.. 2015. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 16(4):237–251. [DOI] [PubMed] [Google Scholar]
- Moyers BT, Morrell PL, McKay JK.. 2018. Genetic costs of domestication and improvement. J Hered. 109(2):103–116. [DOI] [PubMed] [Google Scholar]
- Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, Prins B, Reynolds A, Chia J-M, Ware D, et al. 2011. Genetic structure and domestication history of the grape. Proc Natl Acad Sci U S A. 108(9):3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M, Schatz MC.. 2016. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics 32(19):3021–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, Lugo CSB, Elliott TA, Ware D, Peterson T, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Živković D, Stamatakis A, Alachiotis N.. 2013. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol. 30(9):2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer A, Nizhynska V, Long Q.. 2012. TE-locate: a tool to locate and group transposable element occurrences using paired-end next-generation sequencing data. Biology (Basel) 1(2):395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO.. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28(18):i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuscher S, Furuta T, Bessho-Uehara K, Cosi M, Jena KK, Toyoda A, Fujiyama A, Kurata N, Ashikari M.. 2018. Assembling the genome of the African wild rice Oryza longistaminata by exploiting synteny in closely related Oryza species. Commun Biol. 1(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Robinson JA, Brown C, Kim BY, Lohmueller KE, Wayne RK.. 2018. Purging of strongly deleterious mutations explains long-term persistence and absence of inbreeding depression in island foxes. Curr Biol. 28(21):3487–3494.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP.. 2017. Variant review with the Integrative Genomics Viewer (IGV). Cancer Res. 77(21):e31–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler K, Muyle A, Diez CM, Gaut GRJ, Bousios A, Stitzer MC, Seymour DK, Doebley JF, Liu Q, Gaut BS.. 2019. The genome-wide dynamics of purging during selfing in maize. Nat Plants. 5(9):980–990. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC.. 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 15(6):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M.. 2008. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 40(8):1023–1028. [DOI] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, Sella G.. 2014. The deleterious mutation load is insensitive to recent population history. Nat Genet. 46(3):220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL, et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet. 50(2):285–296. [DOI] [PubMed] [Google Scholar]
- Stuart T, Eichten SR, Cahn J, Karpievitch YV, Borevitz JO, Lister R.. 2016. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife 5:e20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz M-Y, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhou Y, Chen J, Shi J, Zhao H, Zhao H, Song W, Zhang M, Cui Y, Dong X, et al. 2018. Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat Genet. 50(9):1289–1295. [DOI] [PubMed] [Google Scholar]
- Tataru P, Bataillon T.. 2019. polyDFEv2.0: testing for invariance of the distribution of fitness effects within and across species. Bioinformatics 35(16):2868–2869. [DOI] [PubMed] [Google Scholar]
- Tattini L, D’Aurizio R, Magi A.. 2015. Detection of genomic structural variants from next-generation sequencing data. Front Bioeng Biotechnol. 3:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z.. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45(W1):W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell-Mir P, Barteri F, Merenciano M, Gonzalez J, Casacuberta JM, Castanera R.. 2019. A benchmark of transposon insertion detection tools using real data. Mob DNA. 10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voichek Y, Weigel D.. 2020. Identifying genetic variants underlying phenotypic variation in plants without complete genomes. Nat Genet. 52(5):534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vondras AM, Minio A, Blanco-Ulate B, Figueroa-Balderas R, Penn MA, Zhou Y, Seymour D, Ye Z, Liang D, Espinoza LK, et al. 2019. The genomic diversification of grapevine clones. BMC Genomics 20(1):972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Vieira FG, Crawford JE, Chu C, Nielsen R.. 2017. Asian wild rice is a hybrid swarm with extensive gene flow and feralization from domesticated rice. Genome Res. 27(6):1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. 2018. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557(7703):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS.. 2005. The effects of artificial selection on the maize genome. Science 308(5726):1310–1314. [DOI] [PubMed] [Google Scholar]
- Wright SI, Le QH, Schoen DJ, Bureau TE.. 2001. Population dynamics of an Ac-like transposable element in self- and cross-pollinating arabidopsis. Genetics 158(3):1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ.. 2006. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442(7103):705–708. [DOI] [PubMed] [Google Scholar]
- Yao W, Li G, Zhao H, Wang G, Lian X, Xie W.. 2015. Exploring the rice dispensable genome using a metagenome-like assembly strategy. Genome Biol. 16(1):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dong S-S, Xu J-Y, He W-M, Yang T-L.. 2019. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35(10):1786–1788. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liang Z, Cui X, Ji C, Li Y, Zhang P, Liu J, Riaz A, Yao P, Liu M, et al. 2018. N6-methyladenine DNA methylation in Japonica and Indica rice genomes and its association with gene expression, plant development, and stress responses. Mol Plant. 11(12):1492–1508. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, Zhan Q, Lu Y, Zhang L, Huang T, et al. 2018. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet. 50(2):278–284. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Massonnet M, Sanjak JS, Cantu D, Gaut BS.. 2017. Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc Natl Acad Sci U S A. 114(44):11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Minio A, Massonnet M, Solares E, Lv Y, Beridze T, Cantu D, Gaut BS.. 2019. The population genetics of structural variants in grapevine domestication. Nat Plants. 5(9):965–979. [DOI] [PubMed] [Google Scholar]
- Żmieńko A, Samelak A, Kozłowski P, Figlerowicz M.. 2014. Copy number polymorphism in plant genomes. Theor Appl Genet. 127(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene and TE annotations of the reference genome and raw SV calls are available at https://zenodo.org/deposit/3758509. The LASTZ Pipeline for SV detection is available at https://github.com/yiliao1022/lastz_sv_pipeline.