Abstract
All animals are capable of undergoing gametogenesis. The ability of forming haploid cells from diploid cells through meiosis and recombination appeared early in eukaryotes, whereas further gamete differentiation is mostly a metazoan signature. Morphologically, the gametogenic process presents many similarities across animal taxa, but little is known about its conservation at the molecular level. Porifera are the earliest divergent animals and therefore are an ideal phylum to understand evolution of the gametogenic toolkits. Although sponge gametogenesis is well known at the histological level, the molecular toolkits for gamete production are largely unknown. Our goal was to identify the genes and their expression levels which regulate oogenesis and spermatogenesis in five gonochoristic and oviparous species of the genus Geodia, using both RNAseq and proteomic analyses. In the early stages of both female and male gametogenesis, genes involved in germ cell fate and cell-renewal were upregulated. Then, molecular signals involved in retinoic acid pathway could trigger the meiotic processes. During later stages of oogenesis, female sponges expressed genes involved in cell growth, vitellogenesis, and extracellular matrix reassembly, which are conserved elements of oocyte maturation in Metazoa. Likewise, in spermatogenesis, genes regulating the whole meiotic cycle, chromatin compaction, and flagellum axoneme formation, that are common across Metazoa were overexpressed in the sponges. Finally, molecular signals possibly related to sperm capacitation were identified during late stages of spermatogenesis for the first time in Porifera. In conclusion, the activated molecular toolkit during gametogenesis in sponges was remarkably similar to that deployed during gametogenesis in vertebrates.
Keywords: oogenesis, spermatogenesis, evolution, Porifera, transcriptomics, proteomics
Introduction
Sex is a fundamental trait for eukaryote evolution. It allows the cellular and molecular scenario for recombination, underpinning major fitness advantages in eukaryotes (Barton and Charlesworth 1998). Evidences for sexual reproduction (i.e., meiosis, recombination, and syngamy) have been detected in choanoflagellates (Levin and King 2013) and other unicellular eukaryotes (Ramesh et al. 2005; Malik et al. 2007; Logsdon 2008), with the meiotic molecular machinery already in place (Ramesh et al. 2005; Carr et al. 2010). In sexually reproducing animals, gametogenesis is the process by which gametes—ova in females and spermatozoa in males—are produced by cell differentiation of the germ line (McCarrey 2018). Cytologically, the process of gametogenesis is very conserved across all metazoans, including the same steps in both sexes: one round of DNA replication followed by two rounds of meiosis (McCarrey 2018). One of the most important stages of gametogenesis is the first meiotic prophase where pairing of homologous chromosomes and genetic recombination happens before the completion of meiosis I (Haschek et al. 2010). In oogenesis of many animal groups, including mollusks, ascidians, and humans (Russo et al. 1998), a meiotic cell cycle arrest is a very crucial step in which growth and maturation of the oocyte occurs (Telfer and McLaughlin 2007). All the processes after this arrest progress very quickly and cannot be distinguished histologically (McCarrey 2018). In contrast, in spermatogenesis, all phases of gametogenesis are morphologically very distinct and highly conserved in all Metazoa, from sponges to humans (Bellve et al. 1977; Bonilla and Xu 2008; White-Cooper and Bausek 2010). Major processes in spermatogenesis are the decrease in cell size (cytoplasmic loss), chromatin compaction, and flagellum and acrosome formation for sperm capacitation (Auger 2018).
Gametogenesis occurs in gonadal tissues (ovaries/testes) in most animal groups, including early-branching metazoans such as Ctenophora and Cnidaria. Gonads are usually composed of a germinative cell layer with a simple follicle and surrounded by accessory cells (Eckelbarger 1993; Vargas-Ángel et al. 2006; Shikina and Chang 2016). However, Porifera, the earliest splitting metazoan lineage (Dohrmann and Wörheide 2013; Feuda et al. 2017; Zumberge et al. 2018; Laumer et al. 2019), do not have gonads and the female and male gametes, which derive from archaeocytes and choanocytes (Simpson 1984), are widespread in the internal tissue (mesohyl) sometimes enveloped by a follicle layer, with male gametes often grouped in spermatic cysts (e.g., Riesgo, Taylor, et al. 2007; Maldonado and Riesgo 2008; Koutsouveli et al. 2018). In sponges, are the molecular pathways involved in the promotion of gametogenesis different, due to the absence of gonads?
The cytological progression of gametogenesis in Porifera has been described in detail for many sponge groups (see review by Ereskovsky [2010]), highlighting great similarities to other metazoans. Sponge oogenesis consists of two major cellular stages: ameboid-like previtellogenic (PV) oocytes and mature round vitellogenic oocytes. Most of oocyte differentiation and vitellogenesis in sponges occurs during the first meiotic arrest (Maldonado and Riesgo 2008). All the different stages of spermatogenesis (spermatogonia, primary and secondary spermatocytes, spermatids, and spermatozoa) have been observed in sponges (e.g., Riesgo et al. 2008; Lanna and Klautau 2010; Riesgo 2010). Therefore, from an evolutionary point of view, the morphological conservation of oogenesis and spermatogenesis strongly suggests an ancient molecular toolkit. Common molecular markers of gametogenesis (germ cell development, oogenesis, and spermatogenesis) are present in early diverging metazoans such as Placozoa and Cnidaria (Extavour 2009; Eitel et al. 2011; Shikina and Chang 2016), proving that the molecular regulation of gametogenesis appeared very early in animal evolution. Porifera share many genes with the rest of the Metazoa (Srivastava et al. 2010; Riesgo et al. 2014), but regarding the molecular toolkits for gametogenesis, very little is known. For germline determination, the entire complement of germ line markers or the Germline Multipotency Program (GMP) was found in several lineages of sponges (Riesgo et al. 2014; Fierro-Constaín et al. 2017). GMP genes have been expressed not only in somatic multipotent cells but also in gametes of two sponge lineages, indicating that some of those genes may still play a role in the sponge gametogenesis (Leininger et al. 2014; Fierro-Constaín et al. 2017), whereas some others may participate in the development and the maintenance of primordial stem cells (Solana 2013). Interestingly, some genes involved in the oocyte maturation processes, and more precisely in the regulation of the oocyte meiosis arrest, such as mos, are considered to be absent in sponges (Extavour 2009). Using a single cell RNAseq approach in Amphimedon queenslandica, Sebé-Pedrós et al. (2018) identified cells with transcriptomic profiles concordant with spermatozoa, but they did not confirm if those cells were truly sperm. All in all, many uncertainties and gaps remain in our knowledge on the molecular toolkit involved in gametogenesis in sponges.
In the present study, our goal was to identify the molecular toolkit in Porifera during female and male gametogenesis using RNAseq and proteomics. The species of the genus Geodia are appropriate models for studying the molecular aspects associated with gametogenesis, for several reasons. They have a common reproductive strategy due to phylogenetic constrains (Liaci and Sciscioli 1969; Mercurio et al. 2007; Spetland et al. 2007; personal observations) that includes consistent gonochorism (maintaining their sex for life) and oviparity (Liaci and Sciscioli 1969, 1970; Watanabe 1978; Mercurio et al. 2007; Spetland et al. 2007). Besides their consistent reproductive strategy, we selected Geodia spp. for this study because they are dominant species, in terms of size and biomass, of deep-sea boreo-Arctic sponge grounds in the North Atlantic (Klitgaard and Tendal 2004; Cárdenas et al. 2013). These Geodia-dominated grounds provide essential ecosystem services (Maldonado et al. 2016) and due to their vulnerability to anthropogenic activities, they are now considered Vulnerable Marine Ecosystems (UNGA 2006; FAO 2009). Knowledge about the reproduction of these key North Atlantic species will therefore benefit the conservation of these Vulnerable Marine Ecosystems. In particular, we selected these five species of Geodia (order Tetractinellida, family Geodiidae) because they have developed slightly shifted reproductive cycles at the same habitat (Koutsouveli V, unpublished data). So, at time of collection, the different species were at different moments of their life cycles, and by targeting them all we could cover as many developmental stages of gametogenesis as possible. These five species are Geodia hentscheli(Cárdenas et al. 2010), Geodia phlegraei (Sollas 1880), Geodia barretti (Bowerbank 1858), Geodia macandrewii (Bowerbank 1858), and Geodia atlantica (Stephens 1915).
Results and Discussion
In our study, we observed a highly conserved molecular toolkit participating in the physiological process of gametogenesis in all studied Geodia species. Even though some cytological features of gametogenesis have become more complex with evolutionary time, the molecular regulation appears to be very similar from early-branching metazoans to humans.
Cytology of Gametogenesis in Geodia spp
For the subsequent analyses, oocytes were identified as PV, vitellogenic I (Vi_I), II (Vi_II), and III (Vi_III), based on their size and amount of yolk they contained during histological observations. Spermatic cysts were also labeled as SP_I or SP_II according to level of maturation, with SP_I to be present in early spermatogenesis (containing spermatogonia and primary spermatocytes) and SP_II to be in late spermatogenesis (containing mostly secondary spermatocytes, spermatids, and spermatozoa and also few spermatogonia and primary spermatocytes). Accordingly, the individuals of each species were grouped based on the maturation stage of their oocytes and spermatic cysts as assessed at the time of our histological analysis.
Oogenesis
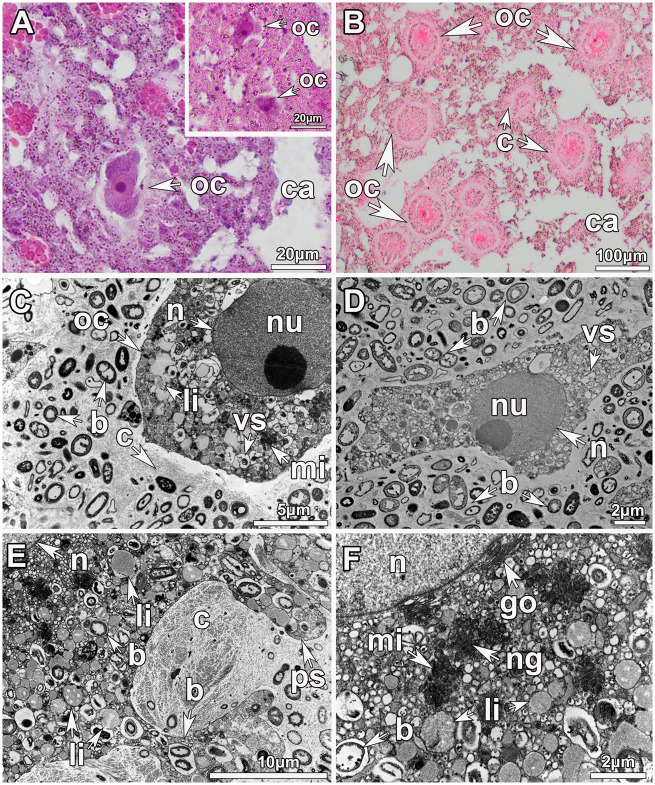
The oocytes within each individual were consistently found in the same developmental stage. PV oocytes of all Geodia spp. were smaller (18 ± 3 μm) (fig. 1A) than the most mature (Vi_III) oocytes (100 μm in largest diameter) (fig. 1B). PV oocytes (fig. 1C and D) had few small lipid droplets (fig. 1C and D), whereas Vi_II oocytes had an ooplasm full of yolk (large lipid droplets) and bacterial symbionts vertically transmitted (fig. 1E). A layer of collagen surrounded the oocyte and became thicker with development (fig. 1C and E). Among the mitochondrial clouds, an electron-dense material identified as nuage was present in Vi_II oocytes (fig. 1F).
Fig. 1.
Morphological features of oocytes in sponge tissues of Geodia species. (A) Light microscopy image showing a PV oocyte (oc) of Geodia phlegraei (≤20 μm in diameter) (insert in the upper right) and a vitellogenic stage I (Vi_I) oocyte in the tissue of Geodia barretti (∼30μm in diameter). (B) Light microscopy image of several vitellogenic oocytes of maturation stage III (Vi_III) in Geodia atlantica close to the canals (ca), ready to be released. A thick layer of collagen (c) surrounds the mature oocytes. (C) Transmission Electron Micrograph (TEM) of a PV oocyte of Geodia hentscheli. The nucleolated (nu) nucleus (n) occupies most of the oocyte. Many vesicles (vs), few lipid droplets (li) as well as mitochondrial clouds (mi) can be seen in the ooplasm. A thin layer of collagen is deposited around the oocyte. Many bacterial (b) symbionts were observed in the mesohyl, none inside the oocyte. (D) TEM image of an ameboid shape PV oocyte of G. hentscheli with a large nucleolated nucleus and the ooplasm full of vesicles but no yolk or lipid droplet. (E) TEM image of a vitellogenic oocyte (Vi_II) of Geodia macandrewii full of lipid yolk (li) and bacterial symbionts in vesicles. The oocyte has pseudopodia (ps) to acquire further bacteria and nutrients for the maternal tissue. A thick layer of collagen surrounds the oocyte. (F) Close-up of (E) of G. macadrewii. Dense accumulations of mitochondria are observed in the ooplasm, close to the nucleus, and among those the nuage (ng) granules. The Golgi apparatus (go) can be seen in the periphery of the nucleus.
Spermatogenesis
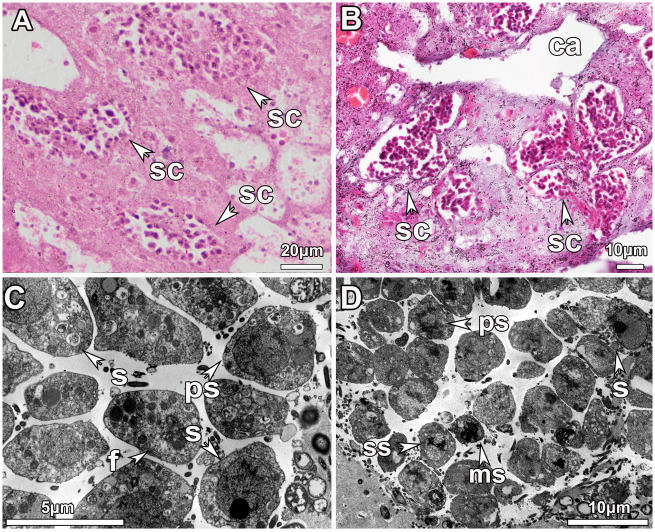
All spermatic cysts were found in the same developmental stage (SP_I or SP_II) consistently within the same individual. Immature spermatic cysts (SP_I) had consistently fewer but bigger spermatic cells (fig. 2A), whereas mature spermatic cysts had a greater amount of dividing smaller spermatic cells (fig. 2B). SP_I spermatic cysts contained exclusively spermatogonia and primary spermatocytes (fig. 2C), whereas SP_II spermatic cysts were composed by gametic cells in mostly later developmental stages; secondary spermatocytes, spermatids, and spermatozoa, but also few spermatogonia and primary spermatocytes (fig. 2D). Along spermatogenesis, a decrease in size and condensation of the chromatin within the nucleus was observed (fig. 2D). Internalization of the choanocyte flagellum occurred at the early spermatic stages (fig. 2C) which then disappeared in the later stages (fig. 2D).
Fig. 2.
Morphological features of spermatic cysts in sponge tissues of Geodia species. (A) Light microscopy image indicating spermatic cyst (sc) in developmental stage I (SP_I) in the mesohyl of Geodia hentscheli. (B) Light microscopy image depicting several spermatic cysts (sc) of developmental stage II (SP_II) accumulated around the canals (ca) in the tissue of Geodia phlegraei. (C) TEM micrograph of a SP_I spermatic cyst from G. hentscheli in which cells at the beginning of gametic development; spermatogonia (s) and primary spermatocytes (ps) were observed. Internalization of the flagellum (f) from the choanocyte occurs at the early stages. (D) TEM micrograph of a SP_II spermatic cyst from G. phlegraei with asynchronous development: spermatogonia (s) in the outer part of the cyst, and primary (ps) to secondary spermatocytes (ss) and mature spermatid/preliminary sperm (ms) in the inner part of the spermatic cyst.
As a summary, PV oocytes were found in G. hentscheli, and G. phlegraei, Vi_I oocytes in G. hentscheli, G. phlegraei and G. barretti, Vi_II in G. macandrewii and Vi_III were observed only in G. atlantica. SP_I spermatic cysts (spermatogonia and spermatocytes) were observed in G. hentscheli and G. phlegraei, and SP_II (spermatogonia, spermatocytes, spermatids, and spermatozoa) were observed in all species, except for G. hentscheli.
Transcriptomic and Proteomic Analysis
Using differential RNAseq, we studied the molecular toolkit upregulated during gametogenesis in all five Geodia species. The amount of raw and filtered reads as also the statistics and completeness metrics of the assembly can be found in supplementary table S1A and B, Supplementary Material online. Our lists of upregulated genes for oogenesis included genes upregulated when compared 1) with males and 2) with nonreproductive specimens; and upregulated genes for spermatogenesis included genes when compared 1) with females and 2) with nonreproductive specimens. In G. hentscheli, 6,107 genes were upregulated in female individuals and 10,491 in male tissues (supplementary tables S2 and S3A, Supplementary Material online). In G. phlegraei, 1,573 genes were upregulated in females and 4,604 genes in males (supplementary tables S2 and S3B, Supplementary Material online). In G. barretti, 7,625 and 9,243 genes were upregulated in female and in male tissues, respectively (supplementary tables S2 and S3C, Supplementary Material online). In G. macandrewii, 541 genes were upregulated in females and 965 genes in males (supplementary tables S2 and S3D, Supplementary Material online). Finally, in G. atlantica, 284 genes were upregulated in females when compared with specimens undergoing spermatogenesis and 107 genes were upregulated in male tissues when compared with female tissues, since we did not find any nonreproductive specimen (supplementary tables S2 and S3E, Supplementary Material online). Roughly between 12% and 69% of these differentially upregulated genes could be identified through BLAST using the Swissprot metazoan database (supplementary table S2, Supplementary Material online). Nonannotated genes could include both genes that are noncoding and others with currently unknown function, including genes that are sponge specific.
Despite studying all the five species, our discussion on oogenesis was essentially focused on the results obtained from G. phlegraei (PV), G. barretti (Vi_I), and G. macandrewii (Vi_II), whereas for spermatogenesis, we mainly discussed the results for G. hentscheli (SP_I) and G. phlegraei (SP_I and SP_II). The reasons for focusing on these species were 1) the higher amount of information obtained from their gene expression pattern and 2) the possibility to cover all the developmental stages along gametogenesis (table 1). Although G. atlantica provided an extra developmental stage for oogenesis (Vi_III), this species was not extensively discussed in our transcriptomic analysis because of its very low gene expression patterns and lack of replicates. Transcriptomic analyses on female and male gametogenesis, including all five species, are provided in supplementary file SF1, Supplementary Material online.
Table 1.
Collection Details of the Specimens Used from Each Geodia Species (G. hentscheli, G. phlegraei, G. barretti, G. macandrewii, and G. atlantica) and Used in the Transcriptomic and Proteomic Analysis.
| Species | Status | Code | Sampling Location | Date |
|---|---|---|---|---|
| G. hentscheli | Oogenesis | PV_1 | Schulz Bank | June 21, 2016 |
| Oogenesis | PV_2 | Schulz Bank | June 21, 2016 | |
| Oogenesis | PV_3 | Schulz Bank | June 21, 2016 | |
| Oogenesis | Vi_I | Schulz Bank | June 21, 2016 | |
| Spermatogenesis | SP_I_1 | Schulz Bank | July 29, 2017 | |
| Spermatogenesis | SP_I_2 | Schulz Bank | July 29, 2017 | |
| No-reproductive | NR_1 | Schulz Bank | June 21, 2016 | |
| No-reproductive | NR_2 | Schulz Bank | July 29, 2017 | |
| G. phlegraei | Oogenesis | PV | Tromsøflaket, Barents Sea | August 3, 2017 |
| Oogenesis | Vi_I | Sula reef | July 23, 2017 | |
| Spermatogenesis | SP_I_1 | Sula reef | July 23, 2017 | |
| Spermatogenesis | SP_I_2* | Tromsøflaket, Barents Sea | August 3, 2017 | |
| Spermatogenesis | SP_II_1 | Tromsøflaket, Barents Sea | August 3, 2017 | |
| Spermatogenesis | SP_II_2 | Tromsøflaket, Barents Sea | August 3, 2017 | |
| No-reproductive | NR | Sula reef | July 23, 2017 | |
| G. barretti | Oogenesis | Vi_I_1 | Sula reef | July 23, 2017 |
| Oogenesis | Vi_I_2 | Tromsøflaket, Barents Sea | August 3, 2017 | |
| Spermatogenesis | SP_II | Tromsøflaket, Barents Sea | August 3, 2017 | |
| No-reproductive | NR_1 | Kosterfjord, West of Yttre Vantenholmen | May 4, 2016 | |
| No-reproductive | NR_2 | Kosterfjord, West of Yttre Vantenholmen | May 4, 2016 | |
| No-reproductive | NR_3 | Kosterfjord, West of Yttre Vantenholmen | May 4, 2016 | |
| Only used for the assembly | Tromsøflaket, Barents Sea | August 3, 2017 | ||
| G. macandrewii | Oogenesis | Vi_II_1* | Skorpeodden, Korsfjord | September 8, 2016 |
| Oogenesis | Vi_II_2 | Sula reef | July 23, 2017 | |
| Spermatogenesis | SP_II | Sula reef | July 23, 2017 | |
| No-reproductive | NR_1 | Tromsøflaket, Barents Sea | August 3, 2017 | |
| No-reproductive | NR_2 | Tromsøflaket, Barents Sea | August 3, 2017 | |
| No-reproductive | NR_3 | Tromsøflaket, Barents Sea | August 3, 2017 | |
| G. atlantica | Oogenesis | Vi_III_1* | Skorpeodden, Korsfjord | September 8, 2016 |
| Oogenesis | Vi_III_2 | Langenuen, Korsfjord | September 9, 2016 | |
| Spermatogenesis | SP_II* | Skorpeodden, Korsfjord | September 8, 2016 | |
| Only used for the assembly | Skorpeodden, Korsfjord | September 8, 2016 | ||
| Only used for the assembly | Sula reef | July 23, 2017 | ||
Note.—The different color indicates the different reproductive status (pink: female; blue: male; gray: nonreproductive). The developmental stage of their gametogenesis is indicated. Asterisks indicate individuals that were used in the proteomic analysis.
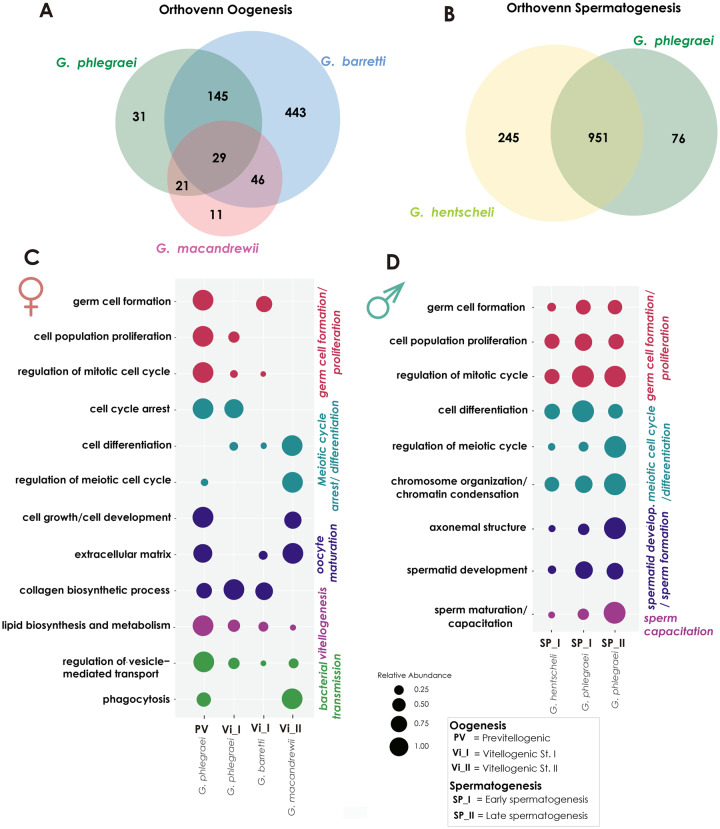
We compared the upregulated genes in female and male individuals across species in order to verify if these species shared any ortholog genes related to gametogenesis. Regarding the upregulated genes in females, we found 29 shared ortholog clusters among G. phlegraei (PV, Vi_I), G. barretti (Vi_I), and G. macandrewii (Vi_II) with Gene Ontology (GO) functions that were related to phagocytosis, extracellular region, regulation of JNK cascade, Wnt signaling pathway, and positive regulation of ATPase activity (fig. 3A and supplementary table S4A, Supplementary Material online). Among the upregulated genes in males, we found 951 shared clusters between G. hentscheli (SP_I) and G. phlegraei (SP_I and SP_II) which were related, among others, to regulation of G2/M transition of mitotic cell cycle, spindle assembly, chromatin organization, sperm axoneme assembly, and spermatid development (fig. 3B and supplementary table S4B, Supplementary Material online). Unique ortholog clusters in G. phlegraei added the GO category spermatogonial cell division (supplementary table S4C, Supplementary Material online). Our analysis suggested that we could include the selected species in our analysis as they shared some gene clusters related to gametogenesis as explained in the following sections and also extract unique information from each of them.
Fig. 3.
Shared upregulated genes during gametogenesis and GO enrichment. Venn diagram depicting the shared ortholog genes which were upregulated during (A) oogenesis across the species Geodia phlegraei, G. barretti, and G. macandrewii and (B) spermatogenesis across the species Geodia hentscheli and G. phlegraei. (C) Bubble graph depicting selected GO-enriched categories related to oogenesis, extracted from the upregulated genes among female individuals with oocytes in different developmental stages, PV (G. phlegraei); Vi_I (G. phlegraei, G. barretti) and Vi_II (G. macandrewii). (D) Bubble graph depicting selected GO-enriched categories related to spermatogenesis, extracted from the upregulated genes among male individuals with spermatic cysts in SP_I (G. hentscheli, G. phlegraei) and SP_II developmental stages (G. phlegraei). GO enrichment analysis was conducted selecting a P value ≤0.05. Each color represents a different group of gene categories according to their function. The size of the circle in each case is relative to the expression level of each gene category. Each gene category was analysed seperately .
In our proteomic analysis, 920 peptide sequences were identified for the G. macandrewii Vi_II female individual and 845 peptides for G. atlantica Vi_III female individual from which, 361 and 246 had an annotation (supplementary fig. S1A and table S5A and B, Supplementary Material online). From the annotated genes above, only 2.2–2.4% of the peptides were annotated for GO categories related to gametogenic processes (supplementary fig. S1A and table S5A and B, Supplementary Material online). In the G. phlegraei male (SP_I) sample, we isolated 1,244 peptides, and in G. atlantica male tissue (SP_II), we identified 811 peptides. From the annotated genes (530 and 256, respectively), only 1.9% and 2.1% had GO terms related to gametogenesis, respectively (supplementary fig. S1B and table S5C and D, Supplementary Material online).
Initiation of Gametogenesis
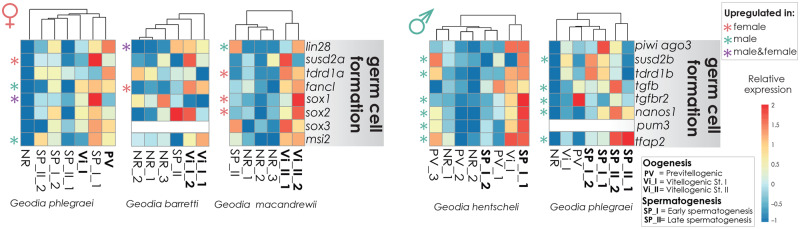
Gametogenesis in metazoans invariably involves an initial step of germline specification (e.g., Matova and Cooley 2001; White-Cooper and Bausek 2010; Saitou and Miyauchi 2016; McCarrey 2018). In our results, GO categories related to germ cell development, maintenance of pluripotency, primordial germ cell proliferation, and gamete generation in many animals were enriched either in females, males, or both (fig. 3C and D and supplementary tables S6, S7, and S8A, Supplementary Material online). Among the upregulated genes involved in primordial germ cell proliferation, we found lin28, sushi domain-containing 2 (susd2), tudor domain-containing 1 (tdrd1), bone morphogenetic protein 6 (bmp6), and E3 ubiquitin-ligase fancl, all of which were previously discussed to be important for germ cell development (fig. 4 and supplementary tables S3 and S8B, Supplementary Material online) (Mahowald 2001; Zhao et al. 2005; West et al. 2009; Eguizabal et al. 2011; Sánchez and Smitz 2012). In addition, we have identified four transcription factors of the Sox family in our studied species (supplementary fig. S2, Supplementary Material online), belonging to the groups B, E/F, and C, and three of those were upregulated during early stages of gametogenesis in our studied species (fig. 4 and supplementary tables S3 and S8B and SF1, Supplementary Material online). Sox proteins play a pivotal role in sex determination, stem cell fate, differentiation, and development (Pangas and Rajkovic 2006). Demosponges have consistently three to four Sox genes, whereas seven were identified in the calcareous sponge Sycon ciliatum (Fortunato et al. 2012), maybe because of possible genome duplication (Fierro-Constaín et al. 2017; Renard et al. 2018). In our phylogenetic reconstruction of the Sox protein family evolution, all our Geodia sequences clustered with Sox sequences from other demosponges (Amphimedon queenslandica and Ephydatia muelleri) (supplementary fig. S2, Supplementary Material online). In S. ciliatum, SoxB and SoxC genes were highly expressed in oocytes and during embryogenesis, whereas SoxF genes were expressed in adult cells (Fortunato et al. 2012). In the ctenophore Mnemiopsis leydi, six Sox genes were identified, and their expression patterns were mostly restricted to adult cells in proliferative areas (Schnitzler et al. 2014). In our case, all three Sox genes seemed to have a widespread expression in both female and male individuals, but Sox1 showed a more restricted expression pattern during early stages of gametogenesis, whereas Sox2 and 3 (SoxB and F) could be less involved in germ cell differentiation (fig. 4 and supplementary SF1, Supplementary Material online). Whether these genes are related to sex determination is possible but not conclusive from our results.
Fig. 4.
Heatmaps depicting the expression level of genes related to germ cell formation during oogenesis and spermatogenesis in individuals of the studied species. The scale for relative expression values increases from blue to red. The asterisks indicate the upregulated genes, pink for upregulated genes found in females, green asterisk for upregulated genes found in males, and purple in both female and male specimens.
Germ cell granules (nuage), which flag the germ cell nature in most organisms, have been previously described in sperm and oocytes of sponges (Lanna and Klautau 2010; Fierro-Constaín et al. 2017; Gonobobleva and Efremova 2017, Riesgo et al. 2018). They have also been discovered in archaeocytes (Isaeva and Akhmadieva 2011), reinforcing the idea that some stem cells in the adult sponge could potentially be predetermined for the germline. Here, we observed potential nuage in oocytes of G. macandrewii (fig. 1F). Many proteins are traditionally associated with nuage structures in animals, including members of the Vasa, Musashi, Argonaute/Piwi, TGF-beta receptors, Nanos, and Pumilio protein families, all of which have a role in the maintenance of the germline (Kobayashi et al. 1996; Cox et al. 1998; Shivdasani and Ingham 2003; Siddall et al. 2006; Seto et al. 2007; Lolicato et al. 2008; Funayama et al. 2010). We found musashi homolog 2 (msi2), piwi-ago3, transforming growth factor beta-1 (tgfb1), TGF-beta receptor (tgfbr1), nanos 1, and pumilio (pum3) and transcription factor AP2 (tfap2) upregulated during gametogenesis in our studied species, especially during the early stages of spermatogenesis in males (fig. 4 and supplementary tables S3 and S8B, Supplementary Material online). Similarly, most of the abovementioned genes were also expressed in the adult multipotent cells and gametes of the calcareous sponge S. ciliatum (Leininger et al. 2014) and the homoscleromorph sponge Oscarella lobularis (Fierro-Constaín et al. 2017), indicating a potential role of those genes in transdifferentiation of somatic stem cells into germ cells in three out of the four Porifera classes. TFAP2 is fundamental in the cnidarian Hydractinia symbiolongicarpus to induce germ cell formation from adult stem cells (DuBuc et al. 2020). These indicate that some of the transcription factors and regulatory genes behind animal germ cell development, maintenance, and self-renewal during gametogenesis appeared very early in evolution and could be functioning in similar ways in Porifera and other Metazoa (e.g., White-Cooper and Bausek 2010).
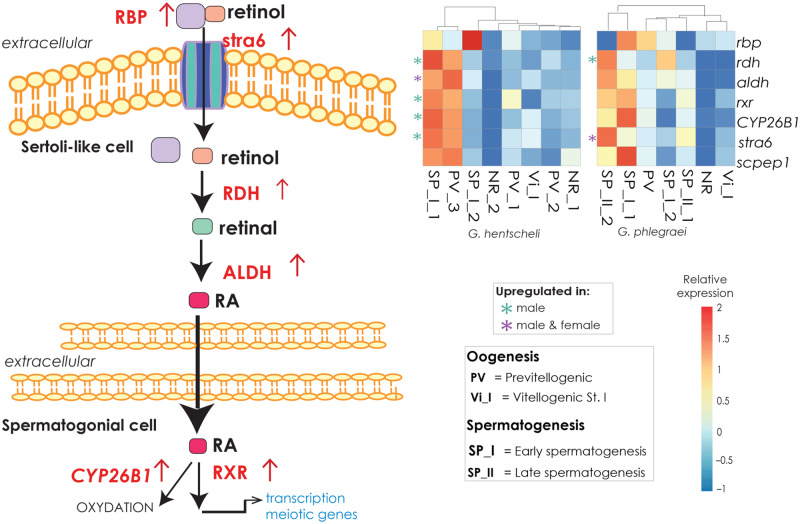
When the expression of regulators maintaining an undifferentiated state is masked by the expression of regulators in favor of differentiation, a germ cell stops dividing mitotically and expresses genes triggering meiosis for further gamete development. In vertebrates and also some invertebrates, meiosis is stimulated by retinoic acid (RA) in both female and male gametogenesis (fig. 5). In male gonads, retinol is transported via retinol binding protein to the stimulated by retinoic acid (STRA) receptor to the Sertoli cell (Berry et al. 2013; Evans et al. 2014). RA synthesis occurs with the help of retinol dehydrogenase (RDH) and aldehyde dehydrogenase (ALDH) (Tong et al. 2013; Arnold et al. 2015). Once synthesized, RA is recognized by its receptors, RAR and RXR in germ cells and stimulates meiosis (Mark et al. 2015; Griswold 2016). The degradation of RA by the enzyme CYP26B1 in the Sertoli or germ cells prevents premature gamete development (Koubova et al. 2006; Feng et al. 2014). In mice, oogenesis is also triggered by STRA receptors, and the mechanism is highly similar to that occurring during spermatogenesis (Anderson et al. 2008). A similar control mechanism could have been already present in the last common ancestor of sponges and the rest of metazoans, since almost all the genes regulating RA synthesis mentioned above were upregulated mainly in males of G. hentscheli, and some of them in G. phlegraei (rdh, aldh, stra6, and rxr) (fig. 5 and supplementary tables S3A and B and S8B, Supplementary Material online). The aldh gene together with the gene Retinoid-inducible serine carboxypeptidase (scpep1), which is also part of the RA biosynthetic pathway, was translated into peptides (supplementary table S5C and D, Supplementary Material online). Regarding the female individuals, although there was a tendency of higher expression for some of these genes in individuals with PV oocytes, only two of those (aldh and stra6) were overexpressed in PV females (fig. 5 and supplementary table S3A and B, Supplementary Material online). Previous studies observed the presence of RA in sponges and its role in morphogenetic events and gemmulation, cell differentiation, and spiculogenesis (Biesalski et al. 1992; Imsiecke et al. 1994; Nikko et al. 2001; Wiens et al. 2003; Müller et al. 2011). Similarly, in our case, RA could induce either the differentiation of choanocytes to spermatogonia (germ cells) and/or the differentiation of spermatogonia to primary gametocytes and initiation of meiosis. Overall, RA pathway activation could be a candidate pathway for the exit of the cells from germ cell fate and initiation of meiosis also in Porifera (fig. 7).
Fig. 5.
The RA pathway. (A) Schematic representation of the RA pathway (modified from Griswold [2016]), which permits a balance between germ cell maintenance and activation of meiosis. Red arrows indicate upregulation of genes in our data. (B) Heatmap of depicting the expression level of the main genes in the RA pathway mainly focused on individuals with gametes in early stages among the studied species (Geodia hentscheli and G. phlegraei). The scale for relative expression values increases from blue to red. The asterisks indicate the upregulated genes in female and or male specimens; the color green was used for upregulated genes in male and purple when found in both female and male individuals. There was no gene exlusively upregulated in female individuals.
Fig. 7.
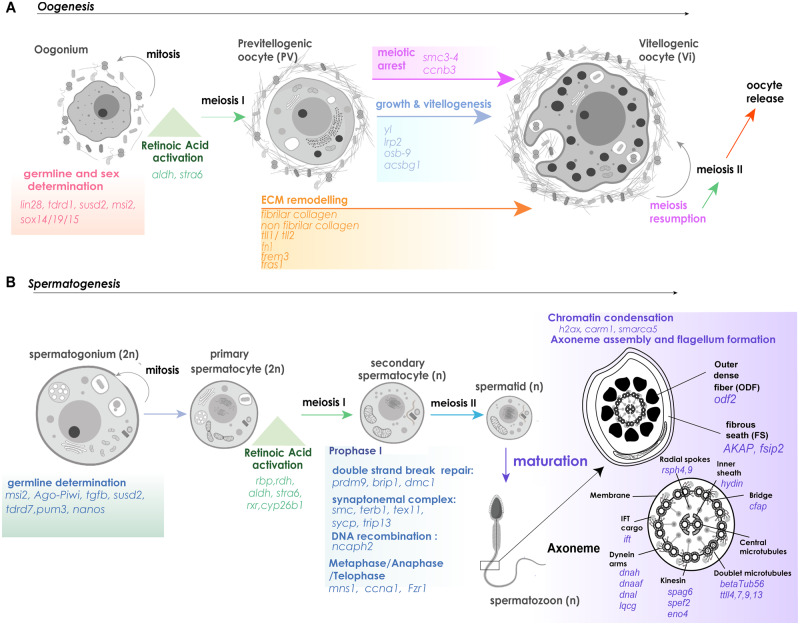
A schematic representation of the molecular toolkits recruited and physiological changes during the progress of (A) oogenesis and (B) spermatogenesis in Geodia spp., based on our histological and transcriptomic results and the theoretical progress of gametogenesis in other Metazoa. The main overexpressed regulatory pathways and genes involved in each developmental stage and transition phase are reported.
Oogenesis
Meiosis and Oocyte Maturation
The oogenesis in Geodia spp. was synchronous within individuals (fig. 1B), which means that once oogonia reached the point of cell cycle arrest and meiosis regulation, there was no further germ cell proliferation, and oogonia entered the phase of differentiation and maturation. Indeed, GO categories related to germline regulation were mainly present at the PV stages in oogenesis, indicating a role at the beginning of gamete formation, whereas those of meiosis and regulation of cell cycle arrest were enriched in later vitellogenic stages (fig. 3C and supplementary tables S6 and S8A, Supplementary Material online). Similarly, in the GO-enriched categories of our proteomic data, “regulation of meiotic nuclear division” and “establishment of meiotic spindle localization,” were enriched in later vitellogenic stages (Vi_II and Vi_III) (supplementary table S9A and B, Supplementary Material online).
The exact expression timing of the molecular complements for meiosis is largely unknown in invertebrates (Kishimoto 2018). There are two cell cycle arrests during meiosis in animals: one that occurs in prophase I (in oogenesis) and is regulated by the expression of kinase cyclin B-associated Cdk1 (cyclin B-Cdk1), and a second arrest that occurs at variable moments across invertebrates but is invariably resumed by the interactions between oocytes and sperm during fertilization (Kishimoto 2018). Genes involved in chromosome stability during meiotic cell cycle arrest, such as structural maintenance of chromosomes protein (smc) (Bickel et al. 2010) and meiosis-specific nuclear structural protein 1 (mns1), were upregulated in females with PV and Vi_I stages (fig. 6A and supplementary tables S3 and S8B, Supplementary Material online). Furthermore, classic genes regulating meiotic divisions were found differentially expressed in females of Vi_II stage (fig. 6A and supplementary tables S3 and S8B, Supplementary Material online), including several G2 mitotic-specific cyclin B genes (ccnb3), which are subunits of kinase cyclin B-associated proteins, important to facilitate progression through meiotic divisions in many organisms (Voronina et al. 2003; Dheilly et al. 2012; Peng et al. 2017; Kishimoto 2018). Mos genes, which are present from placozoans to mammals (Extavour 2009) and have a role in meiotic cell arrest in bilaterians and cnidaria (Amiel et al. 2009), were not found in our study, which corroborates that mos originated after sponges diverged from the metazoan stem (Extavour 2009). This suggests that cell cycle arrest is controlled by other genes in sponges.
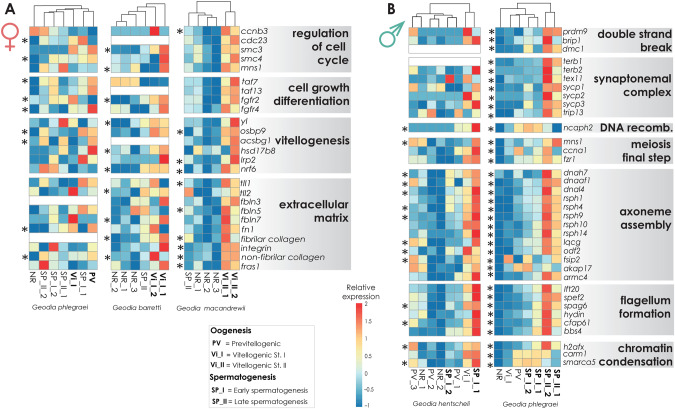
Fig. 6.
(A) Heatmaps depicting the expression level of selected genes related to oogenesis, across individuals with oocytes in different developmental stages: PV, V_I, and V_II (in Geodia phlegraei, G. barretti, and G. macandrewii, respectively). (B) Heatmaps depicting the expression level of selected genes related to spermatogenesis in individuals with spermatid cysts in different developmental stages: SP_I (Geodia hentscheli and G. phlegraei) and SP_II (G. phlegraei). The genes are separated into few big categories according to their function. The scale for relative expression values increases from blue to red. The asterisks indicate the differentially expressed genes in female and male specimens, respectively.
Oocyte maturation is achieved in many animals during cell cycle arrest by the activation of several pathways (i.e., PI3K-AKT, FGF, and Mos-MAPK pathways) and transcription of growth factors (Thisse B and Thisse C 2005; Matus et al. 2007; Sugiura et al. 2007; Ornitz and Itoh 2015; Kishimoto 2018). In our study, females overexpressed genes encoding for transcription of growth factors important during oocyte maturation (Falender et al. 2005; Pangas and Rajkovic 2006; Voronina et al. 2007; Markholt et al. 2012; McGinnis et al. 2013), including transcription initiation factor TFIID subunit gene (taf) and fibroblast growth factor receptor (fgfr), which was also found in the proteomic data (fig. 6A and supplementary tables S3B and S5B, Supplementary Material online). So, in our data, we see signals for meiotic regulation and also meiotic arrest and maturation across all female samples (figs. 3C, 6A, and 7A and supplementary table S8, Supplementary Material online).
Oocyte Growth and Vitellogenesis
Oocyte growth involves a large increase in the volume and many shape modifications, but most importantly during this phase, yolk is accumulated in the oocyte to provide the necessary nutrients for the embryo (Shikina et al. 2013; Reading et al. 2018). All sponges have lecithotrophic larvae, and therefore they entirely rely on the yolk that the oocyte (and then the embryo) contains to develop into a juvenile during the free-swimming stage and early settlement (Maldonado 2006). During the vitellogenesis of Geodia spp., nutrients mainly of lipidic origin are produced and packed by the Golgi apparatus within the ooplasm (fig. 1E and F). Although most of the yolk accumulation in the oocytes was observed at the stages Vi_II and Vi_III in our histological analysis (fig. 1D), GO categories for vitellogenesis started to be enriched at PV stages up to the vitellogenic stages I and II (fig. 3C and supplementary tables S6 and S8A, Supplementary Material online). Indeed, we identified many upregulated genes related to yolk formation in the female individuals which contained vitellogenic oocytes (figs. 6A and 7 and supplementary tables S3 and S8B, Supplementary Material online). Among the most important genes, we found the putative vitellogenin receptor (yl), oxysterol binding protein (obp9), and long-chain fatty acid ligases (acsbg1) (fig. 6A and supplementary tables S3 and S8B, Supplementary Material online). In vertebrates, vitellogenin, a main yolk component in all metazoans, is produced in the liver and is induced by the hormonal control of estradiol 17 (Polzonetti-Magni et al. 2004). Sponges do not have organs and hormonal production is poorly known, but homologs of vitellogenin proteins have been previously characterized in sponges (Riesgo et al. 2014). Furthermore, upregulation of 17-beta-hydroxysteroid dehydrogenase genes (hsd17b8 and hsd17b4) could indicate an active role of those genes in fatty acid metabolism and/or in steroid hormone metabolism during oogenesis in sponges (Mindnich et al. 2004). Another vitellogenin receptor, whose expression level is high in vitellogenic oocytes of teleost fish (Reading et al. 2014), the low-density liporeceptor-related 2 gene (lrp2), was upregulated in our analysis in the vitellogenic oocytes (fig. 6A). HSD17B8 and LRP2, and also other genes related to lipid transport (e.g., nrf-6), are involved in fatty acid biosynthesis during oocyte maturation of the crab Eriocheir sinensis (Chen et al. 2019). Although we cannot speculate on the types of lipids used for yolk formation in our study species, several genes involved in the metabolism of long-chain fatty acids and phospholipids, including phosphatidylcholines, were upregulated during the vitellogenic phase of the female gametes (supplementary table S3, Supplementary Material online). Indeed, fatty acids and phospholipids are abundant in Geodia spp. (Müller et al. 1987; Carballeira and Rodriguez 1991; Djerassi and Lam 1991; Genin et al. 2008). In our proteomic results, we found enriched GO categories, similar to those of transcriptomic data, related to phospholipid, glycolipid, sphingolipid, and glycosphingolipid metabolism in females with vitellogenic oocytes but not in male individuals (supplementary table S9A and B, Supplementary Material online), so this lipid diversity could be possibly linked to yolk formation, although other processes occurring in the female sponge tissue cannot be ruled out.
The role of the extracellular matrix (ECM) in cell growth, differentiation, migration, and maturation of female germ cells is well known (Hay 1991; Rodgers et al. 2003; Guzmán et al. 2014; Viana et al. 2018). Our data support the idea of an indispensable role of the ECM organization molecular machinery in the oocyte development from sponges to late branching metazoans (Morris 1993). In fact, our histological sections showed that gametic cells continuously changed shape, interacting with the surrounding mesohyl, and becoming massively enveloped by a collagenous sheath that increased in size as oogenesis progressed (fig. 1C–F). In all developmental stages, the GO categories of structural morphogenesis, ECM, and collagen production were largely enriched in our RNAseq data, indicating the importance of ECM reorganization during the whole process of oogenesis (figs. 3C, 6A, and 7A and supplementary tables S6 and S8A, Supplementary Material online). In addition, the GO categories “positive regulation of extracellular matrix disassembly” and “positive regulation of extracellular matrix organization” were enriched in the proteomic data set of both female individuals (supplementary fig. S1 and table S9A and B, Supplementary Material online). Among the upregulated genes related to the ECM assembly and reorganization, we found tolloid (tll1/tll2), fibulin (fbln5/fbln7), and fibronectin (fn1), which were significantly overexpressed in Vi_I and Vi_II females (fig. 6A and supplementary tables S3C and D and S8B, Supplementary Material online). These genes have been found to participate in cell growth and maturation in oogenesis of vertebrates (Asem et al. 1992). The most charismatic component of ECM across animals is collagen.
In our female sponges, we found overexpressed genes coding for both fibrillar and nonfibrillar collagens (fig. 6A and supplementary fig. S3 and tables S3 and S8B, Supplementary Material online). Overexpressed genes for fibrillar collagen were found to be from the same group as type I, V, and XI collagens of vertebrates (supplementary fig. S3, Supplementary Material online), but more closely related to collagen type V/XI than I–III, forming a clade with fibrillar collagens of cnidarians (Exposito et al. 1990, 2008, 2010). For nonfibrillar collagens, our sequences for spongin short-chain collagen in Geodia spp. clustered with those of other sponges (supplementary fig. S3, Supplementary Material online), in a separate clade, related to type IV collagens of vertebrates and invertebrates, including sponges (supplementary fig. S3, Supplementary Material online; Exposito et al. 1991; Aouacheria et al. 2006). Genes coding for both collagen families were overexpressed in the sponge E. muelleri during early development of asexual propagules (gemmules) (Exposito et al. 1990, 1991). Fibrillar collagen is the main component of the ECM and in vertebrates fibrillar collagen of types V and XI, together with fibronectins and integrins, initiates the collagen fibrillogenesis in ECM (Kadler et al. 2008). Although the nonfibrillar spongin short-chain collagen does not have an identified function yet (Aouacheria et al. 2006), it could be another component of ECM in sponges, or it could serve for the attachment of the propagules to the substrate (Exposito et al. 2002).
Finally, vesicles with bacterial symbionts were transported from the mesohyl and phagocytized but not digested by the oocyte (fig. 1E). These are the vertically transmitted bacterial symbionts, a transmission previously observed in Geodia cydonium (Sciscioli et al. 1994). GO categories of vesicle transport, phagocytosis, endocytosis, and bacteria recognition were enriched at transcriptomic and translational level (fig. 6A and supplementary fig. S1 and tables S6, S8A, and S9, Supplementary Material online). This vertical transmission is fundamental during oogenesis in many sponge species, reassuring inheritance of the microbial symbionts to the next generation (e.g., Taylor et al. 2007) and offering a possible advantage during juvenile settlement.
According to our data, oogenesis in Porifera develops within relatively few physiological phases, in which oogonia enter meiosis and meiotic cell cycle arrest, where maturation, growth, and vitellogenesis occur. In parallel, variations in structure morphogenesis of the oocytes take place to achieve round shape (fig. 7A). These are also the basic stages of oogenesis in other Metazoa (e.g., Telfer and McLaughlin 2007; Woodruff et al. 2018).
Spermatogenesis
Early Stages of Spermatogenesis
During spermatogenesis, spermatogonia (male germ cells) become primary spermatocytes in which the first meiosis occurs, bringing substantial structural and physiological changes to the gamete cells. From primary to secondary spermatocytes, the number of chromosomes is reduced in half (2n to n) through the first round of meiosis, and then chromatids are separated during the second meiosis to produce spermatids. Then, spermatids experience a massive reduction in size before maturing into spermatozoa. In our study, we identified GO-enriched categories related to cell differentiation, meiotic cycle, and chromatin condensation in male individuals (fig. 3D and supplementary tables S7 and S8A, Supplementary Material online). Although the GO categories related to germ cell formation and proliferation, including “regulation of mitotic cycle,” were similarly enriched in both developmental stages (SP_I and SP_II) (fig. 3D), “regulation of meiotic cycle” was more abundant in individuals with SP_II cysts (fig. 3D). This is something expected from our histological analysis, as males in SP_I stage had spermatic cysts at the beginning of their development, including mainly spermatogonia and primary spermatocytes, whereas in individuals with more mature spermatic cysts, within SP_II spermatic cysts there was germ cell development in parallel with several other stages of sperm production.
During prophase I, homologous chromosomes get closer, the DNA double strands break, the synaptonemal complex is formed, and chromosome synapsis occurs (Haschek et al. 2010). In the pachytene phase, DNA recombination happens (allowed by the synaptonemal complex, a protein structure facilitating crossover of DNA strands), and finally during the diplotene/diakinesis phase, the synaptonemal complex disassembles, and the homologous chromosomes uncoil (Haschek et al. 2010). In particular, we found upregulated genes related to double strand breaks (e.g., histone–lysine N-methyltransferase PRDM9 [prdm9], fanconi anemia group J homolog [brip1], and meiotic recombination DMC1 LIM15 [dmc1]), formation of the synaptonemal complex (e.g., telomere repeats-binding bouquet formation 1-2 [terb1-2], testis-expressed protein 11 [tex11], synaptonemal complex1-3 [sycp1-3], and pachytene checkpoint 2 homolog [trip13]), DNA recombination (e.g., condensin-2 complex subunit H2 [ncaph2]), and overall meiotic process (e.g., meiosis-specific nuclear structural 1, mns1; cyclin-A1 [ccna1]; fizzy-related homolog [fzr1]) in males, mainly with SP_II spermatic cysts (figs. 6B and 7 and supplementary tables S3 and S8B, Supplementary Material online). Overall, most of the genes from the classic set of meiotic genes conserved in eukaryotes (Ramesh et al. 2005; Schurko and Logsdon 2008) were upregulated in males of G. hentscheli and G. phlegraei (msh2,3,4,5,6; hop1,2, mlh1,3; rad50,51; and pms2) (supplementary tables S3A and B and S8B, Supplementary Material online). This set of genes has been also identified in the genome of the protist Giardia and of the choanoflagellate Monosiga ovata (Carr et al. 2010) suggesting meiosis. In sponges, spermatogenesis involves two rounds of meiosis as in the rest of metazoans, and we hypothesize that this set of genes function in meiosis as in other eukaryotes. In addition, the protein tubulin beta-1 chain (BETATUB56D), which plays a role in centriole formation, myosin-9 (MYH9), which participates in spindle formation and positioning in meiotic cycle (Li and Yang 2016), and the inactive peptidyl-prolyl cis-trans isomerase (FKBP6), which allows the chromosome integrity and pairing during meiosis (Crackower et al. 2003) were isolated in the proteomic analysis (supplementary table S5, Supplementary Material online). From the above, we could say that molecular regulation of meiotic cell cycle seemed to be very conserved across metazoans.
Late Stages of Spermatogenesis
In most animals, during the transition from spermatid to spermatozoa, the axoneme of the flagellum is formed. This step is crucial for the maturation, capacitation, and mobility of spermatozoa (Inaba 2011). Although some sponges (e.g., Aplysilla rosea, Halichondria (Halichondria) panicea, and Corticium candelabrum) retain the flagellum during the transition from choanocytes to spermatogonia (Tuzet et al. 1970; Barthel and Detmer 1990; Bergquist 1996; Riesgo, Maldonado, et al. 2007), the most common pattern in sponges is that choanocytes get rid of their flagellum to become spermatogonia and undergo multiple mitoses, and in the final stages of spermatogenesis, spermatids make a flagellum de novo (Maldonado and Riesgo 2008; Ereskovsky 2010). In Geodia spp., the flagellum seems to be resorbed in spermatogonia and rebuilt at later stages (fig. 2C and D). The axoneme assembly is a well-conserved process across animal evolution. The most typical structure is the microtubule organization of nine outer doublet microtubules and two central singlet microtubules (9 + 2) in most eukaryotes (Manton 1953; Inaba 2003). Tubulins and particularly β-tubulins are structural proteins in the axonemal motif (Nielsen et al. 2001; White-Cooper et al. 2009) and the bending of the flagellum is regulated by the axonemal dyneins and the radial spokes, which are both attached to the doublet of microtubule (Porter and Johnson 1989; Smith and Yang 2004; Roberts et al. 2013). We found the complete axonemal toolkit upregulated in the male tissues of our Geodia spp. (figs. 3D, 6B, and 7 and supplementary tables S3, S7, and S8B, Supplementary Material online), including tubulin beta chains, the whole functional complement of dyneins (dynein regulatory complex protein [Iqcg], dynein assembly factor axonemal [dnaaf], dynein heavy [dnah], dynein light [dnal], and radial spoke head [rsph]) (figs. 6B and 7 and supplementary tables S3 and S8, Supplementary Material online). Other genes coding for proteins of the central apparatus including tektin, hydin, and sperm-associated antigen 6, spag6 which participate in axonemal assembly and flagellum motility (Inaba 2011) were also overexpressed in male individuals of our study (figs. 6B and 7 and supplementary tables S3 and S8B, Supplementary Material online).
The main difference between the somatic flagella observed in animal cells and the sperm flagellum is the presence of Outer Dense Fibers (ODFs) and fibrous sheath (FS) (Kierszenbaum 2001; Inaba 2011), which are accessory structures that surround the axoneme to enhance the structural maintenance and motility of the flagellum (Fawcett 1975; Si and Okuno 1993). Although these molecules have been previously detected in sperm flagella of ascidians (Konno et al. 2010), echinoderms, and platyhelminths (White-Cooper et al. 2009), ODF genes may also have a role in centrosome function in other somatic cells (Ishikawa et al. 2005). We identified upregulated genes coding for ODF and FS in males with SP_I and SP_II spermatic cysts (fig. 6B and supplementary tables S3 and S8, Supplementary Material online), so we cannot prove that these genes participate only in the structure of the sperm flagellum, the production of centrosomes during meiosis (Inaba 2011), or the choanocyte flagellum. Although we did not recover any of the above genes in our proteomic analysis, we found other proteins making up enrichment of the GO categories of axonemal structure, which are related to axoneme formation (supplementary table S8, Supplementary Material online). Our findings of the complete axonemal toolkit activated during spermatogenesis, suggest that Geodia spp. generate their flagellum de novo in male gametes and agree with the conserved morphology and movement reported for the sperm flagellum in Porifera (e.g., Reiswig 1983). They are also in concordance with the description of the molecular machinery behind the axonemal structure of the mammalian sperm allowing the formation of competent spermatozoa.
Another important process occurring during spermatogenesis is further DNA condensation. In our histological study, we observed chromatin condensation along the progress of spermatogenesis (fig. 2D), which is a widespread phenomenon across Porifera (e.g., Barthel and Detmer 1990; Riesgo et al. 2008; Lanna and Klautau 2010) and all the other metazoans (Haschek et al. 2010). Specialized histones are involved in this process, particularly H2AX, which replaces the regular histone H2A, and which has a higher affinity for DNA because of its higher arginine to lysine content (Fernandez-Capetillo et al. 2003). The overexpression of h2ax together with the Histone–arginine methyltransferase gene (carm1) in males (figs. 3D and 6B and supplementary tables S3, S7, and S8B, Supplementary Material online) suggests the presence of this high-DNA affinity histones in sponges and demonstrates a conserved mechanism of chromatin condensation across metazoans.
Sperm Capacitation
Sperm capacitation events, from marine invertebrates to mammals, include participation of ion channels (Ca+2 and Na+), increase of the pH, and hyperpolarization of the sperm membrane. The increased intracellular concentration of Ca+2 and cyclic adenosine monophosphate (cAMP), through activation of soluble adenylyl cyclase, and further activation of protein kinase A (PKA) and tyrosine phosphorylation of downstream sperm proteins play a fundamental role in the capacitation events across eukaryotes (fig. 8) (Morisawa and Yoshida 2005; Visconti 2009; Yoshida and Yoshida 2011; Ickowicz et al. 2012). In echinoderms, membrane hyperpolarization is caused by an increase of intracellular cGMP concentration through the activation of GC receptor, and the further efflux of K+ through cGMP/K channels. In ascidians, it is the Ca+2 influx through a Ca+2 channel (i.e., low voltage calcium channel), and further activation of calmodulin (CaM)-dependent pathway, which provokes membrane hyperpolarization and downstream motility events (fig. 8) (Yoshida et al. 1993, 1994; Izumi et al. 1999; Nomura et al. 2004).
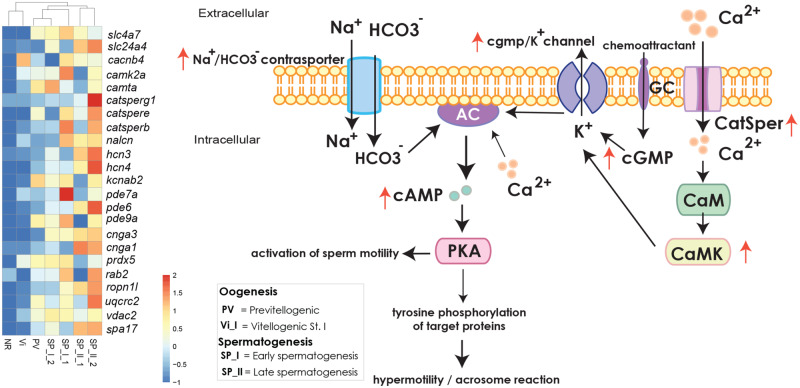
Fig. 8.
Illustration of Ca+2 driven regulation cascades related to sperm motility and capacitation. Bicarbonate, HCO3− is transported via a Na+/HCO3− cotransporter and Ca+2 via Ca+2 channels including the sperm-specific Ca+2 channel, CatSper. Those activate adenylyl cyclase (AC) which increases the levels of cAMP and leads to PKA activation. CaM-dependent increase of Ca+2 as also activation of guanylyl cyclase receptor (GC) and further increase of cGMP into the cell also induce activation of AC and final activation of PKA. Activation of PKA causes flagellar movement. Further phosphorylation of PKA substrates leads to tyrosine phosphorylation and activation of all the final sperm capacitation steps (from sperm motility to axonemal reaction before fertilization) in animals. The illustration is a modified version according to Morisawa and Yoshida (2005), Ickowicz et al. (2012), and Rahman et al. (2017). The red arrows indicate the overexpression of the relevant genes in male specimens of the species Geodia phlegraei as it was the only one with several individuals containing SP_II spermatic cysts. The graph is accompanied by the expression level of the relevant genes in all the studied specimens of G. phlegraei and presented through a heatmap. All the genes indicated were upregulated in males of G. phlegraei. The expression level increases from blue to red color.
In sponges, sperm capacitation has never been studied. However, we expect a basic mechanism of sperm capacitation, similar to that mentioned above, as a GO-enriched category related to sperm capacitation (fig. 3D and supplementary tables S7 and S8A, Supplementary Material online) and genes coding for enzymes and ion channels, part of the regulation cascades of sperm motility, were upregulated in male sponges of our study (G. phlegraei). Various types of calcium channels (voltage-dependent L-type calcium channel [cacnb4], cation channels of sperm relative genes [catsperg, catspere, catsperb]), sodium and potassium channels (sodium hyperpolarization-activated cyclic nucleotide-gated channel [hcn3, hcn4], voltage-gated potassium channel [kbcab2]), and cAMP- and cGMP-related channels (cyclic nucleotide-gated cation channels [cng1,3]) were overexpressed. Several enzymes involved in the metabolism of both cAMP and cGMP were also upregulated, for example, high affinity cAMP-specific 3, 5-cyclic phosphodiesterase (pde7a), specific 3, 5-cyclic phosphodiesterase (pde6), and high affinity cGMP-specific 3, 5-cyclic phosphodiesterase (pde9a) (fig. 8 and supplementary tables S3B and S8B, Supplementary Material online). Finally, genes related to CaM-dependent pathway, such as calcium calmodulin-dependent kinase (camk2a) and Calmodulin-binding transcription activator (camta1), were upregulated in males, indicating that this pathway could also participate in sponge sperm motility. It is definitely difficult to prove, with the current analysis, that all these genes have a role in sperm capacitation in sponges as they have general functions and they could participate in many other physiological processes. However, compared with the marine invertebrates mentioned above and also other animals and plants, in which sperm are immotile or move very slowly inside the male tract (Wolters-Everhardt et al. 1986; Jonge and Barratt 2006) acquiring high motility when induced by female chemoattractants in their surrounding (e.g., sea water) or in the female reproductive tract (Austin 1952; Gervasi and Visconti 2016), the sperm of sponges move very rapidly from the very moment of their release (Hoppe and Reichert 1987; H. Reiswig, personal communication). This suggests that capacitation in terms of enhanced motility of sperm might occur before their release into the seawater and encounter with the eggs. Indeed, presence of upregulated genes in mature sperm (G. phlegraei, Sp_II) coding for proteins, enriched in ejaculated and capacitated sperm of vertebrates, such as Ras-related Rab-2 (rab2), peroxiredoxin-mitochondrial (prdx5), and ropporin-1 (ropn11) (Rahman et al. 2017) (fig. 8 and supplementary tables S3B and S8B, Supplementary Material online), suggests that sperm capacitation happens before release from the sponge tissue. In addition, the role of the mitochondrial cytochrome b-c1 complex subunit 1 (UQCR2) and voltage-dependent anion-selective channel 2, VDAC2, whose genes, uqcr2 and vdac2, were also upregulated in our case (fig. 8 and supplementary table S3B, Supplementary Material online), are crucial regulators of the Ca+2 influx mediating sperm capacitation (Kwon et al. 2013; Shukla et al. 2013; Rahman et al. 2014, 2017).
External chemoattractants have been found in many marine invertebrates but are not known from sponges. Given that the sponge sperm moves rapidly upon release, two scenarios are possible: 1) female chemoattractants could be already filtered by the male sponge, inducing sperm capacitation, and 2) last stages of sperm capacitation would be completed in a further stage since most modifications are posttranslational (Rahman et al. 2017) and therefore do not have changes in expression associated. Overall, it seems that already formed spermatozoa within mature spermatic cysts of G. phlegraei (SP_II) had expressed not only genes related to their maturation (a complete flagellum formation) but also molecular mechanisms responsible for the next and final step, their capacitation. In this sense, sponges seemed to have a similar mechanism regulating sperm capacitation in which ion channels and membrane hyperpolarization induce the enhanced sperm motility.
Summary
In the animal kingdom, sponges are one of the first extant taxa in which female and male sex appeared (Sara 1974; Feuda et al. 2017). Here, we show that most physiological processes and molecular signals responsible for female and male gametogenesis in other metazoans are also present in Porifera, playing a role in the formation of gametes during the reproductive season. In both female and male gametogenesis, we observed upregulated genes related with the germline fate and self-renewal at the onset of the process. Production of RA could potentially regulate the cessation of germ cell multiplication and the entry in the meiotic cycle during the early stages of gametogenesis (fig. 7) also in Porifera as happens in other metazoans (Anderson et al. 2008; Mark et al. 2015), although functional studies are required to confirm this hypothesis.
Our data suggest that, at molecular level, spermatogenesis is more complex than oogenesis in sponges. During spermatogenesis, the progress from a spermatogonium to mature sperm requires the completeness of two meiotic cycles and several physiological, morphological, and functional transformations, whereas oocyte maturation occurs at meiotic arrest, without the necessity of radical cytological changes. On the other hand, many maternal gene transcripts are silenced and expressed later on during embryonic development (Chapman et al. 2014; Reading et al. 2018), which could also explain the consistent pattern of a lesser number of upregulated genes during oogenesis than spermatogenesis in our data.
During oogenesis, we did not observe any clear morphological structures associated with the meiotic process (synaptonemal complex, chromatin compaction), but we found upregulated genes involved in the regulation of both mitotic and meiotic cycles and meiotic arrest (fig. 7). What we did observe was radical changes in cell shape and size with parallel yolk formation, symbiont accumulation, and extracellular collagen formation, which are conserved processes of oocyte maturation in metazoans (except for symbiont acquisition) that occur during the meiotic arrest phase. In spermatogenesis, we observed both the morphological features and the molecular signals related to meiosis completion (e.g., synaptonemal complex, synapsis, and DNA recombination). Although the identity of these genes and proteins was mainly defined according to their function in studies on mice as model organism (Jung et al. 2019), meiosis is a well-conserved process across most of eukaryotes (Loidl 2016). Given the morphological evidence of meiotic processes in Geodia (fig. 2) and other sponge species (e.g., Barthel and Detmer 1990; Gaino et al. 2007; Abdo et al. 2008; Riesgo et al. 2008), we expect these genes to have an almost identical function from Porifera to Chordata. Furthermore, we identified conserved molecular signals associated with sperm cell modification after meiosis, including spermatid generation and sperm maturation. Noteworthy was the overexpression of the molecules regulating the axoneme construction in late spermatogenesis, which allows the formation of a successful sperm flagellum. Sperm flagella have the same typical structure as any flagellum but with some extra accessory structures (ODF, FS) for motility optimization (Kierszenbaum 2001; Inaba 2011). Sponges have other flagellated cells (choanocytes), and some species transform the above cells into spermatozoa retaining the flagellum during the transformation (e.g., Tuzet et al. 1970; Barthel and Detmer 1990; Bergquist 1996; Riesgo, Maldonado, et al. 2007), but Geodia spp. generate the sperm flagellum de novo (figs. 2 and 6B). Interestingly, genes involved in sperm flagellum formation and sperm capacitation in mammals were recovered from our sponge transcriptomes and discussed here for first time, as many of those appeared overexpressed during spermatogenesis (fig. 7).
From cnidarians to chordates, many signaling pathways regulating the onset and correct progress of gametogenesis come from surrounding cells in the gonadal tissues, follicle cells, and Sertoli cells, for instance (e.g., Jørgensen and Lützen 1997; Kiger et al. 2000; Kuznetsov et al. 2001; Kalachev and Reunov 2005; Johnston et al. 2008). Although sponges lack gonads and specialized tissues involved in reproduction, our results show great conservation in the molecular programs regulating the gametogenesis, even in pathways activated in specialized gonadal tissues in other metazoans. This suggests that the main innovations regarding sexual reproduction along animal evolution are probably more related to the cellular specialization and task division than the molecular complements regulating the formation of gamete cells.
A caveat of our study is the large interindividual variability of the gametogenic process. Even though we tried to group individuals according to their similar developmental stage, we could not always obtain consistent intra-specific expression patterns across individuals at the same developmental stage (biological replicates). This could be related to several factors: 1) the individuals could be in slightly different developmental stages than the ones defined, given that it is difficult to distinguish a possible intermediate developmental stage based on morphology alone; 2) the individuals could have different physiological status that could potentially influence the gene expression pattern; and 3) variation of environmental conditions due to different collecting time and spatial scales could have also affected the physiological status. Finally, inter-specific variation is still possible within the same genus as each species could have adopted a slightly different strategy concerning the specific time of molecular regulation for each step of the gametogenesis. However, our study highlights that, overall, Geodia spp. have the same molecular mechanisms to control and conduct gametogenesis in a similar way as gametogenesis occurs in bilaterians. The great number of nonannotated genes in our analysis could also be sponge-specific genes with a specific function on gametogenesis in Porifera. Our study is the most exhaustive incursion into the evolution of the reproductive machinery of these early metazoans, but we only focused on the genus Geodia which is exclusively gonochoristic and oviparous. To further understand the evolution of the molecular toolkit for reproduction in sponges, it would be necessary to address the genomes and transcriptomes of species spanning the four classes and also include hermaphroditic and viviparous species. Indeed, several genes necessary for sex determination and yolk formation have been recovered from the transcriptomes of hermaphroditic but not from oviparous sponges (Riesgo et al. 2014). Understanding how the reproductive toolkits have been modulated during the course of sponge evolution will have profound implications on our understanding of the evolution of sexual reproduction in animals.
Materials and Methods
Sample Collection and Identification of Reproduction
Specimens from the species G. hentscheli, G. phlegraei, G. barretti, G. macandrewii, and G. atlantica were collected from expeditions supported by the SponGES project in the boreo-arctic Northeastern Atlantic in May and September 2016 (Swedish and Norwegian coast), July 2016, and July–August 2017 (Norwegian Sea and Greenland Sea) (table 1). Information about specimens’ collection was archived in PANGAEA data repository (Koutsouveli et al. 2020). Three pieces of tissue were collected from all specimens and preserved 1) in RNAlater Stabilization Solution (Thermo Fisher Scientific) and 2) in 2.5% glutaraldehyde in 0.4 M Phosphate-Buffered Saline (PBS) for histological work. Samples were then stored at 4 °C for 1 day, and then samples in RNAlater were stored at −80 °C until further processing, whereas samples for light microscopy were kept at 4 °C.
Histological Analysis
Tissue pieces collected for histological work were rinsed in a buffer (0.4 M PBS–0.6 M salt), and post fixed in osmium tetroxide 2% in 0.4 M PBS for 2 h. After that, a thorough rinse in distilled water was performed before incubating them overnight in hydrofluoric acid 4% to remove any silica remnant from their skeleton. Samples were then rinsed with distilled water several times and dehydrated in increased amounts of ethanol (50–70–96–100%). Once dehydrated, samples were further processed for light and transmission electron microscopy.
For light microscopy, samples were embedded in paraffin (60 °C melting temperature) overnight at ∼60 °C in an oven. Once the blocks were mounted, sections with thickness 5 μm were cut in a HM325 microtome (ThermoFisher Scientific). After deparaffining with xylene, sections were stained with a standard Harris’ Hematoxylin and Eosin Staining Protocol and studied in Olympus microscope (BX43) with a UC50 camera. Specimens were examined for presence of oocytes or spermatocytes. The specimens with no-reproductive elements in their tissue were considered nonreproductive (table 1). Three fragments (5 mm3 each fragment) from different parts of the sponge tissue were processed for light microscopy and more than 40 sections were obtained in different parts of each fragment to confirm the presence of gametes and the consistency in developmental stage across the body.
For transmission electron microscopy, samples were embedded in LRW resin according to the guidelines of the manufacturer (agar Scientific) overnight. Ultrathin sections of ∼60 nm were obtained with an Ultracut Reichert-Jung ultramicrotome, stained with 2% uranyl acetate/lead citrate (Reynolds 1963), and observed with a Hitachi Transmission Electron Microscope TEM (H-7650) at 80 kV.
Transcriptomic Analysis
RNA Extraction and Library Preparation
Twenty-two samples of five Geodia species were used for the transcriptomic analyses (table 1). Total RNA extraction was performed with a standard TRIzol Reagent (ThermoFisher Scientific) protocol, with an overnight incubation in isopropanol and two consecutive rinses in 100% ethanol of the RNA pellet. Further mRNA purification was done with the Dynabeads mRNA DIRECT kit (ThermoFisher Scientific), starting from the final stage of the protocol, “Elimination of rRNA contamination” and only slight modifications of time incubations. The quantity and overall quality of mRNA were assessed using NanoDrop 2000 (ThermoFisher Scientific) and Qubit RNA Assay kit (ThermoFisher Scientific). Then, cDNA libraries were constructed with either Truseq v2 or Scriptseq v2 kit (Illumina), according to the manufacturer’s instructions, and ∼50 ng of starting mRNA. For all purification steps, we used AMPure XP beads (Beckman Coulter). The cDNA quantity was assessed with a Qubit dsDNA HS Assay kit (ThermoFisher Scientific) and the quality of the libraries was checked with an Agilent Tapestation 2200 system (Agilent Technologies). The libraries were then pooled and further sequenced with an Illumina NextSeq 500 platform at the Natural History Museum of London sequencing facilities (Molecular Core Labs), using a paired-end read strategy (bp length: 2× 150 bp). The raw reads of all the 34 libraries were deposited at Sequence Read Archive with BioProject ID: PRJNA603347 and submission ID: SUB6821624.
Assembly and Annotation
The quality control of the raw reads was conducted with FASTQC (Andrews 2010). The reads were cleaned with Trimmomatic (Bolger et al. 2014). Only the clean paired reads were subsequently used for the de novo assembly for each species with Trinity v2.4.2 (Grabherr et al. 2011). The transcript quantification was conducted using the alignment-based quantification method RSEM (Li and Dewey 2011), supported by the Trinity package, with the script align_and_estimate_abundance.pl. We tested the quality of the assembly by testing the percentage of properly paired-end reads that were aligned to the assembly with bowtie2 (Langmead and Salzberg 2012) and by calculating the transcriptome completeness with Benchmarking Universal Single-Copy Orthologs (Busco V2/3) against metazoan cassettes (Simão et al. 2015). Finally, we calculated the Nx length, GC content, and assembled bases with Trinity.
For the annotation, we used Diamond (Buchfink et al. 2015) against the selection of metazoan proteins in the swissprot database (Boeckmann et al. 2003) accessed in 2018 with a cutoff e-value of 1e-5. The sequences with blast hits were further annotated by Blast2GOPro (Conesa et al. 2005) to retrieve the functional information from the GO terms.
Differential Gene Expression Analysis
We estimated the mapped read abundance using RSEM (Li and Dewey 2011) and then the differential gene expression (DE) analysis was conducted with edgeR (Robinson et al. 2010; McCarthy et al. 2012) with the following parameters: false discovery rate ≤0.01 and 2-fold change differences. We did pairwise comparisons between the two sexes (female vs. male) and female/male versus nonreproductive (NR) along the different species, taking into account that in some cases there were no replicates for some conditions (table 1). In these cases, we used dispersion 0.1 for the conduction of DE analysis with edgeR. The results regarding unreplicated analyses were taken with caution, and only discussed if they were similar to the results obtained in the replicated analyses of the rest of species. The terms “upregulated” and “overexpressed” genes in the Results and Discussion section (see above) refer to genes extracted with either of the two analyses. The difficulty for an intense sampling and appropriate replication for all the reproductive conditions in all species has to be considered, as the studied species come from deep-sea habitats with scarce access over the year. In order to understand how many orthologous upregulated genes were shared among the species during oogenesis and spermatogenesis, we used online platform OrthoVenn, uploading the translated peptide sequences of the upregulated genes in each condition for each species.
The GO terms associated with differentially expressed genes were used to obtain GO enrichment analysis by Fisher’s exact test in Blast2GOPro platform (Conesa et al. 2005) for all the studied species with a threshold of P value 0.05. Genes upregulated from all the pairwise comparisons (for oogenesis: females vs. NR and vs. males, and for spermatogenesis: males vs. NR and vs. females) were used for the analysis in order to understand which genes and GO-enriched categories were expressed during the different developmental stages of oogenesis and spermatogenesis across the different species. The percentage of sequences contained in each GO term was extracted and used for the depiction of a bubble graph in R. In some cases, more than one GO term appeared enriched for the same process, so we joined the relevant categories to have a final summary percentage.
Proteomic Analysis
In this study, we performed a complementary proteomic analysis to support our results derived from the transcriptomic data. We used two female and two male individuals of different species and different developmental stages: one female of G. macandrewii (Vi_II), one female G. atlantica (Vi_III), one male of G. phlegraei (SP_I), and one male of G. atlantica (SP_II). The protein extraction from freeze-dried samples was performed according to the protocol mentioned by Viarengo et al. (1997) and applied previously in sponges (Wanick et al. 2013). The protein extracts were concentrated in a stacking/resolving gel interface. The unseparated protein bands were visualized by Coomassie staining, excised, and cut for protein digestion (Moreno et al. 2014). The dried gel pieces were reswollen in 100 mM Tris–HCl pH 8, 10 mM CaCl2 with 60 ng/μl trypsin or chymotrypsin at 5:1 protein enzyme (w/w) ratio. The tubes were kept in ice for 2 h and incubated at 37 °C (trypsin) or 25 °C (chymotrypsin) for 12 h. Digestion was desalted onto OMIX Pipette tips C18 (Agilent Technologies) until the mass spectrometric analysis.
Protein separation was performed with reverse phase-liquid chromatography RP-LC-MS/MS analysis (dynamic exclusion mode) in an Easy-nLC II system coupled to an ion trap LTQ-Orbitrap-Velos-Pro hybrid mass spectrometer (ThermoFisher Scientific). Peptides were eluted using a 180-min gradient (solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid, 80% acetonitrile in water). Peptide identification from raw data was carried out using PEAKS Studio 10 search engine (Bioinformatics Solutions Inc.) (Han et al. 2011; Zhang et al. 2012). Database search was performed against the transcriptome assembly for each species. False discovery rate for peptide spectrum matches was limited to 0.01. Only proteins with at least two distinct peptides identified from LC/MS/MS analyses were considered reliably identified.
Phylogenetic Analyses
To understand the phylogenetic relationships among Sox genes, we screened for Sox genes in our transcriptomes of all Geodia spp. Three Sox genes were identified in all species, although one of them in G. hentscheli was too small to be included in the analysis. Ninety-three sequences of the high-mobility group domain of the Sox transcription factor (79 amino acids) that were used in Fortunato et al. (2012) and Schnitzler et al. (2014), as well as the Sox sequences from E. muelleri (Jager et al. 2006) were aligned with our 14 sequences retrieved from Geodia spp. using MAFFT v7 (Katoh and Standley 2013). The best evolutionary model was selected with ProtTest-HPC (Darriba et al. 2011) based on the Akaike Information criterion. A phylogenetic hypothesis was obtained with RAxML v8.1.22 (Stamatakis 2014) with LG-GAMMA as parameters of model evolution and 100 replicates for bootstrap sampling. The same pipeline was employed for fibrillar (types I, V, and XI) and nonfibrillar (type IV and spongin short-chain) collagens (separately) recovered as upregulated genes in our transcriptomes and sequences of these types of collagens retrieved from NCBI. For collagens, the full genes were used for the phylogenies. Accession numbers for the sequences used to construct the Sox phylogeny can be found in Fortunato et al. (2012) and Schnitzler et al. (2014), whereas those for fibrillar and nonfibrillar collagen are shown in the trees in supplementary figure S3, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Dr Nathan Kenny for his useful bioinformatic advice and Dr Cristina Díez-Vives for her useful advice in R. Furthermore, we would like to thank María Conejero for her important help in thin sectioning and Kathrin Busch, Asimenia Gavriilidou, Karin Steffen, Dr Francisca Carvalho, Dr Joana Xavier, and Dr Nathan Kenny for their essential help and support during sample collection. Finally, we would like to thank the reviewers for their thorough work and their contribution to the massive improvement of this manuscript. This work was supported by the H2020 EU Framework Programme for Research and Innovation Project SponGES (Deep-sea Sponge Grounds Ecosystems of the North Atlantic: an integrated approach toward their preservation and sustainable exploitation) (Grant Agreement No. 679849). This document reflects only the authors’ view and the Executive Agency for Small and Medium-sized Enterprises (EASME) is not responsible for any use that may be made of the information it contains. This work is dedicated to our beloved collaborator, coordinator of the SponGES project and coauthor of this manuscript, Hans Tore Rapp, who will always stay in our memories.
References
- Abdo DA, Fromont J, McDonald JI.. 2008. Strategies, patterns and environmental cues for reproduction in two temperate haliclonid sponges. Aquat Biol. 1:291–302. [Google Scholar]
- Amiel A, Leclère L, Robert L, Chevalier S, Houliston E.. 2009. Conserved functions for Mos in eumetazoan oocyte maturation revealed by studies in a cnidarian. Curr Biol. 19(4):305–311. [DOI] [PubMed] [Google Scholar]
- Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AMM, Page DC.. 2008. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 105(39):14976–14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available from:http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- Aouacheria A, Geourjon C, Aghajari N, Navratil V, Deléage G, Lethias C, Exposito JY.. 2006. Insights into early extracellular matrix evolution: spongin short chain collagen-related proteins are homologous to basement membrane type IV collagens and form a novel family widely distributed in invertebrates. Mol Biol Evol. 23(12):2288–2302. [DOI] [PubMed] [Google Scholar]
- Arnold SL, Kent T, Hogarth CA, Schlatt S, Prasad B, Haenisch M, Walsh T, Muller CH, Griswold MD, Amory JK, et al. 2015. Importance of ALDH1A enzymes in determining human testicular retinoic acid concentrations. J Lipid Res. 56(2):342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asem EK, Carnegie JA, Tsang BK.. 1992. Fibronectin production by chicken granulosa cells in vitro: effect of follicular development. Acta Endocrinol (Copenh). 127(5):466–470. [DOI] [PubMed] [Google Scholar]
- Auger J. 2018. Spermatogenic cells—structure. Encycl Reprod. 1:53–60. [Google Scholar]
- Austin CR. 1952. The ‘capacitation’ of the mammalian sperm. Nature 170(4321):326. [DOI] [PubMed] [Google Scholar]
- Barthel D, Detmer A.. 1990. The spermatogenesis of Halichondria panicea (Porifera, Demospongiae). Zoomorphology 110(1):9–15. [Google Scholar]
- Barton NH, Charlesworth B.. 1998. Why sex and recombination? Science 281(5385):1986–1990. [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M.. 1977. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 74(1):68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergquist PR. 1996. The marine fauna of New Zealand: porifera, class Demospongiae. Part 5. Dendroceratida and Halisarcida. N Z Oceanogr Inst Mem. 107:1–53. [Google Scholar]
- Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O’Byrne SM, DeSantis D, Klopfenstein M, Feret B, Dennefeld C, Blaner WS, et al. 2013. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem. 288(34):24528–24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel JS, Chen L, Hayward J, Yeap SL, Alkers AE, Chan RC.. 2010. Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 6(7):e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesalski HK, Doepner G, Tzimas G, Gamulin V, Schröder HC, Batel R, Nau H, Müller WE.. 1992. Modulation of myb gene expression in sponges by retinoic acid. Oncogene 7(9):1765–1774. [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I.. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31(1):365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla E, Xu EY.. 2008. Identification and characterization of novel mammalian spermatogenic genes conserved from fly to human. Mol Hum Reprod. 14(3):137–142. [DOI] [PubMed] [Google Scholar]
- Bowerbank JS. 1858. On the anatomy and physiology of the Spongiadae. Part I. On the spicula. Philos Trans R Soc. 148:279–332. [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. [DOI] [PubMed] [Google Scholar]
- Carballeira NM, Rodriguez J.. 1991. Two novel phospholipid fatty acids from the Caribbean sponge Geodia gibberosa. Lipids 26(4):324–326. [DOI] [PubMed] [Google Scholar]
- Cárdenas P, Rapp HT, Klitgaard AB, Best M, Thollesson M, Tendal OS.. 2013. Taxonomy, biogeography and DNA barcodes of Geodia species (Porifera, Demospongiae, Tetractinellida) in the Atlantic boreo-arctic region. Zool J Linn Soc. 169(2):251–311. [Google Scholar]
- Cárdenas P, Rapp HT, Schander C, Tendal OS.. 2010. Molecular taxonomy and phylogeny of the Geodiidae (Porifera, Demospongiae, Astrophorida)—combining phylogenetic and Linnaean classification. Zool Scr. 39(1):89–106. [Google Scholar]
- Carr M, Leadbeater BSC, Baldauf SL.. 2010. Conserved meiotic genes point to sex in the choanoflagellates. J Eukaryot Microbiol. 57(1):56–62. [DOI] [PubMed] [Google Scholar]
- Chapman RW, Reading BJ, Sullivan CV.. 2014. Ovary transcriptome profiling via artificial intelligence reveals a transcriptomic fingerprint predicting egg quality in striped bass, Morone saxatilis. PLoS One 9(5):e96818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang J, Hou X, Yue W, Huang S, Wang C.. 2019. Tissue expression profiles unveil the gene interaction of hepatopancreas, eyestalk, and ovary in the precocious female Chinese mitten crab, Eriocheir sinensis. BMC Genet. 20(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H.. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12(23):3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R.. 2003. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 300(5623):1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada G, Doallo R, Posada D.. 2011. ProtTest-HPC: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly NM, Lelong C, Huvet A, Kellner K, Dubos M-P, Riviere G, Boudry P, Favrel P.. 2012. Gametogenesis in the Pacific Oyster Crassostrea gigas: a microarrays-based analysis identifies sex and stage specific genes. PLoS One 7(5):e36353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djerassi C, Lam WK.. 1991. Sponge phospholipids. Acc Chem Res. 24(3):69–75. [Google Scholar]
- Dohrmann M, Wörheide G.. 2013. Novel scenarios of early animal evolution—is it time to rewrite textbooks? Integr Comp Biol. 53(3):503–511. [DOI] [PubMed] [Google Scholar]
- DuBuc TQ, Schnitzler CE, Chrysostomou E, McMahon ET, Gahan JM, Buggie T, Gornik SG, Hanley S, Barreira SN, Gonzalez P.. 2020. Transcription factor AP2 controls cnidarian germ cell induction. Science 367:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelbarger KJ. 1993. Vitellogenic mechanisms and the allocation of energy to offspring in polychaetes energy. Bull Mar Sci. 1:165–198. [Google Scholar]
- Eguizabal C, Montserrat N, Vassena R, Barragan M, Garreta E, Garcia-Quevedo L, Vidal F, Giorgetti A, Veiga A, Belmonte J.. 2011. Complete meiosis from human induced pluripotent stem cells. Stem Cells 29(8):1186–1195. [DOI] [PubMed] [Google Scholar]
- Eitel M, Guidi L, Hadrys H, Balsamo M, Schierwater B.. 2011. New insights into placozoan sexual reproduction and development. PLoS One 6(5):e19639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereskovsky AV. 2010. The comparative embryology of sponges. Dordrecht (the Netherlands: ): Springer. [Google Scholar]
- Evans E, Hogarth C, Mitchell D, Griswold M.. 2014. Riding the spermatogenic wave: profiling gene expression within neonatal germ and Sertoli cells during a synchronized initial wave of spermatogenesis in mice. Biol Reprod. 90(5):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito JY, Cluzel C, Garrone R, Lethias C.. 2002. Evolution of collagens. Anat Rec. 268(3):302–316. [DOI] [PubMed] [Google Scholar]
- Exposito JY, Larroux C, Cluzel C, Valcourt U, Lethias C, Degnan BM.. 2008. Demosponge and sea anemone fibrillar collagen diversity reveal the early emergence of A/C clades and the maintenance of the modular structure of type V/XI collagens from sponge to human. J Biol Chem. 283(42):28226–28235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito JY, Le Guellec D, Lu Q, Garrone R.. 1991. Short chain collagens in sponges are encoded by a family of closely related genes. J Biol Chem. 266(32):21923–21928. [PubMed] [Google Scholar]
- Exposito JY, Ouazana R, Garrone R.. 1990. Cloning and sequencing of a Porifera partial cDNA coding for a short-chain collagen. Eur J Biochem. 190(2):401–406. [DOI] [PubMed] [Google Scholar]
- Exposito JY, Valcourt U, Cluzel C, Lethias C.. 2010. The fibrillar collagen family. Int J Mol Sci. 11(2):407–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour C. 2009. Oogenesis: making the mos of meiosis. Curr Biol. 19:489–491. [DOI] [PubMed] [Google Scholar]
- Falender AE, Shimada M, Lo YK, Richards JS.. 2005. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev Biol. 288(2):405–419. [DOI] [PubMed] [Google Scholar]
- FAO. 2009. International Guidelines for the Management of Deep-sea Fisheries in the High Seas. Rome: FAO. p. 73. Available from: www.fao.org/docrep/011/i0816t/i0816t00.htm. [Google Scholar]
- Fawcett DW. 1975. The mammalian spermatozoon. Dev Biol. 44(2):394–436. [DOI] [PubMed] [Google Scholar]
- Feng C-W, Bowles J, Koopman P.. 2014. Control of mammalian germ cell entry into meiosis. Mol Cell Endocrinol. 382(1):488–497. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A.. 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4(4):497–508. [DOI] [PubMed] [Google Scholar]
- Feuda R, Dohrmann M, Pett W, Philippe H, Rota-Stabelli O, Lartillot N, Wörheide G, Pisani D.. 2017. Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr Biol. 27(24):3864–3870.e4. [DOI] [PubMed] [Google Scholar]
- Fierro-Constaín L, Schenkelaars Q, Gazave E, Haguenauer A, Rocher C, Ereskovsky A, Borchiellini C, Renard E.. 2017. The conservation of the germline multipotency program, from sponges to vertebrates: a stepping stone to understanding the somatic and germline origins. Genome Biol Evol. 9(3):474–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato S, Adamski M, Bergum B, Guder C, Jordal S, Leininger S, Zwafink C, Rapp HT, Adamska M.. 2012. Genome-wide analysis of the sox family in the calcareous sponge Sycon ciliatum: multiple genes with unique expression patterns. EvoDevo 3(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama N, Nakatsukasa M, Mohri K, Masuda Y, Agata K.. 2010. Piwi expression in archeocytes and choanocytes in demosponges: insights into the stem cell system in demosponges. Evol Dev. 12(3):275–287. [DOI] [PubMed] [Google Scholar]
- Gaino E, Lepore E, Rebora M, Mercurio M, Sciscioli M.. 2007. Some steps of spermatogenesis in Halichondria semitubulosa (Demospongiae, Halichondriidae). Ital J Zool. 74(2):117–122. [Google Scholar]
- Genin E, Wielgosz-Collin G, Njinkoué JM, Velosaotsy NE, Kornprobst JM, Gouygou JP, Vacelet J, Barnathan G.. 2008. New trends in phospholipid class composition of marine sponges. Comp Biochem Physiol B Biochem Mol Biol. 150(4):427–431. [DOI] [PubMed] [Google Scholar]
- Gervasi MG, Visconti PE.. 2016. Chang’s meaning of capacitation: a molecular perspective. Mol Reprod Dev. 83(10):860–874. [DOI] [PubMed] [Google Scholar]
- Gonobobleva EL, Efremova SM.. 2017. Germ cell determinants in the oocytes of freshwater sponges. Russ J Dev Biol. 48(3):231–235. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome assembly from RNA-Seq data. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD. 2016. Spermatogenesis: the commitment to Meiosis. Physiol Rev. 96(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán JM, Luckenbach JA, Yamamoto Y, Swanson P.. 2014. Expression profiles of Fsh-regulated ovarian genes during oogenesis in coho salmon. PLoS One 9(12):e114176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, He L, Xin L, Shan B, Ma B.. 2011. PeaksPTM: mass spectrometry-based identification of peptides with unspecified modifications. J Proteome Res. 10(7):2930–2936. [DOI] [PubMed] [Google Scholar]
- Haschek WM, Rousseaux CG, Wallig MA.. 2010. Male reproductive system In: Haschek WM, Rousseaux CG, Wallig MA, editors. Fundamentals of toxicologic pathology. San Diego (CA: ): Academic Press; p. 553–597. [Google Scholar]
- Hay E. 1991. Cell biology of extracellular matrix. New York: Plenum Press. [Google Scholar]
- Hoppe WF, Reichert M.. 1987. Predictable annual mass release of gametes by the coral reef sponge Neofibularia nolitangere (Porifera: Demospongiae). Mar Biol. 94(2):277–285. [Google Scholar]
- Ickowicz D, Finkelstein M, Breitbart H.. 2012. Mechanism of sperm capacitation and the acrosome reaction: role of protein kinases. Asian J Androl. 14(6):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsiecke G, Borojevic R, Custodio M, Müller WEG.. 1994. Retinoic acid acts as morphogen in freshwater sponges. Invertebr Reprod Dev. 26(2):89–98. [Google Scholar]
- Inaba K. 2003. Molecular architecture of the sperm flagella: molecules for motility and signalling. Zool Sci. 20(9):1043–1056. [DOI] [PubMed] [Google Scholar]
- Inaba K. 2011. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol Hum Reprod. 17(8):524–538. [DOI] [PubMed] [Google Scholar]
- Isaeva VV, Akhmadiev AV.. 2011. Germinal granules in archaeocytes of the sponge Oscarella malakhovi Ereskovsky, 2006. Russ J Mar Biol. 37(3):209–216. [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S.. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 7(5):517–524. [DOI] [PubMed] [Google Scholar]
- Izumi H, Márián T, Inaba K, Oka Y, Morisawa M.. 1999. Membrane hyperpolarization by sperm-activating and -attracting factor increases cAMP level and activates sperm motility in the ascidian Ciona intestinalis. Dev Biol. 213(2):246–256. [DOI] [PubMed] [Google Scholar]
- Jager M, Quéinnec E, Houliston E, Manuel M.. 2006. Expansion of the SOX gene family predated the emergence of the Bilateria. Mol Phylogenet Evol. 39(2):468–477. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Wright WW, DiCandeloro P, Wilson E, Kopf GS, Jelinsky SA.. 2008. Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc Natl Acad Sci U S A. 105(24):8315–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonge CD, Barratt C.. 2006. The cell: production, maturation, fertilization, regeneration. Cambridge: Cambridge University Press. [Google Scholar]
- Jørgensen C, Lützen J.. 1997. Ultrastructure of the non-germinal cells in the testes of ascidians (Urochordata) and their role in the phagocytosis of sperm. Zoomorphology 117(2):103–113. [Google Scholar]
- Jung M, Wells D, Rusch J, Ahmad S, Marchini J, Myers SR, Conrad DF.. 2019. Unified single-cell analysis of testis gene regulation and pathology in five mouse strains. Elife 8:0–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadler KE, Hill A, Canty-Laird EG.. 2008. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr Opin Cell Biol. 20(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalachev AV, Reunov AA. 2005. Resorption of Gametes in the Testes of the Sea Star Asterina pectinifera (Mueller et Troschel, 1842). Russ J Mar Biol. 31(2):119–123. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL. 2001. Spermatid manchette: plugging proteins to zero into the sperm tail. Mol Reprod Dev. 59(4):347–349. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. 2018. MPF-based meiotic cell cycle control: half a century of lessons from starfish oocytes. Proc Jpn Acad Ser B Phys Biol Sci. 94(4):180–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitgaard AB, Tendal OS.. 2004. Distribution and species composition of mass occurrences of large-sized sponges in the northeast Atlantic. Prog Oceanogr. 61(1):57–98. [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T.. 1996. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380(6576):708–711. [DOI] [PubMed] [Google Scholar]
- Konno A, Padma P, Ushimaru Y, Inaba K.. 2010. Multidimensional analysis of uncharacterized sperm proteins in Ciona intestinalis: EST-based analysis and functional immunoscreening of testis-expressed genes. Zool Sci. 27(2):204–215. [DOI] [PubMed] [Google Scholar]
- Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC.. 2006. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A. 103(8):2474–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsouveli V, Cárdenas P, Rapp HT, Riesgo A. 2020. Transcriptomic and histological analysis (Light and Transmission Electron Microscopy) on five Geodia species to identify their reproductive status and the gene expression during gametogenesis. Natural History Museum, PANGAEA.Available from:https://doi.pangaea.de/10.1594/PANGAEA.911273. [Google Scholar]
- Koutsouveli V, Taboada S, Moles J, Cristobo J, Ríos P, Bertran A, Solà J, Avila C, Riesgo A.. 2018. Insights into the reproduction of some Antarctic dendroceratid, poecilosclerid, and haplosclerid demosponges. PLoS One 13(2):e0192267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S, Lyanguzowa M, Bosch TC. 2001. Role of epithelial cells and programmed cell death in Hydra spermatogenesis. Zoology 104(1):25–31. [DOI] [PubMed] [Google Scholar]
- Kiger AA, White-Cooper H, Fuller MT.. 2000. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature 407(6805):750–754. [DOI] [PubMed] [Google Scholar]
- Kwon WS, Park YJ, Mohamed ESA, Pang MG.. 2013. Voltage-dependent anion channels are a key factor of male fertility. Fertil Steril. 99(2):354–361. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanna E, Klautau M.. 2010. Oogenesis and spermatogenesis in Paraleucilla magna (Porifera, Calcarea). Zoomorphology 129(4):249–261. [Google Scholar]
- Laumer CE, Fernandez R, Lemer S, Combosch D, Kocot KM, Riesgo A, Andrade SCS, Sterrer W, Sorensen MV, Giribet G.. 2019. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc Biol Sci. 286(1906):20190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Adamski M, Bergum B, Guder C, Liu J, Laplante M, Bråte J, Hoffmann F, Fortunato S, Jordal S, et al. 2014. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat Commun. 5:1–15. [DOI] [PubMed] [Google Scholar]
- Levin TC, King N.. 2013. Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Curr Biol. 23(21):2176–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, Yang WX.. 2016. Myosin superfamily: the multi-functional and irreplaceable factors in spermatogenesis and testicular tumors. Gene 576(1):195–207. [DOI] [PubMed] [Google Scholar]
- Liaci L, Sciscioli M.. 1970. The sexual cycle of Erylus discophorus (Schmidt) (Porifera, Tetractinellida). Riv Biol. 63:255–270. [PubMed] [Google Scholar]
- Liaci LS, Sciscioli M.. 1969. La riproduzione sessuale di alcuni tetractinellidi (Porifera). Boll Zool. 36(1):61–67. [Google Scholar]
- Logsdon JM. 2008. Evolutionary genetics: sex happens in giardia. Curr Biol. 18:66–68. [DOI] [PubMed] [Google Scholar]
- Loidl J. 2016. Conservation and variability of meiosis across the eukaryotes. Annu Rev Genet. 50(1):293–316. [DOI] [PubMed] [Google Scholar]
- Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, Grimaldi P.. 2008. Potential role of Nanos3 in maintaining the undifferentiated spermatogonia population. Dev Biol. 313(2):725–738. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. 2001. Assembly of the Drosophila germ plasm In: Etkin LD, Jeon KW, editors. Cell lineage and embryo patterning. New York: Academic Press; p. 187–213. [Google Scholar]
- Maldonado M. 2006. The ecology of the sponge larva. Can J Zool. 84(2):175–194. [Google Scholar]
- Maldonado M, Aguilar R, Bannister R, Bell J, Conwa KW, Dayton P, Diaz M, Gutt J, Kelly M, Kenchington E.. 2016. Sponge grounds as key marine habitats: a synthetic review of types, structure, functional roles, and conservation concerns In:Rossi S, Bramanti L, Gori A, Orejas C, editors. Marine animal forests: the ecology of benthic biodiversity hotspots marine animal forests. Switzerland: Springer International Publishing; p. 1–39. [Google Scholar]
- Maldonado M, Riesgo A.. 2008. Reproduction in the phylum Porifera: a synoptic overview. Treballs SCB 59:29–49. [Google Scholar]
- Malik SB, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM.. 2007. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One 3(8):e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manton I. 1953. Number of fibrils in the cilia of the green algae. Nature 171(4350):485–486. [DOI] [PubMed] [Google Scholar]
- Mark M, Teletin M, Vernet N, Ghyselinck NB.. 2015. Role of retinoic acid receptor (RAR) signaling in post-natal male germ cell differentiation. Biochim Biophys Acta 1849(2):84–93. [DOI] [PubMed] [Google Scholar]
- Markholt S, Grøndahl ML, Ernst EH, Andersen CY, Ernst E, Lykke-Hartmann K.. 2012. Global gene analysis of oocytes from early stages in human folliculogenesis shows high expression of novel genes in reproduction. Mol Hum Reprod. 18(2):96–110. [DOI] [PubMed] [Google Scholar]
- Matova N, Cooley L.. 2001. Comparative aspects of animal oogenesis. Dev Biol. 231(2):291–320. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ.. 2007. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 217(2):137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarrey JR. 2018. Gametogenesis overview In: McCarrey JR, Yan W, editors. Encyclopedia of reproduction. Oxford: Elsevier; p. 2–4. [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK.. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40(10):4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis LK, Limback SD, Albertini DF.. 2013. Signaling modalities during oogenesis in mammals. Curr Top Dev Biol. 102:227–242. [DOI] [PubMed] [Google Scholar]
- Mercurio M, Corriero G, Gaino E.. 2007. A 3-year investigation of sexual reproduction in Geodia cydonium (Jameson 1811) (Porifera, Demospongiae) from a semi-enclosed Mediterranean bay. Mar Biol. 151(4):1491–1500. [Google Scholar]
- Mindnich R, Möller G, Adamski J.. 2004. The role of 17 beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 218(1–2):7–20. [DOI] [PubMed] [Google Scholar]
- Moreno ML, Escobar J, Izquierdo-Álvarez A, Gil A, Pérez S, Pereda J, Zapico I, Vento M, Sabater L, Marina A, et al. 2014. Disulfide stress: a novel type of oxidative stress in acute pancreatitis. Free Radic Biol Med. 70:265–277. [DOI] [PubMed] [Google Scholar]
- Morisawa M, Yoshida M.. 2005. Activation of motility and chemotaxis in the spermatozoa: from invertebrates to humans. Reprod Med Biol. 4(2):101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris P. 1993. The developmental role of the extracellular matrix suggests a monophyletic origin of the kingdom animalia. Evolution N Y. 47(1):152–165. [DOI] [PubMed] [Google Scholar]
- Müller WE, Rottmann M, Diehl-Seifert B, Kurelec B, Uhlenbruck G, Schröder HC.. 1987. Role of the aggregation factor in the regulation of phosphoinositide metabolism in sponges. Possible consequences on calcium efflux and on mitogenesis. J Biol Chem. 262(20):9850–9858. [PubMed] [Google Scholar]
- Müller WEG, Binder M, von Lintig J, Guo Y-W, Wang X, Kaandorp JA, Wiens M, Schröder HC.. 2011. Interaction of the retinoic acid signaling pathway with spicule formation in the marine sponge Suberites domuncula through activation of bone morphogenetic protein-1. Biochim Biophys Acta Gen Subj. 1810(12):1178–1194. [DOI] [PubMed] [Google Scholar]
- Nielsen MG, Turner FR, Hutchens JA, Raff EC.. 2001. Axoneme-specific beta-tubulin specialization: a conserved C-terminal motif specifies the central pair. Curr Biol. 11(7):529–533. [DOI] [PubMed] [Google Scholar]
- Nikko E, Van de Vyver G, Richelle-Maurer E.. 2001. Retinoic acid down-regulates the expression of EmH-3 homeobox-containing gene in the freshwater sponge Ephydatia muelleri. Mech Ageing Dev. 122(8):779–794. [DOI] [PubMed] [Google Scholar]
- Nomura M, Yoshida M, Morisawa M.. 2004. Calmodulin/calmodulin-dependent protein kinase II mediates SAAF-induced motility activation of ascidian sperm. Cell Motil Cytoskeleton 59(1):28–37. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N.. 2015. The Fibroblast Growth Factor signaling pathway. Wires Dev Biol. 4(3):215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangas SA, Rajkovic A.. 2006. Transcriptional regulation of early oogenesis: in search of masters. Hum Reprod Update 12(1):65–76. [DOI] [PubMed] [Google Scholar]
- Peng L, Wang L, Yang YF, Zou MM, He WY, Wang Y, Wang Q, Vasseur L, You MS.. 2017. Transcriptome profiling of the Plutella xylostella (Lepidoptera: Plutellidae) ovary reveals genes involved in oogenesis. Gene 637:90–99. [DOI] [PubMed] [Google Scholar]
- Polzonetti-Magni AM, Mosconi G, Soverchia L, Kikuyama S, Carnevali O.. 2004. Multihormonal control of vitellogenesis in lower vertebrates. Int Rev Cytol. 239:1–46. [DOI] [PubMed] [Google Scholar]
- Porter ME, Johnson KA.. 1989. Dynein structure and function. Annu Rev Cell Biol. 5(1):119–151. [DOI] [PubMed] [Google Scholar]
- Rahman MS, Kwon WS, Pang MG.. 2014. Calcium influx and male fertility in the context of the sperm proteome: an update. Biomed Res Int. 2014:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Kwon WS, Pang MG.. 2017. Prediction of male fertility using capacitation-associated proteins in spermatozoa. Mol Reprod Dev. 84(9):749–759. [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik S-B, Logsdon JM.. 2005. A phylogenomic inventory of meiotic genes: evidence for sex in giardia and an early eukaryotic origin of meiosis. Curr Biol. 15(2):185–191. [DOI] [PubMed] [Google Scholar]
- Reading B, Andersen L, Ryu Y-W, Mushirobira Y, Todo T, Hiramatsu N.. 2018. Oogenesis and egg quality in finfish: yolk formation and other factors influencing female fertility. Fishes 3(4):45. [Google Scholar]
- Reading BJ, Schilling J, Molloy KT, Williams VN, Baltzegar DA, Sullivan CV, Glassbrook N, Hiramatsu N, Mizuta H, Luo W, et al. 2014. Lrp13 is a novel vertebrate lipoprotein receptor that binds vitellogenins in teleost fishes. J Lipid Res. 55(11):2287–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiswig HM. 1983. Porifera In: Adiyodi KG, Adiyodi RG. editors. Reproductive biology of invertebrates, Vol. II, spermatogenesis and sperm function. Chichester (United Kingdom: ): Wiley; p 1–21. [Google Scholar]
- Renard E, Leys SP, Wörheide G, Borchiellini C.. 2018. Understanding animal evolution: the added value of sponge transcriptomics and genomics. BioEssays 40(9):1700237. [DOI] [PubMed] [Google Scholar]
- Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 17(1):208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo A. 2010. Phagocytosis of sperm by follicle cells of the carnivorous sponge Asbestopluma occidentalis (Porifera, Demospongiae). Tissue Cell 42(3):198–201. [DOI] [PubMed] [Google Scholar]
- Riesgo A, Cavalcanti FF, Kenny NJ, Ríos P, Cristobo J, Lanna E.. 2018. Integrative systematics of clathrinid sponges: morphological, reproductive and phylogenetic characterisation of a new species of Leucetta from Antarctica (Porifera, Calcarea, Calcinea) with notes on the occurrence of flagellated sperm. Invert Syst. 32(4):827–841. [Google Scholar]
- Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP.. 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 31(5):1102–1120. [DOI] [PubMed] [Google Scholar]
- Riesgo A, Maldonado M, Durfort M.. 2007. Dynamics of gametogenesis, embryogenesis, and larval release in a Mediterranean homosclerophorid demosponge. Mar Freshwater Res. 58(4):398–417. [Google Scholar]
- Riesgo A, Maldonado M, Durfort M.. 2008. Occurrence of somatic cells within the spermatic cysts of demosponges: a discussion of their role. Tissue Cell 40(6):387–396. [DOI] [PubMed] [Google Scholar]
- Riesgo A, Taylor C, Leys SP.. 2007. Reproduction in a carnivorous sponge: the significance of the absence of an aquiferous system to the sponge body plan. Evol Dev. 9(6):618–663. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Stan A.. 2013. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 14(11):713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers RJ, Irving-Rodgers HF, Russell LD.. 2003. Extracellular matrix of the developing ovarian follicle. Reproduction 126(4):415–424. [DOI] [PubMed] [Google Scholar]
- Russo GL, Wilding M, Marino M, Dale B.. 1998. Ins and outs of meiosis in ascidians. Semin Cell Dev Biol. 9(5):559–567. [DOI] [PubMed] [Google Scholar]
- Saitou M, Miyauchi H.. 2016. Gametogenesis from pluripotent stem cells. Cell Stem Cell 18(6):721–735. [DOI] [PubMed] [Google Scholar]
- Sánchez F, Smitz J.. 2012. Molecular control of oogenesis. Biochim Biophys Acta Mol Basis Dis. 1822(12):1896–1912. [DOI] [PubMed] [Google Scholar]
- Sara M. 1974. Sexuality in the Porifera. Boll Zool. 41(4):327–348. [Google Scholar]
- Schnitzler CE, Simmons DK, Pang K, Martindale MQ, Baxevanis AD.. 2014. Expression of multiple Sox genes through embryonic development in the ctenophore Mnemiopsis leidyi is spatially restricted to zones of cell proliferation. EvoDevo 5(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM.. 2008. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays 30(6):579–589. [DOI] [PubMed] [Google Scholar]
- Sciscioli M, Lepore E, Gherardi M, Liaci LS.. 1994. Transfer of symbiotic bacteria in the mature oocyte of Geodia cydonium (Porifera, Demosponsgiae): an ultrastructural study. Cah Biol Mar. 35:471–478. [Google Scholar]
- Sebé-Pedrós A, Chomsky E, Pang K, Lara-Astiaso D, Gaiti F, Mukamel Z, Amit I, Hejnol A, Degnan BM, Tanay A.. 2018. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat Ecol Evol. 2(7):1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC.. 2007. The coming of age for Piwi proteins. Mol Cell 26(5):603–609. [DOI] [PubMed] [Google Scholar]
- Shikina S, Chang CF.. 2016. Sexual reproduction in stony corals and insight into the evolution of oogenesis in cnidaria In: Goffredo S, Dubinsky Z, editors. The cnidaria, past, present and future. Switzerland: Springer International Publishing; p. 248–268. [Google Scholar]
- Shikina S, Chen CJ, Chung YJ, Shao ZF, Liou JY, Tseng HP, Lee YH, Chang CF.. 2013. Yolk formation in a stony coral Euphyllia ancora (Cnidaria, Anthozoa): insight into the evolution of vitellogenesis in nonbilaterian animals. Endocrinology 154(9):3447–3459. [DOI] [PubMed] [Google Scholar]
- Shivdasani AA, Ingham PW.. 2003. Regulation of stem cell maintenance and transit amplifying cell proliferation by TGF-β signaling in Drosophila spermatogenesis. Curr Biol. 13(23):2065–2072. [DOI] [PubMed] [Google Scholar]
- Shukla KK, Kwon WS, Rahman MS, Park YJ, You YA, Pang MG.. 2013. Nutlin-3a decreases male fertility via UQCRC2. PLoS One 8(10):e76959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y, Okuno M.. 1993. The sliding of the fibrous sheath through the axoneme proximally together with microtubule extrusion. Exp Cell Res. 208(1):170–174. [DOI] [PubMed] [Google Scholar]
- Siddall NA, McLaughlin EA, Marriner NL, Hime GR.. 2006. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A. 103(22):8402–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simpson TL. 1984. Gamete, embryo, larval development In: The cell biology of sponges. New York: Springer; p. 341–413. [Google Scholar]
- Smith EF, Yang P.. 2004. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton 57(1):8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J. 2013. Closing the circle of germline and stem cells: the Primordial Stem Cell hypothesis. EvoDevo 4(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollas WJ. 1880. The sponge-fauna of Norway; a Report on the Rev. A.M. Norman’s Collection of Sponges from the Norwegian Coast. Ann Mag Nat Hist Zool Bot Geol. 5(26):130–144. [Google Scholar]
- Spetland F, Rapp HT, Hoffmann F, Tendal OS, 2007. Sexual reproduction of Geodia barretti Bowerbank, 1858 (Porifera, Astrophorida) in two Scandinavian fjords. In: Custódio MR, Hajdu E, Lóbo-Hajdu G, Muricy G, editors. Porifera research: biodiversity, innovation and sustainability. Proceedings of the 7th international sponge symposium; Série Livros 28, Museu Nacional, Rio de Janeiro, Brazil. p. 613–620.
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466(7307):720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens J. 1915. Sponges of the coasts of Ireland. I. The Triaxonia and part of the Tetraxonida. Fish Irel Sci Investig. 1914:1–43. [Google Scholar]
- Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O’Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ.. 2007. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development 134(14):2593–2603. [DOI] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M.. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 71(2):295–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer EE, McLaughlin M.. 2007. Natural history of the mammalian oocyte. Reprod Biomed Online 15(3):288–295. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C.. 2005. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 287(2):390–402. [DOI] [PubMed] [Google Scholar]
- Tong M-H, Yang QE, Davis JC, Griswold MD.. 2013. Retinol dehydrogenase 10 is indispensible for spermatogenesis in juvenile males. Proc Natl Acad Sci U S A. 110(2):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzet O, Garrone R, Pavans de Ceccatty M.. 1970. Observations ultrastructurales sur la spermatogenese chez la demosponge Aplysilla rosea Sch. (Dendroceratide): une metaplaise exemplaire. Ann Sci Nat. 12:27–50. [Google Scholar]
- UNGA. 2006. The Impacts of Fishing on Vulnerable Marine Ecosystems: Actions taken by States and regional fisheries management organizations and arrangements to give effect to paragraphs 66 to 69 of General Assembly Resolution 59/25 on sustainable fisheries, regarding the impacts of fishing on vulnerable marine ecosystems. [Google Scholar]
- Vargas-Ángel B, Colley SB, Hoke SM, Thomas JD.. 2006. The reproductive seasonality and gametogenic cycle of Acropora cervicornis off Broward County, Florida, USA. Coral Reefs 25(1):110–122. [Google Scholar]
- Viana IKS, Gonçalves LAB, Ferreira MAP, Mendes YA, Rocha RM.. 2018. Oocyte growth, follicular complex formation and extracellular-matrix remodeling in ovarian maturation of the imperial zebra pleco fish Hypancistrus zebra. Sci Rep. 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viarengo A, Ponzano E, Dondero F, Fabbri R.. 1997. A simple spectrophotometric method for metallothionein evaluation in marine organisms: an application to Mediterranean and Antarctic molluscs. Mar Environ Res. 44(1):69–84. [Google Scholar]
- Visconti PE. 2009. Understanding the molecular basis of sperm capacitation through kinase design. Proc Natl Acad Sci U S A. 106(3):667–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Lovasco LA, Gyuris A, Baumgartner RA, Parlow AF, Freiman RN.. 2007. Ovarian granulosa cell survival and proliferation requires the gonad-selective TFIID subunit TAF4b. Dev Biol. 303(2):715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Marzluff WF, Wessel GM.. 2003. Cyclin B synthesis is required for sea urchin oocyte maturation. Dev Biol. 256(2):258–275. [DOI] [PubMed] [Google Scholar]
- Wanick RC, de Sousa Barbosa H, Frazão LR, Santelli RE, Arruda MAZ, Coutinho CC.. 2013. Evaluation of differential protein expression in Haliclona aquarius and sponge-associated microorganisms under cadmium stress. Anal Bioanal Chem. 405(24):7661–7670. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. 1978. The development of two species of Tetilla (Demosponge). NSR O U. 29:71–106. [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park I-H, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. 2009. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460(7257):909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H, Bausek N.. 2010. Evolution and spermatogenesis. Philos Trans R Soc B Biol Sci. 365(1546):1465–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H, Doggett K, Ellis RE.. 2009. The evolution of spermatogenesis In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm biology: an evolutionary perspective. Oxford: Elsevier Ltd; p. 151–183. [Google Scholar]
- Wiens M, Batel R, Korzhev M, Müller W.. 2003. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J Exp Biol. 206(18):3261–3271. [DOI] [PubMed] [Google Scholar]
- Wolters-Everhardt E, Dony JM, Lemmens WA, Doesburg WH, De Pont JJ.. 1986. Buffering capacity of human semen. Fertil Steril. 46(1):114–119. [DOI] [PubMed] [Google Scholar]
- Woodruff TK, Lee HC, Lee J, Edmonds M.. 2018. Oocyte maturation during folliculogenesis In: McCarrey JR, Yan W, editors. Encyclopedia of reproduction. Oxford: Elsevier; p. 172–175. [Google Scholar]
- Yoshida M, Inaba K, Ishida K, Morisawa M.. 1994. Calcium and cyclic AMP mediate sperm activation, but Ca2+ alone contributes sperm chemotaxis in the ascidian, Ciona savignyi. Dev Growth Differ. 36(6):589–595. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Inaba K, Morisawa M.. 1993. Sperm chemotaxis during the process of fertilization in the ascidians Ciona savignyi and Ciona intestinalis. Dev Biol. 157(2):497–506. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Yoshida K.. 2011. Sperm chemotaxis and regulation of flagellar movement by Ca2+. Mol Hum Reprod. 17(8):457–465. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B.. 2012. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics 11(4):M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QG, Lu BS, Huang PT.. 2005. Functions of FANCL in primordial germ cell formation and Fanconi anemia. Yi Chuan Xue Bao 32(9):993–1000. [PubMed] [Google Scholar]
- Zumberge JA, Love GD, Cárdenas P, Sperling EA, Gunasekera S, Rohrssen M, Grosjean E, Grotzinger JP, Summons RE.. 2018. Demosponge steroid biomarker 26-methylstigmastane provides evidence for Neoproterozoic animals. Nat Ecol Evol. 2(11):1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.