Abstract
Defining the interaction of Arf GAPs with specific Arfs is important for understanding their functions in the endocytic system. Cell-based approaches have been valuable for identifying Arfs and Arf GAPs active in the endocytic compartment; however, the cell-based assays have some limitations in establishing relationships among the Arfs and ArfGAPs. Here we describe a simple in vitro assay that will provide a means for comparing Arfs as substrates and serve to complement cell-based studies.
INTRODUCTION
The endocytic compartment is a dynamic system of endomembranes through which numerous proteins transit with specific itineraries (D’Souza-Schorey & Chavrier, 2006; Grant & Donaldson, 2009). The function and organization of the compartment is important for many cellular processes. The ADP-ribosylation factors (Arfs) are members of a family of guanine nucleotide-binding proteins that regulate membrane traffic including that in the endocytic compartment (Kahn et al., 2006). Humans have 5 genes encoding Arf proteins. They are divided into 3 classes based on primary structure: Class 1 (Arf1 and 3); class 2 (Arf4 and 5) and; class 3 (Arf6). All three classes of Arfs function in the endocytic compartment (Donaldson & Jackson, 2011; Gillingham & Munro, 2007; Grant & Donaldson, 2009; Hickson et al., 2003; Maldonado-Baez, Williamson, & Donaldson, 2013).
The effects of Arfs are mediated by proteins that bind to Arfs (Donaldson & Jackson, 2011; D’Souza-Schorey & Chavrier, 2006; East & Kahn, 2011; Gillingham & Munro, 2007; Kahn, 2009). Important classes of Arf effector proteins include vesicle coat proteins, such as clathrin adaptors AP-1 and AP-3, and lipid metabolizing proteins, such as phosphatidylinositol 4-phosphate 5-kinase. The proteins that regulate the Arfs, Arf-GTPase-activating proteins (GAPs), also have a critical role in determining the function of the Arfs.
Thirty-one genes encode Arf GAPs in Human, outnumbering Arfs by more than 6-fold (Kahn et al., 2008, 2006). Many of the Arf GAPs have multiple splice variants. In contrast to the Arfs, in which a particular isoform may have more than one site of action, the Arf GAPs may be site specific. Recent investigations have revealed that effects of particular isoforms of Arf on itineraries of proteins that transit the endocytic pathway may be determined, in part, by the site-specific Arf GAP (Chen, Luo, Jian, & Randazzo, 2014).
An understanding of the complex and specific itineraries of proteins that transit the endocytic pathway requires characterization of the Arf-ArfGAP pairs functioning in the compartment. Determining the GAP that is catalytic for a specific Arf is difficult using cell-based assays. There are multiple interactions that could confound the interpretations of experiments using whole cells. For example, ASAP1 binds to FIP3, an Arf6-binding protein. Consequences of the ASAP1-FIP3 interaction are that Arf6 may coimmunoprecipitate and colocalize with ASAP1 due to binding to FIP3 and, potentially, Arf6·GTP levels are affected because Arf6·GTP is stabilized by the complex with FIP3. Furthermore, the elevated Arf6·GTP potentially can affect the activity of Arf exchange factors such as ARNO, thereby affecting Arf1·GTP levels (Cohen et al., 2007; DiNitto et al., 2007). Thus, the effect of the GAP on Arf·GTP levels in cells could be in part independent of GAP catalytic function.
Simple in vitro GAP assays are a valuable complement to cell-based assays in work defining the role of Arfs and Arf GAPs in endocytic traffic. One important factor in GAP assays is the quality of the Arfs used as substrates. Fortunately for Arfologists, full length Arfs from the three major classes can be prepared in native form as we describe here. Also important is that experiments are designed to provide information about relative enzymatic power, which we also describe in this chapter.
1. METHODS
1.1. PREPARATION OF MyrArfs
1.1.1. Background
Arf proteins are Ras-superfamily members. Like other Ras-superfamily members, Arfs are comprised of a nucleotide-binding fold with two motifs, called switch 1 and switch 2, whose conformation is sensitive to nucleotide in the binding pocket. With GTP bound, switch 1 and switch 2 adopt a conformation that interacts with effector proteins and GAPs. Unique to the Arfs is an N-terminal extension of approximately 16 amino acids from the nucleotide-binding fold. The glycine at position 2 in the N-terminus is covalently modified with myristic acid. The N-terminal extension and the C-terminus of Arfs are the most variable between isoforms, and the myristoylated N-terminus may be considered as a third switch motif. In Arf·GDP, the N-terminus associates with Arf between switch 1 and switch 2. In Arf·GTP, the N-terminus is displaced from Arf and associates with membranes. In another Arf family member, Arl2, the amino terminus is part of the interface with the effectors. We have not found the myristate to be critical for interaction with GAPs (in sharp contrast to ArfGEFs) (Chen, Jian, Luo, & Randazzo, 2012; Jian, Gruschus, Sztul, & Randazzo, 2012; Randazzo, 1997). The myristoylated protein is easier to load with nucleotide but myristoylated protein is more difficult to prepare. However, other alterations of the N-terminus are often not tolerated for the GAP assay (Jian, Cavenagh, Gruschus, Randazzo, & Kahn, 2010; Yoon et al., 2004). Deletion of the N-terminus from Arf can reduce activity with some Arf GAPs by 10,000-fold, and substrate specificity that is obvious when examining full-length proteins is lost. Fusion of GST to the N-terminus results in an Arf that binds poorly to nucleotide and poorly interacts with GAPs (Jian et al., 2010). In short, either deletion or fusion of protein to the N-terminus of Arf may yield a protein that is not useful for GAP assays.
The other region of Arf that must be considered when comparing Arf isoforms is the C-terminus, which also varies between Arfs and associates with the membrane surface when Arf is bound to GTP (Liu, Kahn, & Prestegard, 2010). We have found that fusion of short tags or GFP directly to the C-terminus, in general, interferes with GAP activity (Jian et al., 2010). The one exception is a 6His tag, although we have not extensively examined this modification. In cell-based studies, the linker between Arf and GFP has been found to have a critical effect (Dejgaard, Luo, Randazzo, & Presley, submitted for publication). However, we have not extensively examined the various Arf-GFP recombinants for in vitro assays, so we cannot comment on them.
Taking these considerations together, we recommend using full length Arf, either myristoylated or nonmyristoylated, for examination of Arf GAPs. There is no evidence that a His tag will influence interpretation of experiments examining the GAPs, but preparation of the untagged Arfs is straightforward (Randazzo, Weiss, & Kahn, 1995), so the use of the His tag is not necessary. Here we briefly describe the preparation of recombinant Arf1, Arf5, and Arf6 modified with myristic acid. We have not tried Arf3. We have had limited success with Arf4 in that we have not been able to prepare chemical amounts but have been able to prepare enough for GAP assays. We do not describe preparation of Arf3 or Arf4 in this chapter.
1.1.2. Materials
BL21 (DE3) Escherichia coli cotransformed with plasmids for expression of Arf1, Arf5, or Arf6 with yeast N-myristoyltransferase (Randazzo, Terui, et al., 1995; Randazzo, Weiss, et al., 1995).
Luria broth with 100 μg/mL ampicillin and 25 μg/mL kanamycin.
Luria broth/agarose plates with 100 μg/mL ampicillin and 25 μg/mL kanamycin.
Isopropyl β-d-1-thiogalactopyranoside.
Complete protease inhibitor cocktail tablets (Roche, Cat#11697498001).
HiLoad 16/10 phenyl Sepharose HP column (GE Healthcare Life Sciences).
Hiload 26/60 Superdex 75 column (GE Healthcare Life Sciences).
SDS-PAGE apparatus.
Coomassie blue dye.
Cell disruptor (Microfluidics M-110P).
Refrigerated ultracentrifuge with Ti45 fixed-angle rotor (Beckman).
5-mL HiTrap Q HP column (GE Healthcare Life Sciences).
AKTA FPLC (GE Healthcare Life Sciences).
Amicon centrifugal filters (Ultracel 10K).
1.1.3. Buffers
T20N100M1D1: 20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT.
T20N3000M1D1: 20 mM Tris, pH 8.0, 3000 mM NaCl, 1 mM MgCl2, 1 mM DTT.
T20N100M1D1G10: 20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10% Glycerol.
T20N25M1D1G10: 20 mM Tris, pH 8.0, 25 mM NaCl, 1 mM MgCl2, 1 mM DTT, 10% Glycerol.
1.1.4. Expression of MyrArfs
We use the same protocol to express representative Arfs from each class, Arf1, Arf5, and Arf6, modified with myristate, described elsewhere in detail (Chen et al., 2012). Briefly, BL21 (DE3) bacteria are grown on Luria-broth agar culture plates containing 100 μg/mL ampicillin and 25 μg/mL kanamycin. A single colony is used for a 100 mL culture in Luria Broth with 100 μg/mL ampicillin and 25 μg/mL kanamycin. The culture is incubated at 37 °C until the optical density (OD) at 600 nm is 0.6. The bacteria are collected by centrifugation. They can be used immediately or stored overnight at 4 °C. The collected bacteria are used to start a 4 L culture in Luria Broth with 100 μg/mL ampicillin and 25 μg/mL kanamycin at 37 °C. When the OD is 0.6, myristic acid is added to achieve a final concentration of 10 μM. For myrArf6, IPTG is added to achieve a final concentration of 1 mM. Incubation is continued for an additional 3 h at 37 °C. For myrArf1 and myrArf5, IPTG is added to a final concentration of 0.1 mM and incubation is continued at room temperature overnight. Bacteria are collected by centrifugation and can be used immediately or stored at −80 °C.
1.1.5. Purification of myristoylated Arfs
The specific purification protocol depends on the isoform, as we briefly describe here. More complete protocols have been previously described (Chen et al., 2012).
Human myrArf1 (myristoylated Arf1) and myrArf5 protein are purified by a method modified from Randazzo (1997). The pellet from 2 to 4 L of bacteria cell culture that coexpresses Arf and N-myristoyltransferase is lysed using a cell disruptor (M-110P cell disruptor, Microfluidics) in T20N100M1D1 buffer and complete protease inhibitor cocktail tablets (Roche). Cell lysates are clarified by centrifugation at 100,000× g for 60 min at 4 °C. The lysate is then passed through two consecutive 5 mL HiTrapQ HP columns. The material that did not adhere to the column (flow through fraction) is confirmed to contain protein using the BioRad protein assay. The flow through fraction is then adjusted to 3000 mM NaCl by adding solid NaCl and applied to a phenyl-Sepharose HP column with a bed volume of 20–22 mL. The column is developed in a 100 mL gradient from 3000 to 20 mM NaCl. MyrArf1 typically elutes in the center of the gradient. Fractions containing myrArf1 are further purified by size exclusion using a Hiload 26/60 Superdex G75 column in T20N100M1D1G10. The fractions containing MyrArf are combined and concentrated by centrifugal filtration with an Amicon Ultracel 10K to around 1.0 mL (final protein concentration should be 1–2 mg/mL). The purified protein is aliquotted and then snap-frozen in a dry ice/ethanol bath. The protein is stored at −80 °C.
MyrArf6 is purified as described previously (Chen et al., 2012; Jian et al., 2012). MyrArf6 is extracted from BL21(DE3) bacteria cotransformed with expression plasmids for Arf6 and yeast N-myristoyltransferase. First, bacteria are lysed in T20N100M1D1 plus protease inhibitors, and then the pellet is collected and washed in T20N100M1D1. MyrArf6 is extracted from the pellet into T20N25M1D1G10 containing 1% Triton X-100. The protein is precipitated from the detergent extract using ammonium sulfate, dissolved in T20N25M1D1G10 plus 1% Triton X-100, and dialyzed against T20N25M1D1G10 plus 0.1% Triton X-100. After dialysis, the sample is passed through a 5 mL HiTrapQ column preequilibrated with the same buffer as used for dialysis. Like myrArf1 and myrArf5, myrArf6 does not bind to the Q column and is typically recovered in the flow-through fraction. The myrArf6 prepared in the manner is typically in the GTP-bound form. The GTP can be exchanged for [α32P]GTP, which is used as a tracer to follow the conversion of GTP to GDP in the GAP assay.
1.2. GAP ASSAY
1.2.1. Background
The assay described here follows the hydrolysis of GTP bound to Arf using a radioactive tracer, P32, in the α position of GTP. We and others have also used an assay in which the conversion of Arf·GTP to Arf·GDP is followed by a change in tryptophan fluorescence in Arf. However, this indirect assay can be confounded by a number of factors, sometimes leading to dramatically different estimates of enzymatic parameters than those obtained by directly following GTP hydrolysis (Luo, Ha, Hayashi, & Randazzo, 2009; Luo & Randazzo, 2008).
The assay described here involves first exchanging unlabeled nucleotide bound to Arf with [α32P]GTP. Arf·[α32P]GTP is added to the reaction containing variable concentrations of Arf GAP. A plot ln(Arf·GTP0/Arf·GTP)/t versus [ArfGAP], where Arf·GTP0 is Arf·GTP present in the absence of GAP and Arf·GTP is the amount after incubation with Arf GAP, will have a linear portion at low GAP concentrations with a slope equal to the enzymatic power. Alternatively, the amount of GAP required to reach 50% hydrolysis of the GTP on Arf is inversely proportional to enzymatic power and can be used for comparisons. The assay and analysis is described in more detail below. The reaction occurs on a hydrophobic surface. The surface can be provided by large unilamellar vesicles (LUVs). We describe the preparation of LUVs below.
1.2.2. Materials
Lipid hydration buffer:
25 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM DTT (can be stored for 1–2 weeks at 4 °C)
5 × Exchange buffer:
125 mM HEPES, pH 7.4, 5 mM EDTA, pH 8.0, 2.5 mM MgCl2, 500 mM NaCl, 5 mM DTT, 5 mM ATP (store at −20 °C)
5 × GAP reaction buffer:
125 mM HEPES, pH 7.4, 500 mM NaCl, 10 mM MgCl2, 5 mM DTT, 5 mM GTP (store at −20 °C)
Wash buffer:
25 mM Tris·HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT (can be stored for 1–2 weeks at 4 °C)
Lipids and Lipid extruder (Avanti Polar Lipids):
Phosphatidylcholine (PC, chicken egg), Phosphatidylethanolamine (PE, bovine liver), Phosphatidylserine (PS, porcine brain), Phosphatidylinositol (PI, bovine liver), Phosphatidylinositol 4,5-bisphosphate (PIP2, porcine brain), Phosphatidylinositiol 3,4,5-trisphosphate (PIP3), Cholesterol.
Nitrocellulose filters:
Protran BA85, 0.45 μm pore size, 25 mm from Whatman
PEI-cellulose plastic backed thin layer chromatography plates.
Phosphorimager:
StormImager, GE Healthcare Lifesciences.
1.2.3. Preparing large unilamellar vesicles
Mix lipids in chloroform/methanol in a 12 × 75 mm siliconized glass tube at the desired molar ratio in a quantity sufficient for making a 5- to 10-fold concentrate of vesicles to be added to an assay. Typically, the mixture of 40% PC, 25% PE, 15% PS, 10% PI, and 10% cholesterol is used. The amount of PI is reduced and replaced with PIP2 and PIP3 dependent on the Arf GAP. For example, for ASAP1 or AGAP1, 9% PI and 1% PIP2 was used, whereas for ARAP2, 8% PI, 1% PIP2, and 1% PIP3 was used.
Chloroform is evaporated under a gentle stream of nitrogen for 30–60 min at room temperature in a fume hood. Residual chloroform is removed with a lyophilizer with a pressure of <100 μm of mercury for at least 1 h. Add lipid hydration buffer to the dried lipids to achieve a stock concentration at 5 mM. Allow the lipids to hydrate for 10 min at room temperature, then mix to obtain a suspension. Make sure to dislodge the dry lipids from the wall of the glass tube. Freeze and thaw the suspension five times using an ethanol/dry ice bath and a room temperature water bath. Using a lipid extruder, pass the lipid suspension through a Whatman Nulepore Track Etched membrane with a pore size of 1 μm 11 times.
1.2.4. Exchanging GXP for [α32P]GTP
1.2.4.1. Nucleotide exchange reaction for myrArfs
To prepare 100 μL of ~0.25 μM myrArf·GTP, add 52.5 μL water, 20 μL of 5× exchange buffer, 10 μL of 5 mM LUVs, 2.5 μL [α32P]GTP, and 5 μL of 10 μM GTP (specific activity = 25,000—100,000 cpm/pmol). Mix and add 10 μL of 5 μM myrArf. The reaction was incubated for 30–60 min at 30 °C. Add 1.5 μL of 100 mM MgCl2 to adjust Mg2+ to a final concentration of 2 mM and place on ice. The [α32P]GTP-loaded myrArf is stable for hours, but should be used on the same day it is prepared.
1.2.4.2. Nucleotide exchange reaction for non-myrArfs
To load non-myrArfs with [α32P]GTP, the exchange reaction contains 1 μM non-myrArf, 0.1–1 μM [α32P]GTP (specific activity = 25,000–100,000 cpm/pmol), and 0.1% Triton X-100 in 1× exchange buffer. The reaction was incubated for 30–60 min at 30 °C.
1.2.5. GAP assay
Load Arf proteins with [α32P]GTP as described above.
Prepare serial dilutions of the purified Arf GAP.
Typically, a 25-μL GAP reaction was set up on ice as follows. When myrArfs are used as substrates, add 12.5 μL water, 5 μL 5 × GAP reaction buffer, 2.5 μL 5 mM LUVs, and 2.5 μL of the diluted Arf GAP in a tube. When non-myrArfs are used, replace LUVs with mixed micelles containing 360 μM phosphatidic acid with 90 μM PIP2 and/or approximately 10 μM PIP3 in 0.1% Triton X-100 at final concentrations.
To initiate the reaction, add 2.5 μL [α32P]GTP-loaded Arf and vortex to mix.
Immediately incubate at 30 °C for 3 min.
Stop the reaction by adding 2 mL of ice-cold wash buffer.
Nucleotide bound to Arf is trapped on nitrocellulose filters using a vacuum manifold. Wash each filter four times with 2 mL ice-cold wash buffer.
Place each filter into a glass scintillation vial containing 0.75 mL 2 N formic acid to release the nucleotides from Arf.
Apply 20–30 μL of the formic acid extract, 10 μL at a time, to a polyethyleneimine-cellulose plastic-backed thin layer chromatography (TLC) plate.
GDP and GTP are separated by developing the thin layer chromatogram in 1 N formic acid and 1 N LiCl.
Expose the TLC plate to a phosphorimager screen for an appropriate period of time. Determine the relative amounts of [α32P]GDP and [α32P]GTP using a phosphorimager.
1.3. RESULTS AND ANALYSIS OF GAP ASSAYS
For comparing the activity of different Arf GAPs, mutants of an Arf GAP or one Arf GAP under different conditions, for example, PIP2 concentrations, GAP is titrated into the reaction as illustrated in Figure 1. One of two parameters can be used to compare the relative enzymatic activity: the GAP amount that induces hydrolysis of 50% of the GTP on Arf (C50) and the estimated enzymatic power, ln(GTP0/GTP)/t/[GAP].
FIGURE 1. GAP activity measured by the conversion of Arf·[α32P]GTP to Arf·[α32P]GDP.
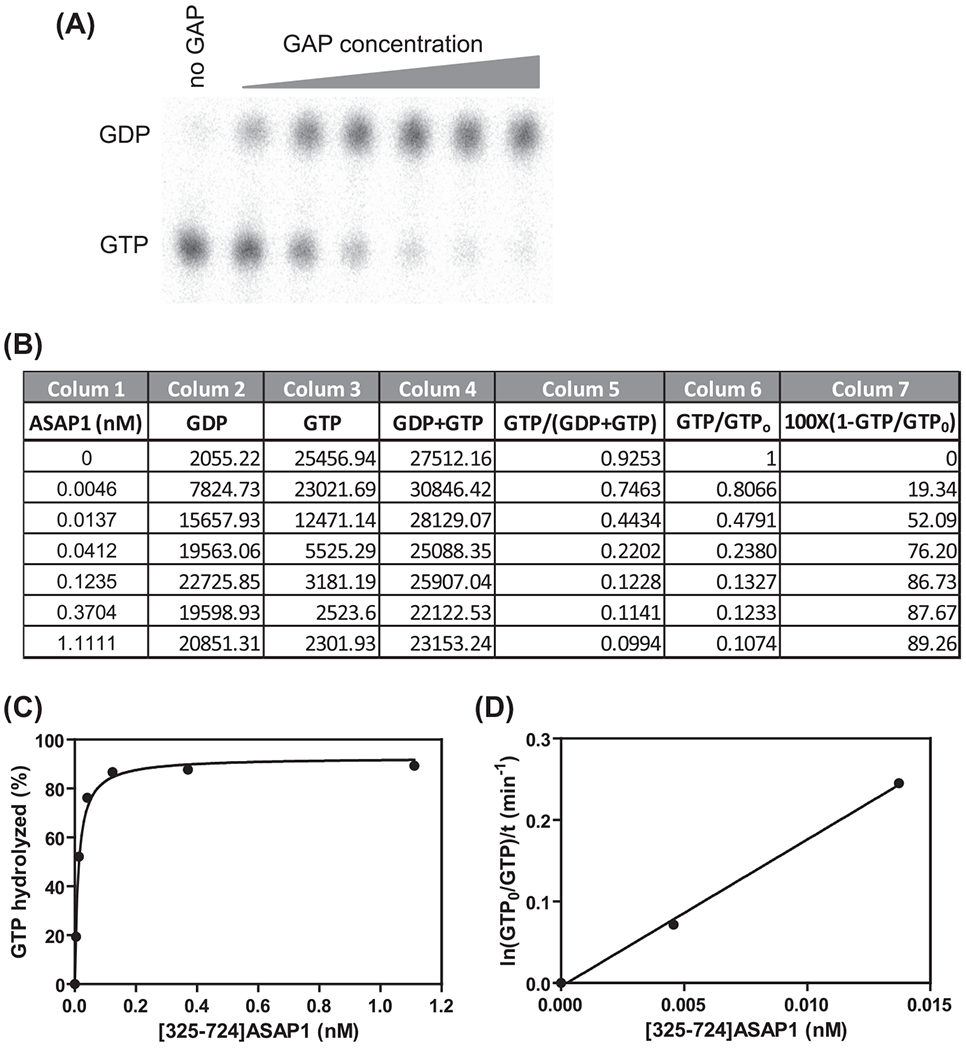
(A) Separation of GTP and GDP by thin layer chromatography. (B) Spreadsheet for calculating the fraction of GTP hydrolyzed from phosphorimager measurements. (C,D) An active fragment of ASAP1 was titrated into the reaction containing LUVs with 2.5% PIP2 and 7.5% PI to determine C50 (C) and enzymatic power (D). (C) The percentage of Arf·GTP hydrolysis was determined in (B), plotted as a function of the centration of ASAP1 and data were fitted to a hyperbolic equation to determine C50. C50 = 0.01 nM. (D) The data were natural log transformed, plotted against the ASAP1 concentration and fitted to a linear function to determine the enzymatic power, which was 18 min−1 nM−1.
In Figure 1(A), the raw results of a typical experiment are shown. Following incubation with the indicated amount of a GAP, radiolabeled nucleotide was extracted from the protein with formic acid and GDP was separated from GTP by thin layer chromatography. Radioactivity in the nucleotides was quantified using a phosphorimager.
1.3.1. Calculation of fractional conversion of Arf·GTP to Arf·GDP and estimation of relative enzymatic activity
Background:
In every experiment, there will be some background GDP signal. The signal does not come from endogenous GTPase activity in Arf as far as we can tell. The signal does not change during incubations of an hour at 30 °C. In the calculations described below, signals are normalized to exclude the background (GTP0, the relative amount of GTP in the reaction containing no GAP).
Normalizing signals:
To improve consistency in the values, calculations are normalized for the total nucleotide signal recovered in each sample. The calculations for an example experiment shown in Figure 1 are illustrated in the spreadsheet in Figure 1(B). The first column of the spreadsheet gives the concentration of the GAP in the reaction. The second column is the signal from GDP and the third column is the signal from GTP (measured using a phosphorimager). GDP and GTP are normalized for total nucleotide signal recovered in the sample. Total nucleotide signal is the sum of columns 2 and 3, calculated in column 4. Normalized signal, i.e., signal as a fraction of total for GTP, is given in column 5.
Calculating fraction of GTP hydrolyzed:
The fraction of GTP hydrolyzed is calculated as the 1–GTP/GTP0. The calculation is illustrated in the spreadsheet as each entry in column 5 is divided by the first entry (column 6) and then subtracted from 1. Percent hydrolysis is obtained by multiplying by 100% (column 7). A plot of GAP concentration versus percent hydrolysis is shown in Figure 1(C).
Estimating relative enzymatic power:
The enzymatic power can be estimated from the linear portion of a plot of ln(GTP0/GTP)/t versus GAP concentration (Randazzo et al., 2013). The slope of the linear part of the curve has the dimensions of min−1 nM−1 and is the enzymatic power. Ln(GTP0/GTP)/t is calculated as the Ln(first row entry in column 5/each row entry column 5)/t (usually use 3 min for the fixed point assay). An example plot is shown in Figure 1(D).
SUMMARY
An assay for Arf GAP activity including the preparation of reagents is described, which provides a complementary approach to cell-based assays examining the isoform-specific interaction of Arfs with Arf GAPs that may function in the endocytic compartment.
ACKNOWLEDGMENT
The work was supported by the intramural program of the National Cancer Institute (Project BC0007365).
REFERENCES
- Chen PW, Jian X, Luo R, & Randazzo PA (2012). Approaches to studying Arf GAPs in cells: in vitro assay with isolated focal adhesions. Current Protocols in Cell Biology. 10.1002/0471143030.cb1713s55 (Chapter 17), Unit 17 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P-W, Luo R, Jian X, & Randazzo PA (2014). The Arf6 GTPase-activating proteins ARAP2 and ACAP1 define distinct endosomal compartments that regulate integrin α5β1 traffic. The Journal of Biological Chemistry, 289(44), 30237–30248. 10.1074/jbc.M114.596155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, & Donaldson JG (2007). Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domain. Molecular Biology of the Cell, 18(6), 2244–2253. 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejgaard SY, Luo R, Randazzo PA & Presley JF “Arf4 is regulated by ArfGAP1 and facilitates sorting of ERGIC53 on pre-Golgi membranes” submitted for publication. [Google Scholar]
- DiNitto JP, Delprato A, Lee MTG, Cronin TC, Huang S, Guilherme A, et al. (2007). Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Molecular Cell, 28(4), 569–583. 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, & Jackson CL (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nature Reviews Molecular Cell Biology, 12(6), 362–375. 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, & Chavrier P (2006). ARF proteins: roles in membrane traffic and beyond. Nature Reviews Molecular Cell Biology, 7(5), 347–358. 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- East MP, & Kahn RA (2011). Models for the functions of Arf GAPs. Seminars in Cell & Developmental Biology, 22(1), 3–9. 10.1016/j.semcdb.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, & Munro S (2007). The small G proteins of the Arf family and their regulators. Annual Review of Cell & Developmental Biology, 23, 579–611. [DOI] [PubMed] [Google Scholar]
- Grant BD, & Donaldson JG (2009). Pathways and mechanisms of endocytic recycling. Nature Reviews Molecular Cell Biology, 10(9), 597–608. 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GRX, Matheson J, Riggs B, Maier VH, Fielding AB, Prekeris R, et al. (2003). Arfophilins are dual Arf/Rab 11 binding proteins that regulate recycling endosome distribution and are related to Drosophila nuclear fallout. Molecular Biology of the Cell, 14(7), 2908–2920. 10.1091/mbc.E03-03-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian XY, Cavenagh M, Gruschus JM, Randazzo PA, & Kahn RA (2010). Modifications to the C-terminus of Arf1 alter cell functions and protein interactions. Traffic, 11(6), 732–742. 10.1111/j.1600-0854.2010.01054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian XY, Gruschus JM, Sztul E, & Randazzo PA (2012). The pleckstrin homology (PH) domain of the Arf exchange factor Brag2 is an allosteric binding site. The Journal of Biological Chemistry, 287(29), 24273–24283. 10.1074/jbc.M112.368084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA (2009). Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Letters, 583(23), 3872–3879. 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM, Nie ZZ, Premont RT, et al. (2008). Consensus nomenclature for the human ArfGAP domain-containing proteins. The Journal of Cell Biology, 182(6), 1039–1044. 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, & Schurmann A (2006). Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. The Journal of Cell Biology, 172(5), 645–650. 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YZ, Kahn RA, & Prestegard JH (2010). Dynamic structure of membrane-anchored Arf-GTP. Nature Structural & Molecular Biology, 17(7), 876–881. 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Ha VL, Hayashi R, & Randazzo PA (2009). Arf GAP2 is positively regulated by coatomer and cargo. Cellular Signalling, 21(7), 1169–1179. 10.1016/j.cellsig.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RB, & Randazzo PA (2008). Kinetic analysis of Arf GAP1 indicates a regulatory role for coatomer. The Journal of Biological Chemistry, 283(32), 21965–21977. 10.1074/jbc.M802268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Baez L, Williamson C, & Donaldson JG (2013). Clathrin-independent endocytosis: a cargo-centric view. Experimental Cell Research, 319(18), 2759–2769. 10.1016/j.yexcr.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA (1997). Resolution of two ADP-ribosylation factor 1 GTPase-activating proteins from rat liver. Biochemical Journal, 324, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA, Jian X, Chen P-W, Zhai P, Soubias O, & Northup JK (2013). Quantitative analysis of guanine nucleotide exchange factors (GEFs) as enzymes. Cellular Logistics, 3(1), e27609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo PA, Terui T, Sturch S, Fales HM, Ferrige AG, & Kahn RA (1995). The myristoylated amino-terminus of ADP-ribosylation factor-1 is a phospholipid-sensitive and GTP-sensitive switch. The Journal of Biological Chemistry, 270(24), 14809–14815. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Weiss O, & Kahn RA (1995). Preparation of recombinant ADP-ribosylation factor. Methods in Enzymology, 257, 128–135. [DOI] [PubMed] [Google Scholar]
- Yoon HY, Jacques K, Nealon B, Stauffer S, Premont RT, & Randazzo PA (2004). Differences between AGAP1, ASAP1 and Arf GAP1 in substrate recognition: interaction with the N-terminus of Arf1. Cellular Signalling, 16(9), 1033–1044. 10.1016/j.cellsig.2004.02.008. [DOI] [PubMed] [Google Scholar]


