Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors with a low survival rate. The identification of mechanisms underlying the development of HCC helps uncover cellular and molecular targets for the diagnosis, prevention, and treatment of HCC. Golgi protein 73 (GP73) level is upregulated in HCC patients and potentially can be a therapeutic target. Despite many studies devoted to GP73 as a marker for HCC early diagnosis, there is little discussion about the function of GP73 in HCC tumorigenesis. Given the poor response to currently available HCC therapies, a better understanding of the role of GP73 in HCC may provide a new therapeutic target for HCC. The current paper summarizes the role of GP73 as a diagnostic marker as well as its roles in liver carcinogenesis. Its roles in other types of cancer are also discussed.
Keywords: Liver cancer, Hepatocellular carcinoma (HCC), Diagnostic markers, Golgi protein 73 (GP73), Alpha-fetoprotein (AFP), Gamma-glutamyl transferase (GGT), Hepatitis C virus (HCV), Hepatitis B virus (HBV)
1. Introduction
Liver cancer is the sixth most common malignancy and the fourth leading cause of cancer-related death with more than 800,000 new cases emerged and 782,000 deaths caused in 2018.1 Hepatocellular carcinoma (HCC) is the terminal step of the sequential progression from chronic hepatitis to fibrosis and cirrhosis and accounts for about 70–85% of primary liver cancer.2 The incidence of HCC is relatively high in China and Western Africa due to aflatoxin exposure and hepatitis B virus (HBV) infection.1,3,4 In the United States, the incidence of HCC is also rising, albeit the pace may have slowed since 2010.5 When it is diagnosed, HCC is usually at advanced stages and current treatment options have poor therapeutic efficacy. However, a 5-year survival rate higher than 70% can be achieved if HCC is diagnosed at an early stage.6 Thus, early diagnosis and treatment play pivotal roles in HCC treatment outcomes.7
One of the most commonly used HCC diagnostic biomarkers is alpha-fetoprotein (AFP). AFP is a fetal-specific glycoprotein produced primarily by the fetal liver. More than 70% of HCC patients have a high serum concentration of AFP due to the tumor excretion. The use of AFP in clinical practice is limited due to low sensitivity at cutoff values maintaining sufficiently high specificity. AFP has 3 different glycoforms, namely AFP-L1, AFP-L2, and AFP-L3, according to their binding capability to lens culinaris agglutinin (LCA). AFP-L1, as the non-LCA-bound fraction, is the major glycoform in the serum of nonmalignant patients; whereas AFP-L3, as the LCA-bound fraction, is the major glycoform found in the serum of HCC patients. Des-gamma-carboxy prothrombin (DCP) is another HCC diagnostic marker. It has been shown that a combination of AFP, AFP-L1, and DCP, is highly sensitive and useful for diagnosing HCC. The related information has been reviewed by others.8–12
In addition to the above-mentioned traditional HCC diagnostic biomarkers, other molecules have been considered and investigated. Those include Golgi protein 73 (GP73), galectins, glypican 3 (GPC3), glutamine synthase, heat shock protein 70, cytokeratin 19, midkine, osteopontin, squamous cell carcinoma antigen, Annexin A2, fibroblast growth factor 3/4, microRNAs, long non-coding RNAs (lncRNAs), circulating tumor cells, cell-free DNA, and others. This review paper focuses on GP73, which is also known as Golgi membrane protein 1 (GOLM1) or Golgi phosphoprotein 2 (GOLPH2) uncovered in the year 2000.13
2. Introduction of GP73
GP73 is a type II transmembrane glycoprotein residing in the Golgi membrane.14 It is only expressed in bile duct epithelial cells, and has low or none detectable expression levels in hepatocytes of a healthy liver.13 However, hepatocyte expression of GP73 is markedly increased in diseased livers regardless of the etiology, whereas the expression level in biliary epithelial cells does not change much. Thus, GP73 expression increases due to hepatitis C virus (HCV) infection, HBV-associated cirrhosis and HCC, alcoholic hepatitis, as well as autoimmune hepatitis.15 The high expression level of GP73 found in HCC patients suggests its role in liver carcinogenesis.16 Moreover, serum GP73 is elevated in HCC patients making it a valuable biomarker for early HCC diagnosis.17–19
2.1. Cell localization and structure of GP73
The GP73 gene has 3042 base pairs containing 9 introns and 10 exons and is localized in human chromosome 9q21.33. GP73 contains 401 amino acids with an estimated molecular weight of 45 kD. However, due to protein modification such as glycosylation, GP73 has a molecular weight of 73 kD based on denatured gel electro-phoresis.13 The gene is cloned from liver cells obtained from an adult giant-cell hepatitis patient.13 The co-localization of GP73 and giantin, a Golgi protein regulating the connections of Golgi stacks, indicates that GP73 is a type II Golgi membrane protein residing in the cis and medial-Golgi cisternae.14
Structurally, GP73 has 5 domains. Domain 1 is a short N-terminal cytoplasmic tail that has 12 amino acids; domain 2 is a transmembrane domain; domain 3 is a coiled-coil structure; domain 4 has a loose structure; and domain 5 is a highly conservative region (Fig. 1).20 There are several possible glycosylation sites at the C-terminal.21,22 Unlike other Golgi-intrinsic proteins, GP73 lacks an oligosaccharide modification function.23 The coiled-coil region has a protein-protein interaction function,24,25 and the N-terminal as well as the transmembrane domain are essential for Golgi localization.20 The endogenous and secreted GP73 forms dimers under normal conditions via forming disulfide bonds within the coiled-coil domain.20
Fig. 1. Schematic diagrams of GP73.
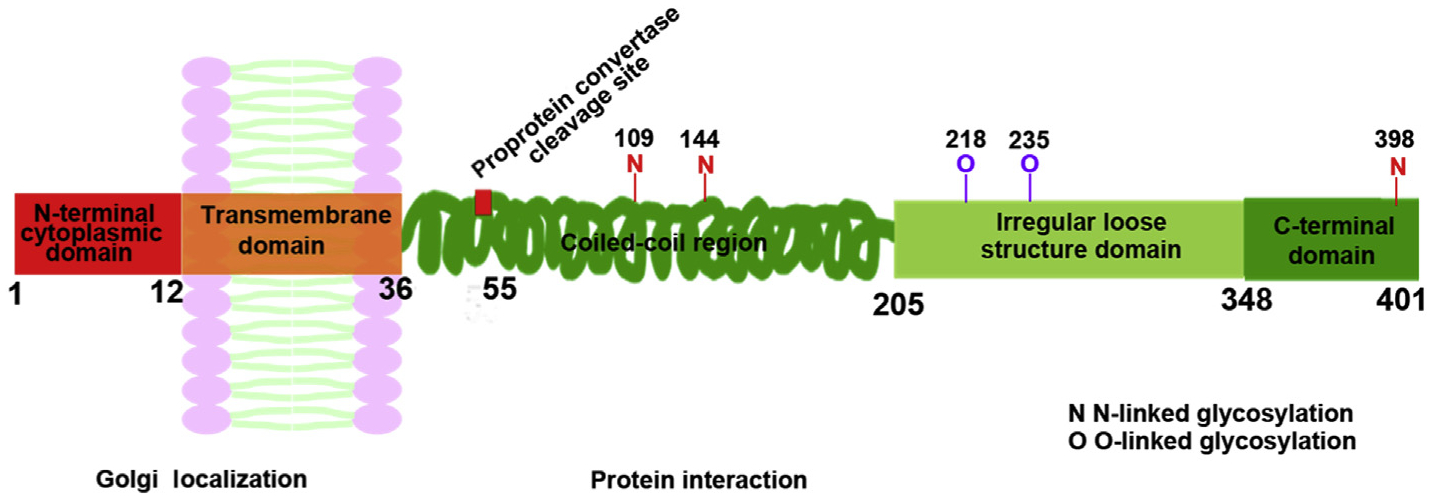
GP73 has 5 domains including a short N-terminal cytoplasmic tail, transmembrane domain, coiled-coil structure, loose structure, and highly conservative domain. Abbreviation: GP73, Golgi protein 73.
GP73 originally was not considered to be a secreted protein, but it is detected in the serum of HCC patients. The evidence of secretion was due to the identification of a proprotein convertase cleavage site at the 55th amino acid.21 Additionally, GP73 has multiple potential glycosylation sites, and more than 75% of GP73 is glycosylated,26 which is then secreted into the blood through intracellular transport. The presence of glycosylation sites allows the utilization of proteomic technology to identify heterogeneous-fucosylated GP73, which has increased sensitivity and specificity for HCC diagnosis.27
2.2. Distribution of GP73
GP73 is highly expressed in the colon, stomach, prostate, and trachea, but has little or no expression in muscle, lymphoid tissue, and leukocytes. At the cellular level, GP73 is mainly found in epithelial cells.13,15,27 Although we are unclear about the actual function of GP73, many studies based on protein modification reveal that GP73 does not have catalytic enzyme activity.15 The Golgi complex has a pivotal role in sorting and modifying proteins exported from the endoplasmic reticulum. GP73 may process proteins synthesized in the rough endoplasmic reticulum and assist in the transportation of protein cargo through the Golgi apparatus. However, GP73 has no homology with those known Golgi glycosyltransferases as well as transporters of nucleotide sugars, nucleotide sulfate, and ATP,28,29 suggesting GP73 may have a unique role.
3. GP73 as a serum marker for HCC diagnosis
3.1. Serum GP73 as a sole diagnostic marker
Many studies demonstrate that GP73 is a biomarker for the diagnosis of early stage HCC with high sensitivity and specificity.19,30–32 In addition, serum GP73 concentration is positively associated with HCC progression and negatively linked with clinical outcomes.18,33–40 A large cohort study consisting of 1690 healthy adults, 337 HBV carriers, and 789 HCC patients revealed that HCC patients have significantly higher serum GP73 levels than healthy people and HBV carriers without noticeable liver disease.19 Another study examined the relationship between serum GP73 concentration and HCC treatment in 9 patients. The data showed that GP73 level decreased after tumor ablation. Additionally, patients with a high serum GP73 level relapsed within 5 years. Thus, serum GP73 level has a monitoring value in HCC patients.41 However, another study reported that serum GP73 is not a suitable biomarker for HCC diagnosis because its level overlaps with the presence of cirrhosis, which affects its diagnostic accuracy.42 In that study, serum GP73 level remained at an elevated level even after tumor resection.42 Nevertheless, in liver transplantation patients, serum GP73 levels predicted tumor recurrence and survival rate, which was demonstrated by studying 60 liver transplantation patients.43
3.2. GP73 is used in combination with other diagnostic markers
The use of GP73 in combination with AFP, gamma-glutamyl transferase (GGT), and dickkopf-1 (DKK-1) can increase the sensitivity and specificity of HCC diagnosis.44–46 The serum level of AFP in HCC patients with cirrhosis decreases dramatically after tumor resection.42 When GP73 is combined with AFP, the sensitivity and the specificity for HCC diagnosis is 89.2% and 85.2%, respectively.39,44–47 Moreover, a combination of GP73 and AFP-L3 is useful to diagnose low serum AFP or AFP-negative HCC cases. Together, sensitivity and specificity are improved when multiple biomarkers are used.44,47–49
Among all the hepatic enzymes, GGT and alkaline phosphatase have served as predictive markers for cancer patients. GGT regulates glutathione metabolism. Its overproduction by the tumor can be a way of self-defense to counteract oxidized waste. However, it has a large overlap with various liver diseases, which limits its diagnosis specificity. Because there was no correlation between GP73 and GGT, they may have complementary roles in the detection of HCC.50
DKK-1 is a Wnt/β-catenin signaling pathway inhibitor, and serum DKK-1 levels are elevated in HCC patients. DKK-1 overexpression promotes the migration and invasion of human HCC cells including Huh7 and Hep3B, whereas DKK-1 silencing inhibits them.51 Overexpression of DKK-1 is also found in human hepatoblastomas.52 It is a promising biomarker for HCC even in AFP-negative patients.52 When GP73 is combined with DDK-1, the sensitivity and specificity for HCC diagnosis is 97.4% and 93.1%, respectively.32 See Table 1.
Table 1.
Diagnosis of HCC using single or combinations of markers.
| Sensitivity (%) | Specificity (%) | Accuracy (%) | |
|---|---|---|---|
| Single marker | |||
| GP73 | 76.34 | 80.09 | 79.61 |
| AFP | 55.6 | 86.7 | 73.4 |
| GGT | 68.4 | 97.1 | 84.8 |
| DKK-1 | 93.6 | 86.9 | 89.08 |
| Combination with GP73 | |||
| GP73 + AFP | 89.2 | 85.2 | 96 |
| GP73 + AFP-L3 | 78.49 | 85.03 | 84.19 |
| GP73 + GGT | 91.1 | 77.1 | 83.2 |
| GP73 + DKK-1 | 97.4 | 93.1 | 94.5 |
Abbreviations: AFP, alpha-fetoprotein; DKK-1, dickkopf-1; GGT, gamma-glutamyl transferase; GP73, Golgi protein 73; HCC, hepatocellular carcinoma.
Taken together, the overall survival rate and disease-free survival rate of HCC patients with elevated GP73 expression are significantly lower than those with low GP73 expression.53 Moreover, increased serum level of GP73 is associated with a degree of liver injuries ranging from hepatitis, fibrosis, and cirrhosis to HCC, which implies that GP73 is secreted by damaged liver cells and activated stellate cells depending on the level of liver damage and fibrosis.32,45
4. Regulation and functional role of GP73
4.1. Induction of GP73
HBV and HCV viral attacks can directly cause the release of pro-inflammatory cytokines including interleukin-6 (IL-6), tumor necrosis factor-alpha (TNF-α), IL-1, and IL-8.54 The serum level of IL-6 in patients with chronic HBV infection increases gradually as the disease progress. IL-6 not only participates in the damage and repair process of hepatic cells, but also promotes the proliferation of hepatic stellate cells.55 Additionally, IL-6 induces the expression of GP73 through gp130 and is associated with phosphorylated signal transducer and activator of transcription 3 (STAT3).56
In cell line models, GP73 is highly expressed in HepG2.2.15 cells, which have active HBV replication, but is not detectable in HepG2T14.1 cells, which do not support HBV replication. GP73 is also undetectable in HBV-free HepG2 cells. Moreover, in immortalized human hepatic adenocarcinoma SK-HEP-1 cells, the level of GP73 is low, interferon-gamma (IFN-γ) treatment can induce GP73, whereas TNF-α reduces it. Additionally, HBV promotes GP73 production in peripheral blood mononuclear cells obtained from healthy people and in macrophages obtained from acute monocytic leukemia cells.57 Taken together, these data suggest that increased GP73 in hepatocytes likely involves hepatotropic viruses or cytokine production.15
Although the expression of GP73 is associated with HBV replication, the expression of GP73 is significantly induced when peripheral blood mononuclear cells and human monocytic THP-1 cells are treated with the supernatants containing replication-competent or non-replicating HBV as well as recombinant hepatitis B surface antigen (HBsAg). However, the expression of GP73 is not affected when those cells are treated with the supernatant neutralized with antibodies specific for HBsAg or recombinant hepatitis B core antigen (HBcAg). In summary, the data suggest that HBsAg promotes GP73 production.57
In HepG2 and Huh7 cells, overexpression of GP73 facilitates HBV replication in a dose-dependent manner, whereas silencing GP73 reduces HBV production. Silencing GP73 also induces NF-κB (p50), which triggers the secretion of antiviral cytokines. Additionally, overexpression of NF-κB (p50) reduces the production of HBsAg and GP73 in HBV-infected HepG2 and Huh7 cells. Thus, by reducing NF-κB signaling and innate immune response, GP73 stimulates HBV replication. Together, there is a distinct positive feedback mechanism between HBV replication and GP73 production. Silencing GP73 can be a potential prevention and treatment of HBV infection.57
Another mechanism by which GP73 can be induced is via hypoxia inducible factors-2alpha (HIF-2α). HIF-2α is an important transcriptional factor making cancer cells adapted to a hypoxic condition.58 It has been shown that HIF-2α and GP73 expression are positively correlated in HCC tissues. In hepatoblastoma HepG2 cells, HIF-2α, but neither HIF-1α nor hepatitis B virus protein X, induces the expression of GP73.59
Mechanistic target of rapamycin complex 1 (mTORC1) also up-regulates GP73 to promote HCC proliferation and migration. The mechanistic target of rapamycin (mTOR) signaling pathway plays a key role in HCC. Hyperactivation of mTORC1 promotes the expression of GP73, which enhances HCC cell proliferation and migration through CD44 and matrix metallopeptidase-7.60
GP73 drives HCC metastasis through activation of epidermal growth factor receptor (EGFR)/receptor tyrosine kinase (RTK) signaling. GP73 binds to EGFR/RTK anchoring on the trans-Golgi network and recycling back to the plasma membrane, leading to activation of the downstream factors, thereby promoting HCC cell migration and invasion.61
4.2. Silencing GP73
Silencing of GP73 induces apoptosis. Knockdown of the GP73 gene leads to reduced Bcl-2 to Bax ratio, increased cytochrome c, as well as reduced Capase-3 in HepG2 cells and Bel7402 cells.62
GP73 is also implicated in autophagy. By removing damaged organelles or misfolded proteins, autophagy has a crucial role in energy balance and maintaining hepatic homeostasis. Autophagy is active at a basic level and can be induced in response to stress including nutrient deficiency, hypoxia, endoplasmic reticulum stress, as well as exposure to pathogens or drugs thereby autophagy serves as a protective mechanism and can prevent the development of cancer. However, when cancer has formed, increased autophagic flux can enable cancer cells to survive and expand. Furthermore, many HCC treatment medications increase autophagy, which is implicated in drug resistance. Moreover, other medications such as Regorafenib has autophagy activation effects. Together, the role of autophagy in HCC treatment remains to be clarified.63
Microtubule-associated protein 1 light chain 3 (LC3) is an autophagosome membrane marker. Silencing of GP73 in HepG2 cells effectively triggers the conversion of LC3-I to LC3-II and the production of LC3-positive vacuoles. In contrast, GP73 upregulation significantly inhibits the formation of starvation-induced LC3-positive structures in HepG2 cells.64 The significance of the finding in HCC treatment remains to be uncovered. Together, GP73 is implicated in prohibiting apoptosis as well as regulating autophagy.
Silencing of GP73 suppresses cell motility, invasion, and epithelial-mesenchymal transition (EMT) by reducing p-retinoblastoma tumor suppressor protein (p-Rb) in HCC.65,66 GP73 decreases expression of N-cadherin and E-cadherin and enhances expression of vimentin.65–67 Hence, GP73 may serve as a molecular target against EMT in HCC metastasis therapy.68
It has been shown that there are several microRNAs that can regulate the expression level of GP73. MiR-141–3p targets the GP73 to reverse EMT leading to reduced HCC progression and metastasis.69 MiR-382 also targets GP73 to inhibit metastasis of HCC. The down-regulation of miR-382, which up-regulates GP73, in HCC predicts poor survival.70 The expression of miR-27b is inversely associated with the level of GP73 in HCC, and thus may have a role in regulating GP73. Moreover, low miR-27b expression is a significant and independent predictor of poor survival in HCC. MiR-27b suppresses the expression of GP73 is therefore a potential prognostic biomarker and treatment target of HCC.71 In addition, the serum level of miR-212 and miR-132, which silences GP73, is reduced in primary liver cancer patients.72 Furthermore, miR-493–5p is significantly downregulated in HCC. Via inhibiting the expression of GP73, miR-493–5p inhibits HCC cell proliferation, arrests cancer cells at the G0/G1 phase, and induces apoptosis.73 See Table 2.
Table 2.
Factors that regulate GP73 expression.
| Regulators | Factors | GP73 expression |
|---|---|---|
| Virus | HBsAg, HCV | Up |
| Cytokines | IL-6, IFN-γ, IL-1, IL-8 | Up |
| TNF-α | Down | |
| Proteins | HIF-2α, mTORC1, | Up |
| NF-κB (p50) | Down | |
| miRNAs | miR-141–3p, miR-382, miR-27b, miR-212, miR-132, miR-493–5p | Down |
Abbreviations: GP73, Golgi protein 73; HBsAg, hepatitis B surface antigen; HCV, hepatitis C virus; HIF-2α, hypoxia inducible factors-2alpha; IFN-γ, interferon-gamma; IL, interleukin; miRNAs, microRNAs; mTORC1, mechanistic target of rapamycin complex 1; NF-κB, nuclear factor-kappa B; TNF-α, tumor necrosis factor-alpha.
4.3. Phenotype of GP73 deficiency mouse models
To investigate biological roles for GP73, there are two GP73 deficiency mouse models that have been generated. The first one is a transgenic mouse model with truncated c-terminal (GP73tr/tr). The lifespan of GP73tr/tr mice is shorter than that of wild-type mice. Microscopically, the GP73tr/tr mice developed severe epithelial abnormalities in the kidney and liver.74 Another model uses the CreloxP system and generates hepatocyte-specific GP73 deficiency. However, these mice have normal growth, behavior, and fertility.75 It would be interesting to investigate HCC development using those knockout mouse models. Taken together, GP73 plays multiple roles in epithelial cell function, but is not vital for normal liver function.
5. GP73 and other cancers
In addition to HCC, GP73 is implicated in carcinogenesis of other types of cancer including melanoma, glioblastoma, prostate cancer, lung cancer, and colorectal cancer.
5.1. GP73 and melanoma
Melanoma is a potentially lethal skin cancer, and its incidence has risen rapidly in the past 50 years.76 A high expression level of GP73 in melanoma is related to the aggressiveness of the disease, reducing the disease-free survival period, as well as reducing cancer-specific overall survival.77 However, elevated levels of GP73 in tumor-associated macrophages is associated with a prolonged disease-free survival period and cancer-specific overall survival, because tumor metastases are not found in patients with high numbers of GP73-positive tumor-associated macrophages. It is speculated that GP73 may be associated with prolonged stabilization of M1 phenotype macrophages, which may result in observed favorable clinical outcome.77
MiR-200a directly targets the GP73 in melanoma to inhibit cell proliferation and metastasis by regulating the phosphoinositide 3-kinase (PI3K)/AKT signaling pathway and EMT. Down-regulated miR-200a, which increases the expression of the GP73, is associated with a poor prognosis of melanoma.78
By whole-exome sequencing, a rare nonsynonymous variant of GP73 is found in melanoma. GP73 is considered a predisposition gene for melanoma by comparing 454 melanoma patients and 396 controls.79 Taken together, these data indicate the oncogenic role of GP73 in melanoma, which might be potential targets for molecular-based treatment.
5.2. GP73 and lung cancer
Lung cancer, including small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), is the second most common cancer and the leading cause of cancer death worldwide.80 GP73 mainly plays roles in NSCLC.81 NSCLC includes adenocarcinoma and squamous cell carcinoma, which comprises about 80% of all diagnosed lung cancers.81 GP73 is upregulated in both adenocarcinoma and squamous cell carcinoma. However, high GP73 expression is negatively associated with overall survival and recurrence-free survival of patients with adenocarcinoma, but not of patients with squamous carcinoma.80,81 GP73 can increase adenocarcinoma aggressiveness by activating matrix metallopeptidase-13.82 A high expression level of GP73 in lung adenocarcinoma is also associated with high-grade malignancy.81
The changes of DNA copy number and methylation might dysregulate the expression of GP73 in lung adenocarcinoma. It has been shown that DNA methyltransferase 1 (DNMT1) inhibits the function of miR-200a, which silences GP73 expression, thereby inhibiting cell proliferation. Thus, DNMT1/miR-200a/GP73 signaling is implicated in lung adenocarcinoma development.81 Taken together, GP73 is an independent predictor of the prognosis of NSCLC.83
5.3. GP73 and pancreatic cancer
Pancreatic cancer is one of the most malignant cancers in the digestive tract, which is difficult to diagnose and treat. The 5-year survival rate is only 5%.84 Thus, appropriate biomarkers for early diagnosis are urgently needed. GP73 is highly expressed in pancreatic ductal adenocarcinoma.85 GP73 overexpression increases the growth and motility of pancreatic ductal adenocarcinoma cells. In contrast, downregulation of GP73 has opposite effects. Such effect is through regulating EMT signaling.84 Additionally, GP73 is downstream of RAS signaling, which promotes the progression of pancreatic ductal adenocarcinoma.85 Furthermore, GP73 interacts with AKT and increases its activity by phosphorylation. Activated AKT promotes the expression of a panel of genes involved in cell growth and apoptosis. Moreover, deactivate AKT eliminates GP73-induced cell growth in pancreatic cancer cells.85
5.4. GP73 and prostate cancer
Prostate cancer is the most commonly diagnosed cancer in men, and GP73 is highly expressed in it.86–91 GP73 has oncogenic effects by promoting proliferation, migration, and invasion of prostate cancer cells such as DU145, PC3, and CWR22Rv1. GP73 also inhibits apoptosis by activating the PI3K/AKT signaling pathway.90 Similar to HCC, miR-27b, a tumor suppressor, silences the expression of GP73 in prostate cancer PC3C and DU145 cells.71,92 Additionally, loss of the tumor-suppressor miR-143/145 cluster enhances prostate cancer cell migration and invasion through upregulating the expression of GP73.93 Thus, GP73 is a biomarker and therapeutic target for prostate cancer.
5.5. GP73 in breast and testicular cancer
Serum level of GP73 is elevated in patients with breast cancer compared with healthy people.94 Therefore, GP73 is hypothesized to be an early indicator of breast cancer. Like lung cancer, GP73 promotes breast cancer cell metastasis by regulating matrix metallopeptidase-13. It also facilitates the growth of breast cancer cells.95
Testicular neoplasms represent the most common malignancy in young adults, and seminoma is the most frequent single histologic subtype. GP73 is highly expressed in seminomas and has been proposed to be an immunohistochemical marker for the assessment of testicular neoplasms.96
6. Conclusion
GP73 as an oncogene, is regulated by the inflammatory signaling pathway, miRNAs, and hyper-activated mTOR signaling pathway. GP73 promotes the occurrence and development of HCC through inhibiting apoptosis and autophagy, as well as promoting EMT (Fig. 2). Although the oncogenic roles of GP73 in HCC have been revealed, a drug that inhibits GP73 has not been developed. Since elevated GP73 is also found in many other types of cancer, GP73 alone may not be a specific biomarker for HCC diagnosis. However, due to its oncogenic effects and elevated expression in many different types of cancer, targeting GP73 will likely have promising effects in cancer prevention and treatment. Additional research is warranted to further understand the functional roles of GP73 in oncogenesis.
Fig. 2. Schematic illustration of the mechanisms by which GP73 expression is regulated.
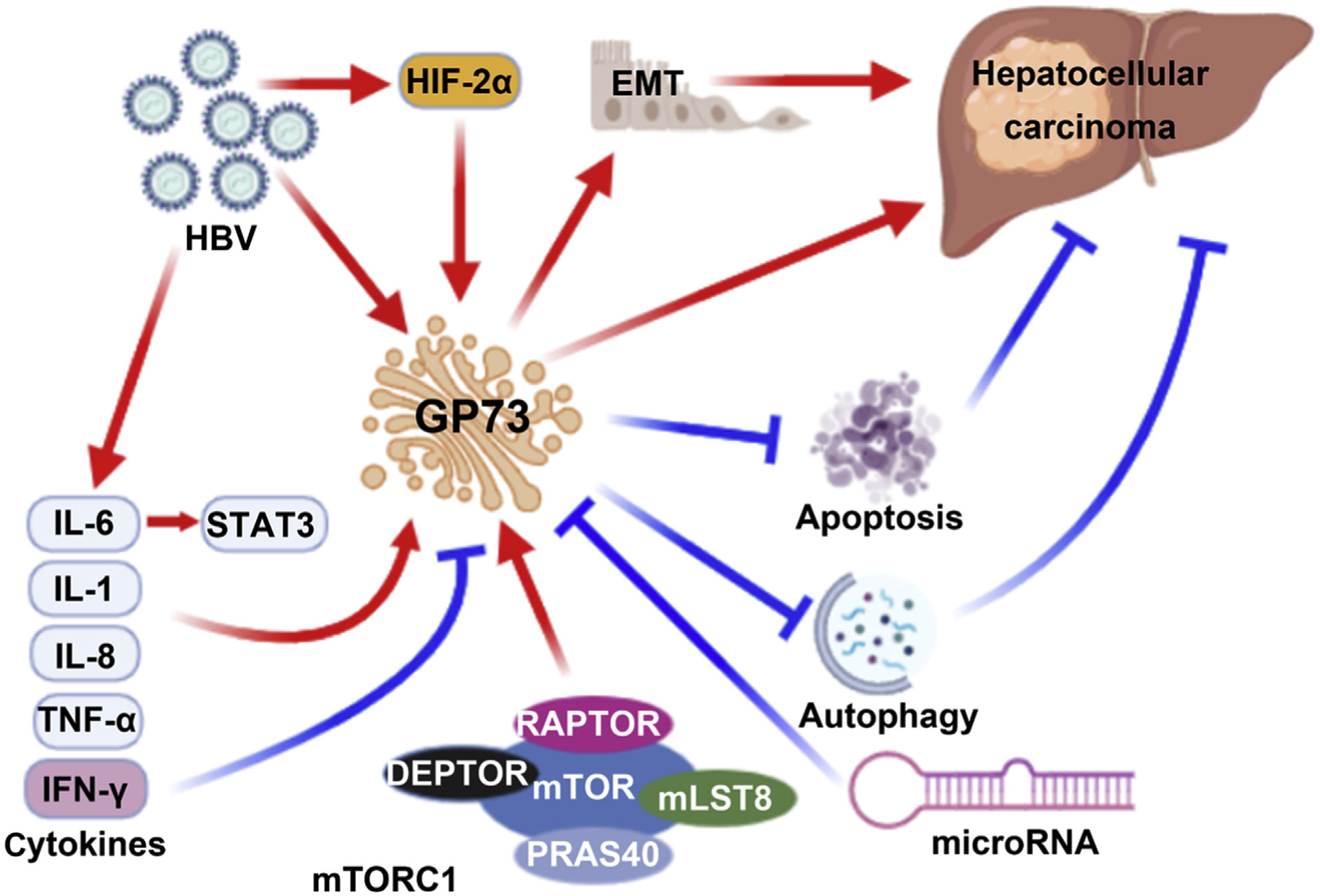
GP73 is regulated by HBsAg, cytokines, hyper-activated mTORC1, HIF-2α, and miRNA. GP73 promotes the occurrence and development of HCC through inhibition of apoptosis and autophagy, as well as promoting EMT. This figure was generated using tools available in the BioRender.com. Abbreviations: DEPTOR, DEP-domain containing mTOR-interacting protein; EMT, epithelial-mesenchymal transition; GP73, Golgi protein 73; HBV, hepatitis B virus; HIF-2α, hypoxia inducible factors-2alpha; IFN-γ, interferon-gamma; IL, interleukin; mLST8, mammalian lethal with SEC13 protein 8; mTOR, mechanistic target of rapamycin; mTORC1, mechanistic target of rapamycin complex 1; PRAS40, prolin-rich Akt substrate of 40 kD; RAPTOR, regulatory-associated protein of mammalian target of rapamycin; STAT3, signal transducer and activator of transcription 3; TNF-α, tumor necrosis factor-alpha.
Acknowledgements
The authors thank Miranda Claire Gilbert and Leyi Wang for editing this manuscript. This study was supported by grants funded by the USA National Institutes of Health (NIH) R01CA222490 to Y.-J. Y. Wan, along with grants from the National Natural Science Foundation of China Project number 81602456 and financial support from the China Scholarship Council (CSC) to Y. Wang.
Footnotes
Declaration of competing interest
The authors declare that they have no conflict of interest.
References
- 1..Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. [DOI] [PubMed] [Google Scholar]
- 3..Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60: 277–300. [DOI] [PubMed] [Google Scholar]
- 4..Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–595. [DOI] [PubMed] [Google Scholar]
- 5..Kim HS, El-Serag HB. The epidemiology of hepatocellular carcinoma in the USA. Curr Gastroenterol Rep. 2019;21:17. [DOI] [PubMed] [Google Scholar]
- 6..Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med. 2014;11, e1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7..Liang R, Liu Z, Piao X, et al. Research progress on GP73 in malignant tumors. Onco Targets Ther. 2018;11:7417–7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8..Wong RJ, Ahmed A, Gish RG. Elevated alpha-fetoprotein: differential diagnosis - hepatocellular carcinoma and other disorders. Clin Liver Dis. 2015;19: 309–323. [DOI] [PubMed] [Google Scholar]
- 9..West CA, Black AP, Mehta AS. Analysis of hepatocellular carcinoma tissue for biomarker discovery In: Hoshida Y, ed. Hepatocellular Carcinoma: Translational Precision Medicine Approaches. Cham (CH): Humana Press; 2019:93–107. [PubMed] [Google Scholar]
- 10..Wang CH, Wey KC, Mo LR, Chang KK, Lin RC, Kuo JJ. Current trends and recent advances in diagnosis, therapy, and prevention of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2015;16:3595–3604. [DOI] [PubMed] [Google Scholar]
- 11..Chen H, Chen S, Li S, et al. Combining des-gamma-carboxyprothrombin and alpha-fetoprotein for hepatocellular carcinoma diagnosing: an update meta-analysis and validation study. Oncotarget. 2017;8:90390–90401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12..Song P, Gao J, Inagaki Y, et al. Biomarkers: evaluation of screening for and early diagnosis of hepatocellular carcinoma in Japan and China. Liver Cancer. 2013;2: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13..Kladney RD, Bulla GA, Guo L, et al. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14..Kim HJ, Lv D, Zhang Y, Peng T, Ma X. Golgi phosphoprotein 2 in physiology and in diseases. Cell Biosci. 2012;2:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15..Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology. 2002;35:1431–1440. [DOI] [PubMed] [Google Scholar]
- 16..Ba MC, Long H, Tang YQ, Cui SZ. GP73 expression and its significance in the diagnosis of hepatocellular carcinoma: a review. Int J Clin Exp Pathol. 2012;5: 874–881. [PMC free article] [PubMed] [Google Scholar]
- 17..Wang L, Yao M, Liu S, et al. Serum Golgi protein 73 as a potential biomarker for hepatic necroinflammation in population with nonalcoholic steatohepatitis. Dis Markers. 2020;2020:6036904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18..Block TM, Comunale MA, Lowman M, et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci U S A. 2005;102:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19..Mao Y, Yang H, Xu H, et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59:1687–1693. [DOI] [PubMed] [Google Scholar]
- 20..Hu L, Li L, Xie H, Gu Y, Peng T. The Golgi localization of GOLPH2 (GP73/GOLM1) is determined by the transmembrane and cytoplasmic sequences. PloS One. 2011;6, e28207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21..Bachert C, Fimmel C, Linstedt AD. Endosomal trafficking and proprotein convertase cleavage of cis Golgi protein GP73 produces marker for hepatocellular carcinoma. Traffic. 2007;8:1415–1423. [DOI] [PubMed] [Google Scholar]
- 22..Fimmel CJ, Wright L. Golgi protein 73 as a biomarker of hepatocellular cancer: development of a quantitative serum assay and expression studies in hepatic and extrahepatic malignancies. Hepatology. 2009;49:1421–1423. [DOI] [PubMed] [Google Scholar]
- 23..Munro S Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24..Lupas A Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 25..Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. [DOI] [PubMed] [Google Scholar]
- 26..Norton PA, Comunale MA, Krakover J, et al. N-linked glycosylation of the liver cancer biomarker GP73. J Cell Biochem. 2008;104:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27..Iftikhar R, Kladney RD, Havlioglu N, et al. Disease- and cell-specific expression of GP73 in human liver disease. Am J Gastroenterol. 2004;99:1087–1095. [DOI] [PubMed] [Google Scholar]
- 28..Kleene R, Berger EG. The molecular and cell biology of glycosyltransferases. Biochim Biophys Acta. 1993;1154:283–325. [DOI] [PubMed] [Google Scholar]
- 29..Abeijon C, Mandon EC, Hirschberg CB. Transporters of nucleotide sugars, nucleotide sulfate and ATP in the Golgi apparatus. Trends Biochem Sci. 1997;22: 203–207. [DOI] [PubMed] [Google Scholar]
- 30..Marrero JA, Romano PR, Nikolaeva O, et al. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol. 2005;43: 1007–1012. [DOI] [PubMed] [Google Scholar]
- 31..Ibrahim GH, Mahmoud MA, Aly NM. Evaluation of circulating Transforming growth factor-beta1, Glypican-3 and Golgi protein-73 mRNAs expression as predictive markers for hepatocellular carcinoma in Egyptian patients. Mol Biol Rep. 2013;40:7069–7075. [DOI] [PubMed] [Google Scholar]
- 32..N Zekri AR, El Kassas M, Salam ESE, et al. The possible role of Dickkopf-1, Golgi protein- 73 and Midkine as predictors of hepatocarcinogenesis: a review and an Egyptian study. Sci Rep. 2020;10:5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33..Wang F, Long Q, Gong Y, et al. Epithelium-Specific ETS (ESE)-1 upregulated GP73 expression in hepatocellular carcinoma cells. Cell Biosci. 2014;4:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34..Chen MH, Jan YH, Chang PM, et al. Expression of GOLM1 correlates with prognosis in human hepatocellular carcinoma. Ann Surg Oncol. 2013;20 Suppl 3:S616–S624. [DOI] [PubMed] [Google Scholar]
- 35..Liu X, Wan X, Li Z, Lin C, Zhan Y, Lu X. Golgi protein 73(GP73), a useful serum marker in liver diseases. Clin Chem Lab Med. 2011;49:1311–1316. [DOI] [PubMed] [Google Scholar]
- 36..Sai W, Wang L, Zheng W, et al. Abnormal expression of Golgi protein 73 in clinical values and their role in HBV-related hepatocellular carcinoma diagnosis and prognosis. Hepat Mon. 2015;15, e32918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37..Yang Y, Xiao L, Mao R, et al. Expression profiles and differential diagnostic value of serum Golgi protein-73 in patients with liver cirrhosis and primary hepatic carcinoma (in Chinese). Zhonghua Gan Zang Bing Za Zhi. 2012;20: 920–924. [DOI] [PubMed] [Google Scholar]
- 38..Yao S, Zhang J, Chen H, et al. Diagnostic value of immunohistochemical staining of GP73, GPC3, DCP, CD34, CD31, and reticulin staining in hepatocellular carcinoma. J Histochem Cytochem. 2013;61:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39..Bao YX, Yang Y, Zhao HR, et al. Clinical significance and diagnostic value of Golgi-protein 73 in patients with early-stage primary hepatocellular carcinoma (in Chinese). Zhonghua Zhong Liu Za Zhi. 2013;35:505–508. [PubMed] [Google Scholar]
- 40..Sai WL, Yao M, Shen SJ, et al. Dynamic expression of hepatic GP73 mRNA and protein and circulating GP73 during hepatocytes malignant transformation. Hepatobiliary Pancreat Dis Int. 2020. [DOI] [PubMed] [Google Scholar]
- 41..Hann HW, Wang M, Hafner J, et al. Analysis of GP73 in patients with HCC as a function of anti-cancer treatment. Cancer Biomark. 2010;7:269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42..Liu T, Yao M, Liu S, et al. Serum Golgi protein 73 is not a suitable diagnostic marker for hepatocellular carcinoma. Oncotarget. 2017;8:16498–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43..Wang Y, Zhang Z, Zou D, et al. Serum Golgi protein 73 is a prognostic biomarker of liver transplantation patients. Int J Clin Exp Pathol. 2017;10: 8626–8632. [PMC free article] [PubMed] [Google Scholar]
- 44..Jia Z, Wang L, Liu C, Yu Z, Chai L, Zhao M. Evaluation of α-fetoprotein-L3 and Golgi protein 73 detection in diagnosis of hepatocellular carcinoma. Contemp Oncol (Pozn). 2014;18:192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45..Tian L, Wang Y, Xu D, et al. Serological AFP/Golgi protein 73 could be a new diagnostic parameter of hepatic diseases. Int J Canc. 2011;129:1923–1931. [DOI] [PubMed] [Google Scholar]
- 46..Shi Y, Chen J, Li L, et al. A study of diagnostic value of golgi protein GP73 and its genetic assay in primary hepatic carcinoma. Technol Cancer Res Treat. 2011;10: 287–294. [DOI] [PubMed] [Google Scholar]
- 47..Shi QF, Pan XD, Zhang WG, Shao WF. Diagnostic value and clinical significance of the Golgi protein-73 in hepatocellular carcinoma (in Chinese). Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2012;26:480–482. [PubMed] [Google Scholar]
- 48..Zhang Z, Zhang Y, Wang Y, Xu L, Xu W. Alpha-fetoprotein-L3 and Golgi protein 73 may serve as candidate biomarkers for diagnosing alpha-fetoprotein-negative hepatocellular carcinoma. Onco Targets Ther. 2015;9:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49..Zhao Y, Wang M, Cui C, et al. Significance of combined tests of serum golgi glycoprotein 73 and other biomarkers in diagnosis of small primary hepatocellular carcinoma. Cancer Biomark. 2015;15:677–683. [DOI] [PubMed] [Google Scholar]
- 50..Hou SC, Xiao MB, Ni RZ, et al. Serum GP73 is complementary to AFP and GGT-II for the diagnosis of hepatocellular carcinoma. Oncol Lett. 2013;6:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51..Kim SU, Park JH, Kim HS, et al. Serum Dickkopf-1 as a biomarker for the diagnosis of hepatocellular carcinoma. Yonsei Med J. 2015;56:1296–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52..Yu B, Yang X, Xu Y, et al. Elevated expression of DKK1 is associated with cytoplasmic/nuclear beta-catenin accumulation and poor prognosis in hepatocellular carcinomas. J Hepatol. 2009;50:948–957. [DOI] [PubMed] [Google Scholar]
- 53..Shan SG, Gao YT, Xu YJ, et al. Gradually increased Golgi protein 73 expression in the progression of benign liver diseases to precancerous lesions and hepatocellular carcinoma correlates with prognosis of patients. Hepatol Res. 2013;43:1199–1210. [DOI] [PubMed] [Google Scholar]
- 54..Ramakrishna G, Rastogi A, Trehanpati N, Sen B, Khosla R, Sarin SK. From cirrhosis to hepatocellular carcinoma: new molecular insights on inflammation and cellular senescence. Liver Cancer. 2013;2:367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55..Geier A, Jahn D, Hermanns HM. Interleukin-6: the dark side of liver regeneration in chronic liver disease. Hepatology. 2017;66:667–668. [DOI] [PubMed] [Google Scholar]
- 56..Liang H, Block TM, Wang M, et al. Interleukin-6 and oncostatin M are elevated in liver disease in conjunction with candidate hepatocellular carcinoma biomarker GP73. Cancer Biomark. 2012;11:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57..Liu L, Zhu J, Yang J, et al. GP73 facilitates hepatitis B virus replication by repressing the NF-κB signaling pathway. J Med Virol. 2020. February 20 10.1002/jmv.25718. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 58..Geis T, Döring C, Popp R, et al. HIF-2alpha-dependent PAI-1 induction contributes to angiogenesis in hepatocellular carcinoma. Exp Cell Res. 2015;331: 46–57. [DOI] [PubMed] [Google Scholar]
- 59..Yang SL, Zeng C, Fang X, et al. Hepatitis B virus upregulates GP73 expression by activating the HIF-2α signaling pathway. Oncol Lett. 2018;15:5264–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60..Chen X, Wang Y, Tao J, et al. mTORC1 up-regulates GP73 to promote proliferation and migration of hepatocellular carcinoma cells and growth of xeno-graft tumors in mice. Gastroenterology. 2015;149:741–752 (e714). [DOI] [PubMed] [Google Scholar]
- 61..Ye QH, Zhu WW, Zhang JB, et al. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30: 444–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62..Zhang YL, Zhang YC, Han W, et al. Effect of GP73 silencing on proliferation and apoptosis in hepatocellular cancer. World J Gastroenterol. 2014;20: 11287–11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63..Yang S, Yang L, Li X, et al. New insights into autophagy in hepatocellular carcinoma: mechanisms and therapeutic strategies. Am J Cancer Res. 2019;9: 1329–1353. [PMC free article] [PubMed] [Google Scholar]
- 64..Zhou YY, Jiang JC, You J, Zhou LF. Effect of Golgi phosphoprotein 2 (GOLPH2/GP73) on autophagy in human hepatocellular carcinoma HepG2 cells. Tumour Biol. 2015;36:3399–3406. [DOI] [PubMed] [Google Scholar]
- 65..Liu Y, Zhang X, Sun T, et al. Knockdown of Golgi phosphoprotein 2 inhibits hepatocellular carcinoma cell proliferation and motility. Oncotarget. 2016;7: 21404–21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66..Jiang K, Li W, Shang S, et al. Aberrant expression of Golgi protein 73 is indicative of a poor outcome in hepatocellular carcinoma. Oncol Rep. 2016;35: 2141–2150. [DOI] [PubMed] [Google Scholar]
- 67..Bao YX, Cao Q, Yang Y, et al. Expression and prognostic significance of golgi-glycoprotein73 (GP73) with epithelial-mesenchymal transition (EMT) related molecules in hepatocellular carcinoma (HCC). Diagn Pathol. 2013;8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68..Yang Y, Liu Q, Zhang H, et al. Silencing of GP73 inhibits invasion and metastasis via suppression of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncol Rep. 2017;37:1182–1188. [DOI] [PubMed] [Google Scholar]
- 69..Hou X, Yang L, Jiang X, et al. Role of microRNA-141–3p in the progression and metastasis of hepatocellular carcinoma cell. Int J Biol Macromol. 2019;128: 331–339. [DOI] [PubMed] [Google Scholar]
- 70..Zhang S, Ge W, Zou G, et al. MiR-382 targets GOLM1 to inhibit metastasis of hepatocellular carcinoma and its down-regulation predicts a poor survival. Am J Cancer Res. 2018;8:120–131. [PMC free article] [PubMed] [Google Scholar]
- 71..Liang H, Ai-Jun J, Ji-Zong Z, et al. Clinicopathological significance of miR-27b targeting Golgi protein 73 in patients with hepatocellular carcinoma. Anti-cancer Drugs. 2019;30:186–194. [DOI] [PubMed] [Google Scholar]
- 72..Sha M, Wang B, Xiao L, Ye J, Wang J, Luan ZY. Expression of miR-212 and miR-132 in serum of patients with primary liver cancer and their targeted regulation of GP73 (in Chinese). Zhonghua Gan Zang Bing Za Zhi. 2017;25:920–926. [DOI] [PubMed] [Google Scholar]
- 73..Zhao J, Xu T, Wang F, Cai W, Chen L. miR-493–5p suppresses hepatocellular carcinoma cell proliferation through targeting GP73. Biomed Pharmacother. 2017;90:744–751. [DOI] [PubMed] [Google Scholar]
- 74..Wright LM, Yong S, Picken MM, Rockey D, Fimmel CJ. Decreased survival and hepato-renal pathology in mice with C-terminally truncated GP73 (GOLPH2). Int J Clin Exp Pathol. 2009;2:34–47.18830387 [Google Scholar]
- 75.Arora G Hepatocyte Specific GP73/GOLM1 Knockout Mice and Human Hepatocellular Carcinoma Cell Lines Provide Insights into the Potential Roles of GP73 HCC Serum Biomarker. 2016.
- 76..Matthews NH, Li WQ, Qureshi AA, Weinstock MA, Cho E. Epidemiology of melanoma In: Ward WH, Farma JM, eds. Cutaneous Melanoma: Etiology and Therapy. Brisbane (AU): Codon Publications; 2017. [PubMed] [Google Scholar]
- 77..Donizy P, Kaczorowski M, Biecek P, Halon A, Szkudlarek T, Matkowski R. Golgi-related proteins GOLPH2 (GP73/GOLM1) and GOLPH3 (GOPP1/MIDAS) in cutaneous melanoma: patterns of expression and prognostic significance. Int J Mol Sci. 2016;17:1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78..Chen WY, Xu YY, Zhang XY. Targeting GOLM1 by microRNA-200a in melanoma suppresses cell proliferation, invasion and migration via regulating PI3K/Akt signaling pathway and epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2019;23:6997–7007. [DOI] [PubMed] [Google Scholar]
- 79..Teerlink CC, Huff C, Stevens J, et al. A nonsynonymous variant in the GOLM1 gene in cutaneous malignant melanoma. J Natl Cancer Inst. 2018;110: 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80..Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103: 463–473. [DOI] [PubMed] [Google Scholar]
- 81..Yang L, Luo P, Song Q, Fei X. DNMT1/miR-200a/GOLM1 signaling pathway regulates lung adenocarcinoma cells proliferation. Biomed Pharmacother. 2018;99:839–847. [DOI] [PubMed] [Google Scholar]
- 82..Aruna Li LM. Overexpression of golgi membrane protein 1 promotes non-small-cell carcinoma aggressiveness by regulating the matrix metal-opeptidase 13. Am J Cancer Res. 2018;8:551–565. [PMC free article] [PubMed] [Google Scholar]
- 83..Zhang Y, Hu W, Wang L, Han B, Lin R, Wei N. Association of GOLPH2 expression with survival in non-small-cell lung cancer: clinical implications and biological validation. Biomark Med. 2017;11:967–977. [DOI] [PubMed] [Google Scholar]
- 84..Song YX, Xu ZC, Li HL, Yang PL, Du JK, Xu J. Overexpression of GP73 promotes cell invasion, migration and metastasis by inducing epithelial-mesenchymal transition in pancreatic cancer. Pancreatology. 2018;18:812–821. [DOI] [PubMed] [Google Scholar]
- 85..Duan J, Li X, Huang S, et al. GOLPH2, a gene downstream of ras signaling, promotes the progression of pancreatic ductal adenocarcinoma. Mol Med Rep. 2018;17:4187–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86..Li W, Wang X, Li B, Lu J, Chen G. Diagnostic significance of overexpression of Golgi membrane protein 1 in prostate cancer. Urology. 2012;80:952–957. [DOI] [PubMed] [Google Scholar]
- 87..Varambally S, Laxman B, Mehra R, et al. Golgi protein GOLM1 is a tissue and urine biomarker of prostate cancer. Neoplasia. 2008;10:1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88..Kristiansen G Immunhistochemische Algorithmen in der Prostatadiagnostik: was gibt es Neues? [Immunohistochemical algorithms in prostate diagnostics: what’s new?](in German). Pathologe. 2009;30:146–153. [DOI] [PubMed] [Google Scholar]
- 89..Kristiansen G, Fritzsche FR, Wassermann K, et al. GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics. Br J Cancer. 2008;99:939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90..Yan G, Ru Y, Wu K, et al. GOLM1 promotes prostate cancer progression through activating PI3K-AKT-mTOR signaling. Prostate. 2018;78:166–177. [DOI] [PubMed] [Google Scholar]
- 91..Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92..Goto Y, Kojima S, Nishikawa R, et al. The microRNA-23b/27b/24–1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget. 2014;5:7748–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93..Kojima S, Enokida H, Yoshino H, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;59:78–87. [DOI] [PubMed] [Google Scholar]
- 94..Zhang A, Cao B. Generation and characterization of an anti-GP73 monoclonal antibody for immunoblotting and sandwich ELISA. J Biomed Res. 2012;26: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95..Zhang R, Zhu Z, Shen W, Li X, Dhoomun DK, Tian Y. Golgi membrane protein 1 (GOLM1) promotes growth and metastasis of breast cancer cells via regulating matrix metalloproteinase-13 (MMP13). Med Sci Monit. 2019;25:847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96..Fritzsche FR, Kristiansen G, Riener MO, Dietel M, Oelrich B. GOLPH2 expression may serve as diagnostic marker in seminomas. BMC Urol. 2010;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]


