Abstract
Long noncoding RNAs (lncRNAs) play a crucial role in progression of bladder urothelial carcinoma (BUC). However, the molecular mechanisms behind this role have not been elucidated yet. Here, we found that the lncRNA MAFG‐AS1, which is highly expressed in BUC, is correlated with aggressive characteristics and poor prognosis of BUC. We demonstrate that MAFG‐AS1 can promote BUC proliferation, invasion, metastasis, and epithelial‐mesenchymal transition in vitro and in vivo. Mechanistically, MAFG‐AS1 direct binding to Hu antigen R (HuR) could recruit ubiquitin‐specific proteinase 5 (USP5) to prevent HuR from degrading by ubiquitination. We further demonstrate that overexpression of MAFG‐AS1 can upregulate the expression of polypyrimidine tract‐binding protein 1 (PTBP1) through promoting its stability mediated by bound HuR. In conclusion, these findings indicate that MAFG‐AS1 promotes the progression of BUC via regulation of the HUR/PTBP1 axis. Targeting MAFG‐AS1 may provide a novel strategy for individualized therapy and a potential biomarker for prognosis of BUC.
Keywords: bladder urothelial carcinoma, HuR, MAFG‐AS1, PTBP1
Our findings indicate that MAFG‐AS1 acts as an oncogene to promote the progression of BUC via regulation of the HUR/PTBP1 axis. Targeting MAFG‐AS1 may provide a novel strategy for individualized therapy and a potential biomarker for prognosis of BUC
Abbreviations
- ARE
AU‐rich elements
- BUC
bladder urothelial carcinoma
- ChIRP
chromatin isolation by RNA purification
- CHX
cycloheximide
- Co‐IP
co‐immunoprecipitation
- FISH
fluorescence in situ hybridization
- HuR
Hu antigen R
- IHC
immunohistochemistry
- IP
immunoprecipitation
- lncRNA
long noncoding RNA
- MIBC
muscle invasive bladder cancer
- miRNA
micro‐RNA
- NMIBC
nonmuscle invasive bladder cancer
- PRM
parallel reaction monitoring
- PTBP1
polypyrimidine tract‐binding protein 1
- RBP
RNA‐binding protein
- RIP
RNA immunoprecipitation
- RT‐qPCR
quantitative real‐time polymerase chain reaction
- TNM
tumor‐node metastasis
- USP5
ubiquitin‐specific processing protease 5
1. INTRODUCTION
As the most prevalent malignant neoplasm that occurs in the urinary system, bladder urothelial carcinoma (BUC) has experienced an increasing incidence and mortality rate in recent years. There are an estimated 430 000 BUC new cases, and 165 000 deaths globally every year. 1 BUC is divided into two classifications: nonmuscle invasive bladder cancer (NMIBC), which accounts for 70‐80% of cases, and muscle invasive bladder cancer (MIBC), which represents the remaining cases. Due to the progressiveness and distant metastasis of NMIBC, nearly 75% of high‐grade NMIBC end with relapse, progression, or death within 5 years. Despite significant improvements in radical cystectomy, neoadjuvant chemotherapy, and radiotherapy for MIBC, the mortality rate is up to 77.6% within 5 years of diagnosis. 2 , 3 Therefore, the progression, extensive metastasis, and recurrence of BUC remain major challenges, and the investigations of molecular mechanisms that contribute to these phenomena is urgently required.
Long noncoding RNAs (lncRNAs) are RNAs longer than 200 nucleotides with no or limited ability to encode proteins, 4 and can promote tumor proliferation, invasion, and migration through various mechanisms to affect tumor development. 5 , 6 , 7 , 8 For example, lncRNAs may act as sponges by absorbing micro‐RNA (miRNA), or regulate neighboring genes in cis by recruiting regulatory factors to the locus and regulating their functions. Moreover, lncRNAs can also bind proteins to promote their functions by preventing their degradation and increasing their stability. In recent years, there has been increasing evidence showing that the interactions between lncRNAs and proteins play an important role in carcinogenesis and metastasis. 9 , 10 , 11 , 12 , 13 , 14 For example, BLACAT2 can bind with WDR5 to promote lymphatic metastasis through upregulating VEGF‐C expression in BUC. 15 Additionally, Wang et al 16 have confirmed that lncRNA EPIC1 can directly bind to Myc and form a positive feedback loop to stimulate tumorigenesis in breast cancer. However, further clarification of the molecular interactions between lncRNAs and proteins is required, and this may contribute to understanding the progressiveness and distant metastasis in BUC.
RNA‐binding proteins (RBPs) are proteins that interact with RNA molecules to regulate the production, maturation, and localization of RNA in cells as well as their translation and degradation. Some studies have reported that dysregulated RBPs can lead to tumorigenesis by affecting the expression of genes. Hu antigen R (HuR), also known as ELAVL1, a member of ELAV‐like protein 1 family, is a ubiquitously expressed RBP that binds to AU‐rich elements (ARE) in the 3′UTR of mRNA. 6 , 17 Reports have shown that HuR could function as an oncogene in tumors, 18 , 19 , 20 , 21 and that increased HuR expression contributes to carcinogenesis and poor prognosis in BUC. 22 , 23 However, the precise mechanisms of HuR involvement in tumorigenesis of BUC remain elusive.
Here, we determined that MAFG‐AS1 is the only lncRNA that is highly expressed in BUC according to both the GSE31189 and TCGA databases. High expression of MAFG‐AS1 is correlated with aggressive cancer characteristics and poor prognosis. Furthermore, we found that MAFG‐AS1 can directly bind to HuR and stabilize it by recruiting the deubiquitinating enzyme, ubiquitin‐specific processing protease 5 (USP5). Moreover, we demonstrate that MAFG‐AS1 promotes malignant phenotypes in BUC by upregulating polypyrimidine tract‐binding protein 1 (PTBP1) via direct binding to and stabilization of HuR. Taken together, we reveal that the MAFG‐AS1/HuR/PTBP1 axis facilitates carcinogenesis and progression in BUC, which could provide new targets and strategies for the therapy of BUC.
2. MATERIALS AND METHODS
2.1. Bioinformatics analysis
lncRNA expression profiles in human BUC tissues were derived from the Cancer RNA‐Seq Nexus, 24 TANRIC, 25 and Xena databases. GEPIA and UALCAN 26 databases were used for survival analysis. GEO (GSE31189, GSE87304, GSE13507, GSE31684, GSE124305), ChIPBase 2.0, and starBase 2.0 databases were used to predict the correlation between RBPs and lncRNAs. catRAPID, 27 starBase 2.0, HDOCK, 28 and POSTAR2.0 databases were used to predict the possible binding sites of RBPs on lncRNA. Gene ontology (GO) analysis was conducted using the Metascape database. A list of RNA degradation genes was downloaded from the Molecular Signatures Databases 29 (KEGG_RNA_DEGRADATION, M963). Gene set enrichment analysis (GSEA) was performed using the GSEA software. All databases used are listed in Table S7.
2.2. Tissues and cell lines
For MAFG‐AS1 expression profiling analysis, a cohort of BUC tissues was derived from 102 patients with BUC who had undergone radical cystectomy at the Sun Yat‐sen University Cancer Center (Guangzhou, Guangdong, China) and Third Xiangya Hospital of Central South University (Changsha, Hunan, China) from 2007 to 2015. The tumor‐node metastasis (TNM), grade, and stage were classified with the World Health Organization and American Joint Committee on Cancer. The clinicopathologic characteristics of these patients are summarized in Table S1. An additional panel of six formalin‐fixed, paraffin‐embedded tissues, snap‐frozen fresh BUC tissues, and matched adjacent normal bladder tissues were obtained from patients who underwent radial cystectomy at the Third Xiangya Hospital of Central South University between 2016 and 2018. These clinical specimens were previously approved for research use by the patients and the ethics committee. The fresh tissue samples were quantified by quantitative real‐time polymerase chain reaction (RT‐qPCR) and/or Western blotting as described below. We purchased human BUC cells (5637, BIU87, EJ, RT4, and T24 cells) from ATCC (Rockville, MD), and cultured in DMEM (Invitrogen, CA), added with 10% fetal bovine serum (FBS) (GIBCO, NY), 1 mmol/L glutamine, and 1% penicillin/streptomycin.
Highlights
lncRNA MAFG‐AS1 is correlated with aggressive characteristics of BUC;
BUC patients with high levels of MAFG‐AS1 have poor prognosis;
MAFG‐AS1 direct binding to HuR could recruit USP5 to prevent HuR from degrading by ubiquitination;
MAFG‐AS1 promotes malignant phenotypes by upregulating of PTBP1 via direct binding to and stabilization of HuR in BUC.
2.3. Cell transfection
Plasmids for overexpression MAFG‐AS1 were purchased from GeneChem Biotechnology (Shanghai, China). Three MAFG‐AS1‐specific si‐RNAs (si‐RNA1, si‐RNA2, and si‐RNA3), si‐HuR, si‐PTBP1, si‐USP5, si‐UCHL5, si‐COPS6, si‐PSMD14, si‐OTUB1, and shRNAs targeting MAFG‐AS1, HuR, PTBP1, and negative control were purchased from GeneChem Biotechnology. The T24 and RT4 cells were transfected the recombinant plasmid using Lipofectamine 3000 (Invitrogen). RT‐qPCR was performed to determine the transfection efficiency after 48‐72 hours of transfection.
2.4. RT‐qPCR
Total RNA was extracted from BUC tissues and cells using TRIzol (Invitrogen) and the purity of the RNA was assessed spectrophotometrically (A260/A280 > 1.8). Total RNA (500 ng) was subjected into cDNA using the MMLV reverse transcriptase enzyme (Promega, Madison, WI). RT‐qPCR was performed using SYBR Green PCR Master Mix (Life Technologies Corporation, CA). Gene expression was normalized to that GAPDH mRNA or U1 snRNA. The reactions were performed independently and in triplicate. The 2−ΔΔCt method was applied to calculate relative expression levels of genes. All primers used are listed in Table S8.
2.5. Wound‐healing and transwell assays
The migration capacity of BUC cells was measured by wound‐healing assays as described previously. 30 Briefly, BUC cells were cultivated in a six‐well plate. The cellular monolayers were wounded by scratching with a 20 μL pipette tip when cultures reached >90% confluency. Images were photographed at 0, 24, and 48 hours after wounding. The invasiveness of cells assessed using a transwell assay. BUC cells (5 × 104) were plated into a transwell plate of 8 μm pores (Corning Inc, Corning, NY). The two layers of the transwell contained two different media compositions: DMEM medium containing 1% FBS (upper transwell chamber), and DMEM containing 15% FBS (lower transwell chamber). The invasive cells were fixed with 95% ethanol, stained with hematoxylin (after 24 hours incubation), and then counted using an inverted microscope.
2.6. MTT assay
Transfected BUC cells were inoculated in 96‐well plates and incubated for 24, 48, 72, and 96 hours at 37°C and 5% CO2, then MTT (50 μL) (Sigma Chemicals, MO) was added to each well and incubated for an additional 4 hours. Supernatants were removed from cells by aspiration and DMSO was added. Finally, the absorbance at 570 nm was measured.
2.7. Clone formation assay
BUC cells, 48 hours posttransfection were inoculated into a six‐well plate and cultured at 37°C, 5% CO2, and saturated humidity for 2‐3 weeks. Cell colonies were fixed with 4% paraformaldehyde and stained with hematoxylin. Fixed and stained cells were counted to quantify clone formation. If the number of cloned cells added up to 50, it was counted as a clone.
2.8. Western blotting
Western blotting was performed as described previously. 31 , 32 Briefly, tissues and cells were lysed on ice for 30 minutes using RIPA lysis buffer (Beyotime, Shanghai, China) containing 10% protease inhibitor cocktail (Roche, IN), and the protein concentration was quantified with a BCA Protein Assay Kit (Thermo Scientific). Subsequently, the protein samples were separated on a 10% SDS‐polyacrylamide gel and transferred to PVDF membrane. The membranes were blocked in 5% skimmed milk powder in PBS and incubated overnight anti‐HuR (Proteintech, 11910‐1‐AP), anti‐PTBP1 (Abcam, ab5642), anti‐UCHL5 (Abcam, ab124931), anti‐USP5 (Proteintech, 10473‐1‐AP), anti‐COPS6 (Abcam, ab77299), anti‐PMSD14 (Proteintech, 12059‐1‐AP), anti‐OTUB1 (Abcam, ab233160), and anti‐GAPDH (Abcam, ab125247) at 4°C with. This was followed by 30‐60 minutes incubation with secondary antibody. ECL was used to visualize the signals of protein bands and quantified using the optical density analysis software Quantity One (Bio‐Rad). GAPDH protein levels were used as internal control.
2.9. Xenograft mouse model
Five‐week‐old female BALB/c nude mice weighing 18‐22 g was purchased from Shanghai Laboratory Animal Center (SLAC, Shanghai, China). Briefly, 1 × 106 of overexpressing MAFG‐AS1 T24/RT4 cells, sh‐MAFG‐AS1 T24/RT4 cells, MAFG‐AS1 + sh‐HuR T24/RT4 cells, and MAFG‐AS1 + sh‐PTBP1 T24/RT4 cells were injected subcutaneously into mice. Tumor volumes were calculated using the equation (length × width2)/2 every 3 days. Nude mice were euthanized at day 25. For lung metastasis models, 2 × 106 of T24/RT4 cells were injected into mice via the tail vein. The tumors and lungs were stained with hematoxylin and eosin (H&E) at day 45. The number of lung metastatic nodules in each mouse was calculated. All experimental procedures were approved by the University Ethics Committee.
2.10. Immunohistochemistry (IHC)
IHC staining was performed following similar methods to those previously published. 31 Cancer tissue sections were deparaffinized, then blocked with 1% PBA. The prepared sections were incubated with anti‐HuR (Proteintech, 11910‐1‐AP), anti‐PTBP1 (Abcam, ab5642), and anti‐Ki67 (Genetex, GTX 16667) at 4°C overnight. The incubated polymer enhancers were then added with a biotin‐labeled secondary antibody. Next, the sections were stained with a DAB staining solution, and counterstained with hematoxylin. Two independent pathologists assessed the IHC scores in a blinded manner; the details of the scoring methodology have been previously published. 31
2.11. Fluorescence in situ hybridization (FISH)
The Cy3‐labeled MAFG‐AS1 probe and U6 RNA were designed and synthesized by RiboBio (Guangzhou, China). FISH kit (RiboBio) was used to implement the FISH experiments as per the manufacturer's instructions.
2.12. Immunoprecipitation (IP)
2.12.1. RNA immunoprecipitation (RIP)
The RIP assay was performed using the EZ‐Magna RIP kit (Millipore, MA). Briefly, collected and incubated BUC cells (1 × 107) with RIP lysis buffer. The precleared lysates were used for RIP with anti‐HuR (Proteintech, 11910‐1‐AP) and rabbit isotype control IgG antibodies. RNA was isolated and purified using an acid phenol/chloroform method.
2.12.2. Co‐immunoprecipitation (Co‐IP)
The growth medium of adherent cells in a 10 cm cell culture dish was removed by aspiration and washed once with PBS. The cells were then lysed with 1 mL IP buffer, mixed with 2 μg anti‐HuR (Proteintech, 11910‐1‐AP) and incubated with rotation overnight at 4°C. Protein A/G Agarose was fully resuspended in 20 μL buffer A and shaken slowly. IP complex was washed in IP buffer five times and 20 μL of 1 × SDS‐PAGE loading buffer was added and mixed. The pellet was vortexed and then centrifuged to concentrate the sample at the bottom of the tube. Western blot analysis was performed using antiubiquitin (Proteintech, 10201‐2‐AP) and anti‐USP5 (Proteintech, 10473‐1‐AP).
2.13. Chromatin isolation by RNA purification
In total, 1 × 109 to 5 × 109 cells were used for the chromatin isolation by RNA purification‐mass spectrometry (ChIRP‐MS) experiment. Briefly, after harvesting, cross‐linking, and lysis of cells, the DNA was sheared into small fragments through sonication. Lysed cells were transfected with a biotin‐labeled MAFG‐AS1 probe or NC probe (Table S4). ChIRP was performed with the following modifications 33 : (a) cross‐linking cells in 3% formaldehyde for 30 minutes, and then quenched with 0.125 M glycine for 5 minutes; (b) for hybridization, 100 pmol probe was mixed with chromatin (1 mL) in Falcon tube (15 mL) and incubated at 37°C for 4 hours with shaking, stripped the beads from the buffer with a DynaMag‐2 magnet; (c) for MS, the beads were removed twice from precleared lysates using a magnetic stand; (d) for RNase control, the lysates were first pooled and mixed into two equal parts. Before hybridization, samples were incubated at 37°C for 30 minutes with mixing. Then, RNA extraction and protein elution were carried out as previously described. 34 Finally, protein samples were size‐separated by a nano‐UPLC liquid phase system (EASY‐nLC1200) and used for MS.
2.14. Parallel reaction monitoring (PRM)
PRM‐MS analysis was as previously described. 35 Tryptic polypeptides (5 μL) from each sample were dissolved in solution A (0.1% formic acid‐acetonitrile). Peptides were separated on a reversed‐phase chromatographic column (Reprosil‐Pur 120 C18‐AQ, 1.9 μm, Dr. Math) using the following gradient system at a flow rate of 300 nL/min: 92 minutes from 8% to 35% buffer B (98% acetonitrile and 0.1% formic acid), 20 minutes from 35% to 45% buffer B, 2 minutes from 45% to 100% buffer B, 2 minutes at 100% buffer B, and 2 minutes at 2% buffer B. PRM data were collected by MS and imported into Skyline (version 3.6.1) for transition extraction.
2.15. Stability and α‐amanitin treatment
We inoculated MAFG‐AS1 shRNA, NC shRNA, with T24/RT4 cells overexpressing MAFG‐AS1 and empty plasmid. Then treated with 50 μg/Ml α‐amanita toxin, 36 and harvested after 24 hours. The cells were processed for RNA purification and RT‐qPCR after 6, 12, 18, 24, and 48 hours of treatment. Three independent samples were collected for each data point.
2.16. Statistical analysis
GraphPad Prism 8.0.2 was used for all statistical analyses, and all experiments were repeated three times. Data are expressed as mean ± standard deviation (SD). Significant differences between the two groups were analyzed using Student's t‐tests, whereas significant differences between more than two groups were analyzed using one‐way ANOVA followed by Dunnett's test. Survival curves were plotted using the Kaplan‐Meier method and compared with log‐rank tests. P‐value < .05 was considered to be a significant difference.
3. RESULTS
3.1. MAFG‐AS1 is highly expressed and negatively correlated with prognosis in BUC
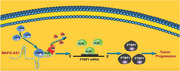
The level of MAFG‐AS1 was higher in BUC tissues than that in adjacent normal tissues using RT‐qPCR (Figure S1C). Moreover, analysis of both GEO (GSE31189) and TCGA databases revealed that MAFG‐AS1 is the only lncRNA that is significantly upregulated in both databases (Figure 1A). MAFG‐AS1 was significantly upregulated in BUC patients in clinical stages II, III, and IV compared with adjacent normal tissues (Figures 1B and 1C). Interestingly, MAFG‐AS1 was upregulated in multiple cancers, for instance, breast cancer and colorectal adenocarcinoma (Figure S1A). We analyzed the expression of MAFG‐AS1 to determine its potential clinical and prognostic significance in a cohort of 102 BUC tissues. We found that the higher levels of MAFG‐AS1 were correlated with advanced T and N stages (Table S1). Moreover, BUC patients with highly expressed MAFG‐AS1 were associated with poorer prognosis by Kaplan‐Meier survival analysis (Figure 1D). In univariate analyses, MAFG‐AS1 levels, T status, and N status were notably associated with survival of BUC patients (Table S2). Further multivariate analysis revealed that MAFG‐AS1 and N status were independent prognostic indicators for survival (Table S3). Additionally, the expression level of MAFG‐AS1 in BUC patients with stage III‐IV BUC was higher than that in stage II patients (Figure S1B). The relative expression of MAFG‐AS1 in BUC patients at different stages was further verified by analyzing the TANRIC database. As shown in Figure 1F, the expression level of MAFG‐AS1 was markedly higher in advanced clinical stages (III‐IV, N = 292), advanced T stages (T3‐4, N = 269), advanced N stages (N1‐2, N = 174), and advanced M stages (M1‐2, N = 219) of BUC patient tissues, compared to those with earlier clinical stages (I‐II, n = 132), T stages (T1‐2, n = 124), N stage (N0, n = 246), and M stage (M0, N = 204), respectively. Furthermore, the GEPIA dataset indicated that patients with high MAFG‐AS1 expression levels not only had a shorter overall survival but also a shorter disease‐free survival (Figure 1E; Figure S1D). Altogether, these data suggested that MAFG‐AS1 was highly expressed in BUC, which associated with aggressive characteristics and poor prognosis, further supporting the oncogenic role of MAFG‐AS1 in BUC.
FIGURE 1.
MAFG‐AS1 is highly expressed and negatively correlated with prognosis in bladder urothelial carcinoma (BUC). A, Screening significantly dysregulated lncRNAs in BUC. B, Expression level of MAFG‐AS1 in BUC patients with each stage; data from Cancer RNA‐Seq Nexus. C, The expression of MAFG‐AS1 in cancer and normal tissues of BUC; data from GES31189 dataset and TANRIC database. D and E, The relationship between MAFG‐AS1 and the prognosis of BUC. F, The expression of MAFG‐AS1 in different stages of BUC patients. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
3.2. MAFG‐AS1 can promote proliferation, invasion, metastasis, and EMT of BUC cells
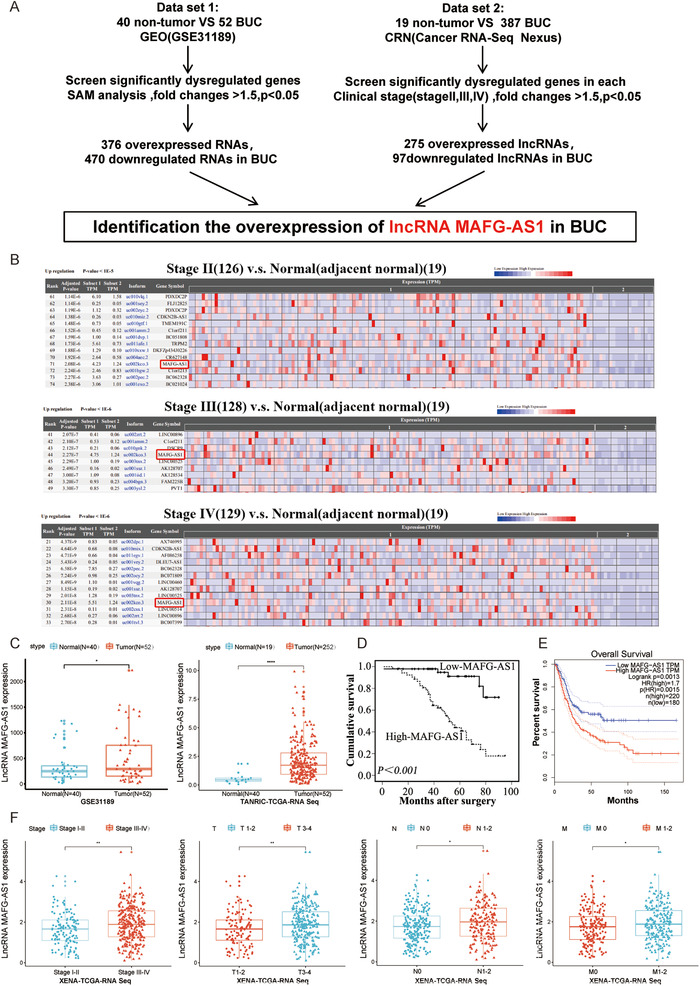
Since the upregulated MAFG‐AS1 is closely related to the prognosis of patients, we further explored the function of MAFG‐AS1 in BUC. We examined the expression of MAFG‐AS1 in five BUC cell lines and found that the expression of MAFG‐AS1 was relatively higher in T24 cells compared with RT4 cells through RT‐qPCR (Figure 2A). Subsequently, RT4 and T24 BUC cell lines were established to stably overexpress MAFG‐AS1 and siRNA targeting MAFG‐AS1 (Figures 2B and 2C). The proliferative capacities of cells were strikingly suppressed after knockdown of MAFG‐AS1 by clone formation and MTT assays (Figures 2D and 2E). The influence of MAFG‐AS1 on invasion and metastasis was investigated by transwell and wound‐healing assay. Similarly, the invasion and metastatic ability of BUC cells was weakened after knocking down MAFG‐AS1 (Figures 2F and 2G). On the contrary, overexpression of MAFG‐AS1 dramatically elevated cell proliferation, migration, and invasion in BUC cells (Figure 2D‐G). Meanwhile, E‐cadherin, the epithelial marker, was increased after silencing MAFG‐AS1, while the expression of vimentin, the mesenchymal marker, was increased in MAFG‐AS1‐overexpressing cells, indicating that MAFG‐AS1 contributed to epithelial‐mesenchymal transition (EMT) progression in BUC (Figure 2H). Additional studies were conducted to verify the effects of MAFG‐AS1 on subcutaneous tumor formation and distant lung metastasis in BUC cells. Tumor volume was significantly reduced in the sh‐MAFG‐AS1 group compared with the control group (Figure 2I). The positive rate of proliferation index Ki67 (Figure 2J) and the number of lung metastatic nodules (Figure 2K) were significantly reduced after silencing MAFG‐AS1, while overexpression of MAFG‐AS1 exerted promoting effects on the tumor volume (Figure 2I), proliferative ability (Figure 2J), and lung metastatic capacities (Figure 2K). These results indicated that upregulation of MAFG‐AS1 considerably promoted progression of BUC both in vivo and in vitro.
FIGURE 2.
MAFG‐AS1 can promote the proliferation, invasion, metastasis, and epithelial‐mesenchymal transition (EMT) of bladder urothelial carcinoma (BUC) cells. A, The expression level of MAFG‐AS1 in different BUC cell lines (BIU87, 5637, T24, EJ, and RT4) was detected by RT‐qPCR. B and C, The expression level of MAFG‐AS1 in T24/RT4 cells was detected by RT‐qPCR after transfection of si‐RNA and overexpression plasmid. D and E, MTT (D) and clone formation assays (E) showed the proliferation of T24 and RT4 cells with knockdown or overexpression of MAFG‐AS1. F and G, Wound‐healing (F) and transwell assays (G) were performed to evaluate cell migration and invasion in T24 and RT4 cells treated with knockdown or overexpression of MAFG‐AS1. H, Western blot analysis showed the expression level of EMT‐related proteins. I, Images of tumor xenografts from nude mice. J, Immunohistochemistry to detect the expression level of Ki67 in tissues. K, Representative images of lung metastatic foci after hematoxylin and eosin (H&E) staining. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
3.3. MAFG‐AS1 binds directly to HuR
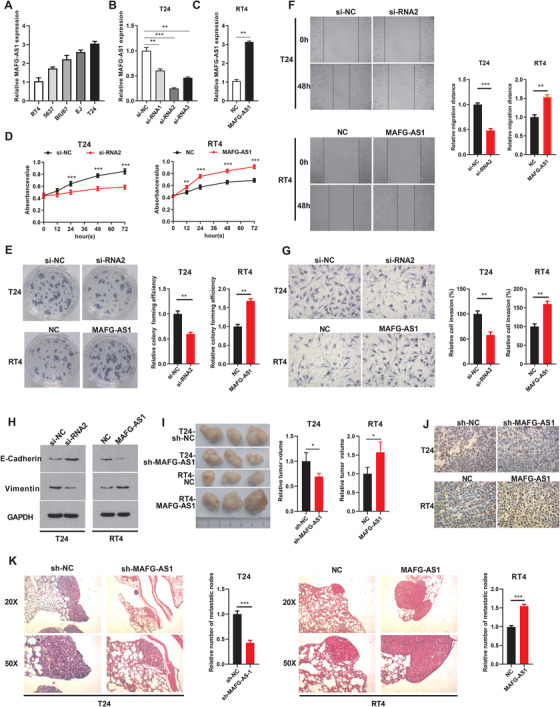
To identify the location of MAFG‐AS1, RNA FISH was performed. The results revealed that MAFG‐AS1 was enriched in not only cytoplasm but also in nucleus of T24 and RT4 cells (Figure 3A). Taking into account that lncRNAs can bind with proteins to promote tumor development, we then screened for the binding protein of MAFG‐AS1 using ChIRP‐MS assay (Figure 3B). The results showed that the specific probe group of MAFG‐AS1 bound to 26 proteins (Figure 3C; Table S5). We then performed GO analysis based on only these 26 genes. The GO analysis showed that these genes were related to the regulation of mRNA processing (Figure 3D). GSEA suggested that MAFG‐AS1 is involved in RNA degradation (Figure 3E).
FIGURE 3.
MAFG‐AS1 binds directly to Hu antigen R (HuR). A, FISH for detecting the localization of MAFG‐AS1 in T24 and RT4 cells. B, Schematic diagram of the ChIRP experiment. C, ChIRP‐MS detects the binding proteins of MAFG‐AS1. D, Gene ontology (GO) analysis results. E, The GSEA analysis results (187 gene sets were used for the GSEA). F, HDOCK database predicts that MAFG‐AS1 binds to HuR. G, PRM assay validates proteins that MAFG‐AS1 may bind to. H, RIP assay to verify that HuR binds to MAFG‐AS1. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001. DAPI, 4ʹ,6‐diamidino‐2‐phenylindole; T24‐Lnc, T24‐MAFG‐AS1; T24‐Ctrl, T24‐negative control
HuR, a member of ELAVL Family, is an RBP involved in a variety of biological processes including RNA degradation. The HDOCK and catRAPID web servers predicted that HuR has binding sites for MAFG‐AS1 (Figure 3F; Figure S4A). Furthermore, ChIRP‐MS data were further verified using PRM, which detected a strong signal of HuR in the protein pulled down by the MAFG‐AS1‐specific probe (Figure 3G; Table S6). To confirm the binding between MAFG‐AS1 and HuR, RIP was performed and showed that MAFG‐AS1 was evidently enriched in HuR group compared to the IgG control group (Figure 3H), indicating that MAFG‐AS1 could bind to HuR directly. However, the consequences induced by this binding and whether the oncogenic role of MAFG‐AS1 is associated with this interaction remain unknown.
3.4. MAFG‐AS1 can stabilize HuR by recruiting the deubiquitinating enzyme USP5
To identify the specific relationship between MAFG‐AS1 and HuR, we conducted Western blotting in BUC cell lines (Figure 4A). Moreover, the expression of HuR was substantially upregulated in cancer tissues compared to noncancerous tissues (Figure 4B), which was further verified by IHC (Figure 4C). It was found that MAFG‐AS1 was positively correlated with HuR using the ChIPBase 2.0 database (Figure S3A). Knockdown of MAFG‐AS1 resulted in a notable reduction of HuR; however, HuR expression level was significantly increased when MAFG‐AS1 was overexpressed in T24 and RT4 cells (Figure 4D). Therefore, we hypothesized that MAFG‐AS1 might affect the stability of HuR. To verify this hypothesis, we treated T24 and RT4 cells with cycloheximide (CHX), an intracellular protein synthesis inhibitor, with or without proteasome inhibitor MG132. The results showed that HuR protein degradation was effectively slowed down in the MG132‐treated group, suggesting that HuR protein was affected by the ubiquitinated protease system (Figure 4E). Then, MAFG‐AS1 was knocked down in T24 and RT4 cells with CHX. Similarly, knockdown of MAFG‐AS1 reduced the stability of HuR compared with the control group (Figure 4F).
FIGURE 4.
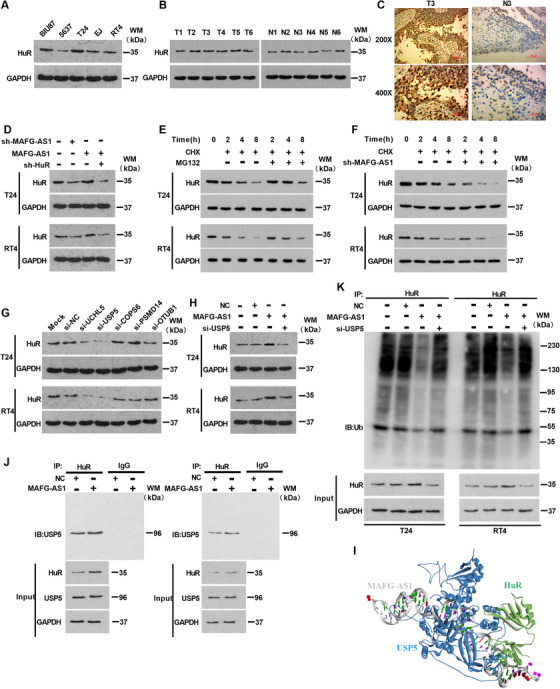
MAFG‐AS1 can stabilize Hu antigen R (HuR) by recruiting the deubiquitinating enzyme USP5. A, Western blot analysis of HuR expression in bladder urothelial carcinoma (BUC) cells. B, Western blot analysis of HuR expression in cancer and normal tissues of bladder. C, Immunohistochemical detection of HuR expression in cancer and normal tissues of bladder. D, Western blotting was used to detect the expression level of HuR in each experimental group. E and F, T24 and RT4 cell lines were cultured with cycloheximide (10 μg/mL), adding MG132 (20 μM) or transfected with sh‐MAFG‐AS1, and cell lysates were analyzed by Western blotting. G, Detection of HuR protein expression by Western blotting after knockdown of deubiquitinating enzyme in T24 and RT4 cell lines. H, Western blotting shows the expression level of HuR in T24 and RT4 cells after transfected with MAFG‐AS1 or MAFG‐AS1 and si‐USP5. I, The HDOCK website predicts that HuR can bind to USP5. J, USP5 expression was determined by Co‐IP using HuR‐antibody to precipitate binding proteins. K, The level of ubiquitination of HuR was detected by Co‐IP after overexpression of MAFG‐AS1 or overexpression of MAFG‐AS1 and knocking down USP5 in T24/RT4 cells. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
It is generally recognized that ubiquitination is a posttranslational modification that can regulate proteins’ functions, such as protein stability, subcellular localization, and activity. lncRNAs have been reported to participate in protein degradation through the ubiquitin proteasome system. Interestingly, it was predicted that there were multiple ubiquitinated sites in HuR using PhosphoSitePlus website (https://www.phosphosite.org//). Therefore, we inferred that MAFG‐AS1 increased the stability of HuR via inhibition of ubiquitination. Next, we selected five deubiquitinating enzymes (UCHL5, USP5, COPS6, PSMD14, and OTUB1) that were positively correlated with MAFG‐AS1 or associated with poor prognosis in BUC using the UALCAN database (Figure S2A‐J). Among the selected enzymes, knockdown of USP5 repressed the protein level of HuR the most (Figure 4G). Additionally, MAFG‐AS1 was positively correlated with USP5 according to the GEO (GSE31684, GSE124305) and ChIPBase 2.0 database (Figure S3C,H,I). Western blotting analysis exhibited that HuR expression could be promoted by overexpression of MAFG‐AS1, but the effects could be weakened by cotransfection with MAFG‐AS1 and si‐USP5 (Figure 4H). Surprisingly, we found that HuR could bind to USP5 directly through analyzing both HDOCK and catRAPID (Figure S4D). Furthermore, MAFG‐AS1 tended to bind with USP5 (Figure 4I; Figure S4B,C). We performed Co‐IP assays using anti‐HuR as a precipitating antibody to identify the relationship between USP5 and HuR in T24 and RT4 cells. As shown in Figure 4J, HuR directly binds to USP5. Moreover, the ubiquitination level of HuR was decreased in the MAFG‐AS1‐overexpressing cells, which could be reversed by cotransfection with MAFG‐AS1 and si‐USP5 in T24 and RT4 cells (Figure 4K). Taken together, MAFG‐AS1 can stabilize HuR by recruitment of USP5, a deubiquitinating enzyme, thereby protecting HuR from degradation through ubiquitination.
3.5. MAFG‐AS1 could promote PTBP1 translation by increasing its expression through enhancing HuR stability
To illustrate what effects could be induced by the binding between MAFG‐AS1 and HuR, we analyzed several databases (such as catRAPID, starBase 2.0, and POSTAR2.0) to find out downstream effects of HuR. It was predicted that HuR, as an RBP, is highly likely to bind to PTBP1 (Figure 5D). PTBP1 was highly expressed in T24 cells with high metastatic invasion (Figure 5A), while RT4 cells with low metastasis levels exhibited decreased PTBP1 expression. Importantly, PTBP1 was upregulated in BUC tissues (Figure 5B). These results were further verified by IHC experiments (Figure 5C). Furthermore, HuR was coexpressed with PTBP1 at the transcriptional level based on data (GSE87304, GSE13507, GSE31684, GSE124305) from GEO and ChIPBase 2.0 databases (Figure 5E; Figure S3B,D‐F). The RIP assay indicated that the PTBP1 mRNA could interact with HuR (Figure 5F). Therefore, we assumed that MAFG‐AS1 could promote the expression of PTBP1 by enhancing HuR stability. The expression of PTBP1 mRNA was increased significantly when MAFG‐AS1 was overexpressed in T24 and RT4 cells (Figure 5G), and knockdown of HuR significantly inhibited MAFG‐AS1 promotion of PTBP1 expression. Meanwhile, the expression of PTBP1 protein increased when MAFG‐AS1 was overexpressed in T24 and RT4 cells, (Figure 5H), which could be reduced by knocking down USP5 or HuR (Figure 5I).
FIGURE 5.
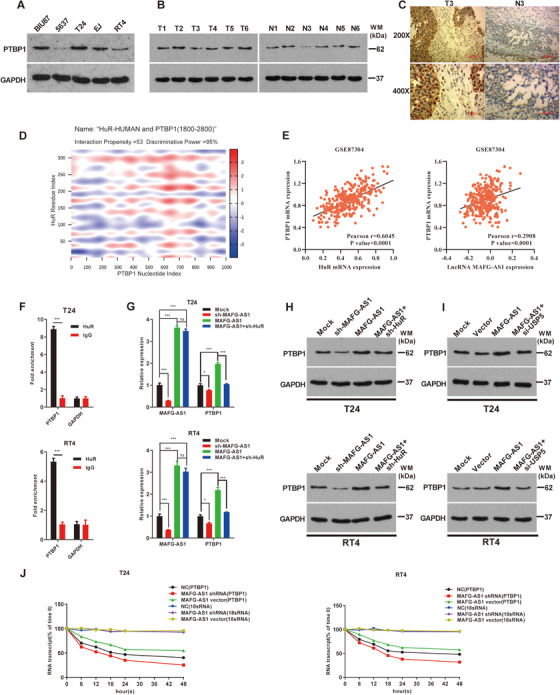
MAFG‐AS1 could promote PTBP1 translation by increasing its expression through enhancing Hu antigen R (HuR) stability. A, The expression of PTBP1 protein was detected by Western blot in bladder urothelial carcinoma (BUC) cell lines (BIU87, 5637, T24, EJ, and RT4). B, Western blot assay detected the expression of HuR in tumor and adjacent tissues. C, Immunohistochemical detection of PTBP1 protein expression in tumor tissues and adjacent tissues of BUC patients. D, The catRAPID database predicted that HuR is much more likely to bind to PTBP1 mRNA (discriminative power [DP] ranges from 0% (unpredictability) to 100% (predictability); DP values above 50% indicate that the interaction is likely to take place, whereas DPs above 75% represent high‐confidence predictions). E, HuR is positively correlated with PTBP1 in BUC based on the GEO dataset (GSE124305, GSE87304). F, RIP assay was conducted to detect whether HuR protein can bind to PTBP1 mRNA. G, The effect of MAFG‐AS1 and HuR on PTBP1 mRNA levels was detected by RT‐qPCR. H and I, Western blot detected the expression of PTBP1 protein. J, The stability of PTBP1 mRNA was measured by RNA stability assay in each experimental group. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
We intended to verify whether MAFG‐AS1 regulates PTBP1 mRNA stability. Thus, transfected of MAFG‐AS1‐vector or sh‐MAFG‐AS1 into T24 and RT4 cells, with treatment of α‐amanitin, to inhibit transcription; 18S ribosomal RNA was used as an internal reference. Increased PTBP1 mRNA level was observed after overexpression of MAFG‐AS1. Conversely, MAFG‐AS1 knockdown shortened the half‐life of PTBP1 mRNA (Figure 5J).
Overall, the results concluded that MAFG‐AS1 could promote the expression of PTBP1 by promoting HuR stability.
3.6. MAFG‐AS1/HuR/PTBP1 axis can promote proliferation, invasion, and EMT of BUC in vivo and in vitro
We have identified that MAFG‐AS1 promoted PTBP1 translation by increasing its stability through enhancing the expression of HuR in BUC. Next, to examine the biological role of that the MAFG‐AS1/HuR/PTBP1 axis in cell proliferation, invasion, and metastasis of BUC, we conducted MTT, clone formation, transwell, and wound‐healing assays, respectively. The results indicated that overexpression of MAFG‐A1 dramatically increased cell viability (Figures 6A and 6B), invasion, and metastasis of BUC cells (Figures 6C and 6D). Additionally, an epithelial marker E‐cadherin protein level was markedly decreased, while the level of mesenchymal marker, vimentin, was significantly increased when MAFG‐AS1 was overexpressed in T24 and RT4 cells (Figure 6E). By contrast, ectopic overexpression of MAFG‐AS1 and knockdown of HuR or PTBP1 partly inhibited the promoting effects on proliferation, invasion, metastasis, and EMT of BUC cells induced by MAFG‐AS1. Subsequently, we tested the role of MAFG‐AS1 on BUC in vivo experiments. After knocking down MAFG‐AS1, the tumor‐forming ability of the cells was decreased (Figure 7A), the EMT process was inhibited (Figure 7B); in sharp contrast, overexpression of MAFG‐AS1 exerted a significant promoting effect on tumor growth and EMT process (Figures 7A and 7B). In addition, it was found that the tumorigenic ability of MAFG‐AS1 could be weakened by knockdown of HuR or PTBP1. Consistently, EMT process was inhibited with cotransfection of MAFG‐AS1 and knockdown of HuR or PTBP1 compared with overexpression of MAFG‐AS1 (Figures 7A and 7B). IHC to detect tumor proliferation‐related indicators Ki67 also confirmed this result (Figure 7C). Therefore, the findings of this study uncover a novel molecular mechanism in the progression of BUC via MAFG‐AS1/HuR/PTBP1 axis, which promotes cell proliferation, invasion, and EMT in BUC.
FIGURE 6.
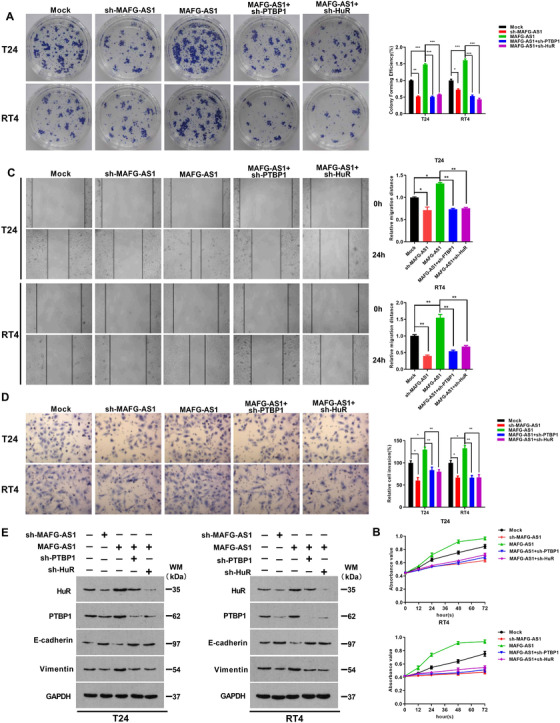
MAFG‐AS1/HuR/PTBP1 axis can promote proliferation, invasion, metastasis, and epithelial‐mesenchymal transition (EMT) of bladder urothelial carcinoma (BUC) in vivo. A and B, Colony formation assay (A) and MTT assay (B) were used to examine the effect of MAFG‐AS1/HuR/PTBP1 axis on cell proliferation. C and D, The effect of MAFG‐AS1/HuR/PTBP1 axis on cell metastasis and invasion was examined by wound‐healing assay (C) and transwell assay (D). E, The expression of HuR, PTBP1, E‐cadherin, and vimentin was examined by Western blot. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
FIGURE 7.
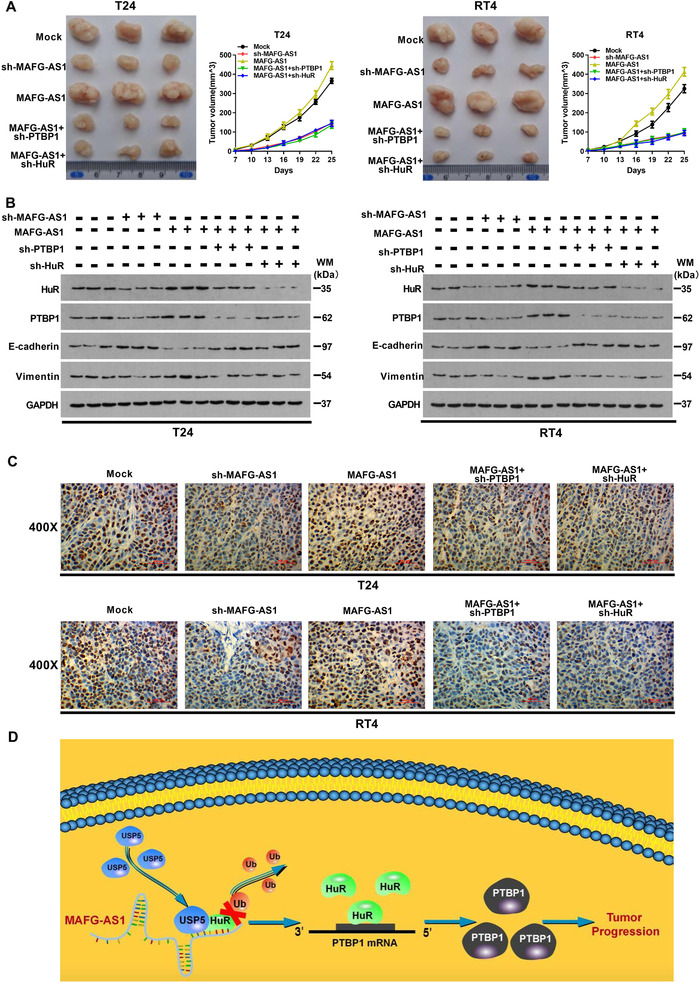
MAFG‐AS1/HuR/PTBP1 axis can promote proliferation, invasion, and epithelial‐mesenchymal transition (EMT) of bladder urothelial carcinoma (BUC) in vitro. A, Image of mouse xenograft in vivo assay and volume of tumor in each group. B, Western blot was used to detect the expression level of HuR, PTBP1, and EMT‐related proteins in tumor tissues of mice. C, Immunohistochemistry was used to detect the expression level of Ki67 in each group. D, Schematic diagram of MAFG‐AS1/HuR/PTBP1 axis promotes tumor progression in BUC. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
4. DISCUSSION
Emerging and accumulating studies reveal that a number of lncRNAs can regulate the genesis and development of tumors through a variety of mechanisms. MAFG‐AS1 is a newly discovered oncogenic lncRNA in several cancers. 37 , 38 However, the role of MAFG‐AS1 in BUC remains poorly understood. Here, we found that MAFG‐AS1 expression is upregulated in BUC tissues based on TCGA and GEO (GSE31189) databases. Then, in vitro and in vivo experiments revealed that MAFG‐AS1 could promote cell proliferation, invasion, metastasis, and EMT progression in BUC. Further analysis found that increased MAFG‐AS1 in BUC patients was correlated with poor prognosis and advanced TNM stage. Therefore, MAFG‐AS1 may be a prognostic biomarker for BUC.
HuR, an RBP, belongs to the human/embryo lethal abnormal vision (Hu/ELAV) RBP family, and regulates mRNA stability, translation, and miRNA production, 39 and its activity is mainly regulated by nuclear translocation, 40 phosphorylation, 41 and ubiquitination. 42 Recently, studies have shown that lncRNAs participate in regulation of HuR function. lincRNA‐UFC1 19 can directly bind to HuR protein and stabilize its expression, while lncRNA OCC‐1 can promote its ubiquitination at the posttranslational level to degrade the protein level of HuR. 43 In this study, it was predicted that MAFG‐AS1 could bind to HuR through HDOCK, and verified by PRM and RIP analysis. Importantly, MAFG‐AS1 was positively correlated with HuR at the transcriptional level according to ChIPBase 2.0 and starBase 2.0. Furthermore, overexpression of MAFG‐AS1 remarkably enhanced HuR protein levels, indicating that MAFG‐AS1 is highly likely to stabilize the expression of HuR protein upon binding. Ubiquitination is involved in the stability of proteins and eventually leads to their specific degradation. Therefore, we identified several deubiquitinating enzymes via several online databases, such as UALCAN, ChIPBase 2.0, and GEPIA. We identified that USP5 was positively correlated with HuR and MAFG‐AS1 as well with poor prognosis in BUC. USP5 is a deubiquitinating enzyme that has been reported to facilitate tumorigenesis in multiple cancers. 44 , 45 Furthermore, we predicted, using HDOCK and catRAPID, that there are binding sites between USP5, HuR, and MAFG‐AS1. This analysis suggested that MAFG‐AS1 is likely to promote tumorigenesis and progression of BUC by inhibiting the ubiquitination of HuR via recruiting USP5. It was further validated that USP5 directly binds to HuR by Co‐IP assays. We observed that overexpression of MAFG‐AS1 could upregulate HuR protein level in T24 and RT4 cells, which could be partly inhibited by silencing USP5. These data proved that MAFG‐AS1 could directly bind to HuR and stabilize it by recruiting the USP5 deubiquitinating enzyme in BUC. Our results provided a reasonable answer to the question of why HuR is overexpressed in BUC raised in previous studies. However, further investigation is needed to identify whether HuR is an independent prognostic factor.
HuR could confer transcriptional stability and promote translation by binding to ARE of the 3′ untranslated region of mRNA in cells. 46 , 47 As a result, we hypothesized that HuR might play a role in BUC by regulating certain key mRNAs. PTBP1, a known hnRNPI, could promote the progression of multiple tumors. 48 , 49 Moreover, it was found that highly expressed PTBP1 facilitates genesis and development of tumors, and was associated with poor prognosis as well as lymph node metastasis in BUC. 50 , 51 As yet, no studies have explored the molecular mechanism involved in the high expression of PTBP1 in BUC. Interestingly, we found that HuR is positively correlated with PTBP1 based on the ChIPBase and GEO database, which is consistent with previous results. 52 Stability tests with RIP and RNA revealed that HuR promotes the translational level of PTBP1 by binding and stabilizing its mRNA. Furthermore, function recovery assays showed that overexpression of MAFG‐AS1 and knockdown of HuR or PTBP1 reduced cell proliferation, metastasis, and inhibited EMT process in BUC. We confirmed that MAFG‐AS1 facilitates carcinogenesis and progression in BUC by upregulating PTBP1 via directly binding and stabilizing HuR. Our results also suggest that PTBP1 serves as an oncogene in BUC, and clarified the possible molecular mechanism of overexpressed PTBP1.
In conclusion, we confirmed that MAFG‐AS1 is markedly upregulated in BUC tissues, and higher expressed MAFG‐AS1 is associated with progression of BUC and poor survival of BUC patients. On the molecular level, MAFG‐AS1 could promote cell proliferation, metastasis, and EMT process via the MAFG‐AS1/HuR/PTBP1 axis in BUC. We reveal that MAFG‐AS1 may be a novel target and a potential biomarker for BUC treatment. Targeting MAFG‐AS1/HuR/PTBP1 axis can provide new therapeutic avenues for clinical treatment of BUC.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHOR CONTRIBUTIONS
Mengqing Xiao and Ke Cao designed the study, analyzed and interpreted the data, and wrote the manuscript. Jianye Liu, Liang Xiang, Kai Zhao, Dong He, Qinghai Zeng, Yuxing Zhu, and Yeyu Zhang contributed to data acquisition, analysis, and interpretation. Jianye Liu and Qun Zhang provided clinical database compilation and analysis. Hao Bo and Xingyu Chen performed all the bioinformatics analysis. Yan Liu, Xiaoming Liu, Lian Gong, Ying Bao, Yi Hu, Yaxin Cheng, Liping Deng, and Rongrong Zhu carried out the experiments. Dan Xie, Minhua Deng, Xiaowei Xing, Ming Zhou, Wei Xiong, Yanhong Zhou, Jianda Zhou, and Xiaohui Li provided technical expertise and support. All the authors have seen and approved the final version of the manuscript, and agreed to submit the manuscript for consideration of publication in this journal.
ETHICS STATEMENT
This study was approved by the Third Xiangya Hospital of Central South University, and informed consent was obtained from all the patients. All the experiments in this study were conducted following the corresponding regulations and guidelines.
Supporting information
Supplementary Figure S1 MAFG‐AS1 is highly expressed and negatively correlated with prognosis in BUC. A, GEPIA was used to analyze the expression of MAFG‐AS1 in most types of cancer. B, High MAFG‐AS1 expression was associated with advanced clinical stages using GEPIA database. C, Expression of MAFG‐AS1 was identified by RT‐qPCR in cancer and normal tissues of bladder. D, Patients with high MAFG‐AS1 expression had shorter DFS in BUCs; data from GEPIA. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
Supplementary Figure S2 Potential deubiquitinating enzymes that may be recruited by MAFG‐AS1. A‐E, Five deubiquitinating enzymes such as UCHL5 (A), USP5 (B), COPS6 (C), PSMD14 (D), and OTUB1 (E) were selected using UALCAN and ChIPBase 2.0 database. F‐J, The transfection efficiency of si‐UCHL5 (F), si‐USP5 (G), si‐COPS6 (H), si‐PSMD14 (I), and si‐OTUB1 (J) was shown by Western blot in T24 and RT4 cells
Supplementary Figure S3 Correlation between MAFG‐AS1, USP5, and HuR was predicted by CHIPBASE2.0 and GEO datasets in BUC. A, The correlation between MAFG‐AS1 and HuR in BUC was predicted by ChIPBase 2.0. B‐F, The correlation between HuR and PTBP1 in BUC was predicted by ChIPBase 2.0 (B), GSE83586 (C), GSE13507 (D), GSE31684 (E), and GSE124305 (F). G, The correlation between HuR and USP5 was predicted by ChIPBase 2.0 in BUC. H and I, The correlation between MAFG‐AS1 and USP5 was predicted by GSE124305 (H) and GSE87304 (I) in BUC.
Supplementary Figure S4 Bioinformatics analysis based on catRAPID and HDOCK databases. A, The potential binding sites between MAFG‐AS1 and HuR were detected by catRAPID databases. B and C, The potential binding sites between MAFG‐AS1 and USP5 were detected using catRAPID (B) and HDOCK (C) databases. D, The potential site between HuR and USP5 was detected by HDOCK database.
TableS1
TableS2
TableS3
TableS4
TableS5
TableS6
TableS7
ACKNOWLEDGMENTS
We thank all the authors for their excellent work. This work was supported by the National Natural Science Foundation of China (81874137), the Outstanding Youth Foundation of Hunan Province (2018JJ1047), the Huxiang Young Talent Project (2016RS3022), the Hunan Province Science and Technology Talent Promotion Project (2019TJ‐Q10), the Project of Scientific Research Plan of Health and Family Planning Commission of Hunan Province (c20180476), and the Independent Exploration and Innovation Project of Central South University, Grant/Award Number: 2018zzts938.
Xiao M, Liu J, Xiang L, et al. MAFG‐AS1 promotes tumor progression via regulation of the HuR/PTBP1 axis in bladder urothelial carcinoma. Clin Transl Med. 2020;10:e241 10.1002/ctm2.241
DATA AVAILABILITY STATEMENT
All the data generated or analyzed during this study are included in this published article. The datasets supporting the conclusions of this article are included within the article and its additional supporting files.
REFERENCES
- 1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96‐108. [DOI] [PubMed] [Google Scholar]
- 2. Giacalone NJ, Shipley WU, Clayman RH, et al. Long‐term outcomes after bladder‐preserving tri‐modality therapy for patients with muscle‐invasive bladder cancer: an updated analysis of the Massachusetts General Hospital experience. Eur Urol. 2017;71(6):952‐960. [DOI] [PubMed] [Google Scholar]
- 3. Kulkarni G, Hermanns T, Wei Y, et al. Propensity score analysis of radical cystectomy versus bladder‐sparing trimodal therapy in the setting of a multidisciplinary bladder cancer clinic. J Clin Oncol. 2017;35(20):2299‐2305. [DOI] [PubMed] [Google Scholar]
- 4. Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50(12):1705‐1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen X, Li A, Sun BF, et al. 5‐Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21(8):978‐990. [DOI] [PubMed] [Google Scholar]
- 6. Lebedeva S, Jens M, Theil K, et al. Transcriptome‐wide analysis of regulatory interactions of the RNA‐binding protein HuR. Mol Cell. 2011;43(3):340‐352. [DOI] [PubMed] [Google Scholar]
- 7. Zhai W, Zhu R, Ma J, et al. A positive feed‐forward loop between LncRNA‐URRCC and EGFL7/P‐AKT/FOXO3 signaling promotes proliferation and metastasis of clear cell renal cell carcinoma. Mol Cancer. 2019;18(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lan T, Yuan K, Yan X, et al. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res. 2019;79(13):3220‐3234. [DOI] [PubMed] [Google Scholar]
- 9. Stellos K, Gatsiou A, Stamatelopoulos K, et al. Adenosine‐to‐inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR‐mediated post‐transcriptional regulation. Nat Med. 2016;22(10):1140‐1150. [DOI] [PubMed] [Google Scholar]
- 10. Xu S, Wang P, Zhang J, et al. Ai‐lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA‐RNA and RNA‐protein interactions in human cancer. Mol Cancer. 2019;18(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Z, Yao Z, Wang L, et al. Activation of ferritinophagy is required for the RNA‐binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14(12):2083‐2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang T, Guo C, Xia T, et al. LncCCAT1 promotes breast cancer stem cell function through activating WNT/β‐catenin signaling. Theranostics. 2019;9(24):7384‐7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z, Chen X, Lu B, et al. Up‐regulated LINC01234 promotes non‐small‐cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng S, Yang L, Zou Y, et al. Long non‐coding RNA HUMT hypomethylation promotes lymphangiogenesis and metastasis via activating FOXK1 transcription in triple‐negative breast cancer. J Hematol Oncol. 2020;13(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He W, Zhong G, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer‐associated lymphangiogenesis and lymphatic metastasis. J Clin Invest. 2018;128(2):861‐875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell‐cycle progression in cancer. Cancer Cell. 2018;33(4):e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. López D, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA‐binding protein HuR. Proc Natl Acad Sci U S A. 2004;101(9):2987‐2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barbisan F, Mazzucchelli R, Santinelli A, et al. Overexpression of ELAV‐like protein HuR is associated with increased COX‐2 expression in atrophy, high‐grade prostatic intraepithelial neoplasia, and incidental prostate cancer in cystoprostatectomies. Eur Urol. 2009;56(1):105‐112. [DOI] [PubMed] [Google Scholar]
- 19. Cao C, Sun J, Zhang D, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β‐catenin in HCC cells. Gastroenterology. 2015;148(2):415‐426.e418. [DOI] [PubMed] [Google Scholar]
- 20. Kang MJ, Ryu BK, Lee MG, et al. NF‐kappaB activates transcription of the RNA‐binding factor HuR, via PI3K‐AKT signaling, to promote gastric tumorigenesis. Gastroenterology. 2008;135(6):e2031‐e2033. [DOI] [PubMed] [Google Scholar]
- 21. Young LE, Sanduja S, Bemis‐Standoli K, Pena EA, Price RL, Dixon DA. The mRNA‐binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136(5):1669‐1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang PI, Li WM, Wang YH, et al. HuR cytoplasmic expression is associated with increased cyclin A expression and poor outcome with upper urinary tract urothelial carcinoma. BMC Cancer. 2012;12:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miyata Y, Watanabe S, Sagara Y, et al. High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS One. 2013;8(3):e59095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li JR, Sun CH, Li W, et al. Cancer RNA‐Seq Nexus: a database of phenotype‐specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016;44(D1):D944‐D951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J, Han L, Roebuck P, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728‐3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large‐scale prediction of protein‐RNA interactions. Bioinformatics. 2013;29(22):2928‐2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: a web server for protein‐protein and protein‐DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365‐W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu Y, He D, Bo H, et al. The MRVI1‐AS1/ATF3 signaling loop sensitizes nasopharyngeal cancer cells to paclitaxel by regulating the Hippo‐TAZ pathway. Oncogene. 2019;38(32):6065‐6081. [DOI] [PubMed] [Google Scholar]
- 31. Zeng Q, Liu J, Cao P, et al. Inhibition of REDD1 sensitizes bladder urothelial carcinoma to paclitaxel by inhibiting autophagy. Clin Cancer Res. 2018;24(2):445‐459. [DOI] [PubMed] [Google Scholar]
- 32. Liu JY, Zeng QH, Cao PG, et al. SPAG5 promotes proliferation and suppresses apoptosis in bladder urothelial carcinoma by upregulating Wnt3 via activating the AKT/mTOR pathway and predicts poorer survival. Oncogene. 2018;37(29):3937‐3952. [DOI] [PubMed] [Google Scholar]
- 33. Chu C, Quinn J, Chang H. Chromatin isolation by RNA purification (ChIRP). J Vis Exp. 2012;(61):3912 10.3791/3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chu C, Zhang Q, da Rocha S, et al. Systematic discovery of Xist RNA‐binding proteins. Cell. 2015;161(2):404‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11(11):1475‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med. 2008;14(7):723‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jia YC, Wang JY, Liu YY, Li B, Guo H, Zang AM. LncRNA MAFG‐AS1 facilitates the migration and invasion of NSCLC cell via sponging miR‐339‐5p from MMP15. Cell Biol Int. 2019;43(4):384‐393. [DOI] [PubMed] [Google Scholar]
- 38. Sui Y, Lin G, Zheng Y, Huang W. LncRNA MAFG‐AS1 boosts the proliferation of lung adenocarcinoma cells via regulating miR‐744‐5p/MAFG axis. Eur J Pharmacol. 2019;859:172465. [DOI] [PubMed] [Google Scholar]
- 39. Ke Y, Han Y, Guo X, et al. PARP1 promotes gene expression at the post‐transcriptiona level by modulating the RNA‐binding protein HuR. Nat Commun. 2017;8:14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao L, Li XX, Chung HK, et al. RNA‐binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via post‐transcriptional control of CNPY3. Gastroenterology. 2019;157(3):731‐743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Battaglia‐Hsu SF, Ghemrawi R, Coelho D, et al. Inherited disorders of cobalamin metabolism disrupt nucleocytoplasmic transport of mRNA through impaired methylation/phosphorylation of ELAVL1/HuR. Nucleic Acids Res. 2018;46(15):7844‐7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abdelmohsen K, Srikantan S, Yang X, et al. Ubiquitin‐mediated proteolysis of HuR by heat shock. EMBO J. 2009;28(9):1271‐1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC‐1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46(11):5809‐5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu X, Huang A, Cui X, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9(14):4208‐4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Z, Tong J, Tang X, et al. The ubiquitin ligase HERC4 mediates c‐Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood. 2016;127(13):1676‐1686. [DOI] [PubMed] [Google Scholar]
- 46. Ghosh M, Aguila HL, Michaud J, et al. Essential role of the RNA‐binding protein HuR in progenitor cell survival in mice. J Clin Invest. 2009;119(12):3530‐3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav‐like protein. J Biol Chem. 1996;271(14):8144‐8151. [DOI] [PubMed] [Google Scholar]
- 48. Georgilis A, Klotz S, Hanley CJ, et al. PTBP1‐mediated alternative splicing regulates the inflammatory secretome and the pro‐tumorigenic effects of senescent cells. Cancer Cell. 2018;34(1):e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sayed ME, Yuan L, Robin JD, et al. NOVA1 directs PTBP1 to hTERT pre‐mRNA and promotes telomerase activity in cancer cells. Oncogene. 2019;38(16):2937‐2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bielli P, Panzeri V, Lattanzio R, et al. The splicing factor PTBP1 promotes expression of oncogenic splice variants and predicts poor prognosis in patients with non‐muscle‐invasive bladder cancer. Clin Cancer Res. 2018;24(21):5422‐5432. [DOI] [PubMed] [Google Scholar]
- 51. Xie R, Chen X, Chen Z, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31‐44. [DOI] [PubMed] [Google Scholar]
- 52. Jiang D, Zhang Y, Yang L, et al. Long noncoding RNA HCG22 suppresses proliferation and metastasis of bladder cancer cells by regulation of PTBP1. J Cell Physiol. 2020;235(2):1711‐1722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 MAFG‐AS1 is highly expressed and negatively correlated with prognosis in BUC. A, GEPIA was used to analyze the expression of MAFG‐AS1 in most types of cancer. B, High MAFG‐AS1 expression was associated with advanced clinical stages using GEPIA database. C, Expression of MAFG‐AS1 was identified by RT‐qPCR in cancer and normal tissues of bladder. D, Patients with high MAFG‐AS1 expression had shorter DFS in BUCs; data from GEPIA. Bars represent standard deviation, ns P > .05, *P < .05, **P < .01, ***P < .001
Supplementary Figure S2 Potential deubiquitinating enzymes that may be recruited by MAFG‐AS1. A‐E, Five deubiquitinating enzymes such as UCHL5 (A), USP5 (B), COPS6 (C), PSMD14 (D), and OTUB1 (E) were selected using UALCAN and ChIPBase 2.0 database. F‐J, The transfection efficiency of si‐UCHL5 (F), si‐USP5 (G), si‐COPS6 (H), si‐PSMD14 (I), and si‐OTUB1 (J) was shown by Western blot in T24 and RT4 cells
Supplementary Figure S3 Correlation between MAFG‐AS1, USP5, and HuR was predicted by CHIPBASE2.0 and GEO datasets in BUC. A, The correlation between MAFG‐AS1 and HuR in BUC was predicted by ChIPBase 2.0. B‐F, The correlation between HuR and PTBP1 in BUC was predicted by ChIPBase 2.0 (B), GSE83586 (C), GSE13507 (D), GSE31684 (E), and GSE124305 (F). G, The correlation between HuR and USP5 was predicted by ChIPBase 2.0 in BUC. H and I, The correlation between MAFG‐AS1 and USP5 was predicted by GSE124305 (H) and GSE87304 (I) in BUC.
Supplementary Figure S4 Bioinformatics analysis based on catRAPID and HDOCK databases. A, The potential binding sites between MAFG‐AS1 and HuR were detected by catRAPID databases. B and C, The potential binding sites between MAFG‐AS1 and USP5 were detected using catRAPID (B) and HDOCK (C) databases. D, The potential site between HuR and USP5 was detected by HDOCK database.
TableS1
TableS2
TableS3
TableS4
TableS5
TableS6
TableS7
Data Availability Statement
All the data generated or analyzed during this study are included in this published article. The datasets supporting the conclusions of this article are included within the article and its additional supporting files.


