Abstract
The SARS-CoV-2 outbreak, began in late 2019, has caused a worldwide pandemic and shows no signs of slowing. Glucocorticoids (GCs), including dexamethasone (DEX), have been widely used as effective anti-inflammatory and immunosuppressant drugs. In this study, seven GCs had no obvious effect on cell viability of angiotensin converting enzyme 2 (ACE2) high expressed HEK293T cells when concentrations were under 10 μM. Molecular docking results revealed that DEX occupied with active binding site of ACE2 of SARS-CoV-2 spike protein. Surface plasmon resonance (SPR) results showed that KD value between DEX and ACE2 was (9.03 ± 0.78) e−6 M. Cell membrane chromatography (CMC) results uncovered that DEX had a chromatographic retention. DEX was found out to inhibiting the viropexis into ACE2h cells using SARS-CoV-2 spike pseudotyped virus. Therefore, DEX inhibits the entrance of SARS-CoV-2 spike pseudotyped virus into cell by binding to ACE2.
Keywords: Dexamethasone, SARS-CoV-2, ACE2
1. Introduction
In the late of 2019, unknown cases of pneumonia have outbreak in Wuhan, China (Wang et al., 2020). The pathogen has been identified as a novel enveloped RNA betacoronavirus2 that has currently been named 2019-novel corona virus (SARS-CoV-2), which has a phylogenetic similarity to SARS-CoV (Lu et al., 2020). The virus is transmitted by droplets and aerosols (Guo et al., 2020). The disease caused by these coronaviruses has pneumonialike symptoms, such as fever and dry cough, and leads to progressive respiratory failure and even death (Wang et al., 2020; Weiss and Murdoch, 2020).
GC is the most important regulatory hormone in the body's stress response (Stahn and Buttgereit, 2008), possessing anti-inflammatory (Ayroldi et al., 2012; Coutinho and Chapman, 2011), anti-toxic, anti-allergic (McKeever et al., 2018), anti-shock (Venkatesh et al., 2018), non-specific immunosuppression and antifebrile effects, which can prevent the occurrence of immunological inflammatory reactions and pathological immune reactions (Hardy et al., 2020). GCs were widely used during the outbreaks of severe acute respiratory syndrome (SARS)-CoV (Stockman et al., 2006) and Middle East respiratory syndrome (MERS)-CoV (Arabi et al., 2018). Since the outbreak of the SARS-CoV-2 in 2020, physicians have tried many drugs to treat it (Tu et al., 2020; Wu et al., 2020). As a result, GCs are being used in patients with SARS-CoV-2 (Johnson and Vinetz, 2020; Villar et al., 2020) in addition to other therapeutics recommended by WHO and health authorities in many countries (Guan et al., 2020). Clinical studies by British scholars have shown that DEX can have a certain therapeutic effect on SARS-CoV-2 patients (Selvaraj et al., 2020). However, how DEX performs its therapeutic effects remains unclear. Meanwhile, the curative effect of GCs on SARS-CoV-2 is still controversial (Lewis et al., 2019; Russell et al., 2020). Not only that, patients who were given corticosteroids were more likely to require mechanical ventilation in MERS infection (Arabi et al., 2018), diabetes in SARS-CoV infection (Li et al., 2004) and avascular necrosis in SARS-CoV-2 (Mehra et al., 2020) as complications associated with corticosteroid treatment.
The renin–angiotensin–aldosterone system (RAAS) is a significant system managing vasoactive peptides. ACE2, an enzyme that physiologically counters RAAS activation but also functions as a receptor for both SARS viruses (Yan et al., 2020). Therefore, studying whether the compound blocks or antagonizes the ACE2 receptor in epithelial cells can help to screen and identify potential drugs that can treat coronavirus disease 2019 (COVID-19) (Liu et al., 2020). Renhong Yan et al. reported the structural basis for the recognition of SARS-CoV-2 by full-length human ACE2 (Yan et al., 2020). They present cryo–electron microscopy structures of full-length human ACE2, and uncovered the receptor binding domain (RBD) of SARS-CoV-2 spike protein is recognized by the extracellular peptidase domain of ACE2 mainly through polar residues.
In the present study, SPR and molecular docking were used to detect the binding characteristics of GCs with ACE2 and the RBD of SARS-CoV-2 spike protein. The inhibition effect of GCs on ACE2h cells invaded by pseudotyped virus was determined by fluorescence.
2. Materials and methods
2.1. Drugs and reagents
DEX, betamethasone (BET), hydrocortisone (HYD), fluorohydrocortisone (FLD), triamcinolone acetonide (TRI), methyl prednisolone (MET), fluocinolone (FLO) were purchased from Dalian Meilun Co., Ltd. Cell Counting Kit were purchased from 7 Sea Pharmatech Co., Ltd (Shanghai, China). SPR related consumables were purchased from Nicoya (ON, Canada). SARS-CoV-2 spike pseudotyped virus (PSC001) from Sino Biological Co., Ltd (Beijing, China). The Luciferase luminescence value reached 106 RLU (Relative Light Unit) 48–72 h after pseudovirus infected ACE2h cells. 1010 virus copies/mL. Spike S1 protein content of SARS-COV-2 was 860 ng/mL. VSVG (GM-0220PC) was purchased from Genomedotech, Shanghai. Dulbecco's Modification of Eagle's Medium (DMEM) with high glucose (Cat. No. SH30022.01), and fetal bovine serum (Cat. No. 16140071) were from HyClone (Logan, UT, USA). Penicillin–streptomycin solution was obtained from Xi'an Hat Biotechnology Co., Ltd (Xi'an, China). Protease inhibitor and phosphatase inhibitor cocktails were purchased from Roche Diagnostic (Mannheim, Germany). The 5 × loading buffer was purchased from Thermo Fisher Scientific, lnc. (MA, USA), and SDS-PAGE was from Pioneer Biotech Co., Ltd (Xi'an, China). Polyvinylidene fluoride membranes were from Hangzhou Microna Membrane Technology Co., Ltd (Hangzhou, China). Tween-20 was provided by Shaanxi Pioneer Biotech Co., Ltd (Xi'an, China). Enhanced Chemiluminescence (ECL) kit was from Proteintech Group, lnc (Rosemont, USA).
2.2. Cell culture
HEK293T cells was from ATCC, ACE2 high expressing-HEK293T cells (ACE2h cells) were constructed by Genomeditech (Shanghai, China). ACE2h cells were kept in DMEM high glucose medium containing 10% FBS, 1% penicillin-streptomycin, 4 μg/mL puromycin and cultured at 37 °C containing 5% of CO2.
2.3. Cytotoxicity assay
Cell viability was determined following the instruction from the company. Briefly, ACE2h cells were seeded into 96-well plates at a density of 5 × 104 cells per well and then treated with different concentrations of GCs (0, 1, 5, 10, 25, 50, 100, 200, and 400 μM) for 24 h, then 10 μL of Cell Counting Kit solution was added to each well followed by 2 h of incubation. The relative cell viability was assessed by the detection of the absorbance at 450 nm using a microplate reader (Bio-Rad, Carlsbad, CA, USA). The survival rate of ACE2h cells was calculated as the following formula:
2.4. Real-time PCR
RNA was extracted from ACE2h cells. Briefly, after washing cells with PBS, Trizol was used in extracting RNA from cells. Then, by adding chloroform, isopropyl alcohol in turn and centrifugation, RNA appeared at the bottom of the tube. The solvent was removed with ethanol and then redissolved in DEPC water. gDNA wiper and reverse transcription were performed using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Japan) under manufactory's instruction. TB Green Premix Ex Taq II (Takara, Japan) and Bio-Rad CFX connect was used in real-time PCR.
2.5. Western blotting
Total proteins from different cells were extracted in ice-cold condition by using RIPA lysis buffer containing 10% protease inhibitor and a phosphatase inhibitor cocktail. The protein in the cell lysates was denatured by boiling the samples for 5 min with a 5 × loading sample buffer, equal amounts of protein were separated on a 10% gel using SDS-PAGE. The separated proteins were transferred onto polyvinylidene fluoride membranes and blocked by constant stirring with 5% non-fat milk in Tris-buffered saline containing Tween-20. The membranes were then incubated overnight at 4 °C with the following primary antibodies: anti-ACE2 (1:500, EPR4435, Abcam), and anti-GAPDH (1:2000, a#2118, CST). The membranes were washed thrice with TBST and then incubated with secondary antibodies (at a dilution of 1:20,000 in TBST) for 1 h at 37 °C. The membranes were washed another three times with TBST for 10 min and developed using ECL kit. Image-Pro Plus 5.1 software (Rockville, MD, USA) was used to quantify the protein levels.
2.6. Docking studies
Molecular docking studies were carried out with SYBYL-X 2.0 version. The small molecules and the X-ray crystal structure of protein (PDB code: 6M0J) were imported. Water molecules were removed and hydrogen was added. Tripos force field and Pullman charge were applied to minimize. GCs were depicted by SYBYL/Sketch module (Tripos Inc.), optimized by Powell's method with Tripos force field with convergence criterion at 0.005 kcal/(Å mol), and assigned with Gasteiger–Hückel method.
2.7. Surface plasmon resonance assay
For assessment of surface plasmon resonance, ACE2 protein (20 μg/mL), bought from Sino Biological Inc., was fixed on a carboxyl sensor chip (Nicoya, Canada) by capture-coupling, then GCs at 6.25, 12.5, 25, 50 and 100 μM were injected sequentially into the chamber in PBS running buffer. The interaction of ACE2 with GCs was detected by using Open SPRTM (Nicoya Lifesciences, Waterloo, Canada) at 25 °C. The binding time and disassociation time were both 250 s, the flow rate was 20 μL/s, and the chip was regenerated with hydrochloric acid (pH 2.0). A one-to-one diffusion-corrected model was fitted to the wavelength shifts corresponding to the different drug concentration. The data were retrieved and analyzed by using TraceDrawer.
2.8. Cell membrane chromatography
The protocol of ACE2h/CMC column preparation is as follows (Ma et al., 2017). In brief, a certain extent number (~1 × 107) of well-grown ACE2h cells were gathered and washed three times by precooled physiological saline. Then ruptured with 30 min ultrasonic in 50 mM Tris-HCl hypotonic solution (pH 7.4) and 3 min homogenization. The suspension of the ruptured cells was then centrifuged for 10 min at 1000×g, the raw cell membrane was obtained in the supernatant and washed twice. The refined cell membrane was resuspended in ice-cold physiological saline solution at 5 mL. 50 mg silica gel beads were activated at 105 °C for 30 min for better adsorption with cell membrane. The ACE2h cell membrane stationary phase (CMSP) was prepared with 5 mL cell membrane suspension and 50 mg activated silica gel beads under vortex at 4 °C negative pressure for 5 min. The suspension of CMSP was then stirred at 4 °C for 30 min and subsequently stood at 4 °C overnight for further adsorption. The prepared CMSP was packed into the commercial stainless-steel column (10 mm × 2.0 mm id) with water at 1 mL/min flow rate for 10 min. The CMC assay was performed using a SHIMADZU LC-2010AHT high-performance liquid chromatography (HPLC). The CMC column was preequilibrated for 1 h before sample injection, and then pH and FC were analyzed using the CMC columns. Flow rate 0.2 ml/min; column temperature 37 °C; mobile phase 1 mM phosphate-buffered saline, pH 7.2.
2.9. The detection of SARS-CoV-2 spike pseudotyped virus entry into ACE2h cells
5 × 104 of ACE2h cells in 100 μL DMEM per well were seeded into white 96-well plates. The cells were cultured in a 37 °C incubator containing 5% CO2 for 2 h. 50 μL medium was aspirated carefully from 96 wells, 50 μL medium containing corresponding dose of the medicine was added and incubated for 2 h. The final concentration of the drugs in each well was 10 μM. Then 5 μL of SARS-CoV-2 spike pseudotyped virus was added (Sino Biological, PSC001), and incubated in the 37 °C incubator containing 5% CO2 for 4 h followed with adding 100 μL of complemented DMEM per well. After 6–8 h of further infection, the culture medium containing the virus was sucked away and replaced by 200 μL of fresh DMEM, and incubated continuously at 37 °C for 48 h, the culture medium was aspirated and 20 μL of cell lysate was add from the Luciferase Assay System (Promega, E1500) to each well, then 100 μL of luminescence solution was added to wells before the luciferase luminescence detection, chemiluminescence was detected by a microplate reader under 560 nm, the exposure time was 1 s. The same method was used to verify whether DEX inhibits VSVG from entering ACE2h cells.
2.10. Statistical analysis
Data are presented as the mean ± standard error of mean (SD) and were statistically analyzed using analysis of variance (ANOVA). Two-tailed tests were used for comparisons between two groups, and differences were considered statistically significant at p < 0.05.
3. Results
3.1. The effect of GCs on ACE2h cells viability
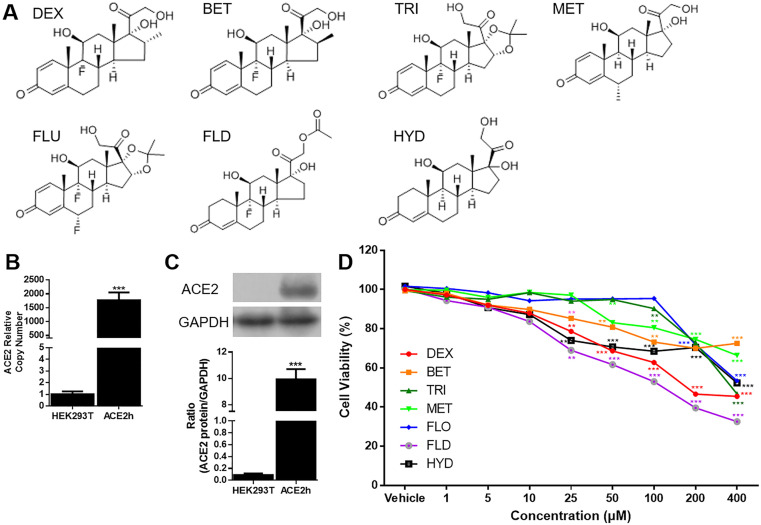
All the seven GCs shared the same basic steroidal structure, including C4-5 double bond, C3 ketone group and C17 side chain. Differences, including chirality in the remaining sites, gave rise to different potency and metabolic properties; As showed in Fig. 1 B and C, the ACE2 mRNA and protein were both overexpressed detected by real-time PCR and western blotting, indicating that ACE2h cell line were constructed successfully. All of the GCs showed no significant inhibition of cell activity when the concentration was less than 10 μM. When concentrations raised to 100 μM, FLO and TRI still have no effect on cell activity while cell viability remain 62.73 ± 2.68% (DEX), 73.2 ± 5.35% (BET), 80.5 ± 6.55% (MET), 68.52 ± 5.98% (HYD), and 52.98 ± 5.19% (FLD), respectively. When concentration reached 400 μM, BET showed the least impact on cell viability. Thereafter, the experiments were carried out at concentration no more than 10 μM in vitro.
Fig. 1.
Structure and cytotoxicity of GCs on ACE2h cells. A. Structure of 7 GCs; B. ACE2 mRNA was overexpressed in ACE2h cells compared to HEK293T cells. The experiments were repeat three times. Data are presented as mean ± S.D.; C. ACE2 was overexpressed in ACE2h cells compared to HEK293T cells. The experiments were repeat three times. Data are presented as mean ± S.D. (***p < 0.001, compared with HEK293T); (***p < 0.001, compared with HEK293T); D. Cytotoxicity of GCs on ACE2h cells under concentration from 0 to 400 μM. The experiments were repeat three times. Data are presented as mean ± S.D. (*p < 0.05, **p < 0.01, ***p < 0.001 compared to vehicle).
3.2. Molecular docking results of GCs and ACE2
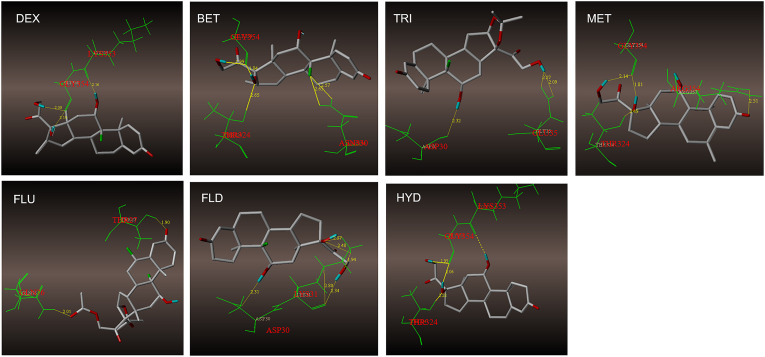
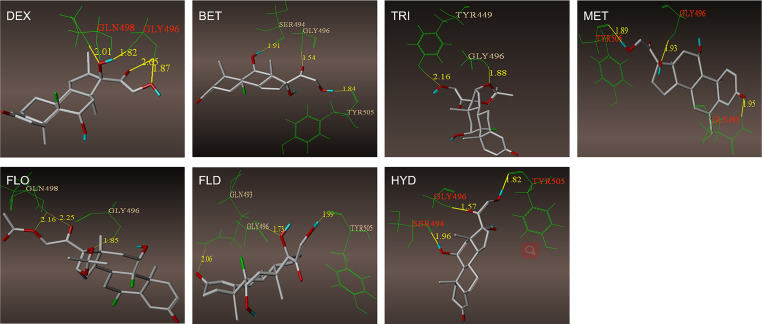
Currently, it is widely believed that SARS-CoV-2 infects cells via binding ACE2 protein with RBD of SARS-CoV-2 spike protein on the cell membrane surface. Therefore, it was of great significance to investigate whether GCs could interact with these proteins. Molecular docking was used to analyze the binding sites of GCs on ACE2 and RBD. As showed in Fig. 2 , the binding sites and hydrogen bonding of 7 GCs with ACE2 were presented. Binding character of GCs with RBD of SARS-CoV-2 spike protein were presented in Supplementary Fig. 1. It was assumed that seven GCs can bind to ACE2 and RBD, DEX can interact with active amino acid residue of both proteins, LYS353 for ACE2 and GLN498 for RBD (Table 1 ). Although other GCs can also bind with active residue, such as TRI and FLD bind with ASP30 in ACE2, and FLO with GLN498 in RBD, they cannot interact with both proteins.
Fig. 2.
The binding sites and hydrogen bonds of 7 GCs with ACE2. A. GCs bind with ACE2 by several amino residues. DEX, TRI, MET, FLD and HYD bind to active binding sites. B. GCs bind with RBD by several amino residues. DEX, and FLO bind to active binding sites.
Table 1.
Amino acid residues that form hydrogen bonds between GCs and ACE2 and RBD of 2019-nCoV spike protein.
| GC | ACE2 | RBD of 2019-nCoV spike protein |
|---|---|---|
| DEX | LYS353a, GLY354 | GLY496, GLN498a |
| BET | THR324, GLY354, ASN330 | SER494, GLY496, TYR505 |
| TRI | ASP30a, GLN35 | TYR449, GLY496 |
| MET | THR324, GLY354, ARG357a | GLN493, GLY496, TYR505 |
| FLO | THR27, GLN76 | GLY496, GLN498a |
| FLD | ASP30a | GLN493, GLY496 |
| HYD | THR324, LYS353, GLY354 | SER494, GLY496, TYR505 |
Amino acid residues are the active sites of ACE2 binding to RBD during virus invasion.
3.3. The binding character of GCs with ACE2
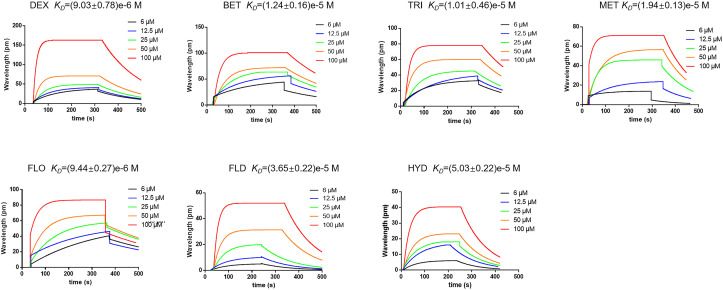
SPR were used to evaluate the binding characteristics. Seven GCs have a certain degree of binding. DEX showed the best affinity with ACE2 than other GCs, the binding constant K D value was (9.03 ± 0.78)e−6 M (Fig. 3 ).
Fig. 3.
Interaction between GCs and ACE2. GCs have a certain ability to combine with ACE2 detected by SPR method. DEX showed better affinity with ACE2 than other 6 GCs, the KD value is (9.03 ± 0.78)e−6 M.
3.4. Chromatographic retention of GCs on ACE2h-CMC column
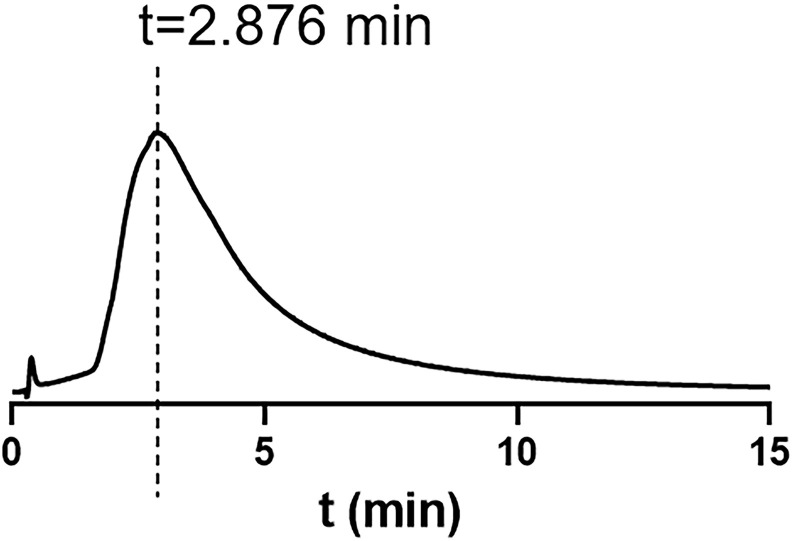
Considering that the interaction between ACE2 and GCs was detected by SPR in a non-bioactive state, we further investigated the interaction by CMC which was a bio affinity chromatography always used in screening bioactive components in complex mixture and analyzing receptor-ligand interaction (Ma et al., 2017). ACE2h-CMC was used in analyzing potential compounds target on ACE2. As showed in Fig. 4 , DEX was retained on the ACE2h-CMC column due to the interaction between receptor and ligand while other GCs were not, indicating that DEX could bind with ACE2 in vitro. The retention time of DEX was 2.876 min. Based on these results, DEX could bind with ACE2 and may have an effect on virus viropexis.
Fig. 4.
DEX was retained on the ACE2h-CMC column while other GCs were not. The retention time of DEX was 2.876 min.
3.5. DEX suppressed the entry of SARS-CoV-2 spike pseudotyped virus into ACEh cells
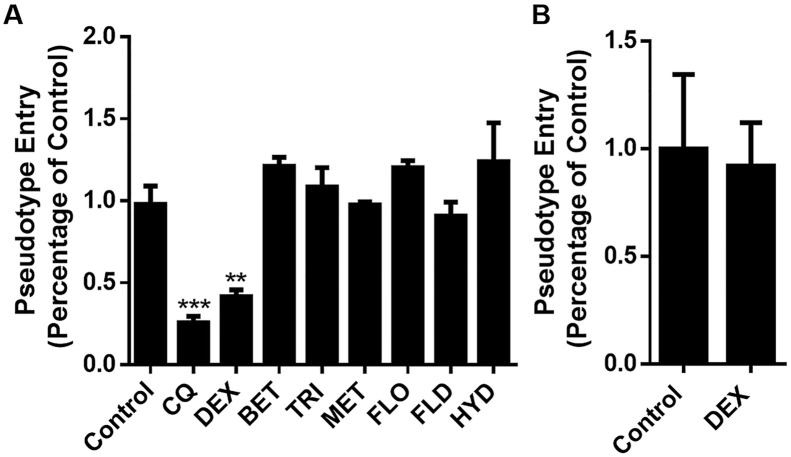
ACEh cells infected by SARS-CoV-2 spike pseudotyped virus were considered as control, the luciferase luminescence value of control was defined as 1. Chloroquine (CQ) was used as a positive control (Colson et al., 2020). Under the treatment of DEX (10 μM), the SARS-CoV-2 spike pseudotyped virus entrance ratio was reduced to 41 ± 7.38, while treated by the same dosage of other 6 GCs, the ratios were 121 ± 9.35% (BET), 108 ± 16.7% (TRI), 97 ± 3.18% (MET), 120 ± 6.13% (FLO), 90 ± 14.6% (FLD), and 124 ± 4.09% (HYD), respectively (Fig. 5 A). The ability of the SARS-CoV-2 spike pseudotyped virus to enter the ACE2h cells were reduced significantly under the treatment of DEX. To confirm whether DEX have a general effect on pseudovirus gene expression, we performed the mock infection with VSVG. The results showed that the entry of VSVG into ACEh cells was not inhibited by DEX (Fig. 5B).
Fig. 5.
Effect of GCs on the entrance of SARS-CoV-2 spike pseudotyped virus into ACE2h cells. A. DEX inhibit the entrance of SARS-CoV-2 spike pseudotyped virus into ACE2h cells; B. DEX had no effect on pseudovirus gene expression. The experiments were repeated three times. Data are presented as mean ± S.D. **p < 0.01, ***p < 0.001, compared with group 0.
4. Discussion
SARS-CoV-2 is globally prevalent in 2020, and there are currently no specific drugs against the virus. ACE2 is the target receptor of SARS-CoV-2 virus, DEX have shown certain efficacy in clinical use (Ledford, 2020; Selvaraj et al., 2020). Our study had investigated the binding character of 7 GCs by molecular docking, SPR, and ACE2h CMC assay, and confirmed that DEX could bind to ACE2 and inhibit the entry of pseudovirus through binding to its receptor ACE2.
Among the 7 GCs we've investigated, MET and FLO showed little cytotoxicity when concentration was under 100 μM. BET and MET had least influence on cell viability when concentration was up to 400 μM. Consistent with structural pharmacology and clinical observation, when there are C9 F and no C1,2 double bond, side effects of GC are more obvious (Pereira and Freire de Carvalho, 2011). Although DEX, FLD, and HYD had relatively more cytotoxicity, GCs had no obvious influence on cell viability when concentration were less than 10 μM. Thereafter, the experiments were carried out at concentration no more than 10 μM in vitro. Firstly, as showed in Fig. 2 and supplementary Fig. 1, molecular docking was used to simulate the combination of 7 GCs with ACE2 and RBD. Although all 7 GCs bind to the proteins, DEX can bind to the active sites, GLN498 of ACE2, showing good combination potential (Yan et al., 2020). Next, SPR was used to investigate the binding characteristics of GCs to ACE2 protein. SPR results further proved that DEX interacted with ACE2 better than other GCs. Finally, the bioactive CMC method was carried out to evaluate the interaction ability between GCs and ACE2. CMC is a method which investigates the ligand-receptor interaction by chromatography on the premise of not destroying the senior structure of the receptor on the cell membrane (Fu et al., 2019; He et al., 2007). Due to the existence of spatial structure as well as biological activity of receptor such as ACE2, this method can ensure the authenticity of the measured affinity as much as possible. DEX was retained on ACE2h-CMC column. So far, we believed that DEX can interacted with ACE2. Although the K D between ACE2 and spike protein was much smaller than that between ACE2 and DEX, DEX was preincubated before adding spike protein pseudovirus in this experiment. Plenty of DEX was allowed to bind to ACE2 freely. Subsequently, the spike protein pseudovirus added were more difficult to bind to ACE2, thus inhibiting the invasion of pseudovirus. Virus entry into cells is a critical step in the process of virus infection (Gao et al., 2020). However, novel coronavirus research is greatly limited by the need to achieve laboratory safety level 3 or above for direct research using virus strains. Pseudotyped virus refers to a retrovirus that can integrate the membrane glycoproteins of a different kind of virus to form an external viral membrane, while the genome retains the genomic characteristics of the retrovirus itself (Zhao et al., 2013). The SARS-CoV-2 pseudotyped virus proccess protein required for infection, and can be used for simulating the viropexis of SARS-CoV-2 (Nie et al., 2020). By using SARS-CoV-2 pseudovirus as an infection model to assess the anti-virus effect of GCs (Wang et al., 2020), we confirmed that DEX had the ability to suppress the entrance of SARS-CoV-2 spike pseudotyped virus into ACE2h cells. Additionally, VSVG mock infection was carried out to prove that there was no general effect on pseudovirus gene expression, as showed in Fig. 5B.
Additionally, we evaluated the binding character of DEX and RBD by molecular docking. DEX bind to LYS353, a active residue of RBD. More interestingly, when the virus infects the body, the two amino acid residues (GLN498 and LYS353) of ACE2 and RBD which may interact with DEX by molecular docking, happen to form hydrogen bonds with each other. Perhaps this synergy is what gives DEX its superior antiviral effect.
Based on our findings, we looked at the dosages of DEX for different corona viruses treatments. At present, DEX is still used clinically for its anti-inflammatory activity (Johnson and Vinetz, 2020). However, there was a dramatic drop in doses. For SARS-CoV-1, over 100 mg/day DEX has been used in order to acquire less fatality, shorter hospitalization days (Chen et al., 2006). As for SARS-CoV-2, low dose dexamethasone (6 mg once daily, 10 days) reduces deaths in patients hospitalised with COVID-19 who need ventilation, according to preliminary results from the recovery trial (Mahase, 2020). At such low dose, DEX has little anti-inflammatory effect. Besides, an excellent result has been obtained from a short-term use (4 mg three times daily for two days) (Selvaraj et al., 2020). Low doses and short periods of DEX use can greatly avoid the serious side effects of glucocorticoid pulse therapy, such as femoral head necrosis, hyperglycemia, psychosis, and avascular necrosis (Arabi et al., 2018; Stockman et al., 2006). Meanwhile, we hypothesized that the significant decrease in dose and duration of DEX is due to the ability to inhibit viral invasion of cells. The ability of DEX to resist SARS-CoV-2 in vivo needs further study.
In conclusion, we found that DEX inhibits the entrance of SARS-CoV-2 spike pseudotyped virus into ACE2h cells by preventing ACE2 from binding to the spike protein.
CRediT authorship contribution statement
Yongjing Zhang: Conceptualization, Methodology, Writing - original draft. Shiling Hu: Investigation, Software. Jue Wang: Data curation, Investigation. Zhuoyin Xue: Investigation. Cheng Wang: Software. Nan Wang: Writing - review & editing, Supervision, Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by National Natural Science Foundation of China [Grant number: 81930096], China Postdoctoral Science Foundation [Grant number: 2019M653672], and Fundamental Research Funds for the Central University (Grant number: xjj2018168).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2020.12.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., Saudi Critical Care Trial G. Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018;197(6):757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- Ayroldi E., Cannarile L., Migliorati G., Nocentini G., Delfino D.V., Riccardi C. Mechanisms of the anti-inflammatory effects of glucocorticoids: genomic and nongenomic interference with MAPK signaling pathways. Faseb. J. 2012;26(12):4805–4820. doi: 10.1096/fj.12-216382. [DOI] [PubMed] [Google Scholar]
- Chen R.C., Tang X.P., Tan S.Y., Liang B.L., Wan Z.Y., Fang J.Q., Zhong N. Treatment of severe acute respiratory syndrome with glucosteroids: the Guangzhou experience. Chest. 2006;129(6):1441–1452. doi: 10.1378/chest.129.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55(4):105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho A.E., Chapman K.E. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol. Cell. Endocrinol. 2011;335(1):2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Lv Y., Jia Q., Lin Y., Han S. Dual-mixed/CMC model for screening target components from traditional Chinese medicines simultaneously acting on EGFR & FGFR4 receptors. Talanta. 2019;192:248–254. doi: 10.1016/j.talanta.2018.09.053. [DOI] [PubMed] [Google Scholar]
- Gao M., Yang L., Chen X., Deng Y., Yang S., Xu H.…Gao X. A study on infectivity of asymptomatic SARS-CoV-2 carriers. Respir. Med. 2020;169:106026. doi: 10.1016/j.rmed.2020.106026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., China Medical Treatment Expert Group for C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J.…Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.S., Raza K., Cooper M.S. Therapeutic glucocorticoids: mechanisms of actions in rheumatic diseases. Nat. Rev. Rheumatol. 2020;16(3):133–144. doi: 10.1038/s41584-020-0371-y. [DOI] [PubMed] [Google Scholar]
- He L.C., Wang S.C., Yang G.D., Zhang Y.M., Wang C.H., Yuan B.X., Hou X.F. Progress in cell membrane chromatography. Drug Discov Ther. 2007;1(2):104–107. [PubMed] [Google Scholar]
- Johnson R.M., Vinetz J.M. Dexamethasone in the management of covid -19. BMJ. 2020;370:m2648. doi: 10.1136/bmj.m2648. [DOI] [PubMed] [Google Scholar]
- Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582(7813):469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- Lewis S.R., Pritchard M.W., Thomas C.M., Smith A.F. Pharmacological agents for adults with acute respiratory distress syndrome. Cochrane Database Syst. Rev. 2019;7:CD004477. doi: 10.1002/14651858.CD004477.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.M., Wang S.X., Gao H.S., Wang J.G., Wei C.S., Chen L.M., Su B. [Factors of avascular necrosis of femoral head and osteoporosis in SARS patients' convalescence] Zhonghua Yixue Zazhi. 2004;84(16):1348–1353. [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Yang L., Lv Y., Fu J., Zhang Y., He L. Determine equilibrium dissociation constant of drug-membrane receptor affinity using the cell membrane chromatography relative standard method. J. Chromatogr. A. 2017;1503:12–20. doi: 10.1016/j.chroma.2017.04.053. [DOI] [PubMed] [Google Scholar]
- Mahase E. Covid-19: low dose steroid cuts death in ventilated patients by one third, trial finds. BMJ. 2020;369:m2422. doi: 10.1136/bmj.m2422. [DOI] [PubMed] [Google Scholar]
- McKeever T., Mortimer K., Wilson A., Walker S., Brightling C., Skeggs A., Harrison T. Quadrupling inhaled glucocorticoid dose to abort asthma exacerbations. N. Engl. J. Med. 2018;378(10):902–910. doi: 10.1056/NEJMoa1714257. [DOI] [PubMed] [Google Scholar]
- Mehra M.R., Desai S.S., Kuy S., Henry T.D., Patel A.N. Retraction: cardiovascular disease, drug therapy, and mortality in covid-19. N. Engl. J. Med. 2020;382(26):2582. doi: 10.1056/NEJMoa2007621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microb. Infect. 2020;9(1):680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R.M., Freire de Carvalho J. Glucocorticoid-induced myopathy. Joint Bone Spine. 2011;78(1):41–44. doi: 10.1016/j.jbspin.2010.02.025. [DOI] [PubMed] [Google Scholar]
- Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/s0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj V., Dapaah-Afriyie K., Finn A., Flanigan T.P. Short-term dexamethasone in sars-CoV-2 patients. R. I. Med. J. 2020;103(6):39–43. [PubMed] [Google Scholar]
- Stahn C., Buttgereit F. Genomic and nongenomic effects of glucocorticoids. Nat. Clin. Pract. Rheumatol. 2008;4(10):525–533. doi: 10.1038/ncprheum0898. [DOI] [PubMed] [Google Scholar]
- Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3(9):e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Chiou S.H. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21(7) doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B., Finfer S., Cohen J., Rajbhandari D., Arabi Y., Bellomo R. The Australian-New Zealand intensive care society clinical trials, G. adjunctive glucocorticoid therapy in patients with septic shock. N Engl J Med. 2018;378(9):797–808. doi: 10.1056/NEJMoa1705835. [DOI] [PubMed] [Google Scholar]
- Villar J., Ferrando C., Martínez D., Ambrós A., Muñoz T., Soler J.A., González-Martín J.M. Dexamethasone treatment for the acute respiratory distress syndrome: a multicentre, randomised controlled trial. Lancet Respir Med. 2020;8(3):267–276. doi: 10.1016/s2213-2600(19)30417-5. [DOI] [PubMed] [Google Scholar]
- Wang Nan, Han Shengli, Liu Rui, Meng Liesu, He Huaizhen, Zhang Yongjing, Wang Cheng, Lv Yanni, Wang Jue, Li Xiaowei. Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of 2019-nCoV Spike pseudotyped virus. Phytomedicine. 2020 doi: 10.1016/j.phymed.2020.153333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J.…Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P., Murdoch D.R. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395(10229):1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Li W., Shi X., Chen Z., Jiang B., Liu J.…Li L. Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID-19) J. Intern. Med. 2020;288(1):128–138. doi: 10.1111/joim.13063. [DOI] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Du L., Ma C., Li Y., Li L., Poon V.K.…Zhou Y. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol. J. 2013;10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]