Abstract
Coca is the natural source of cocaine as well as a sacred and medicinal plant farmed by South American Amerindians and mestizos. The coca crop comprises four closely related varieties classified into two species (Amazonian and Huánuco varieties within Erythroxylum coca Lam., and Colombian and Trujillo varieties within Erythroxylum novogranatense (D. Morris) Hieron.) but our understanding of the domestication and evolutionary history of these taxa is nominal. In this study, we use genomic data from natural history collections to estimate the geographic origins and genetic diversity of this economically and culturally important crop in the context of its wild relatives. Our phylogeographic analyses clearly demonstrate the four varieties of coca comprise two or three exclusive groups nested within the diverse lineages of the widespread, wild species Erythroxylum gracilipes; establishing a new and robust hypothesis of domestication wherein coca originated two or three times from this wild progenitor. The Colombian and Trujillo coca varieties are descended from a single, ancient domestication event in northwestern South America. Huánuco coca was domesticated more recently, possibly in southeastern Peru. Amazonian coca either shares a common domesticated ancestor with Huánuco coca, or it was the product of a third and most recent independent domestication event in the western Amazon basin. This chronology of coca domestication reveals different Holocene peoples in South America were able to independently transform the same natural resource to serve their needs; in this case, a workaday stimulant. [Erythroxylum; Erythroxylaceae; Holocene; Museomics; Neotropics; phylogeography; plant domestication; target-sequence capture.]
Called the “Divine Leaf” by the Inka, coca has been cultivated for over 8000 years and is the most culturally significant pharmaceutical plant in South America; it is also the source of the alkaloid cocaine—an insecticide and local anesthetic that has had the largest impact in Western Medicine of any Neotropical phytochemical (Schultes 1979; Plowman 1986; Nathanson et al. 1993; Dillehay et al. 2010; Restrepo et al. 2019). In parts of Colombia, Ecuador, Peru, Bolivia, and Brazil, the traditional varieties of coca are still cultivated as they have been since Pre-Columbian times (Fig. 1; Plowman 1984; Plowman and Hensold 2004), and today over 5 million South Americans chew the leaves for their mild stimulant and medicinal effects (Conzelman and White 2016). However, prohibition has pushed illicit cultivation for cocaine, now at its highest level since 2000, into lowland forests in Colombia, Peru, Brazil, and even southern Mexico, causing deforestation and civil destabilization in these regions (Plowman 1984; Dávalos et al. 2011; Bewley-Taylor 2016; Casale and Mallette 2016; United Nations 2019).
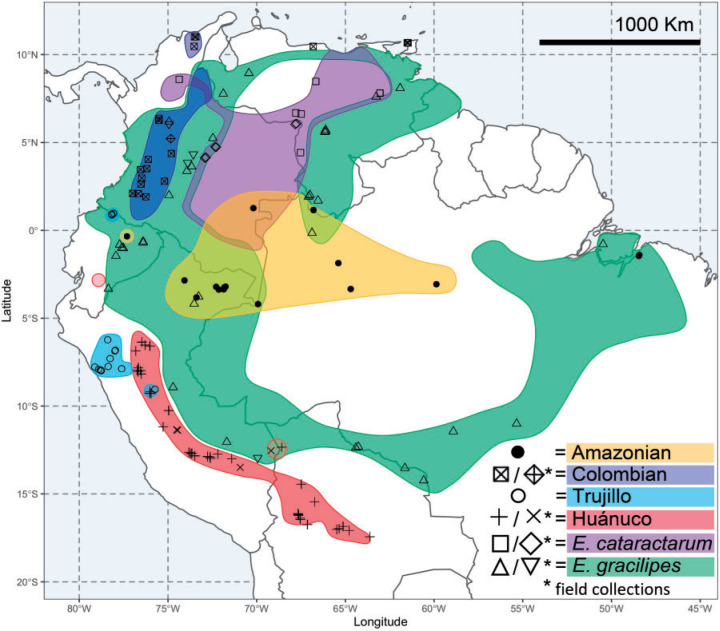
Figure 1.
Map of taxon distributions and sampling localities. Polygons indicate modern areas of cultivation for the four coca varieties sensu Plowman and Hensold (2004); omitted are areas of illicit cultivation (esp. southern Colombia) and garden plots (incl. botanical gardens). Symbols indicate location of sampled herbarium specimens and rotated symbols indicate field collections by D. M. W.
To understand the identity of the coca crop, we sequenced 424 nuclear genes from tissue samples collected almost entirely from historical museum collections (90% of the total) and completed the first investigation of the genetic structure of the four coca varieties and their closest wild relatives (Erythroxylum spp.). With over 10,000 exsiccatae including  950 cocas, the Field Museum (F) holds the world’s largest Neotropical Erythroxylum collection. We utilized this resource to maximize geographic coverage in our sampling of dozens of individuals of the coca varieties and their wild relatives, exemplifying the utility of well-curated museum collections in phylogeographic and population-genomics research. For diverse and poorly studied tropical plant taxa, and especially for drug plants that require complex logistics to collect, ship, and store, museum collections can provide an increasingly important role in systematic research (Rowe et al. 2011; Hart et al. 2016; Forrest et al. 2019). Yet, many studies using historical samples have been limited to organelles or utilized relatively undegraded DNA (Staats et al. 2013; Beck and Semple 2015).
950 cocas, the Field Museum (F) holds the world’s largest Neotropical Erythroxylum collection. We utilized this resource to maximize geographic coverage in our sampling of dozens of individuals of the coca varieties and their wild relatives, exemplifying the utility of well-curated museum collections in phylogeographic and population-genomics research. For diverse and poorly studied tropical plant taxa, and especially for drug plants that require complex logistics to collect, ship, and store, museum collections can provide an increasingly important role in systematic research (Rowe et al. 2011; Hart et al. 2016; Forrest et al. 2019). Yet, many studies using historical samples have been limited to organelles or utilized relatively undegraded DNA (Staats et al. 2013; Beck and Semple 2015).
There are four taxonomic varieties of coca grown in separate geographic areas in South America (Fig. 1; Plowman 1979a,1979b). They are morphologically similar but can be distinguished by several traits, most notably leaf shape and venation patterns (Supplementary Table S1 available on Dryad at https://doi.org/10.5061/dryad.cvdncjt1n; Plowman 1979a; Bohm et al. 1982; Rury and Plowman 1983). Huánuco (or Bolivian) coca (Erythroxylum coca Lam.) is grown in the moist, montane forest on the eastern slopes of the Andes in Peru and Bolivia. This is the most abundant crop for traditional and indigenous coca leaf consumption and was the world’s primary source of cocaine hydrochloride from its discovery in 1865 until 2000 (Gootenberg 2008; United Nations 2019). Amazonian coca (E. coca var. ipadu Plowman) is grown in discrete localities throughout the Amazon basin, and its leaves are consumed as a pulverized powder with other additives (Plowman 1981). Until extensive eradication began about 30 years ago, Colombian coca (Erythroxylum novogranatense (D. Morris) Hieron.) was grown throughout the drier inter-Andean valleys in Colombia, but now it is restricted to the Cauca region and the Sierra Nevada de Santa Marta (Bohm et al. 1982). Lastly, Trujillo coca (E. novogranatense var. truxillense (Rusby) Plowman) is grown in the arid valleys of northwestern Peru for traditional coca leaf use and as a flavoring agent in Coca-Cola® (Gootenberg 2008). We also sampled from a disjunct population of what is believed to be Trujillo coca cultivated on the Colombia/Ecuador border but were unable to sample the disjunct Huánuco coca from southern Ecuador (Plowman 1984).
The primary hypothesis of coca domestication was described by Plowman (Plowman 1979b; Bohm et al. 1982); he posited that Huánuco coca was domesticated in the eastern Andes of Bolivia or Peru from a wild, ancestral (and presumably extinct) form of E. coca. Then, because the two taxa can form infertile hybrids, he believed Huánuco coca was taken north to dry Andean valleys in Peru or Ecuador, where it developed into Trujillo coca. Next, three pieces of evidence led him to postulate Colombian coca was derived from Trujillo: first, all coca macrofossils  1500 years old are of the Trujillo morphotype, providing evidence that Trujillo coca is older than Colombian (Plowman 1984; Dillehay et al. 2010). Second, Trujillo
1500 years old are of the Trujillo morphotype, providing evidence that Trujillo coca is older than Colombian (Plowman 1984; Dillehay et al. 2010). Second, Trujillo  Colombian F1 hybrids are fertile but Trujillo
Colombian F1 hybrids are fertile but Trujillo  Huánuco hybrids are not, so Colombian coca has been interpreted as a derived variety with acquired interspecific hybrid incompatibility (Bohm et al. 1982). Finally, Colombian coca is the only variety that is self-compatible, which is generally a derived trait unlikely to give rise to self-incompatibility (Plowman 1986; Goldberg et al. 2010). Lastly, Huánuco coca was taken east and transformed into Amazonian coca.
Huánuco hybrids are not, so Colombian coca has been interpreted as a derived variety with acquired interspecific hybrid incompatibility (Bohm et al. 1982). Finally, Colombian coca is the only variety that is self-compatible, which is generally a derived trait unlikely to give rise to self-incompatibility (Plowman 1986; Goldberg et al. 2010). Lastly, Huánuco coca was taken east and transformed into Amazonian coca.
However, archaeological evidence does not directly support Plowman’s linear-series hypothesis beginning with Huánuco coca because Trujillo coca has the most extensive Pre-Columbian archaeological record (Mortimer 1901; Plowman 1984), and a recent discovery of Trujillo morphotype leaves in northern Peru pushed coca consumption from 4000 to over 8000 years BP, making it one of oldest cultivated plants in the Americas (Dillehay et al. 2010; Larson et al. 2014). Coca paraphernalia and bountiful artworks provide the only archaeological evidence of coca use in Colombia, the earliest is from the Yotoco culture (100–1200 CE; Reichel-Dolmatoff and Schrimpff 2005). The earliest Huánuco coca remains are endocarps from a Late Intermediate period (1000–1476 CE) site in Junín, Peru, but evidence of coca trade in Bolivia pushes this date to 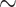 1700 years BP (Plowman 1984; Carter and Mamani 1986; Hastorf 1987; Valdez et al. 2015). On the basis of linguistic and ethnographic similarities across its range, Amazonian coca is presumed to be the most recently developed (Plowman 1981).
1700 years BP (Plowman 1984; Carter and Mamani 1986; Hastorf 1987; Valdez et al. 2015). On the basis of linguistic and ethnographic similarities across its range, Amazonian coca is presumed to be the most recently developed (Plowman 1981).
A more recent hypothesis from Johnson et al. (2005), is that E. coca (Amazonian and Huánuco) and E. novogranatense (Colombian and Trujillo) are sister species resulting from the domestication of a common ancestor (Johnson et al. 2005; Emche et al. 2011). However, their analysis, based on flavonoid profiles and amplified fragment-length polymorphisms, did not include the closest wild relatives and thus could not properly evaluate the independent origins of the coca crops. Our previous study (White et al. 2019) revealed that the closest wild relatives of the coca varieties are the cocaine-producing, wild species Erythroxylum cataractarum Spruce ex Peyr. and Erythroxylum gracilipes Peyr. (Aynilian et al. 1974; Plowman and Rivier 1983; Bieri et al. 2006; Islam 2011). Erythroxylum gracilipes occurs throughout most of the Amazon basin and can be distinguished from the cocas by its larger leaves (11–18 cm vs. 2.5–11 cm) with acuminate apices. Erythroxylum cataractarum of the Llanos of Colombia and Venezuela, is morphologically very similar to E. novogranatense, but with stiffer branchlets, thicker membranaceous to sub-chartaceous leaves, and stipules with more prominent apical setae (Fig. 1; Supplementary Table S1 available on Dryad).
The first goal of this project is to establish the genealogical relationships and genetic structure of a severely understudied crop. By sampling both the coca crops and their closest wild relatives, this is the first study capable of inferring the number of times coca was domesticated from a wild progenitor and also where and when these events occurred. Second, we want to expand our toolkit for “unlocking” genetic data from the “treasure chests” of biological diversity stored in museums and biological collections worldwide (Särkinen et al. 2012; Jones and Good 2016). By sequencing 424 genes from over 130 herbarium collections across six taxa, we have demonstrated the efficacy of exon-capture DNA sequencing with highly degraded DNA from museum collections.
Materials and Methods
More detailed text is available in the Supplementary Material file on Dryad. We extracted genomic DNA from 154 Erythroxylum samples, 138 being herbarium specimens, (Supplementary Table S2 available on Dryad), and used a custom set of RNA probes (White et al. 2019) to sequence 424 nuclear genes. Cleaned reads were de novo assembled into contigs and mapped to the concatenated exon sequences for each gene using HybPiper (Johnson et al. 2016). We mapped the cleaned reads back to the supercontig consensus sequences for each gene and followed the seqcap_pop pipeline (Faircloth 2015; Harvey et al. 2016) and the GATK Best Practices workflow (https://software.broadinstitute.org/gatk/best-practices/workflow?id=11145) to call, annotate, and filter single nucleotide polymorphisms (SNPs).
We aligned gene sequences using MAFFT and inferred gene trees with RAxML before reconstructing a summary tree from 424 gene trees with ASTRAL-III (Katoh et al. 2002; Stamatakis 2014; Zhang et al. 2018). We used IQ-TREE 2 (Hoang et al. 2018; Minh et al. 2020) to infer a maximum likelihood tree from the concatenated SNP data set (“ML-SNP”) and to generate SNP-based (sCF) and gene tree-based (gCF) concordance factors for both the summary tree and the ML-SNP topologies. We then dropped 10 individuals showing admixture or “trans-taxonomic” cluster assignment and used the SNP data set to infer species trees with SVDquartets (Chifman and Kubatko 2014) as well as reduced-taxon summary trees with ASTRAL-III. To assess alternative domestication scenarios, we used ASTRAL-III to estimate quartet support and the probability of a constrained species tree topology.
We used our final data set of 6263 SNPs in a genetic cluster analysis using the snapclust program from the adegenet v.2.13 R package (Jombart and Ahmed 2011; Hoang et al. 2018). This method uses a fast maximum likelihood and geometric approach to assign genotypes to a set number of populations determined from goodness-of-fit statistics (Beugin et al. 2018). For principal components analysis (PCA), we used the ipyrad analysis toolkit (Eaton and Overcast 2020). Using all SNPs and the nine-population assignments, we calculated weighted 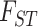 to understand population differentiation (Hudson et al. 1992). We evaluated genetic diversity by the number of private, novel alleles within a population (vcftools and vcf-contrast; Danecek et al. 2011), observed heterozygosity and nucleotide diversity (hierfstat R package; Goudet 2005), and allelic richness (the average number of alleles per SNP with rarefaction correction; diveRsity R package; Keenan et al. 2013). We estimated the degree of admixture between taxa using treemix v1.13 (Pickrell and Pritchard 2012). We dropped gracilipes2-4, leaving six populations, and performed stairway plot analyses (Liu and Fu 2015) to infer effective population size through time.
to understand population differentiation (Hudson et al. 1992). We evaluated genetic diversity by the number of private, novel alleles within a population (vcftools and vcf-contrast; Danecek et al. 2011), observed heterozygosity and nucleotide diversity (hierfstat R package; Goudet 2005), and allelic richness (the average number of alleles per SNP with rarefaction correction; diveRsity R package; Keenan et al. 2013). We estimated the degree of admixture between taxa using treemix v1.13 (Pickrell and Pritchard 2012). We dropped gracilipes2-4, leaving six populations, and performed stairway plot analyses (Liu and Fu 2015) to infer effective population size through time.
We applied an approximate Bayesian computation (ABC) approach to statistically test four alternative domestication scenarios: Plowman’s linear series, Johnson’s sister species, two origins, and three origins; the latter two hypotheses being derived from our genetic data. All E. gracilipes samples were grouped as a single population (see Supplementary Material available on Dryad) and E. cataractarum was excluded. This is a powerful approach for estimating coalescent parameters and comparing complex evolutionary scenarios (Beaumont et al. 2002; Gerbault et al. 2014). It was conducted in three steps using DIYABC v.2.1.0 (Cornuet et al. 2014). First, we generated 40 million simulated data sets under the coalescent based on our four topological scenarios. Second, we selected the subset of data sets closest to the observed (SNP) data according to summary statistics instead of the Bayesian likelihood computation (Nei’s genetic diversity, 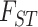 , and Nei’s distance using all data Nei 1972, 1987; Weir and Cockerham 1984). Third, the posterior distributions of coalescent parameters were estimated from the subset using a local linear regression procedure and the posterior probabilities of our four scenarios were compared by calculating the relative proportion of simulated data sets for each scenario present among the 500 data sets closest to the observed data set, as well as logistic regression of each scenario probability based on deviations between observed and simulated summary statistics (Beaumont 2008). We estimated confidence in our final scenario choices by calculating posterior- and prior-based error and scenario-specific type 1 and type 2 errors from additional pseudo-observed data sets generated under each scenario.
, and Nei’s distance using all data Nei 1972, 1987; Weir and Cockerham 1984). Third, the posterior distributions of coalescent parameters were estimated from the subset using a local linear regression procedure and the posterior probabilities of our four scenarios were compared by calculating the relative proportion of simulated data sets for each scenario present among the 500 data sets closest to the observed data set, as well as logistic regression of each scenario probability based on deviations between observed and simulated summary statistics (Beaumont 2008). We estimated confidence in our final scenario choices by calculating posterior- and prior-based error and scenario-specific type 1 and type 2 errors from additional pseudo-observed data sets generated under each scenario.
Results and Discussion
Our study demonstrates that a genomic approach can be applied to a population genetic project focusing on a very recent diversification history using primarily historical museum collections. Our results support a novel and robust hypothesis of multiple independent origins of different coca varieties from E. gracilipes, a widespread, wild species comprised of at least two main clades.
Museum Genomics: Target Capture, Assembly, and Gene Alignment
Hybridization-based exon-capture is an efficient method for sequencing select loci from genomic DNA isolated from herbarium specimens (Hart et al. 2016; Villaverde et al. 2018). We generated 270 GB of sequence data and deposited reads for all 154 samples in the NCBI Short Read Archive under BioProject PRJNA485502. Samples were collected between 1900 and 2016 with a median collection date of 1981 (Supplementary Fig. S1 and Table S2 available on Dryad). These samples presented a spectrum of DNA quantity and degradation, but these two factors had little correlation with the success of our exon capture and target assembly. A linear model fit between the genomic DNA maximum fragment size and the number of genes recovered was nearly flat (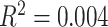 ); the polynomial regression model fit the relationship with an R
); the polynomial regression model fit the relationship with an R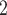 of 0.024 and was also mostly flat (Supplementary Fig. S1c available on Dryad). Samples with low input DNA amounts (less than 200 ng) were scattered throughout the range of observations of gene recovery. Plants collected in the moist tropics require extra care to dry, and DNA quality thus appears to depend more on initial drying and preservation quality rather than age, corroborating (Forrest et al., 2019) (Supplementary Fig. S1a, b available on Dryad).
of 0.024 and was also mostly flat (Supplementary Fig. S1c available on Dryad). Samples with low input DNA amounts (less than 200 ng) were scattered throughout the range of observations of gene recovery. Plants collected in the moist tropics require extra care to dry, and DNA quality thus appears to depend more on initial drying and preservation quality rather than age, corroborating (Forrest et al., 2019) (Supplementary Fig. S1a, b available on Dryad).
Not surprisingly, we observed a positive correlation between the log number of reads and the number of genes with good target coverage, defined as 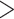 75% of target length (linear
75% of target length (linear 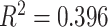 , polynomial
, polynomial 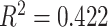 Supplementary Fig. S1d available on Dryad). This demonstrates that we recovered
Supplementary Fig. S1d available on Dryad). This demonstrates that we recovered 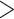 90% of exons at
90% of exons at 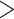 75% of their length with 4
75% of their length with 4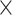 coverage per site at about 10
coverage per site at about 10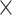 sequencing depth, and this depth provides a good target for sequencing effort. However, our number of reads per sample is quite variable, spanning three orders of magnitude, thus highlighting the variability incurred by combining museum samples of varying qualities together in large sequencing pools. Dilution of RNA baits and smaller numbers of samples per hybridization pool prior to exon-capture will help minimize amplification bias and overall variability of sequence reads per sample.
sequencing depth, and this depth provides a good target for sequencing effort. However, our number of reads per sample is quite variable, spanning three orders of magnitude, thus highlighting the variability incurred by combining museum samples of varying qualities together in large sequencing pools. Dilution of RNA baits and smaller numbers of samples per hybridization pool prior to exon-capture will help minimize amplification bias and overall variability of sequence reads per sample.
We removed 10 of our 154 samples from the analysis because they failed to assemble at least 90 genes with good coverage (Supplementary Fig. S2 available on Dryad). Across the 424 genes, the average assembled contig length per sample ranged from 819 bp to 8279 bp with a median of 2160 bp of sequence per sample and the 424 genes had an average of 141 samples per gene. The length of concatenated exon targets was 740,646 bp (median 1746 bp). Our assembled supercontigs (exon+intron) had an average concatenated length of 1,484,570 bp, revealing that we sequenced about 740 kb of intron and flanking sequence. Gene alignments ranged from 1749–57,834 bp in length (median 7290) with 30–93% missing data (median 59%). Cleaned trimAL alignments ranged from 129 to 8700 bp in length (median 1677) with 0.05–51.73% missing data (median 9.5%).
While previous studies have included key historical collections within broader biodiversity investigations (e.g., McCormack et al. 2016; Prosser et al. 2016; Greiman et al. 2018), our study has shown modern phylogeographic analyses can rely almost entirely on historical museum collections to generate deep and useful genomic data sets for phylogeographic research. Historical museum collections can also alleviate geographic sampling shortfalls, which can lead to erroneous inferences or oversplitting geographic populations (e.g., Jackson et al. 2017; Linck et al. 2019). This applies to systems where field collection is challenging or even dangerous but also in most biological systems due to the ever-increasing difficulty to obtain permits (Prathapan et al. 2018).
Genetic Structure of Coca and Wild Relatives
For a crop of such cultural and economic significance, our scientific understanding of the origin and evolution of coca is minimal. This is not only due to the obstacles to research imposed by its international prohibition but also the botanically challenging nature of Erythroxylum, a morphologically complex and speciose clade of tropical shrubs and treelets. Genetic sampling from the world’s largest Erythroxylum collection at the Field Museum has reshaped our fundamental understanding of the systematics of this clade (White et al. 2019).
The maximum likelihood gene trees inferred from the full gene alignments, cleaned trimAL alignments, or 212 alignments with the least missing data produced nearly identical ASTRAL summary trees, the only differences were among tips within the main clades and do not affect our presented results. Of these, the ASTRAL summary tree (AST; Fig. 2a; Supplementary Fig. S3 available on Dryad) inferred from the full gene alignments had the best local posterior probability along backbone nodes. To measure support for AST versus ML-SNP topology (topological differences are described in the Supplementary Text available on Dryad), we calculated SNP-based concordance factors (sCF; Supplementary Figs. S4 and S5 available on Dryad) and gene-tree concordance factors (gCF; Supplementary Figs. S6 and S7 available on Dryad) and found AST to be the best-supported phylogeny across our sequence and SNP data sets.
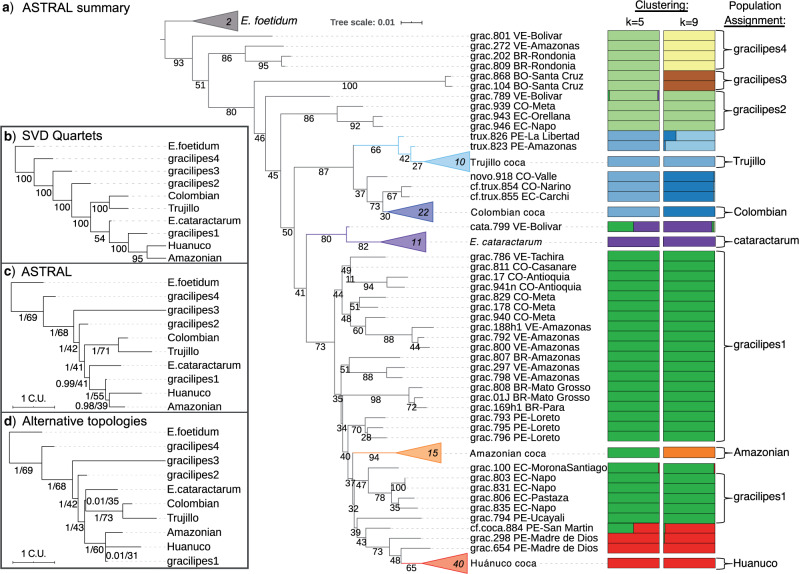
Figure 2.
Phylogeny and genetic clustering of coca varieties and wild relatives. a) ASTRAL-III summary (AST) tree topology with branch lengths optimized using the ML-SNP data set; scale indicates substitutions per site and branch support values denote concordance factors of SNPs; collapsed clades are marked with the number of samples; tip names indicate taxon, sample ID, and geographic provenance indicated by the two letter country code and political subdivision. Probability assignment of individuals to five or nine genetic clusters are indicated in bar plots along with the population assignment of ‘pure’ individuals for species trees and population genetic statistics. b) SVDquartets species tree inferred from SNP data with bootstrap support. c) ASTRAL-III species tree inferred from 424 gene trees with local posterior probability (LPP) and percent quartet support, respectively; branch lengths scaled by coalescent units. d) Constrained ASTRAL-III species trees indicating LPP and percent quartet support for an E. cataractarum 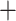 Trujillo/Colombian clade or a Huánuco
Trujillo/Colombian clade or a Huánuco 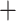 gracilipes1 clade.
gracilipes1 clade.
The AST phylogeny reveals the 37 E. gracilipes samples form a series of nested clades within which arise separate monophyletic lineages comprising E. cataractarum, E. novogranatense (Trujillo and Colombian cocas), Amazonian coca, and Huánuco coca (Fig. 2a). The goodness-of-fit estimators for the number of genetic clusters indicated the most significant information content in 5 and 9 clusters (Supplementary Fig. S8 available on Dryad; clustering results described in Supplementary Text available on Dryad).
The earliest diverging lineages of E. gracilipes are geographically distributed around the Amazon Basin and form a single cluster at 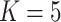 and three clusters at
and three clusters at 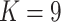 (gracilipes2, gracilipes3, gracilipes4; Fig. 2a). Next, the E. novogranatense lineage is divided into two clades and two clusters (
(gracilipes2, gracilipes3, gracilipes4; Fig. 2a). Next, the E. novogranatense lineage is divided into two clades and two clusters (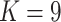 ) that define the Trujillo and Colombian coca varieties. The exceptions are samples trux.854 and trux.855 from the small, disjunct population on Río Chical at the Colombia/Ecuador border, revealing this population has a genealogical history closer to Colombian coca. This population could represent an early Colombian landrace derived from Trujillo coca, or it could be an admixed variety with combined Colombian and Trujillo ancestry. Additional sampling from Ecuador and Venezuela should improve our understanding of the history of this clade.
) that define the Trujillo and Colombian coca varieties. The exceptions are samples trux.854 and trux.855 from the small, disjunct population on Río Chical at the Colombia/Ecuador border, revealing this population has a genealogical history closer to Colombian coca. This population could represent an early Colombian landrace derived from Trujillo coca, or it could be an admixed variety with combined Colombian and Trujillo ancestry. Additional sampling from Ecuador and Venezuela should improve our understanding of the history of this clade.
The remaining E. gracilipes samples form a single cluster, gracilipes1, and belong to well-supported clades with geographic structure (Fig. 2a). Within gracilipes1, 15 samples of Amazonian coca form a clade with perfect posterior probability (Supplementary Fig. S3 available on Dryad), most closely related to gracilipes1 samples from Ecuador and northern Peru. Finally, three gracilipes1 samples from the Ucayali and Madre de Dios Departments of Peru form a nested series leading to the well-supported Huánuco coca clade (Fig. 2a; Supplementary Fig. S3 available on Dryad). The fourth sample in this series, “cf.coca.884,” is described as cultivated on its herbarium voucher, but its exceptional height (3 m), leaf morphology (apex and venation), and clustering results indicate admixture with E. gracilipes (Image in Supplementary material available on Dryad).
The E. cataractarum clade diverges as sister to gracilipes1 and comprises a distinct genetic cluster (Fig. 2a). However, our ML-SNP (Supplementary Figs. S5 and S7 available on Dryad) and SVDquartets tree (Fig. 2b) stirred a prior suspicion that E. cataractarum could actually be sister to the E. novogranatense lineage (White et al. 2019). We tested support among our gene trees for this alternative placement and found it received lower quartet support (35% vs. 41%) and posterior probability (0.01 vs. 0.99). To further evaluate the evolutionary history generating this discordance, we used Treemix to model genomic admixture and gene flow across a tree representing the nine populations. This inferred gene flow from Colombian coca into E. cataractarum and, secondarily, from Amazonian coca into E. cataractarum. The third most significant migration edge was from E. cataractarum into gracilipes4 (Supplementary Fig. S8 available on Dryad). These grow in the vicinity of E. cataractarum (Figs. 1 and 2a), and thus it is sensible that seeds and/or pollen from nearby Amazonian and Colombian coca farms have naturally introduced alleles into E. cataractarum. This result explains the phylogenomic discordance of the E. cataractarum clade and also indicates that it may not have a role in the evolution of coca, though the reverse appears to be true.
Multiple Origins Hypothesis
Our phylogenomic and clustering results indicate E. gracilipes is a paraphyletic taxon with respect to the Amazonian, Huánuco, and Colombian/Trujillo coca lineages (Fig. 2). This structure elucidates a novel hypothesis of multiple origins of domestication of coca from progenitor E. gracilipes (i.e. gracilipes1 and possibly gracilipes2), refuting both Plowman’s linear-series hypothesis beginning with Huánuco coca (Plowman 1979b) and the sister-species hypotheses suggested by Johnson and colleagues (Johnson et al. 2005; Emche et al. 2011).
While there is a clear separation of the E. novogranatense lineage (Colombian and Trujillo) from the E. coca lineage (Amazonian and Huánuco) and thus at least two domestication events, the separation of the Amazonian and Huánuco varieties, which would suggest three origins of domestication, is more tenuous. The first evidence of independent origins of Amazonian and Huánuco coca comes from the phylogeny and clustering results (Fig. 2), where they are separated by samples of gracilipes1 from central and southern Peru and a clade of Ecuadorean gracilipes1. While the placement of this Ecuadorean clade is not statistically robust, three gracilipes1 samples from Peru are firmly supported as basal to the Huánuco clade (Fig. 2a; Supplementary Figs. S2–S7 available on Dryad). The second basis for three domestications is that Amazonian coca clusters with gracilipes1 at 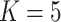 and forms its own cluster in
and forms its own cluster in 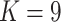 , suggesting closer proximity to gracilipes1 than Huánuco coca.
, suggesting closer proximity to gracilipes1 than Huánuco coca.
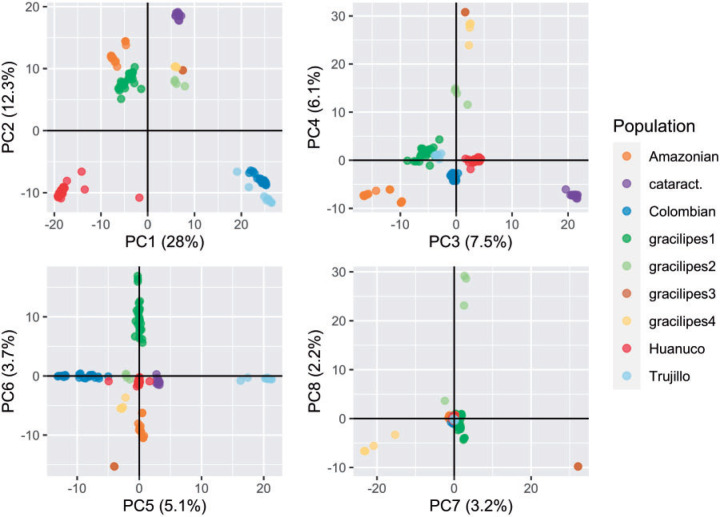
To further evaluate this separation and the progenitor-derivative relationships under our three-domestication hypothesis, we conducted a principal components analysis and calculated population genetic statistics across individuals from the nine populations. The PCA separates individuals into populations in agreement with the clustering analysis (Fig. 3). The nine populations, including Huánuco and Amazonian, cluster independently of one another in our bivariate plots describing the first eight components (explaining 65.9% of SNP variance; Fig. 3); further supporting the evolutionary separation of these two varieties.
Figure 3.
Genetic variation of coca varieties and wild relatives. The first eight principal components are presented in four bivariate plots.
Wright’s index of fixation (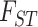 ) showed all pairs showed moderate to high levels of differentiation (range 0.25–0.78; Table 2). By our measures of genetic distance, Amazonian and Huánuco cocas are more similar to E. gracilipes than they are to each other, and Amazonian and gracilipes1 are the least differentiated taxa across all populations (Table 2). Amazonian and Huánuco show a similar degree of differentiation as Colombian and Trujillo (0.55 vs. 0.52, respectively). Statistics for the number of private alleles (PA; not weighted by sample size), average allelic richness per SNP (AR; weighted by sample size), observed heterozygosity (Ho), and genetic diversity (Hs) are presented in Table 1. Amazonian has the highest genetic diversity among the cocas (AR, Hs). Together, these statistics all corroborate the separation of Huánuco and Amazonian coca and thus a three-origin hypothesis of coca domestication. Corroborating expectations of genetic bottlenecks during domestication events (Gross and Olsen 2010), the coca varieties have lower genetic diversity than the wild taxa (PA, AR, Hs; see Supplementary Material available on Dryad for discussion of Ho).
) showed all pairs showed moderate to high levels of differentiation (range 0.25–0.78; Table 2). By our measures of genetic distance, Amazonian and Huánuco cocas are more similar to E. gracilipes than they are to each other, and Amazonian and gracilipes1 are the least differentiated taxa across all populations (Table 2). Amazonian and Huánuco show a similar degree of differentiation as Colombian and Trujillo (0.55 vs. 0.52, respectively). Statistics for the number of private alleles (PA; not weighted by sample size), average allelic richness per SNP (AR; weighted by sample size), observed heterozygosity (Ho), and genetic diversity (Hs) are presented in Table 1. Amazonian has the highest genetic diversity among the cocas (AR, Hs). Together, these statistics all corroborate the separation of Huánuco and Amazonian coca and thus a three-origin hypothesis of coca domestication. Corroborating expectations of genetic bottlenecks during domestication events (Gross and Olsen 2010), the coca varieties have lower genetic diversity than the wild taxa (PA, AR, Hs; see Supplementary Material available on Dryad for discussion of Ho).
Table 2.
Population pairwise 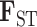
| gracilipes4 | gracilipes3 | gracilipes2 | gracilipes1 | cataract. | Colombian | Trujillo | Amazonian | |
|---|---|---|---|---|---|---|---|---|
| gracilipes3 | 0.68 | — | — | — | — | — | — | — |
| gracilipes2 | 0.42 | 0.62 | — | — | — | — | — | — |
| gracilipes1 | 0.46 | 0.6 | 0.32 | — | — | — | — | — |
| cataract. | 0.6 | 0.73 | 0.51 | 0.41 | — | — | — | — |
| Colombian | 0.66 | 0.76 | 0.56 | 0.49 | 0.6 | — | — | — |
| Trujillo | 0.79 | 0.91 | 0.71 | 0.55 | 0.71 | 0.52 | — | — |
| Amazonian | 0.61 | 0.73 | 0.51 | 0.25 | 0.54 | 0.61 | 0.7 | — |
| Huanuco | 0.74 | 0.83 | 0.67 | 0.4 | 0.69 | 0.71 | 0.78 | 0.55 |
Table 1.
Genetic diversity of coca varieties and wild relatives
| Population | PA | AR | Hs | Ho |
|---|---|---|---|---|
| gracilipes4 | 211 | 1.088 | 0.0734 | 0.028 |
| gracilipes3 | 131 | 1.014 | 0.118 | 0.067 |
| gracilipes2 | 95 | 1.096 | 0.066 | 0.029 |
| gracilipes1 | 467 | 1.116 | 0.083 | 0.023 |
| cataract. | 170 | 1.083 | 0.080 | 0.031 |
| Colombian | 35 | 1.064 | 0.046 | 0.027 |
| Trujillo | 7 | 1.021 | 0.015 | 0.011 |
| Amazonian | 65 | 1.083 | 0.068 | 0.060 |
| Huanuco | 55 | 1.045 | 0.050 | 0.026 |
Populations, as defined in Figure 3, are in rows. Statistics show the number of private alleles (PA), average, rarefaction-corrected allelic richness per SNP (AR), genetic diversity (Hs), and observed heterozygosity (Ho).
Testing Alternative Domestication Scenarios
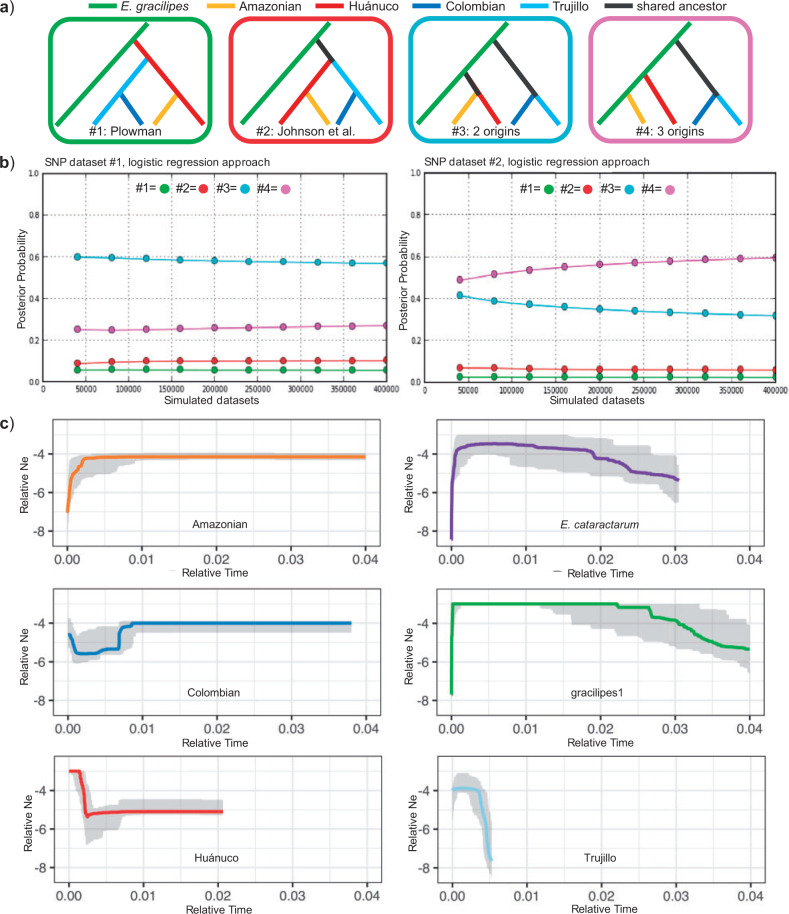
We used coalescent simulations under an approximate Bayesian computation (ABC) framework to estimate statistical support for Plowman’s linear series (Scenario 1; Plowman 1979b), Johnson’s sister species (Scenario 2; Johnson et al. 2005), a two-origin hypothesis combining Huánuco and Amazonian coca (Scenario 3), and a three-origin hypothesis (Scenario 4; Fig. 4a). Simulations generated under Scenario 3 and evaluated using the logistic regression approach received the highest posterior probability of matching the observed summary statistics from our first SNP data set, but Scenario 4 was more similar to our second SNP data set (Fig. 4b). Using the direct proportions of simulated data sets closest to our observed data sets, Scenario 3 best represented both SNP data sets (Supplementary Figs. S10 and S11 available on Dryad). The ABC results consistently showed Plowman’s linear-series and Johnson’s sister-species hypotheses are unlikely historical scenarios explaining the current genetic diversity and divergence of these taxa (Fig. 4b; Supplementary Figs. S10 and S11 available on Dryad).
Figure 4.
ABC tests and effective population size through time. a) Historical scenarios representing four domestication hypotheses. Scenarios 1–4 are drawn within colored boxes and the legend at top identifies the branch color for each taxon. b) Posterior probability of historical scenarios based on multinomial logistic regression procedure. c) Stairway plots reconstructing changes in effective population size through time for each of the four coca varieties, gracilipes1, and E. cataractarum. Solid lines are mean population size across 200 bootstraps and the 95% confidence interval is shown in gray.
Based on several analyses, we can be confident that ABC could discriminate these scenarios and estimate their probabilities. Principal components analyses show the posterior distributions for Scenarios 3 and 4 were able to accurately replicate the observed data (Supplementary Figs. S10c and S11c available on Dryad). The posterior predictive error rates specific to confidence in scenario choice ranged from 0.146 to 0.206 among direct and logistic estimates from the two data sets (Supplementary Table S5 available on Dryad). The global error computed over the whole prior distribution (parameters and scenarios) ranged from 0.243 to 0.307 (Supplementary Table S5 available on Dryad). We summarized calculations of scenario-based prior error rates in confusion matrices and estimates of type I and type II error (Supplementary Tables S6–S9 available on Dryad). We found the simulated scenarios received the highest probability in all cases, but the Type I error, the probability a correct scenario was not chosen, was high for Scenario 1 (0.39–0.48) and Scenario 2 (0.33–0.42; Scenario 3  0.17–0.23; Scenario 4
0.17–0.23; Scenario 4  0.08–0.18). However, the Type I error is significantly lower if we only analyze confusions among Scenario 3, Scenario 4, and Scenario 1 or Scenario 2 (range
0.08–0.18). However, the Type I error is significantly lower if we only analyze confusions among Scenario 3, Scenario 4, and Scenario 1 or Scenario 2 (range  0.07–0.20), which are the meaningful comparisons with respect to our main results. Type II error, the probability a scenario was chosen when data were generated under an alternate scenario, ranged from 0.07 to 0.12 for Scenario 1, 0.15–0.16 for Scenario 2, 0.09–0.11 for Scenario 3, and 0.03–0.04 for Scenario 4. Model check results are presented in Supplementary Figs. S10 and S11 available on Dryad and prior and posterior distributions of the demographic parameters are presented in Supplementary Figs. S12 and S13 available on Dryad.
0.07–0.20), which are the meaningful comparisons with respect to our main results. Type II error, the probability a scenario was chosen when data were generated under an alternate scenario, ranged from 0.07 to 0.12 for Scenario 1, 0.15–0.16 for Scenario 2, 0.09–0.11 for Scenario 3, and 0.03–0.04 for Scenario 4. Model check results are presented in Supplementary Figs. S10 and S11 available on Dryad and prior and posterior distributions of the demographic parameters are presented in Supplementary Figs. S12 and S13 available on Dryad.
We chose not to evaluate support for two or three origins from additional SNP data sets because the multiple domestication hypothesis is a high-confidence conclusion given our genetic and geographic sampling. Our posterior samples converged on parameter estimations and posterior support for the domestication scenarios agrees with the other analyses. In addition, there are limitations in the ABC approach, the largest being that many demographic phenomena beyond our topological scenarios have influenced the allele frequencies and summary statistics our results are based upon (Sunnåker et al. 2013). For instance, the remarkably high observed heterozygosity of Amazonian coca could be explained by the accumulation of somatic mutations during the mostly clonal propagation of this crop, as is observed in other mostly asexual populations (Table 1; Stoeckel and Masson 2014). More sampling of E. gracilipes, delimitation of gracilipes1–4 as distinct taxa (see below), and distinct demographic models of each domestication event will more accurately bear on this history.
Timing and Locations of Domestication
We estimated the relative age of each coca variety by explicitly modeling the change in population sizes through time using site frequency spectra derived from our SNP data (Fig. 4c; Supplementary Fig. S14 available on Dryad). We did not estimate generation times or extrapolate mutation rates from other studies and therefore we cannot estimate the absolute timing of domestication events.
The stairway plots of gracilipes1 and E. cataractarum show these taxa have experienced a gradual population size increase followed by a very recent and drastic decrease (Fig. 4c). All coca varieties except Trujillo are marked by a stable historical population size that is smaller than gracilipes1 through much of its history. Colombian and Amazonian have about the same size, but Huánuco is smaller. Colombian coca shows the earliest departure from this stability and experienced a population decrease followed by a more recent increase, consistent with a domestication bottleneck. For Trujillo coca, population sizes have increased and then leveled off. Huánuco and Amazonian coca depart from the background population size more recently and at about the same time, although confidence intervals show changes in Amazonian could be more recent. Amazonian shows a pattern of recent population decrease whereas Huánuco coca appears to have experienced a bottleneck—a small decrease in population size followed by a large increase, then leveling off (Fig. 4c). In accordance with expectations based on historical coca farming practices, Huánuco coca is inferred to have the largest effective population size at present and Amazonian has the smallest. If we remove singletons, the model reconstructs generally similar population size histories, but without the recent population decreases in gracilipes1 and E. cataractarum, and without a rebound after the bottleneck for Colombian coca (Supplementary Fig. S14 available on Dryad). The relative coalescence times from our ABC parameter estimations also show the Trujillo/Colombian domestication was oldest and, in the case of three origins, that Amazonian coca coalesced with E. gracilipes the fewest generations ago (Supplementary Figs. S12b and S13b available on Dryad).
In addition to the Colombian coca stairway plot, the long branch subtending the E. novogranatense clade (Colombian and Trujillo; Fig. 2a) and the 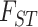 statistics (Table 2) also corroborate the hypothesis that this is the oldest coca crop. Archaeological leaf fragments and chewing paraphernalia reveal coca culture was well established 8000 years BP (Plowman 1984; Dillehay et al. 2010). Evidence from the archaeological record, mating systems, and hybrid crosses (see Bohm et al. 1982) led Plowman to believe that Colombian coca was derived from Trujillo. Under neutral evolutionary models, we would expect higher genetic diversity in progenitor taxa (Gross and Olsen 2010; Feng et al. 2020), but we see Trujillo has fewer private alleles and lower allelic richness, genetic diversity, and observed heterozygosity than Colombian coca (Table 1). However, these statistics are also influenced by different demographic histories (see Mortimer 1901; Plowman 1984; Gootenberg 2008), so we maintain the working hypothesis established by Bohm et al. (1982) that Trujillo coca or a common ancestor was the progenitor of Colombian coca. Thus, a likely region of this domestication event was in Ecuador or northern Peru, near the current range of cultivation.
statistics (Table 2) also corroborate the hypothesis that this is the oldest coca crop. Archaeological leaf fragments and chewing paraphernalia reveal coca culture was well established 8000 years BP (Plowman 1984; Dillehay et al. 2010). Evidence from the archaeological record, mating systems, and hybrid crosses (see Bohm et al. 1982) led Plowman to believe that Colombian coca was derived from Trujillo. Under neutral evolutionary models, we would expect higher genetic diversity in progenitor taxa (Gross and Olsen 2010; Feng et al. 2020), but we see Trujillo has fewer private alleles and lower allelic richness, genetic diversity, and observed heterozygosity than Colombian coca (Table 1). However, these statistics are also influenced by different demographic histories (see Mortimer 1901; Plowman 1984; Gootenberg 2008), so we maintain the working hypothesis established by Bohm et al. (1982) that Trujillo coca or a common ancestor was the progenitor of Colombian coca. Thus, a likely region of this domestication event was in Ecuador or northern Peru, near the current range of cultivation.
Our phylogenetic and clustering analyses indicate Huánuco coca was domesticated in the eastern Andean foothills of southern Peru. The two gracilipes1 samples from Madre de Dios, Peru (grac.298, grac.654) cluster with and are placed at the base of the Huánuco coca clade (Fig. 2a). Neither of these samples was collected in proximity ( 100 km) to coca farms, but this does not rule out the possibility of postdomestication gene flow from coca farms in this region into gracilipes1. A remarkable sample (cf.coca.884) from central Peru is described as cultivated but is morphologically intermediate and admixed between with gracilipes1 (Image in Supplementary material available on Dryad). A morphologically wild-type E. gracilipes from the montane forest in Ecuador (grac.100) was also inferred to be slightly admixed with Huánuco coca, but this is likely due to postdomestication gene flow because the Ecuadorean gracilipes1 forms a clade separate from Huánuco coca (Fig. 2a).
100 km) to coca farms, but this does not rule out the possibility of postdomestication gene flow from coca farms in this region into gracilipes1. A remarkable sample (cf.coca.884) from central Peru is described as cultivated but is morphologically intermediate and admixed between with gracilipes1 (Image in Supplementary material available on Dryad). A morphologically wild-type E. gracilipes from the montane forest in Ecuador (grac.100) was also inferred to be slightly admixed with Huánuco coca, but this is likely due to postdomestication gene flow because the Ecuadorean gracilipes1 forms a clade separate from Huánuco coca (Fig. 2a).
The Amazonian clade is most closely related to gracilipes1 individuals from Loreto, Peru, and Amazonian Ecuador (Fig. 2; Supplementary Figs. S5 and S7 available on Dryad). At  , Amazonian coca is not distinct from gracilipes1, but it does emerge as a separate genetic cluster when nine clusters are defined (Fig. 2a). Lastly, the clustering and
, Amazonian coca is not distinct from gracilipes1, but it does emerge as a separate genetic cluster when nine clusters are defined (Fig. 2a). Lastly, the clustering and 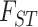 results, as well as the fact that Amazonian coca has the highest genetic diversity of any coca variety (Table 1), support the hypothesis that Amazonian is the most recently domesticated coca crop. This is in line with linguistic and ethnographic observations that indigenous agricultural practices, preparation, consumption, and terminology surrounding coca are remarkably consistent across the Amazon basin, and thus thought to have been more recently dispersed throughout its current range (Plowman 1986). Thus, Amazonian coca was either recently derived from Huánuco coca or it was independently domesticated from gracilipes1 in Amazonian Ecuador or northern Peru.
results, as well as the fact that Amazonian coca has the highest genetic diversity of any coca variety (Table 1), support the hypothesis that Amazonian is the most recently domesticated coca crop. This is in line with linguistic and ethnographic observations that indigenous agricultural practices, preparation, consumption, and terminology surrounding coca are remarkably consistent across the Amazon basin, and thus thought to have been more recently dispersed throughout its current range (Plowman 1986). Thus, Amazonian coca was either recently derived from Huánuco coca or it was independently domesticated from gracilipes1 in Amazonian Ecuador or northern Peru.
Erythroxylum gracilipes: Defining the ‘Mother of Coca’
Although E. gracilipes is one of the few wild species to have been hypothesized as a wild ancestor of the coca crops (Macbride 1949; see White et al. 2019), the taxonomic and ecological understanding of this wild species is minimal. This phylogeographic analysis reveals E. gracilipes is comprised of at least two main clades, gracilipes1 and gracilipes2–4, representing two or more distinct taxa. Amazonian and Huánuco coca are clearly derived from gracilipes1, but E. cataractarum and E. novogranatense could be derived from gracilipes1 or gracilipes2. Given our new understanding of this phylogenetic structure, focused morphological, ecological, and phylogeographic analyses are needed to inform species delimitation and taxonomic revision of E. gracilipes, thus clarifying the taxonomic identity of the progenitor(s) of coca.
While taxonomic revision could lump E. coca and E. novogranatense as varieties within paraphyletic E. gracilipes in order to reflect the evolutionary relationships of these taxa (Baum 2009), we believe they should instead retain their species status because they appear to be morphologically and genetically distinct, independently evolving, monophyletic lineages (Fig. 2a; Table 2; Supplementary Fig. S9 available on Dryad; De Queiroz 2007). Following additional sampling and analysis, gracilipes2–4 should probably be reclassified as one or more distinct species. One of our gracilipes1 samples in this study, grac.798, was collected at the same locality as the E. gracilipes type specimen (Spruce 3068; San Carlos, Amazonas, Venezuela), so gracilipes1 will likely retain the E. gracilipes epithet (Turland et al. 2018). Lastly, the monophyly of E. cataractarum and lack of gene flow with gracilipes1–4 supports the conservation of this epithet. Our results establish the hypothesis that E. cataractarum formed by peripatric speciation via ecological adaptation to gallery forests in the Colombian and Venezuelan Llanos.
Domestication in this system has resulted in plants with smaller, rounder, and softer leaves (lacking the sclerosed spongy mesophyll cells of E. gracilipes; Rury 1981) and erect to virgate branches (Supplementary Table S1 available on Dryad). Ethnobotanical knowledge is also scarce, the only credible record we are aware of reports the leaves are consumed for rheumatism and relaxation by Amerindians in the upper Rio Napo in Ecuador (Friedman et al. 1993). With the knowledge that E. gracilipes is a diverse and complex clade from which all coca varieties evolved, we hope this study will invigorate new botanical collections and systematic and ethnobotanical investigation of this wild taxon.
Conclusions
Our study has demonstrated that modern biotechnologies have finally permitted the efficient sequencing of genomic DNA from the hundreds of millions of preserved specimens sitting in wait in biological collections around the world. While the technology is available, this project also shows the importance of taxonomically complete collections for the efficacy and productivity of systematic and biodiversity investigations. Museum genomics projects can access a wide geographic and temporal scale, but might be best suited to initial, broad investigations like this one before more geographically or taxonomically focused evolutionary studies are conducted.
Harvard ethnobotanist R. E. Schultes called coca the most important South American narcotic plant due to its prevalence and significance for indigenous cultures, as well as the revolutionary role of cocaine in Western medicine (Schultes 1979). Our genetic blueprint of the domestication of coca reveals that different Amerindian peoples have continuously adopted wild E. gracilipes into cultivation as a mild stimulant and medicine. Under the broad context of crop origins and centers of civilization, this contradicts the traditional Vavilovian view of few, distinct centers of origins for crops with extensive dispersal networks (Vavilov and Löve 2009) and instead corroborates localized and widespread domestication practices (Harlan 1971). This pervasive ingenuity has resulted in four unique coca crops: coca cultivation and culture has flourished for over 8000 years in northwestern South America following the domestication of E. novogranatense in that region. Erythroxylum gracilipes was also domesticated into Huánuco coca in Andes/Amazon region of Peru and Bolivia, where it has grown into a sacred commodity and cultural symbol unparalleled in the world of crop plants. The coca grown today by indigenous tribes in the Amazon basin was possibly brought down from the Andes but could also represent a third, and most recent, independent origin of coca. The results of this study have reframed our understanding of the history and diversity of this crop and have directly informed the next generation of research into where, when, and how coca was domesticated from E. gracilipes.
Acknowledgments
We thank the following herbaria for use of material: F, HUEFS, MOL. Thanks to E. Gardner and M. Johnson for guidance with library preparation and Hyb-Seq analyses, J. Walsh for assistance with DNA isolation, and the following people for help in the field and lab: A. Daza, M. Huinga, J. Janovec, F. Parra, E. Vosburgh, and J. Wells. Thanks to Proyecto Khoka in Medellín for collaboration. Specimen collection in Colombia was performed under the general collecting permit Resoluciones ANLA 1177 9 octubre 2014 and 1386 29 octubre 2015 awarded to Universidad de los Andes.
Supplementary material
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.cvdncjt1n.
Funding
This work was supported by National Science Foundation grants to DMW (GRFP-0907994) and RJMG (DEB-1354975), the Grainger Bioinformatics Center and the Pritzker Laboratory for Molecular Systematics and Evolution at the Field Museum. Thank you to the Ree Lab and our anonymous reviewers for helpful comments with the manuscript.
References
- Aynilian G.H., Duke J.A., Gentner W.A., Farnsworth N.R. 1974. Cocaine content of Erythroxylum species. J. Pharm. Sci. 63:1938–1939. [DOI] [PubMed] [Google Scholar]
- Baum D.A. 2009. Species as ranked taxa. Syst. Biol. 58:74–86. [DOI] [PubMed] [Google Scholar]
- Beaumont M.A. 2008. Joint determination of topology, divergence time, and immigration in population trees In: Matsumura S., Forster P., Renfrew C., editors. Simulation, genetics, and human prehistory. Cambridge: McDonald Institute for Archaeological Research; p. 135–154. [Google Scholar]
- Beaumont M.A., Zhang W., Balding D.J. 2002. Approximate Bayesian computation in population genetics. Genetics 162:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J.B., Semple J.C. 2015. Next-generation sampling: pairing genomics with Herbarium specimens provides species-level signal in solidago (Asteraceae). Appl. Plant Sci. 3:1500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beugin M.-P., Gayet T., Pontier D., Devillard S., Jombart T. 2018. A fast likelihood solution to the genetic clustering problem. Methods Ecol. Evol. 9:1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley-Taylor D.R. 2016. Coca and cocaine: the evolution of international control In: Gootenberg P., editor. Roadmaps to regulation: coca, cocaine, and derivatives. Oxford:The Beckley Foundation; p. 1–13. [Google Scholar]
- Bieri S., Brachet A., Veuthey J.-L., Christen P. 2006. Cocaine distribution in wild Erythroxylum species. J. Ethnopharmacol. 103:439–447. [DOI] [PubMed] [Google Scholar]
- Bohm B.A., Ganders F.R., Plowman T. 1982. Biosystematics and evolution of cultivated coca (Erythroxylaceae). Syst. Bot. 7:121–133. [Google Scholar]
- Carter W.E., Mamani M. 1986. Coca in Bolivia. La Paz: Juventud. [Google Scholar]
- Casale J.F., Mallette J.R. 2016. Illicit coca grown in Mexico: an alkaloid and isotope profile unlike coca grown in South America. Forensic Chem. 1:1–5. [Google Scholar]
- Chifman J., Kubatko L. 2014. Quartet inference from SNP data under the coalescent model. Bioinformatics 30:3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelman C.S., White D.M. 2016. The botanical science and cultural value of Coca leaf in South America In: Gootenberg P., editor. Roadmaps to regulation: coca, cocaine, and derivatives. Oxford: The Beckley Foundation. [Google Scholar]
- Cornuet J.M., Pudlo P., Veyssier J., Dehne-Garcia A., Gautier M., Leblois R., Marin J.M., Estoup A. 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30:187–1189. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávalos L.M., Bejarano A.C., Hall M.A., Correa H.L., Corthals A., Espejo O.J. 2011. Forests and drugs: coca-driven deforestation in tropical biodiversity hotspots. Environ. Sci. Technol. 45:1219–1277. [DOI] [PubMed] [Google Scholar]
- De Queiroz K. 2007. Species concepts and species delimitation. Syst. Biol. 56:879–886. [DOI] [PubMed] [Google Scholar]
- Dillehay T.D., Rossen J., Ugent D., Karathanasis A., Vásquez V., Netherly C.P.J. 2010. Early Holocene coca chewing in northern Peru. Antiquity 84:939–953. [Google Scholar]
- Eaton D.A.R., Overcast I. 2020. ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36:2592–2594. [DOI] [PubMed] [Google Scholar]
- Emche S.D., Zhang D., Islam M.B., Bailey B.A., Meinhardt L.W. 2011. AFLP phylogeny of 36 Erythroxylum species. Trop. Plant. Biol. 4:126–133. [Google Scholar]
- Faircloth B.C. 2015. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32:786–788. [DOI] [PubMed] [Google Scholar]
- Feng L.-Y., Liu J., Gao C.-W., Wu H.-B., Li G.-H., Gao L.-Z. 2020. Higher genomic variation in wild than cultivated rubber trees, Hevea brasiliensis, revealed by comparative analyses of chloroplast genomes. Front. Ecol. Evol. 8:237. doi:10.3389/fevo.2020.00237. [Google Scholar]
- Forrest L.L., Hart M.L., Hughes M., Wilson H.P., Chung K.-F., Tseng Y.-H., Kidner C.A. 2019. The limits of Hyb-Seq for herbarium specimens: impact of preservation techniques. Front. Ecol. Evol. 7:439. doi:10.3389/fevo.2019.00439. [Google Scholar]
- Friedman J., Bolotin D., Rios M., Mendosa P., Cohen Y., Balick M.J. 1993. A novel method for identification and domestication of indigenous useful plants in Amazonian Ecuador. In: Janick J., Simon J.E., editors. New crops. New York: Wiley. [Google Scholar]
- Gerbault P., Allaby R.G., Boivin N., Rudzinski A., Grimaldi I.M., Pires J.C., Climer Vigueira C., Dobney K., Gremillion K.J., Barton L., Arroyo-Kalin M., Purugganan M.D., Rubio de Casas R., Bollongino R., Burger J., Fuller D.Q., Bradley D.G., Balding D.J., Richerson P.J., Gilbert M.T.P., Larson G., Thomas M.G. 2014. Storytelling and story testing in domestication. Proc. Natl. Acad. Sci. USA 111:6159–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E.E., Kohn J.R., Lande R., Robertson K.A., Smith S.A., Igiæ B. 2010. Species selection maintains self-incompatibility. Science 330:493–495. [DOI] [PubMed] [Google Scholar]
- Gootenberg P. 2008. Andean cocaine: the making of a global drug. Chapel Hill: University of North Carolina Press. [Google Scholar]
- Goudet J. 2005. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Resour. 5:184–186. [Google Scholar]
- Greiman S.E., Cook J.A., Tkach V.V., Hoberg E.P., Menning D.M., Hope A.G., Sonsthagen S.A., Talbot S.L. 2018. Museum metabarcoding: a novel method revealing gut helminth communities of small mammals across space and time. Int. J. Parasitol. 48:1061–1070. [DOI] [PubMed] [Google Scholar]
- Gross B.L., Olsen K.M. 2010. Genetic perspectives on crop domestication. Trends Plant Sci. 15:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan J.R. 1971. Agricultural origins: centers and noncenters. Science 174:468–474. [DOI] [PubMed] [Google Scholar]
- Hart M.L., Forrest L.L., Nicholls J.A., Kidner C.A. 2016. Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 65:1081–1092. [Google Scholar]
- Harvey M.G., Smith B.T., Glenn T.C., Faircloth B.C., Brumfield R.T. 2016. Sequence capture versus restriction site associated DNA sequencing for shallow systematics. Syst. Biol. 65:910–924. [DOI] [PubMed] [Google Scholar]
- Hastorf C.A. 1987. Archaeological evidence of coca (Erythroxylum coca, erythroxylaceae) in the upper mantaro valley, Peru. Econ Bot. 41:292–301. [Google Scholar]
- Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R., Slatkin M., Maddison W.P. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.B. 2011. Tracing the evolutionary history of coca (Erythroxylum) [thesis]. University of Colorado at Boulder. [Google Scholar]
- Jackson N.D., Carstens B.C., Morales A.E., O’Meara B.C. 2017. Species delimitation with gene flow. Syst. Biol. 66:799–812. [DOI] [PubMed] [Google Scholar]
- Johnson E.L., Zhang D., Emche S.D. 2005. Inter- and intra-specific variation among five Erythroxylum taxa assessed by AFLP. Ann. Bot. 95:601–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.G., Gardner E.M., Liu Y., Medina R., Goffinet B., Shaw A.J., Zerega N.J.C., Wickett N.J. 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4:1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., Ahmed I. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.R., Good J.M. 2016. Targeted capture in evolutionary and ecological genomics. Mol. Ecol. 25:185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan K., McGinnity P., Cross T.F., Crozier W.W., Prodöhl P.A. 2013. diveRsity: an R package for the estimation of population genetics parameters and their associated errors. Methods Ecol. Evol. 4:782–788. [Google Scholar]
- Larson G., Piperno D.R., Allaby R.G., Purugganan M.D., Andersson L., Arroyo-Kalin M., Barton L., Climer Vigueira C., Denham T., Dobney K., Doust A.N., Gepts P., Gilbert M.T.P., Gremillion K.J., Lucas L., Lukens L., Marshall F.B., Olsen K.M., Pires J.C., Richerson P.J., Rubio de Casas R., Sanjur O.I., Thomas M.G., Fuller D.Q. 2014. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. USA 111:6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck E., Epperly K., Van Els P., Spellman G.M., Bryson R.W., McCormack J.E., Canales-Del-Castillo R., Klicka J. 2019. Dense geographic and genomic sampling reveals paraphyly and a cryptic lineage in a classic sibling species complex. Syst. Biol. 68:956–966. [DOI] [PubMed] [Google Scholar]
- Liu X., Fu Y.-X. 2015. Exploring population size changes using SNP frequency spectra. Nat. Genetics 47:555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbride J.F. 1949. Erythroxylaceae. Flora of Peru. Chicago, IL, USA: Field Museum of Natural History; p. 632–647. [Google Scholar]
- McCormack J.E., Tsai W.L.E., Faircloth B.C. 2016. Sequence capture of ultraconserved elements from bird museum specimens. Mol. Ecol. Resour. 16:1189–1203. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer W.G. 1901. History of coca: the divine plant of the Incas. New York: Vail. [Google Scholar]
- Nathanson J.A., Hunnicutt E.J., Kantham L., Scavone C. 1993. Cocaine as a naturally occurring insecticide. Proc. Natl. Acad. Sci. USA 90:9645–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1972. Genetic distance between populations. Am. Nat. 106:283–292. [Google Scholar]
- Nei M. 1987. Molecular evolutionary genetics. New York:Columbia University Press. [Google Scholar]
- Pickrell J.K., Pritchard J.K. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8(11):e1002967. doi:10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman T. 1979a. The identity of Amazonian and Trujillo coca. Bot. Mus. Leafl. Harv. Univ. 27:45–68. [Google Scholar]
- Plowman T. 1979b. Botanical perspectives on coca. J. Psychedelic Drugs. 11:103–117. [DOI] [PubMed] [Google Scholar]
- Plowman T. 1981. Amazonian coca. J. Ethnopharmacol. 3:195–225. [DOI] [PubMed] [Google Scholar]
- Plowman T. 1984. The origin, evolution, and diffusion of coca, Erythroxylum spp., in South and Central America. Pap. Peabody Mus. Archaeol. Ethnogr. 76:125–163. [Google Scholar]
- Plowman T. 1986. Coca chewing and the botanical origins of coca (Erythroxylum spp.) in Latin America In: Pacini D., Franquemont C., editors. Coca and cocaine: effects on people and policy in Latin America. Cornell University: Cultural Survival, Inc./LASP; p. 5–34. [Google Scholar]
- Plowman T., Hensold N. 2004. Names, types, and distribution of neotropical species of Erythroxylum (Erythroxylaceae). Brittonia 56:1–53. [Google Scholar]
- Plowman T., Rivier L. 1983. Cocaine and cinnamoylcocaine content of Erythroxylum species. Ann. Bot. 51:641–659. [Google Scholar]
- Prathapan K.D., Pethiyagoda R., Bawa K.S., Raven P.H., Rajan P.D., Countries 172 co-signatories from 35. 2018. When the cure kills—CBD limits biodiversity research. Science 360:1405–1406. [DOI] [PubMed] [Google Scholar]
- Prosser S.W.J., deWaard J.R., Miller S.E., Hebert P.D.N. 2016. DNA barcodes from century-old type specimens using next-generation sequencing. Mol. Ecol. Resour. 16:487–497. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reichel-Dolmatoff G., Schrimpff R. 2005. Goldwork and shamanism: an iconographic study of the Gold Museum of the Banco de la República, Colombia. Villegas Asociados. [Google Scholar]
- Restrepo D.A., Saenz E., Jara-Muñoz O.A., Calixto-Botía I.F., Rodríguez-Suárez S., Zuleta P., Chavez B.G., Sanchez J.A., D’Auria J.C. 2019. Erythroxylum in focus: an interdisciplinary review of an overlooked genus. Molecules 24:3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe K.C., Singhal S., Macmanes M.D., Ayroles J.F., Morelli T.L., Rubidge E.M., Bi K., Moritz C.C. 2011. Museum genomics: low-cost and high-accuracy genetic data from historical specimens. Mol. Ecol. Resour. 11:1082–1092. [DOI] [PubMed] [Google Scholar]
- Rury P.M. 1981. Systematic anatomy of Erythroxylum P. Browne: practical and evolutionary implications for the cultivated cocas. J. Ethnopharmacol. 3:229–263. [DOI] [PubMed] [Google Scholar]
- Rury P.M., Plowman T. 1983. Morphological studies of archaeological and recent coca leaves (Erythroxylum spp.). Bot. Mus. Leafl. Harv. Univ. 29:297–341. [Google Scholar]
- Särkinen T., Staats M., Richardson J.E., Cowan R.S., Bakker F.T. 2012. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS One 7:e43808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes R.E. 1979. Evolution of the identification of the major South American narcotic plants. J. Psychedelic Drugs 11:119–134. [DOI] [PubMed] [Google Scholar]
- Staats M., Erkens R.H.J., van de Vossenberg B., Wieringa J.J., Kraaijeveld K., Stielow B., Geml J., Richardson J.E., Bakker F.T. 2013. Genomic treasure troves: complete genome sequencing of herbarium and insect museum specimens. PLoS ONE 8(7):e69189. doi:10.1371/journal.pone.0069189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel S., Masson J.-P. 2014. The exact distributions of FIS under partial asexuality in small finite populations with mutation. PLoS One 9:e85228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnåker M., Busetto A.G., Numminen E., Corander J., Foll M., Dessimoz C. 2013. Approximate Bayesian computation. PLoS Comput. Biol. 9:e1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turland N.J., Wiersema J.H., Barrie F.R., Greuter W., Hawksworth D.L., Herendeen P.S., Knapp S., Kusber W.-H., Li D.-Z., Marhold K., May T.W., McNeill J., Monro A.M., Prado J., Price M.J., Smith G.F. 2018. International code of nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Glashütten: Koeltz Botanical Books. [Google Scholar]
- United Nations. 2019. World drug report. Sales No. E.19.XI.8 Available at: https://wdr.unodc.org/wdr2019/. [Google Scholar]
- Valdez L.M., Taboada J., Valdez J.E. 2015. Ancient use of coca leaves in the Peruvian central highlands. J. Anthropol. Res. 71:231–258. [Google Scholar]
- Vavilov N.I., Löve D. 2009. Origin and geography of cultivated plants. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Villaverde T., Pokorny L., Olsson S., Rincón-Barrado M., Johnson M.G., Gardner E.M., Wickett N.J., Molero J., Riina R., Sanmartín I. 2018. Bridging the micro- and macroevolutionary levels in phylogenomics: Hyb-Seq solves relationships from populations to species and above. New Phytol. 220:636–650. [DOI] [PubMed] [Google Scholar]
- Weir B.S., Cockerham C.C. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- White D.M., Islam M.B., Mason-Gamer R.J. 2019. Phylogenetic inference in section Archerythroxylum informs taxonomy, biogeography, and the domestication of coca (Erythroxylum species). Am. J. Bot. 106:154–165. [DOI] [PubMed] [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19:153. [DOI] [PMC free article] [PubMed] [Google Scholar]