Abstract
Nanobodies have been progressively replacing traditional antibodies in various immunological methods. However, the use of nanobodies as capture antibodies is greatly hampered by their poor performance after passive adsorption to polystyrene microplates, and this restricts the full use of double nanobodies in sandwich ELISAs. Herein, using the human soluble epoxide hydrolase (sEH) as a model analyte, we found both the immobilization format and blocking agent have a significant influence on the performance of capture nanobodies immobilized on polystyrene and the subsequent development of double nanobody sandwich ELISAs. We first conducted epitope mapping for pairing nanobodies and then prepared a horseradish peroxidase labeled nanobody using a mild conjugation procedure as detection antibody throughout the work. The resulting sandwich ELISA using capture nanobody (A9, 1.25 μg/mL) after passive adsorption and BSA as blocking agent generated a moderate sensitivity of 0.0164 OD•mL/ng and a LOD of 0.74 ng/mL. However, the introduction of streptavidin as a linker to the capture nanobody at the same working concentration demonstrated a dramatic 16-fold increase in sensitivity (0.262 OD•mL/ng) and 25-fold decrease in the LOD for sEH (0.03 ng/mL). The streptavidin bridged double nanobody ELISA was then applied successfully to tests for recovery, cross-reactivity, and real samples. Meanwhile, we accidentally found that blocking with skim milk could severely damage the performance of the capture nanobody by an order of magnitude, compared to BSA. This work provides guideline to retain the high effectiveness of capture nanobody and thus to further develop double nanobody ELISA for various analytes.
Keywords: Nanobody, Soluble epoxide hydrolase, Double Nanobody, Sandwich ELISA, Polystyrene
Graphical Abstarct
INTRODUCTION
Rapid and sensitive detection technologies are critical for decision-making procedures in many fields, including clinic, food safety, and environmental monitoring. Immunoassay is one of the most powerful of all immunochemistry techniques and has been extensively used to detect and quantitate numerous analytes in many applications.1 Taking in vitro diagnostics (IVD) for example, it possessed the highest market share of the IVD market (37%) with sales of $18.6 billion achieved in 2012 and $26.9 billion predicted by 2024.2,3 Among immunoassays, enzyme linked immunosorbent assay (ELISA) has been most used due to its overwhelming overall advantages of high sensitivity, excellent selectivity, simplicity, fast speed, high throughput, low cost, safety, and general applicability.3,4 The key of an excellent ELISA is the availability of antibodies with high affinity, specificity, and batch-to-batch consistency. However, both academic and industrial communities have been suffering from reproducibility crisis of antibodies caused by the limited supply and large variance between batches of the polyclonal antibodies (pAbs) or the deterioration and loss of hybridomas of monoclonal antibodies (mAbs) in storage.5 Thus, there is a strong demand that “all antibody reagents should be sequenced and then produced from the genetic code obtained”.6
A nanobody, also termed VHH, is an antibody with a single variable domain derived from heavy-chain only antibodies in camelids or cartilaginous fish.7–9 This recombinant antibody is a promising solution to the reproducibility crisis, since it can be easily sequenced and resynthesized. Moreover, nanobodies have received increasing interest owing to their small size, monoclonal nature, genetic manipulability, high thermostability, superior solubility, ease of clone storage and expression in diverse expression platforms, and cost effectiveness for both discovery and continuous production.4,10 It can not only address the issues of limited supply and batch-to-batch variation of pAb, but also the risk of hybridomas deterioration associated with costly liquid nitrogen cryogenic storage and the non-monospecific problem of the classical mAb caused by the frequent expression of additional functional variable regions by hydridomas.11 It is therefore no wonder that nanobodies are progressively replacing conventional IgG based antibodies in various immunological techniques, especially in immunoassay. Our group recently developed many successful immunoassays using nanobodies as detection antibody for various analytes, including small molecules such as 3-phenoxybenzoic acid,10 triclocarban,12 tetrabromobisphenol A,13 ochratoxin,14 triazophos,15 fipronil,16 as well as large proteins such as soluble epoxide hydrolase4,17 and cystic fibrosis transmembrane conductance regulator (CFTR) inhibitory factor.18 However, the use of nanobody as capture antibody in classical sandwich ELISAs relying only on fully sequenced nanobodies for both capturing and reporting is greatly hampered by its poor performance after the passive adsorption of the small-size based nanobody on polystyrene microplates. The fundamental study on the mechanism behind the deterioration of the capture nanobody is lacking, and the physical and functional behavior of the capture nanobody immobilized on polystyrene remains unexplored. It is important to understand the potential cause for the substantial loss of functionality of capture nanobodies adsorbed on polystyrene microplates and hereby further improve their performance in ELISAs.
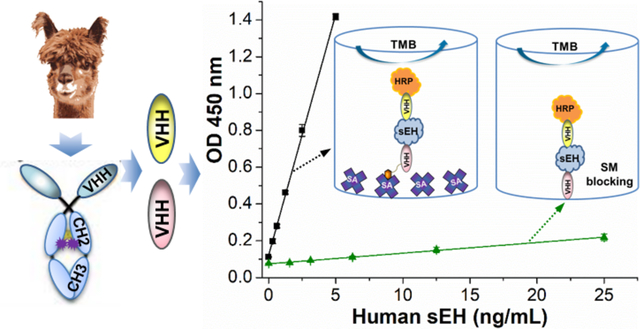
To tackle this problem, we developed two sandwich ELISA formats using nanobodies as capture antibodies and compared them to that using pAb as capture antibody on polystyrene microplates. Human soluble epoxide hydrolase (sEH) was chosen as the model analyte considering its increasing significance as a biomarker of many diseases,19–25 and the availability of corresponding nanobodies in the laboratory. The basic purpose of this study was to examine the physical and functional behavior of capture nanobody immobilized on polystyrene and thus establish a guideline for developing double nanobody ELISA. First, we conducted epitope mapping for pairing nanobodies and then prepared a horseradish peroxidase (HRP) labeled nanobody (HRP-A1) as detection antibody using a mild conjugation method. Second, we compared the performance of a sandwich ELISA for sEH detection using nanobody (format B, Figure 1b) and polyclonal Ab (format A, Figure 1a) passively adsorbed as capture Ab. Third, we evaluated the effect of streptavidin as bridge for immobilizing nanobody on polystyrene (format C, Figure 1c) and compared several ways of blocking in this format. Fourth, three ELISA (format A, B, and C) were compared in a quantitative way at the same working concentration of capture antibody. Fifth, bovine serum albumin (BSA) blocking was compared with skim milk blocking on polystyrene plate with nanobody passively adsorbed (format B). Finally, streptavidin bridged double nanobody ELISAs were applied to tests for recovery and cross-reactivity and the analysis of real samples with various sEH levels.
Figure 1.
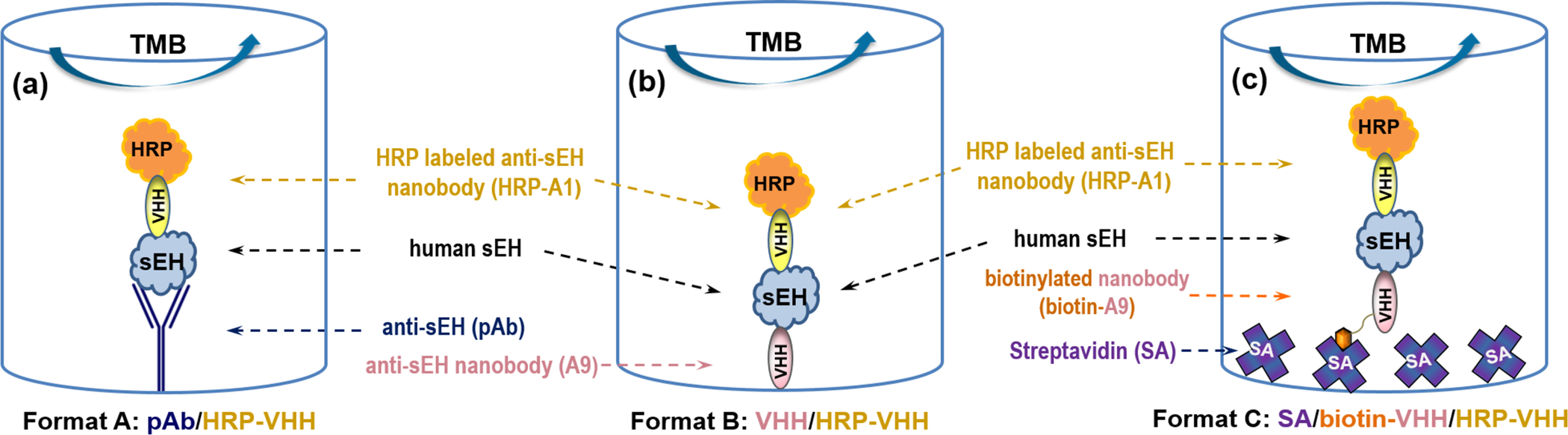
Schematic comparison of three different sandwich ELISA formats for human sEH detection using the same horseradish peroxidase (HRP) labeled anti-sEH nanobody conjugate (HRP-A1) as detection antibody. (a) format A (pAb/HRP-A1): polyclonal antibody passively adsorbed, (b) format B: (A9/HRP-A1): nanobody passively adsorbed, (c) format C (SA/biotin-A9/HRP-A1): nanobody biotinylated and bound to streptavidin passively adsorbed, as capture antibody, respectively.
EXPERIMENTAL SECTION
Materials
Recombinant human sEH, anti-human sEH rabbit pAb and eight nanobodies (A1, A9, B1, B3, B4, B6, B7, and B15) were produced as described in our previous work.17,26 Horseradish peroxidase (HRP, Cat. No. P6782), sodium periodate (311448), Sulfo-NHS-LC-biotin (B1022), and streptavidin (S4762) were purchased from Sigma-Aldrich. BSA (BP1600–100) and skim milk powder (1.15363.0500) were purchased from Fisher Scientific and EMD Millipore, respectively. High-binding polystyrene microplates (Nunc Maxisorp, 442404) were purchased from Thermo Fisher Scientific Inc. A sensitive 3,3′,5,5′-tetramethylbenzidine (TMB) substrate for color development was prepared as detailed in Supporting Information (SI). Eight anti-human sEH VHHs were biotinylated against amine groups at a 10:1 molar ratio of Sulfo-NHS-LC-Biotin to nanobody as previously described.4
Epitope Mapping
Modified from the assay for epitope mapping of mAbs on cells,27 competition assays based on indirect immunoassay were herein proposed to determine the epitope specificity of eight anti-human sEH nanobodies we obtained previously. As shown in Figure S1a, unconjugated nanobodies were incubated with biotinylated nanobodies for competitive binding to human sEH coated on microplate. Percentage of inhibition (PI) was defined as a ratio between the optical density (OD) of well with unconjugated nanobody in excess and the OD obtained for each biotinylated nanobody alone (Figure S1b). Inhibition was regarded significant when the PI was higher than 50%.27
Preparation of HRP Labeled Nanobody Conjugate (HRP-A1)
As illustrated in Figure 2, HRP enzyme was labeled to nanobody (A1) through periodate oxidation (Maraprade reaction) but in a mild condition (pH 5.4, 10 mM NaIO4, 30 min, ice bath (1.2 °C)) with high working concentration of HRP. Fractions without unconjugated monomer after size exclusion chromatography (SEC) were collected and further processed for downstream use as detection antibody. Detailed preparation and characterization of the HRP labeled nanobody conjugate was available in SI.
Figure 2.
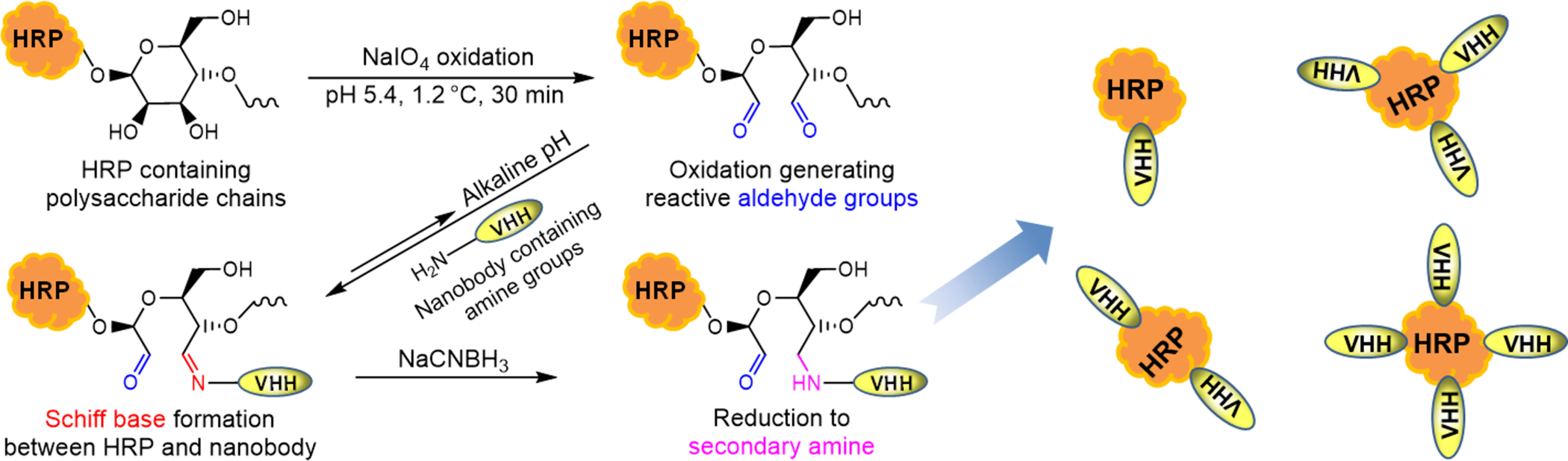
Schematic preparation of the enzyme HRP labeled nanobody A1 conjugate (HRP-A1) based on a mild periodate oxidation (Malaprade reaction). HRP is a heme-containing glycoprotein that may be oxidized with sodium periodate. The resulting reactive aldehyde residues may be conjugated to nanobody with Schiff base formed and further stabilized by sodium cyanoborohydride.
Comparison of Polyclonal Ab and Nanobody as Capture Antibody in ELISA
Anti-human sEH antiserum (1:2000 dilution) or affinity-purified pAb (3 μg/mL) through protein A column was coated in 0.05 M pH 9.6 carbonate-bicarbonate buffer (CB) overnight at 4 °C on a high-binding Nunc microplate (100 μL/well). Meanwhile, nanobody A9 (20 μg/mL) was coated in CB or phosphate buffered saline (PBS) on the same plate. After washing, 3% (w/v) skim milk/PBS (250 μL/well) was added to block the plate for 1 h and subsequently washed. Then, serial concentrations of human sEH standards prepared in PBS containing 0.1 mg/mL BSA were added to the plate (100 μL/well), followed immediately by HRP-A1 in PBS (1:1600, 100 μL/well). After another 1 h incubation, the immunoreaction was ended with the final washing step. TMB substrate (100 μL/well) was then added to the plate and incubated for 15 min. The color development was then terminated with 1 M sulfuric acid (100 μL/well) and the OD was recorded within 10 min using a microplate reader at 450 nm. All incubations unless otherwise specified were performed with shaking (600 rpm) on a microplate shaker at room temperature, and three washings with PBS containing 0.05% Tween-20 (PBST, 300 μL/well) were conducted in each washing step using a plate washer.
Development of SBdNb ELISA (format C) for sEH Detection
Streptavidin (2.5 μg/mL) was coated in CB overnight at 4 °C on Nunc microplate (100 μL/well). After washing, the plates were blocked with 2% BSA (250 μL/well) for 1 h and then washed. Biotinylated nanobody A9 (BioA9, 1.25 μg/mL, 100 μL) were added to each well. After 1 h incubation and then washing, serial human sEH standards (100 μL/well) were added to the plate and then followed immediately by addition of varying detection antibody HRP-A1 (1:1600-1:12800, 100 μL/well) in PBS or 10% SM/PBS. After another 1 h incubation and then the final washing, color was developed for 15 min with TMB and stopped with sulfuric acid. OD was recorded at 450 nm. Prior optimization of streptavidin and BioA9 is detailed in Figure S4.
Comparison of Three Formats at the Same Working Concentration of Capture Antibodies
The SBdNb ELISA and other two formats (A and B) were compared simultaneously with two Nunc microplates used. Streptavidin (2.5 μg/mL) were coated in CB overnight at 4 °C on plate I and the SBdNb ELISA was run on this plate as the same as previously described. For plate II, pAb (1.25 μg/mL in CB), A9 (1.25 μg/mL in PBS), and A9 (20 μg/mL in PBS) were coated overnight at 4 °C (100 μL/well) on columns 1–3, 4–6, and 7–12, respectively. After washing, the plate II was blocked with 2% BSA (columns 1–9) or 3% SM (columns 10–12) at 250 μL per well and this blocking was conducted simultaneously with the addition of BioA9 (1.25 μg/mL) on plate I. After 1 h incubation, both plates were washed and then added with human sEH standards plus HRP-A1 (1:6400) in one step. After another 1 h incubation, both plates were simultaneously washed, incubated with TMB, and finished with OD reading as previously described.
BSA vs SM in Blocking of Microplate with Nanobody Passively Adsorbed
Varying concentrations of nanobody A9 (0.625–20 μg/mL) was directly coated in PBS overnight at 4 °C on Nunc microplate. After washing, both plates were simultaneously blocked with 2% BSA or 3% SM (half plate for each blocking agents, 250 μL/well) for 1 h. Subsequently, downstream steps were performed the same as that for format A as aforementioned.
Cross-reactivity
The cross-reactivity of SBdNb ELISA was evaluated by its sensitivity for an interferent relative to that for the target analyte. A group of epoxide hydrolases were tested.
Matrix Effects
SBdNb ELISA was applied to brain tissue samples from sEH knockout (KO) mice. A simple dilution protocol was followed to evaluate the matrix effect. Briefly, the brain tissue samples were diluted with standard diluent (PBS containing 0.1 mg/mL BSA) to 1:150, 1:450 and 1:1350. Subsequently, spiked samples were obtained by spiking a series of human sEH solutions into samples of various dilutions and analyzed with the SBdNb ELISA.
Real Sample Analysis
S9 fractions from pooled (4–50 individuals) human tissues of six kinds purchased from Xenotech LLC (Lenexa, KS), PMBCs samples from patients without or with type 2 diabetes, and post-mortem brain samples from patients without or with multiple sclerosis were serially diluted with standard diluent and analyzed with the SBdNb ELISA. Dilutions resulting in OD reading in the linear range of the calibration curve were adopted to calculate the human sEH level in the samples.
RESULTS AND DISCUSSION
Epitope Mapping and the Preparation of HRP Labeled Nanobody Conjugate (HRP-A1)
The classical sandwich ELISA captures the analytes using the capture antibody immobilized on the microplate and subsequently detects them with an enzyme labeled detection antibody. Theoretically, double nanobody based sandwich ELISAs in such a classical and simple format (as shown in Figure 1b) can be developed and could fully replace the classical sandwich format using double Abs of pAb or mAb. As is true with antibodies, it is ideal to use a pair of nanobodies recognizing different epitopes in sandwich ELISA. This procedure utilizes formation of a sandwich immunocomplex resulting in increased selectivity. As illustrated in Figure S1, the 8 nanobodies were divided into six groups of mutually noncompetitive antibodies as follows: group I (A1, B4, B7), group II (A9), group III (B1), group IV (B3), group V (B6), and group VI (B15). This also means that the six groups of nanobodies recognize six different epitopes of human sEH. Considering their high yield of expression and good affinity, A1 from group I and A9 from group II were chosen as the pair in this work for double nanobody ELISA development. A1 is used to conjugate enzyme label since it has been well validated as a detection antibody after biotinylation elsewhere.4
As illustrated in Figure 2, HRP enzyme was labeled to nanobody (A1) through periodate oxidation (Maraprade reaction) but in a mild condition (pH 5.4, 10 mM NaIO4, 30 min, ice bath (1.2 °C)). This revised working condition is much milder than that of periodate oxidation in the classical literature for preparing HRP labeled IgG Ab.28 It cannot only better retain the enzyme activity of the oxidized HRP, but it also avoids the formation of HRP polymer caused by overoxidation. In addition, unlike IgG, the unique small size of nanobody allowed a clearer separation of HRP, antibody, and the conjugates in the resulting mixture through SEC and thereby a harvest of the conjugates with unconjugated monomer eliminated. This is very meaningful since the free antibody can suppress the specific signal of the conjugates and the unconjugated enzyme can cause increased nonspecific background. They together may greatly reduce the signal to noise ratio. The synthesis, purification, and characterization of HRP labeled nanobody conjugates are detailed in SI. Eventually, the resultant purified HRP-A1 conjugates possess an average of 3 nanobodies conjugated per HRP according to the densitometry on SDS-PAGE (Figure S2c) or 1.8 nanobodies per HRP calculated from its adsorption at 280 nm and 403 nm. As shown in Figure S3, the optimal working concentration of the HRP-A1 conjugate obtained is 1:1600 for sandwich ELISA (format A) in terms of sufficient signal and low background, using antiserum at1:2000 (the same coating as ref4) as capture antibody and 3% skim milk (SM) for plate blocking. The sensitivity obtained in this condition is around one fourth of that in the ultrasensitive PolyHRP ELISA previously published,4 indicating the high potency of the HRP labeled nanobody.
Comparison of ELISA Formats Using Polyclonal Ab and Nanobody as Capture Antibody
To fully replace traditional antibodies in immunoassay, it is crucial that nanobodies can demonstrate superior or at least comparable performance to classical polyclonal antibodies of IgG type as both detection and capture antibodies. The previous studies as aforementioned indicated satisfactory performance of nanobodies as detection antibody with the extra advantages of excellent solubility and superior penetration. However, performance comparison between IgG and nanobody as capture antibody in a quantitative way is rarely seen. Thus, we evaluated their capture performance of the pAb and nanobody reagents by comparing the ELISA format A and B as shown in Figure 1. Yet, for comparison, it is important to have a proper parameter as an index of the performance. For the study of pAb and mAb, Butler et al. defined and introduced the parameter of antigen capture capacity (AgCC) as a reflection of antibody activity or affinity. AgCC equals [Ag]b/[Ag]t. It is expressed as percent where [Ag]b is the amount of Ag bound in the linear binding region (region of capture Ab in excess) at high dilution of antigen. [Ag]t is the total amount of antigen added. They used iodinated antigen for tracing and quantification. This parameter tends to be constant in the linear binding region.29 In our work, we adopted sensitivity (slope) in linear range as a direct measure of the immunoactivity of nanobody immobilized on polystyrene. This parameter eliminates the obscurity of AgCC, making quantitative comparisons simple and straightforward. Figure 3 shows the signal responses of two ELISA (format A and B) run under same conditions for human sEH detection. For format B, 20 μg/mL nanobody coated in 10 mM phosphate buffered saline (PBS, pH 7.4) buffer generated 38% higher assay sensitivity (0.0116 OD•mL/ng) than that coated in CB (0.0084 OD•mL/ng), indicating PBS to be a superior coating buffer to CB for nanobody A9. This is however much lower than that of format A. Format A using affinity purified polyclonal antibody (pAb, 3 μg/mL, purified through protein A column with a yield of 6 mg pAb per mL of antiserum) and antiserum (1:2000 dilution) as capture antibodies demonstrated a sensitivity of 0.1650 and 0.0443 OD•mL/ng, respectively. It is astonishing that 20 μg/mL nanobody (17 kDa, monovalent) just gave just 7% assay sensitivity of 3 μg/mL pAb (150 kDa, divalent) as capture antibody, though the former contains 29.4-fold (or 294-fold supposing antigen-specific pAb accounts for only 10% IgG1) antigen-binding sites as the latter does. It was unclear in that microenvironment what exactly caused the dramatic deterioration of capture nanobody passively adsorbed on the high-binding polystyrene microplate.
Figure 3.
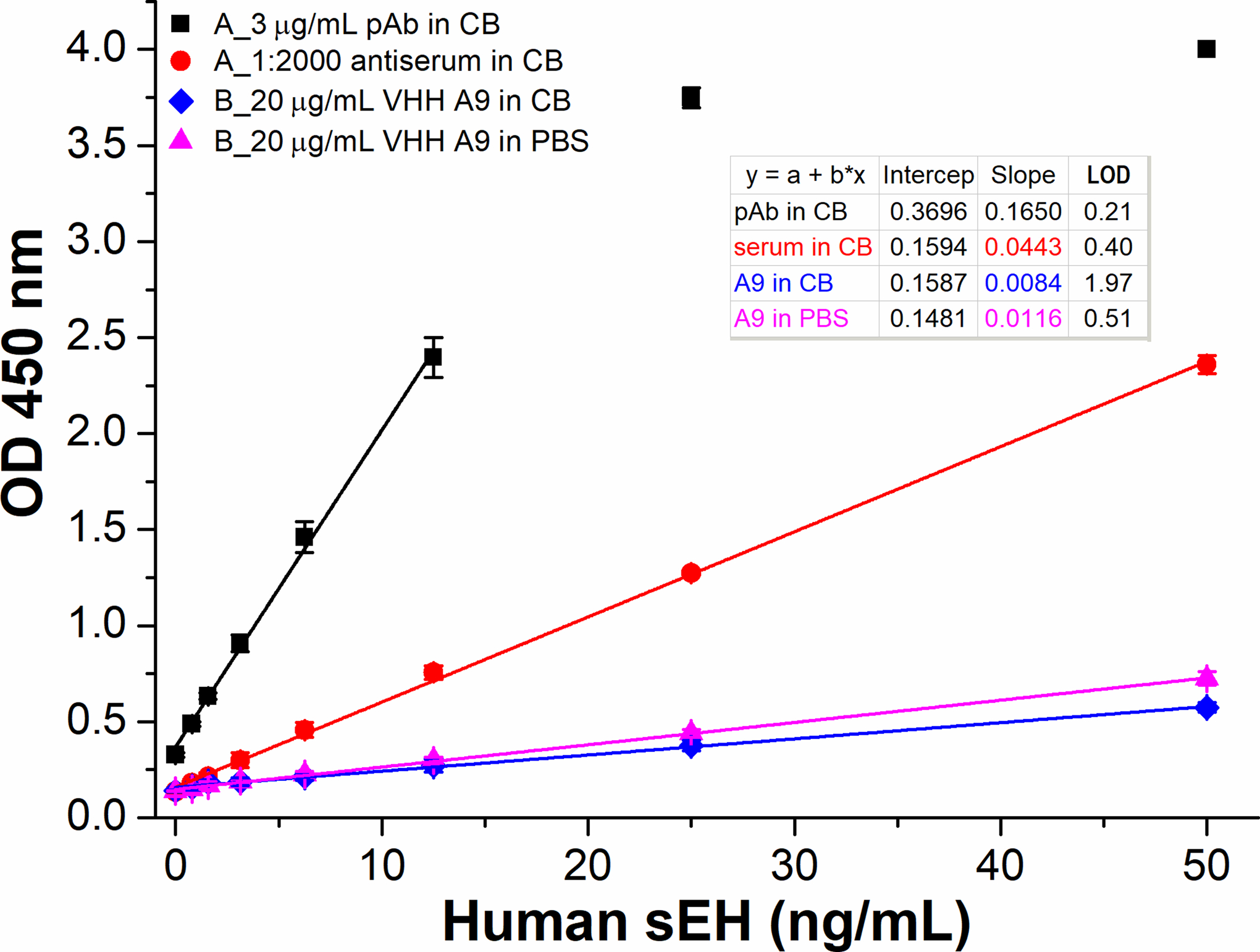
Signal responses of four ELISAs in two formats (A and B) run on the same plate using HRP-VHH A1 (1:1600 dilution) as the detection antibody. For format A, anti-sEH affinity purified polyclonal antibody (pAb, 3 μg/mL) or antiserum (1:2000 dilution) was used as the capture antibody through passive adsorption on high-binding polystyrene microplate. For format B, nanobody A9 (20 μg/mL) coated in CB or PBS buffer was used as the capture antibody after passive adsorption. The four ELISAs were performed on the same plate using 3% skim milk (SM) for blocking after the coating step. The same reagents involved for different formats are used from the same batch prepared. All same or similar steps were performed at the same time under the same conditions. Error bars indicate standard deviations (n = 3). All coefficient of determination R2>0.99.
Development of Streptavidin Bridged Double Nanobody ELISA
Butler et al found only < 3% of the binding sites of mAb and about 5–10% of those of pAb were capable of capturing antigens after passive adsorption on polystyrene. They also surprisingly found over 70% capture capability could be preserved when the antibodies were immobilized via a streptavidin bridge, i.e., the protein-(strept)avidin-biotin-capture (PABC) system.30,31 Thus, we tried a similar strategy to evaluate the possibility of improving the capture performance of a nanobody on polystyrene. However, a direct coating of streptavidin rather than PABC was used. As shown in Figure 4a, the streptavidin bridged double nanobody ELISA (SBdNb ELISA, format C) demonstrated a dramatic OD signal enhancement with sensitivity of (0.4447 OD•mL/ng) achieved at the working concentrations of streptavidin, biotin-A9 and HRP-A1 being 2.5 μg/mL, 1.25 μg/mL and 1:1600 dilution, respectively. This improved sensitivity is 38-fold of the format B (Figure 3) but with just 1/16 the amount of capture nanobody used. This huge increase in sensitivity demonstrated the excellent capability of the streptavidin bridge in retaining the substantial functional activity of the immobilized capture nanobody. We thought there might be two roles of the streptavidin in the preservation of nanobody functionality. One is that the streptavidin works as an insulating gasket which prevents nanobody from contacting polystyrene and the consequent denaturation. The other is that it acts as a giant booster seat or steppingstone anchored in the deep polystyrene groove for better exposure of capture nanobody to antigen, for the polystyrene surface is not atomically flat and cavities up to 5 μm and from 10 to 40 nm in depth were observed in Nunc Maxisorp plate with atomic force microscope.32 Yet, it is unclear if these or other mechanisms account for the dramatic role of streptavidin in this protection phenomenon. In addition, it is meaningful to optimize the working concentrations of the assay so as to conserve reagents while maintaining the performance. As shown in Figure S4, 2.5 μg/mL was chosen for streptavidin coated in CB and 1.25 μg/mL for biotin-A9 in PBS after optimization by varying the working concentrations of streptavidin (10, 5, 2.5, 1.25, and 0.625 μg/mL) and biotin-A9 (5, 2.5, 1.25, and 0.625 μg/mL). For the streptavidin coated solid phase, SM should not be used as the blocking agent since it contains endogenous biotins which may occupy the biotin-binding sites of streptavidin immobilized and consequently prevent biotinylated antibody from binding. This interference was first observed in immunoblotting using streptavidin for coating and SM for blocking.33 It was also confirmed in SBdNb ELISA that the SM blocking gave a very weak signal response while the BSA blocking showed the opposite (see Figure S4a,f). Therefore, avoidance of SM blocking is crucial to SBdNb ELISA. As an alternative to SM, BSA blocking allowed high sensitivity but also high background (OD>0.3) when HRP-A1 at 1:1600 was used. This increased background is attributed to the weaker blocking efficacy of BSA compared to SM.34 The latter contains small caseins, enabling deeper penetration and thus more efficient blocking of the plate. Besides sensitivity, limit of detection (LOD) is another important index related to detectability. It is the lowest quantity of an analyte that can be distinguished from the absence of that analyte (a blank value) with a specified confidence level (generally 99%)35 and hence is a statistical parameter. The LOD = 3SB/k refers to the calculated analyte concentration corresponding to signal response of the blank plus three times its standard deviation and is used with unit of ng/mL throughout this work, where k is the slope or sensitivity obtained in a linear regression analysis and SB is the standard deviation of the blank. The LOD is not only determined by the sensitivity but also the standard deviation of the blank. Therefore, higher sensitivity may mean increased odds of lower LOD but cannot guarantee it, as is shown in Figure 3–5 of this paper. Although the background signal above does not determine the sensitivity and detectability of an assay, clean or weak background is always preferred by the end users. To decrease the background, normally caused by nonspecific adsorption of the tracer, decreasing the working concentration of HRP-A1 or adding blocking proteins in tracer was performed. As illustrated in Figure 4a, decreasing HRP-A1 led to a decrease of both background and sensitivity. For HRP-A1 diluted in 1:6400 and 1:12800, acceptable clean background of OD around 0.1 was achieved with sufficient sensitivity comparable to the PolyHRP ELISA previously published;4 and the linearity can be extended to 10 ng/mL (data not shown) and the LOD is 0.03 and 0.06 ng/mL, respectively. On the other hand, using 10% SM in PBS as the diluent of HRP-A1 also reduced the background significantly (Figure 4b), because SM is a potent blocker that outcompetes the HRP-A1 for nonspecific binding sites. BSA was not adopted since it showed a modest effect as a simultaneous blocking agent in other ELISAs developed in our lab (data not shown). Similar phenomena revealing the efficacy of SM and inefficacy of BSA for simultaneous blocking was previously observed in immunohistology as well.36 Despite decreased background due to SM, the sensitivities (Figure 4b) also dropped remarkably with only around 1/3 remaining compared to that of corresponding dilutions of HRP-A1 in PBS illustrated in Figure 4a. The decrease in sensitivity likely is due to the SM-containing endogenous biotin,33 which probably displaced 2/3 of previously bound biotinylated capture nanobody (biotin-A9) from streptavidin-coated plate. It is worth noting that the high affinity of streptavidin for free biotin (Ka =1015 M−1) actually does not hold for biotinylated macromolecules probably because of the steric hindrance caused by the linked proteins. Vincent and Samuel found that the affinity of biotinylated macromolecules to streptavidin decreased orders of magnitude compared to that of free biotin to streptavidin.37 Thus, the prevalent idea that the binding between streptavidin and biotinylated biomolecules is almost irreversible should be treated with caution. This also means that risk of competition or displacement from second biotinylated probe (especially if it is in excess) with the first one can happen when free streptavidin is used to connect two biotinylated probes, e.g., PABC system aforementioned. Therefore, it is important to use direct coating of streptavidin and avoid SM in SBdNb ELISA. Considering the balance among background, sensitivity and cost efficiency, SBdNb ELISA with HRP-A1 in 1:6400 dilution illustrated in Figure 4a was chosen as the optimal condition and used for downstream selectivity, recovery, and real sample tests.
Figure 4.
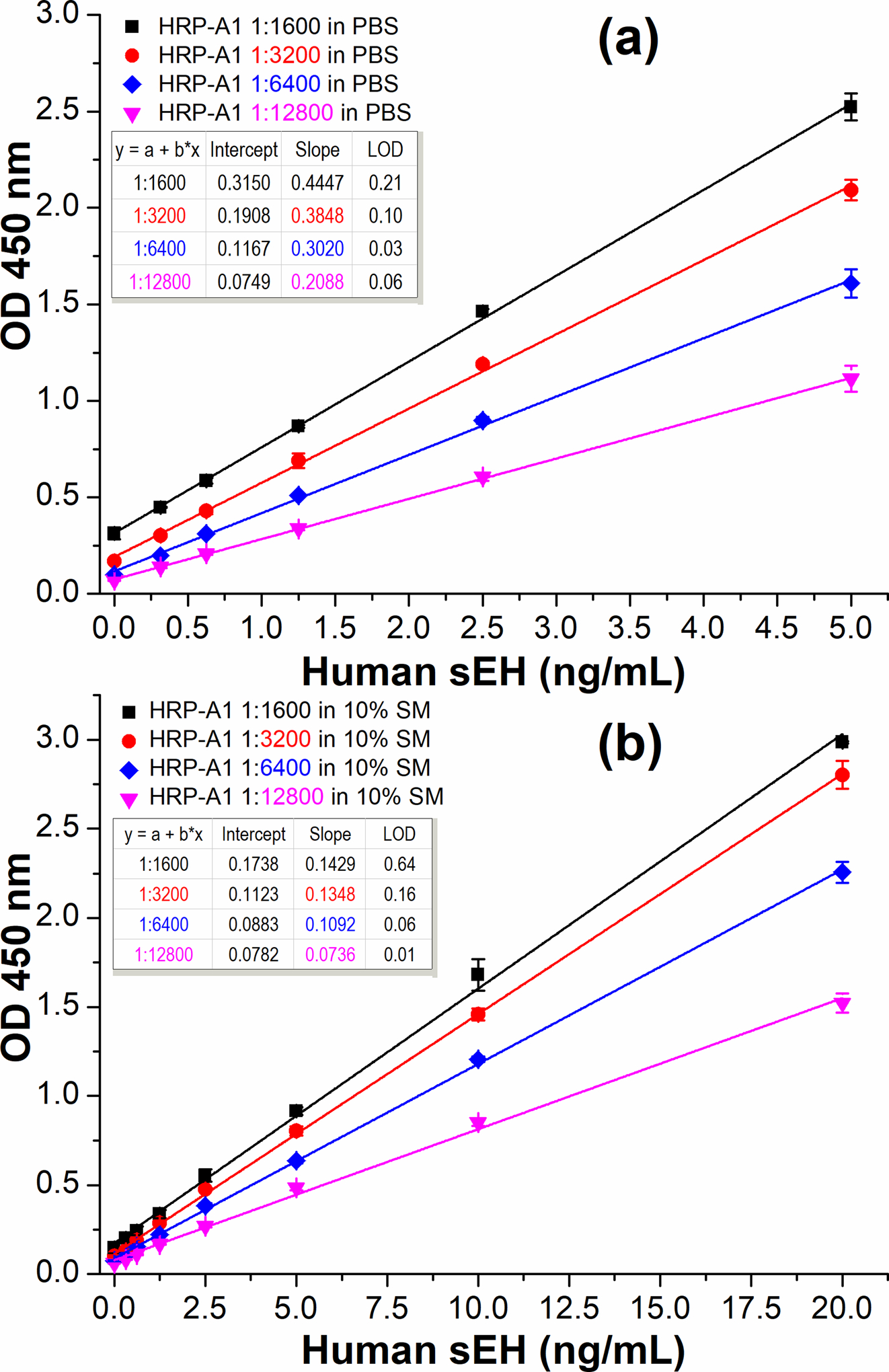
Calibration curves of SBdNb ELISAs (format C) with varying concentrations of detection antibody (HRP-A1, 1:1600–1:12800 dilution) in PBS (a) or 10% SM/PBS (b). Other conditions: streptavidin, 2.5 μg/mL coated in CB overnight at 4 °C; blocking buffer, 2% BSA/PBS 1 h; biotin-A9, 1.25 μg/mL 1 h; HRP-A1, 1h; TMB 15 min. All same or similar steps were performed under the same conditions (e.g., incubation time, reagents). Error bars indicate standard deviations (n = 3). All coefficient of determination R2>0.99.
Figure 5.
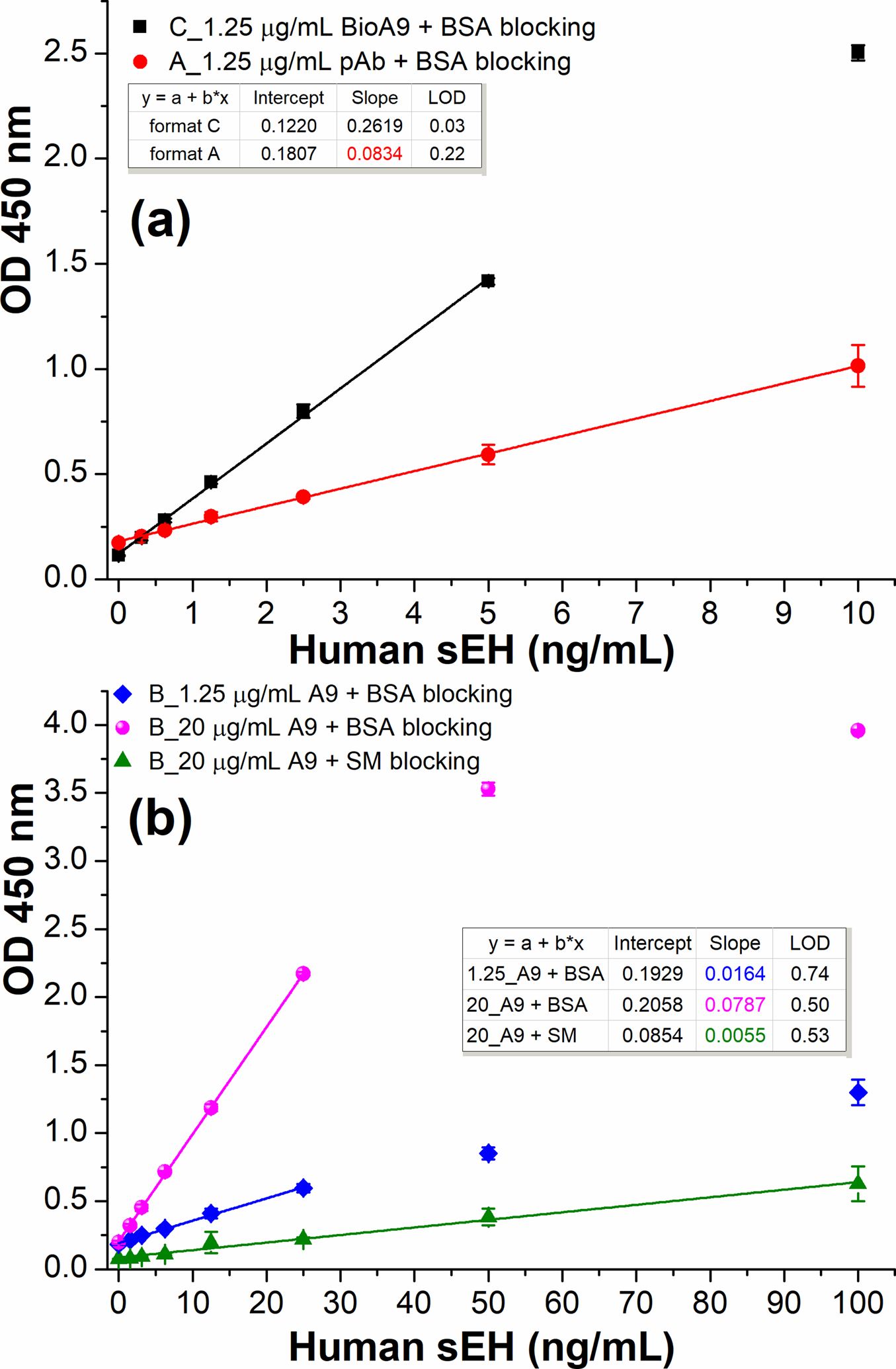
Comparison of three ELISA formats using HRP-A1 (1:6400 dilution in PBS) as detection antibody. (a) format C (SBdNb ELISA, square): 2.5 μg/mL streptavidin, 2% BSA/PBS blocking, 1.25 μg/mL biotin-A9; format A (circle): 1.25 μg/mL anti-sEH pAb, 2% BSA/PBS blocking. (b) format B with varying nanobody A9 coated in PBS and different blocking agents: diamond, 1.25 μg/mL A9 plus 2% BSA/PBS blocking; sphere, 20 μg/mL A9 plus 2% BSA/PBS blocking; up-triangle, 20 μg/mL A9 plus 3% SM/PBS blocking. All same or similar steps were performed under the same conditions. Error bars indicate standard deviations (n = 3). All R2>0.99 except the up-triangle of Fig. 5b (R2=0.9878).
Comparison of SBdNb ELISA and Other Two ELISA Formats
As previously described, the SBdNb ELISA demonstrated excellent performance thanks to the streptavidin bridge. To precisely evaluate the influence of antibody type and streptavidin bridge on the functionality of capture antibodies, it is meaningful to compare SBdNb ELISA (format C) with other formats at the same working concentration of the capture antibody. Figure 5a shows the signal responses of formats A and C using the same concentration (1.25 μg/mL) of pAb and biotinylated nanobody (BioA9) as capture antibodies, respectively. Format C generated a sensitivity (0.2619 OD•mL/ng) 3.1-fold of that of format A (0.0834 OD•mL/ng), with the advantage of nanobody containing more antigen-binding sites probably revealed at the same mass compared to pAb. Moreover, the format C gave a lower background than format A. This is likely due to the single domain nature of nanobody devoid of Fc portion. In addition, the SBdNb ELISA demonstrated the magic of streptavidin bridge when compared to format B with 1.25 μg/mL nanobody A9 coated directly. Its sensitivity is 16-fold of that of format B (Figure 5b, diamond, 0.0164 OD•mL/ng). In other words, less than 6% nanobody adsorbed passively on polystyrene is still effective for antigen binding. Thus, we can conclude that the procedure used for immobilizing nanobody is crucial to the functionality of capture nanobody. Moreover, it is worth noting that format B (Figure 5b, diamond) has 141% sensitivity as the ELISA (format B) described in Figure 3 where PBS was used as the coating buffer, though the working concentrations of capture nanobody and HRP-A1 in the latter is 16-fold and 4-fold higher than that of the former, respectively. This unexpected increased sensitivity is likely due to the switch of blocking agent from 3% SM/PBS to 2% BSA/PBS. It seems that SM not only reduced the nonspecific background but also shielded substantial specific signal in format B. This was further confirmed by the parallel ELISAs of format B using 20 μg/mL A9 for direct coating (passive adsorption) followed by 2% BSA or 3% SM for blocking (see Figure 5b). The SM blocking based direct double nanobody ELISA demonstrated only 7% sensitivity of the BSA blocking based assay. That means that at least 93% of the capture nanobody adsorbed lost its functionality or effectiveness because of SM blocking. There is the question of whether SM disabled more capture nanobody (e.g., through changing its conformation or masking paratopes) due to its nature alone or if it stripped more adsorbed nanobody off polystyrene than BSA did during blocking. The answer to this remains unclear and further investigation is needed.
Systematic Comparison of BSA Blocking with SM Blocking in Format B
The radical difference in blocking the plate with BSA vs SM was previously demonstrated with a high coating concentration of nanobody (20 μg/mL). To further evaluate this difference, varying coating concentrations (0.625–20 μg/mL) of capture nanobody A9 was passively adsorbed and later blocked with BSA or SM in parallel for each on the same high-binding polystyrene microplate (Nunc Maxisorp). As illustrated in Figure 6, sensitivity increased with increasing coating concentrations of capture nanobody in both blocking conditions. For BSA blocking, the saturation of sensitivity occurred at 5 μg/mL of capture A9 (Figure 6a). Whereas for SM blocking shown in Figure 6b, a huge drop in sensitivity by an order of magnitude was observed in various coating concentrations of nanobodies compared to the BSA blocking based situation. Thus, we can conclude that capture nanobody passively adsorbed could retain good functionality with BSA blocking, and SM blocking should be avoided due to its severe damage to the efficacy of capture nanobody. On the other hand, the format B demonstrated a 1/4 sensitivity of the optimized SBdNb ELISA when the saturating 5 μg/mL of A9 was used as the optimal coating concentration, though the former used 4-fold more nanobody. This sensitivity is actually sufficient for detecting many real samples where the sEH abundance is not too low. Compared to SBdNb ELISA, the format B based on passive adsorption of capture nanobody saved steps of streptavidin bridge and the biotinylation of nanobody, however, at the cost of dramatic loss of antibody efficiency, i.e., with no more than 1/16 functionality per nanobody amount remaining.
Figure 6.
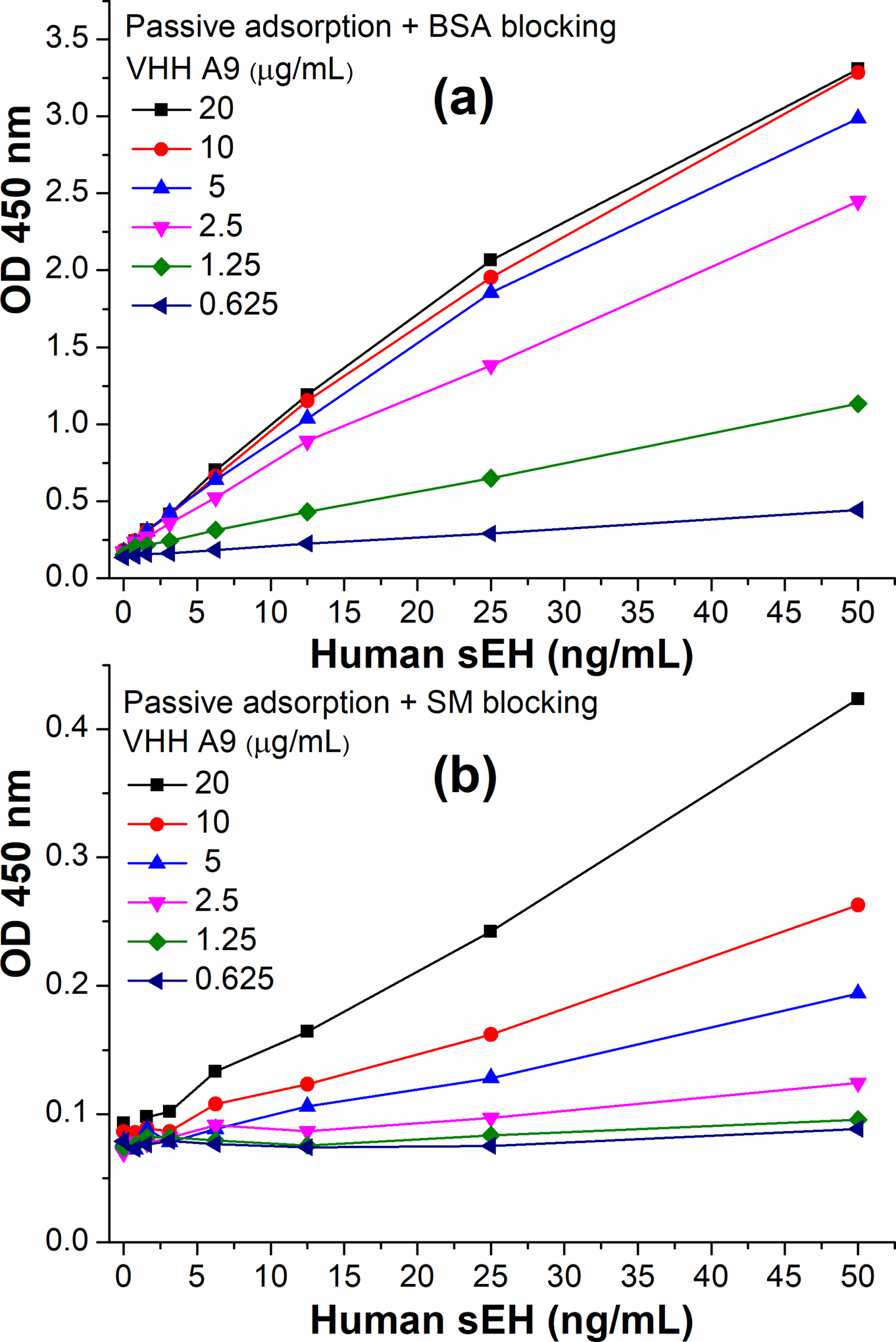
Comparison of ELISA format B using different blocking reagents with varying working concentrations of capture nanobody A9 (0.625–20 μg/mL). Direct coating of A9 (format A) through passive adsorption was followed by 2% BSA blocking (a) or 3% SM blocking (b). HRP-A1 (1:6400 dilution in PBS) was used as detection antibody.
Validation of SBdNb ELISA
Based on the aforementioned comparison of three ELISA formats, SBdNb ELISA (format C) turned out to be the superior double nanobody ELISA in terms of sensitivity as well as reagents cost-effectiveness and thus was used for further validation. As detailed in Table S1, the SBdNb ELISA demonstrated excellent selectivity for the target, allowing specific detection of only the bioactive sEH with negligible interference from epoxide hydrolases in the same enzyme family, such as human mEH and human EH3. Moreover, the matrix effects were evaluated through spike-and-recovery tests against spiked samples at varying dilutions. As detailed in Table S2, the background decreased and the net signal increased with increasing dilutions of the sample matrix of sEH free mouse brain tissue. The recovery was 75–98%, 78–90% and 78–97% for the spiked samples prepared at 1:150, 1:450 and 1:1350 dilution of the sample matrix, respectively. The corresponding overall recovery was 75%, 84% and 91%, respectively. These data support the acceptance of SBdNb ELISA for sEH detection in biological samples. Furthermore, a successful analytical approach must satisfy the need for detection of real samples with high accuracy.38 The SBdNb ELISA was used to analyze the sEH in the S9 fractions of pooled (4–50 persons) human tissue samples described in SI. The resultant data were compared with that obtained through the enzyme activity based radiometric assay, Western-blot, conventional ELISA, ELISA NbS7/NbS43, and PolyHRP ELISA (Table S3). We can find that the data by SBdNb ELISA is comparable to that by other fiver methods. Moreover, the SBdNb ELISA showed good correlation to the radioactive assay in these tissue samples with correlation coefficient r > 0.99, suggesting the sEH protein level through SBdNb ELISA is a good indication of its enzyme activity. Also, as specified in Table S4 and S5, the SBdNb ELISA successfully measured peripheral blood mononuclear cells (PBMCs) samples from patients without or with type 2 diabetes, and brain samples from patients without or with multiple sclerosis, respectively. Generally, PBMCs have very low abundance of sEH and thus require assays of high sensitivity. The results obtained through SBdNb ELISA well disclosed the association between sEH overexpression and the pathogenesis of type 2 diabetes, as well as the lack of a correlation between sEH and the progression of multiple sclerosis. This further indicates the significance of SBdNb ELISA as an important diagnostic tool for human health.
Further Overall Discussion
Interestingly, Zhu et al. reported a streptavidin-biotin-based directional double nanobody sandwich ELISA for detection of influenza H5N1. The capture nanobody was biotinylated in vivo at the opposite terminal of antigen-binding site and later bound to a commercial streptavidin-coated microplate for a directional immobilization with the paratope facing outward. The resultant directional ELISA was claimed to show more sensitivity than the undirectional conventional ELISA involving passive adsorption of capture nanobody and BSA blocking.39 However, the sensitivities of both ELISAs are moderate and seem to be comparable. A confirmative comparison is difficult to make in view of the limited quality of the linear fitting. Also, Rossotti et al. developed a sensitive double nanobody based indirect sandwich ELISA for sEH detection using avidin as a bridge on polystyrene plates. This format was adopted as the adaptation to a screening platform they proposed, where the output clones were first biotinylated in vivo in 96-well culture plates and then bound to avidin-coated microplates for immobilization. Clones with high affinity were ranked according to their reactivity with the HRP labeled antigen.40 Actually, their avidin bridged indirect sandwich ELISA is similar to our SBdNb ELISA. We therefore inferred that the high sensitivity of the assay is not only due to nanobodies of high affinity obtained alone, but also due to the bioactivity enhancement of the avidin bridge. Despite emergence of some early research on developing double nanobody based ELISA, field has mainly focused on the analytical aspects. The exploration of other factors contributing to the performance of capture nanobodies is rare in the literature.
CONCLUSION
We developed an improved double nanobody sandwich ELISA for human sEH detection. Epitope mapping was successfully performed to pair nanobodies for sandwich immunoassay, and HRP labeled nanobody conjugate as detection antibody was favorably prepared through an improved periodate oxidation in a much mild but efficient conjugation manner. Compared to the double nanobody ELISA with capture nanobody passively adsorbed on polystyrene microplate, the streptavidin bridged double nanobody ELISA demonstrated a 16-fold higher sensitivity and 25-fold lower LOD of the former, with the sensitivity improved from 0.0164 to 0.2619 OD•mL/ng and LOD from 0.74 to 0.03 ng/mL for human sEH detection. The enhanced ELISA was then successfully applied to tests for recovery and cross-reactivity and the analysis of real samples. Meanwhile, the blocking with skim milk was unexpectedly found to severely damage the performance of capture nanobody passively adsorbed by an order of magnitude, compared to that with BSA. Thus, understanding of the physical and functional behavior of capture nanobodies immobilized on polystyrene related to both the immobilization format and blocking agent provides a general guideline for developing double nanobody ELISAs for various targets.
Supplementary Material
ACKNOWLEDGMENT
This work was financially supported by National Institute of Health (NIEHS Superfund P42ES04699 and RIVER Award R35 ES030443-01). D.L. received partial support from National Natural Science Foundation of China (81402722) and The International Postdoctoral Exchange Fellowship (06/2014-06/2015) by The Office of China Postdoctoral Council. A.T. and R.N. received support from National Multiple Sclerosis Society (PP-1606-24495 and RG 829-P-40). We would like to thank Ms. Catherine Elizabeth Clennan for allowing the use of her llama artwork in the abstract graphic.
Footnotes
The authors declare no competing financial interest.
Supporting Information Available
The Supporting Information is available free of charge at http://pubs.acs.org.
Supplementary experimental section; epitope mapping of nanobodies; characterization of the HRP labeled nanobody; optimization of HRP-A1 working concentration; optimization of SA and BioA9; selectivity of SBdNb ELISA; recovery test; S9 fraction of human tissues analysis; analysis of PBMCs from diabetic patients; analysis of brain samples from MS patients;
REFERENCES
- (1).Harlow E; Lane D Antibodies: A Laboratory Manual; Cold Spring Harbor Laboratory: New York, 1988, p 291. [Google Scholar]
- (2).Huckle D In The Immunoassay Handbook (Fourth Edition), Wild D, Ed.; Elsevier: Oxford, 2013, pp 517–531. [Google Scholar]
- (3).VynZ Research https://www.globenewswire.com/news-release/2019/07/09/1879881/0/en/Global-Immunoassay-Market-is-Set-to-Reach-USD-26-9-Billion-by-2024-Growing-at-a-CAGR-of-5-6-During-the-Forecast-Period-VynZ-Research.html (accessed Mar 12, 2020).
- (4).Li D; Cui Y; Morisseau C; Gee SJ; Bever CS; Liu X; Wu J; Hammock BD; Ying Y Nanobody based immunoassay for human soluble epoxide hydrolase detection using polymeric horseradish peroxidase (PolyHRP) for signal enhancement: the rediscovery of PolyHRP? Anal. Chem 2017, 89, 6248–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Baker M Reproducibility crisis: Blame it on the antibodies http://www.nature.com/news/reproducibility-crisis-blame-it-on-the-antibodies-1.17586 (accessed Mar 12, 2020). [DOI] [PubMed]
- (6).Arnaud CH http://cen.acs.org/articles/93/web/2015/02/Call-Antibody-Standardization.html (accessed Mar 12, 2020).
- (7).Greenberg AS; Avila D; Hughes M; Hughes A; McKinney EC; Flajnik MF A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [DOI] [PubMed] [Google Scholar]
- (8).Hamers-Casterman C; Atarhouch T; Muyldermans S; Robinson G; Hammers C; Songa EB; Bendahman N; Hammers R Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [DOI] [PubMed] [Google Scholar]
- (9).Steeland S; Vandenbroucke RE; Libert C Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discovery Today 2016, 21, 1076–1113. [DOI] [PubMed] [Google Scholar]
- (10).Kim H-J; McCoy MR; Majkova Z; Dechant JE; Gee SJ; Tabares-da Rosa S; González-Sapienza GG; Hammock BD Isolation of Alpaca Anti-Hapten Heavy Chain Single Domain Antibodies for Development of Sensitive Immunoassay. Anal. Chem 2012, 84, 1165–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bradbury AR; Trinklein ND; Thie H; Wilkinson IC; Tandon AK; Anderson S; Bladen CL; Jones B; Aldred SF; Bestagno M When monoclonal antibodies are not monospecific: hybridomas frequently express additional functional variable regions. MAbs 2018, 10, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tabares-da Rosa S; Rossotti M; Carleiza C; Carrión F; Pritsch O; Ahn KC; Last JA; Hammock BD; González-Sapienza G Competitive Selection from Single Domain Antibody Libraries Allows Isolation of High-Affinity Antihapten Antibodies That Are Not Favored in the llama Immune Response. Anal. Chem 2011, 83, 7213–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang J; Bever CR; Majkova Z; Dechant JE; Yang J; Gee SJ; Xu T; Hammock BD Heterologous antigen selection of camelid heavy chain single domain antibodies against tetrabromobisphenol A. Anal. Chem 2014, 86, 8296–8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Liu X; Xu Y; Wan DB; Xiong YH; He ZY; Wang XX; Gee SJ; Ryu D; Hammock BD Development of a nanobody-alkaline phosphatase fusion protein and its application in a highly sensitive direct competitive fluorescence enzyme immunoassay for detection of ochratoxin a in cereal. Anal. Chem 2015, 87, 1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wang K; Liu Z; Ding G; Li J; Vasylieva N; Li QX; Li D; Gee SJ; Hammock BD; Xu T Development of a one-step immunoassay for triazophos using camel single-domain antibody–alkaline phosphatase fusion protein. Anal. Bioanal. Chem 2019, 411, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang K; Vasylieva N; Wan D; Eads DA; Yang J; Tretten T; Barnych B; Li J; Li QX; Gee SJ; Hammock BD; Xu T Quantitative Detection of Fipronil and Fipronil-Sulfone in Sera of Black-Tailed Prairie Dogs and Rats after Oral Exposure to Fipronil by Camel Single-Domain Antibody-Based Immunoassays. Anal. Chem 2019, 91, 1532–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cui Y; Li D; Morisseau C; Dong J-X; Yang J; Wan D; Rossotti M; Gee S; González-Sapienza G; Hammock B Heavy chain single-domain antibodies to detect native human soluble epoxide hydrolase. Anal. Bioanal. Chem 2015, 407, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vasylieva N; Kitamura S; Dong J; Barnych B; Hvorecny KL; Madden DR; Gee SJ; Wolan DW; Morisseau C; Hammock BD Nanobody-based binding assay for the discovery of potent inhibitors of CFTR inhibitory factor (Cif). Anal. Chim. Acta 2019, 1057, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Morisseau C; Hammock BD Impact of Soluble Epoxide Hydrolase and Epoxyeicosanoids on Human Health. Annu. Rev. Pharmacool. Toxicol 2013, 53, 37–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yao L; Cao B; Cheng Q; Cai W; Ye C; Liang J; Liu W; Tan L; Yan M; Li B; He J; Hwang SH; Zhang X; Wang C; Ai D; Hammock BD; Zhu Y Inhibition of Soluble Epoxide Hydrolase Ameliorates Hyperhomocysteinemia-Induced Hepatic Steatosis by Enhancing β-oxidation of Fatty Acid in Mice. Am. J. Physiol. Gastrointest. Liver Physiol 2019, 316, G527–G538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Imig JD; Hammock BD Soluble epoxide hydrolase as a therapeutic target for cardiovascular diseases. Nat. Rev. Drug Discovery 2009, 8, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Morisseau C; Hammock BD Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu. Rev. Pharmacool. Toxicol 2005, 45, 311–333. [DOI] [PubMed] [Google Scholar]
- (23).Lee KSS; Yang J; Niu J; Ng CJ; Wagner KM; Dong H; Kodani SD; Wan D; Morisseau C; Hammock BD Drug-Target Residence Time Affects in Vivo Target Occupancy through Multiple Pathways. ACS Cent. Sci 2019, 5, 1614–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ren Q; Ma M; Yang J; Nonaka R; Yamaguchi A; Ishikawa K.-i.; Kobayashi K; Murayama S; Hwang SH; Saiki S Soluble epoxide hydrolase plays a key role in the pathogenesis of Parkinson’s disease. PNAS 2018, 115, E5815–E5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Borlongan CV Fatty acid chemical mediator provides insights into the pathology and treatment of Parkinson’s disease. PNAS 2018, 115, 6322–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yu Z; Davis BB; Morisseau C; Hammock BD; Olson JL; Kroetz DL; Weiss RH Vascular localization of soluble epoxide hydrolase in the human kidney. Am. J. Physiol. Renal Physiol 2004, 286, F720–F726. [DOI] [PubMed] [Google Scholar]
- (27).LASTRA PDL; BERG VD; BULLIDO; ALMAZÁN; DOMÍNGUEZ; LLANES; MORGAN. Epitope mapping of 10 monoclonal antibodies against the pig analogue of human membrane cofactor protein (MCP). Immunology 1999, 96, 663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wilson MB, and Nakane PK Recent development in the periodate methods of conjugating horseradish peroxidase (HRPO) to antibodies. Immunofluorescence and Related Staining Techniques, Elsevier/North Holland Biomedical Press, Amsterdam 1978, 215–224. [Google Scholar]
- (29).Joshi KS; Hoffmann LG; Butler JE The immunochemistry of sandwich ELISAs—V. The capture antibody performance of polyclonal antibody-enriched fractions prepared by various methods. Mol. Immunol 1992, 29, 971–981. [DOI] [PubMed] [Google Scholar]
- (30).Butler JE; Ni L; Nessler R; Joshi KS; Suter M; Rosenberg B; Chang J; Brown WR; Cantarero LA The physical and functional behavior of capture antibodies adsorbed on polystyrene. J. Immunol. Methods 1992, 150, 77–90. [DOI] [PubMed] [Google Scholar]
- (31).Suter M; Butler JE The immunochemistry of sandwich ELISAs. II. A novel system prevents the denaturation of capture antibodies. Immunol. Lett 1986, 13, 313–316. [DOI] [PubMed] [Google Scholar]
- (32).Näreoja T; Määttänen A; Peltonen J; Hänninen PE; Härmä H Impact of surface defects and denaturation of capture surface proteins on nonspecific binding in immunoassays using antibody-coated polystyrene nanoparticle labels. J. Immunol. Methods 2009, 347, 24–30. [DOI] [PubMed] [Google Scholar]
- (33).Hoffman WL; Jump AA Inhibition of the streptavidin-biotin interaction by milk. Anal. Biochem 1989, 181, 318–320. [DOI] [PubMed] [Google Scholar]
- (34).Vogt RF; Phillips DL; Henderson LO; Whitfield W; Spierto FW Quantitative Differences among Various Proteins as Blocking-Agents for Elisa Microtiter Plates. J. Immunol. Methods 1987, 101, 43–50. [DOI] [PubMed] [Google Scholar]
- (35).MacDougall D; Crummett WB; et al. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem 1980, 52, 2242–2249. [Google Scholar]
- (36).Duhamel RC; Johnson DA Use of nonfat dry milk to block nonspecific nuclear and membrane staining by avidin conjugates. J. Histochem. Cytochem 1985, 33, 711–714. [DOI] [PubMed] [Google Scholar]
- (37).Vincent P; Samuel D A comparison of the binding of biotin and biotinylated macromolecular ligands to an anti-biotin monoclonal antibody and to streptavidin. J. Immunol. Methods 1993, 165, 177–182. [DOI] [PubMed] [Google Scholar]
- (38).Hou L; Tang Y; Xu M; Gao Z; Tang D Tyramine-Based Enzymatic Conjugate Repeats for Ultrasensitive Immunoassay Accompanying Tyramine Signal Amplification with Enzymatic Biocatalytic Precipitation. Anal. Chem 2014, 86, 8352–8358. [DOI] [PubMed] [Google Scholar]
- (39).Zhu M; Gong X; Hu Y; Ou W; Wan Y Streptavidin-biotin-based directional double Nanobody sandwich ELISA for clinical rapid and sensitive detection of influenza H5N1. J. Transl. Med 2014, 12, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Rossotti MA; Pirez M; Gonzalez-Techera A; Cui Y; Bever CS; Lee KSS; Morisseau C; Leizagoyen C; Gee S; Hammock BD; González-Sapienza G Method for Sorting and Pairwise Selection of Nanobodies for the Development of Highly Sensitive Sandwich Immunoassays. Anal. Chem 2015, 87, 11907–11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


