Abstract
Peptide hormones are attractive as injectable therapeutics and imaging agents, but they often require extensive modification by mutagenesis and/or chemical synthesis to prevent rapid in vivo degradation. Alternatively, the single atom, O-to-S modification of peptide backbone thioamidation has the potential to selectively perturb interactions with proteases while preserving interactions with other proteins, such as target receptors. Here, we use the validated diabetes therapeutic, glucagon-like peptide-1 (GLP-1), and the target of clinical investigation, gastric inhibitory polypeptide (GIP), as proof-of-principle peptides to demonstrate the value of thioamide substitution. In GLP-1 and GIP, a single thioamide near the scissile bond renders these peptides up to 750-fold more stable than the corresponding oxopeptides toward cleavage by dipeptidyl peptidase 4, the principle regulator of their in vivo stability. These stabilized analogs are nearly equipotent with their parent peptide in cyclic AMP activation assays, but the GLP-1 thiopeptides have much lower β-arrestin potency, making them novel agonists with altered signaling bias. Initial tests show that a thioamide GLP-1 analog is biologically active in rats, with an in vivo potency for glycemic control surpassing that of native GLP-1. Taken together, these experiments demonstrate the potential for thioamides to modulate specific protein interactions to increase proteolytic stability or tune activation of different signaling pathways.
Graphical Abstract
INTRODUCTION
In recent years, there has been significant growth in the development of peptide therapeutics (“biologics”) and imaging agents.1–3 Peptide-based therapies and diagnostics are typically based on natural bioactive peptides such as hormones or neuropeptides, and this makes the identification of a ‘lead compound’ much easier than the identification of a lead compound for a small molecule.4–5 However, peptides undergo rapid proteolysis and renal clearance in vivo, leading to unfavorable pharmacokinetics.6 Much time in the development of peptide biologics is spent on modifying the peptide at cleavage sites to reduce proteolysis, while maintaining activity.4, 7–9 The most popular strategies involve unnatural amino acids (D-, β-, or α,α-dimethyl amino acids) used specifically to overcome proteolysis, or “staples” that stabilize secondary structure and consequently reduce proteolysis.10–11 Since these substitutions may have significant undesirable effects on peptide conformation and bioactivity, the identification of appropriate modification sites takes time and effort. Here, we show that thioamide substitution, a single atom O-to-S change, at the cleavage site of a peptide can decrease the rate of proteolysis while preserving pharmacodynamic activity in cell or animal assays.12 We demonstrate this in two therapeutically-relevant peptide substrates of dipeptidyl peptidase 4 (DPP-4): glucagon-like peptide-1 (GLP-1) and gastric inhibitory polypeptide (GIP). These peptides were chosen for proof-of-concept because they are validated therapeutic or diagnostic targets, yet there is still room to improve upon the state-of-the-art. Indeed, we find that our GLP-1 analogs have alterations in the relative activation of second messenger pathways associated with the GLP-1 receptor (GLP-1R), known as signaling bias.13 The stability and novel signaling bias of these thiopeptides could make them valuable tools for investigating GLP-1 signaling in vivo and separating pathways responsible for desirable glycemic control effects from those of undesirable effects such as nausea.
Aside from various insulin forms, one of the most highly used classes of injectable peptide biologics derives from the incretin hormone GLP-1.14 The GLP-17–36 fragment, referred to simply as GLP-1, stimulates insulin, inhibits gastric emptying, and reduces food intake.15 (Fig. 1) However, GLP-1 is inactivated by DPP-4 cleavage with a half-life of less than 2 min.16 DPP-4 preferentially cleaves after Pro or Ala residues penultimate to the N-terminus and functions as the principal determinant of the circulating half-life for GLP-1.17 Therapeutic approaches for enhancing insulin-secreting activity include both degradation-resistant GLP-1R agonists and inhibitors of DPP-4 activity. Several stabilized GLP-1 mimetics are currently prescribed to Type II diabetes patients as injectables taken between twice daily and once weekly, such as exenatide (Byetta®), liraglutide (Victoza®), lixisenatide (Adlyxin™), dulaglutide (Trulicity®), and albiglutide (Tanzeum®).18 Despite the appeal of orally bioavailable small molecule DPP-4 inhibitors such as sitagliptin (Januvia®), there remains a need for injectable GLP-1 mimetics because DPP-4 inhibitors fail to produce some desirable effects of the peptides, such as appetite suppression and weight loss.19–20
Figure 1.
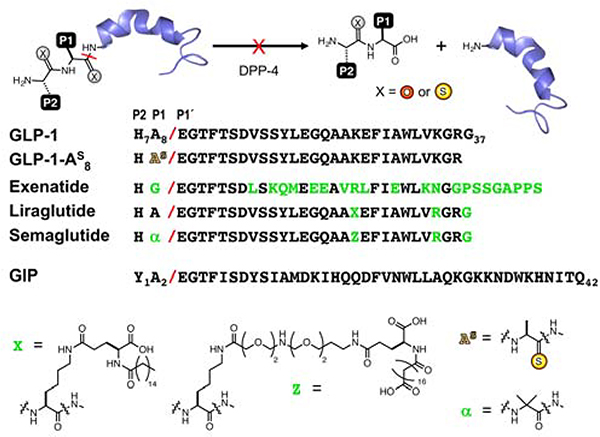
Thioamides Prevent Peptide Inactivation by Proteolysis. Native peptides are inactivated by DPP-4 cleavage at the scissile bond, indicated with a red slash (P2, P1 and P1’ positions are numbered relative to the scissile bond, by convention). Sequences of GLP-1, GLP-1 analogs, and GIP are shown. GLP-1 analogs exenatide, liraglutide, and semaglutide are stabilized by extensive mutation, sidechain fatty acid modification (X or Z), or a combination of fatty acid modification and aminoisobutyric acid (α) incorporation, respectively. Other GLP-1 stabilization strategies are described in the text. Thioamide substitution is indicated by an “S” superscript.
DPP-4 substrates include not only GLP-1, but over 25 peptides with diverse signaling functions, including: glucagon, GIP,17, 21 vasoactive intestinal peptide,22 pancreatic polypeptide,22 peptide YY and neuropeptide Y,23 oxyntomodulin,24 brain natriuretic peptide,25 enterostatin,26 growth-hormone-releasing hormone,27 substance P,28–30 and several cytokines.31–32 All of these DPP-4 substrates have half-lives of less than an hour (in most cases, less than 15 min) and many have potentially useful therapeutic activity. Like GLP-1, GIP acts as a glucose-lowering agent and has been studied extensively as a diabetes treatment.33–35 There is substantial current interest in GLP-1/GIP chimera peptides which can activate both GLP-1R and the GIP receptor (GIPR).36 Unlike GLP-1, GIP currently has no stabilized versions available for therapeutic purposes.
Peptide stabilization strategies previously applied to GLP-1 primarily involve restricting DPP-4 access to the cleavable bond. Exenatide has only natural amino acid changes in its peptide sequence, whereas liraglutide includes a fatty acid modified sidechain (X in Fig. 1), which increases serum albumin binding to reduce both protease access and kidney clearance.37 Albiglutide features genetic fusion of two copies of GLP-1 (modified with a Gly mutation at the scissile bond) to human albumin protein to achieve the same effect.38–39 Other peptides, such as taspoglutide,40 block access to cleavable bonds and stabilize conformations with aminoisobutyric acid (Aib or α in Fig. 1). Semaglutide, a molecule in clinical trials, combines fatty acid modification (Z) with Aib (Fig. 1) to make a GLP-1 analog that requires only once weekly injection.41–42 Gellman and coworkers substituted Aib at Ala8 and Val16, and also made extensive β-amino acid modifications throughout the GLP-1 sequence.43 While there are many other modifications applied to this important problem, these examples represent the major strategies currently employed.
Peptide stabilization by using multiple modifications at non-obvious positions requires extensive structure-activity relationship studies to determine the best locations for these modifications. In contrast, thioamides can be easily introduced at or near the scissile bond. They are also fully compatible with emerging recent trends in developing dual receptor (e.g., GLP-1 and GIP) chimeric agonists by merging sequences.33 While thioamides are not expected to alter renal clearance, this can be addressed by addition of fatty acids as in semaglutide. In the case of imaging reagents, clearance of peptides not bound to receptors is actually beneficial as it will reduce background signal.44
Peptides containing a thioamide substitution near the scissile bond have previously been investigated as potential competitive inhibitors of a variety of proteases.45–52 In most cases, thioamide substitution weakened the affinity of the peptide for its target protease, negating its use as a competitive inhibitor. A study of DPP-4 itself by Fischer and coworkers found that thioamide substitution at the P2 position of Ala-Pro-pNA substrates, where pNA is p-nitroaniline, yielded a nearly 1000-fold decrease in the kcat/Km value relative to the oxoamide.50 Their mechanistic studies revealed that the ~3 kcal/mol higher cis/trans rotational barrier of the thioamide played an important role in lowering kcat. However, since thioamide substitution significantly affected Km as well, subsequent studies of thiopeptide-based DPP-4 inhibitors found them to have higher KIs than the corresponding oxopeptides.53
These results provide a valuable precedent for our applications. While a decrease in DPP-4 affinity prevented thioamides from being useful in inhibitors, it is helpful to us, since we are designing our thiopeptides to evade proteolysis entirely, and wish to avoid unintended inhibition of DPP-4 activity towards its many other important substrates. Thus, there is tantalizing precedent for thioamide stability effects coming from inhibitor design, but to our knowledge, there has been only limited investigation of its utility in stabilizing short signaling peptides (~6 amino acids) with few published examples of trials in animals.54–58
Recent improvements in the synthesis of thioamide-containing peptides and proteins now permit the generation of hormone analogs greater than 30 amino acids, such as those reported here.59–60 (Note: The thioamide position is represented by a superscript “S”, thioalanine is shown in Fig. 1 as an example.) This enables an unrestricted exploration of the effects of thioamides in peptides with significant biological activity to demonstrate the potential for using thioamides to stabilize peptides for in vivo experiments in a general and straightforward manner.
RESULTS
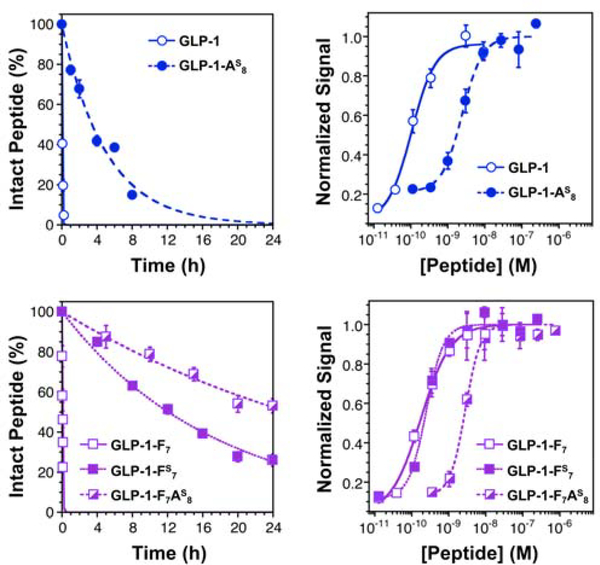
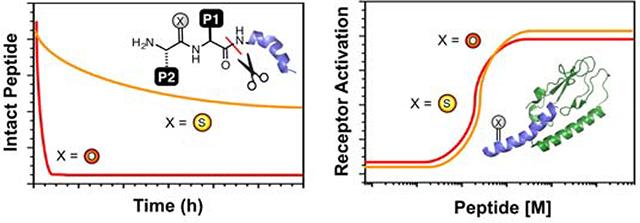
GLP-1 was investigated first due to the precedent for therapeutic relevance of stabilized analogs and the existence of established bioactivity assays.18 GLP-1, GLP-1-F7, and thioamide variants GLP-1-AS8, GLP-1-F7AS8, GLP-1-FS7, and GLP-1-ES9 were generated with a combination of automated and manual synthesis using our established protocols.59, 61–62 These peptides were purified by high performance liquid chromatography (HPLC), and characterized by matrix-assisted laser desorption ionization mass spectrometry (MALDI MS). The in vitro stability of the GLP-1 peptides was measured by incubation in buffer alone and with DPP-4 for different time periods. The reaction was quenched by the addition of HCl and the products analyzed by HPLC to determine the amount of intact peptide. MALDI MS was used to confirm the identities of any HPLC peaks. The in vitro half-life of GLP-1-AS8 (P1) is 3.4 h under conditions where the half-life of native GLP-1 is 2 min. (Fig. 2, Top Left and Table 1). In fact, GLP-1-AS8 appears to be inert to cleavage by DPP-4. The degradation that does occur takes place through an intramolecular reaction that results from the presence of the N-terminal His residue in GLP-1-AS8. (see Supporting Information, Figs. S5–S8) On the other hand, GLP-1-ES9 (P1’) is stable in buffer, but has a half-life of <5 min in the presence of DPP-4. (see Table 1 and Supporting Information, Fig. S3) Unfortunately, GLP-1-HS7 (P1) could not be synthesized in sufficient yields for testing with DPP-4.
Figure 2.
Thioamide Substitution Stabilizes GLP-1 Analogs Without Disrupting Activity. Top Left: In vitro proteolysis data demonstrating that GLP-1-AS8 is cleaved by 2.5 ng/mL DPP-4 more slowly than the respective oxopeptide (GLP-1). Top Right: Dose response curves for GLP-1 and GLP-1-AS8. Bottom Left: In vitro proteolysis data demonstrating that GLP-1-F7AS8 and GLP-1-FS7 are cleaved by 2.5 ng/mL DPP-4 more slowly than their respective oxopeptide (GLP-1-F7). Bottom Right: Dose response curves for GLP-1-F7, GLP-1-F7AS8, and GLP-1-FS7. All cellular responses were determined using DiscoveRx GLP-1R reporter cells, which detect cAMP production following GLP-1R activation in an enzyme-coupled assay. See Supporting Information for details of both assays, see a list of half-lives and EC50 values in Table 1. Bars represent standard error.
Table 1.
Peptide In Vitro Half-lives and Cellular cAMP Activation
| Peptide | Half-life (min or h)a,b | EC50 (pM)a |
|---|---|---|
| GLP-1 | 1.9 ± 0.3 min | 207 ± 42 |
| GLP-1-AS8 | 03.44 ± 0.26 h | 2440 ± 398 |
| GLP-1-ES9 | < 5 min | |
| GLP-1-F7 | 2.6 ± 0.1 min | 171 ± 25 |
| GLP-1-FS7 | 12.09 ± 0.38 h | 244 ± 29 |
| GLP-1-F7AS8 | 25.73 ± 1.06 h | 2715 ± 172 |
| GIP | 5.5 ± 0.1 min | 161 ± 19 |
| GIP-YS1 | 67.68 ± 3.47 h | 430 ± 46 |
| GIP-AS2 | 60.20 ± 2.86 h | 6522 ± 351 |
Values represent mean ± standard error.
Cleavage rates per μg DPP-4 are reported in Table S5 in the Supporting Information.
Since GLP-1 and GLP-1-F7 have previously been shown to be essentially equivalent in potency,63 we chose to use GLP-1-F7 as host system to study thioamide effects at P2 (GLP-1-F7AS8) and P1 (GLP-1-FS7). We hypothesized that GLP-1-F7AS8 would not be prone to the degradation observed for GLP-1-AS8, since it lacked the imidazole moiety of His7, which could act as a catalyst in auto-degradation. Indeed, both GLP-1-F7AS8 and GLP-1-FS7 were stable in buffer (see Supporting Information, Fig. S4). Like the parent GLP-1 peptide, GLP-1-F7 has a half-life of about 2 min in the presence of 2.5 ng/mL DPP-4 (Table 1). Under the same conditions, the thiopeptides GLP-1-F7AS8 (P1) and GLP-1-FS7 (P2) have half-lives of 26 h and 12 h. (Fig. 2, Bottom Left and Table 1) Thus, thioamide substitution at specific sites near the scissile bond of GLP-1 analogs can confer a >500-fold increase in DPP-4 proteolytic stability. Half-lives of this duration are long enough that DPP-4 cleavage will no longer be the limiting factor for pharmacokinetics.18
To determine whether thiopeptides act as competitive inhibitors of DPP-4, various concentrations of a thioamide-modified GLP-1 were mixed with 100 μM Ala-Pro-pNA, a commercial chromogenic substrate of DPP-4. After addition of catalytic amounts of DPP-4, the spectral change resulting from release of pNA was used to monitor DPP-4 activity. We found that GLP-1-AS8, GLP-1-F7AS8, and GLP-1-FS7 do not inhibit DPP-4 at concentrations up to 100 μM, consistent with the idea that their stability to DPP-4 cleavage derives from an effect on binding rather than catalysis. (see Supporting Information, Fig. S12)
To be valuable, the thioamide substitutions must also be non-disruptive to the peptide’s structure and activity. Circular dichroism (CD) measurements demonstrated that all of our thioamide GLP-1 analogs were comparable in helicity to the corresponding oxopeptides. (see Supporting Information, Fig. S1) To measure the GLP-1R EC50s of each peptide, we used engineered DiscoveRx cAMP Hunter™ eXpress CHO-K1 cell lines with an enzyme-coupled cAMP reporter. We find that GLP-1-AS8 is 11-fold less potent than GLP-1. (Fig. 2, Top Right and Table 1) In the same assay, GLP-1-F7 is essentially equipotent with GLP-1, with an EC50 of 171 pM. (Fig. 2, Bottom Right and Table 1) GLP-1-F7AS8 is 16-fold less potent than GLP-1-F7, showing that the decreased potency of GLP-1-AS8 was probably not due to the His-catalyzed side reaction (the assay duration is 30 min, the auto-degradation half-life of GLP-1-AS8 is 3.4 h). Most significantly, GLP-1-FS7 was found to be only two-fold less potent than GLP-1 or GLP-1-F7, with an EC50 of 244 pM. (Fig. 2, Bottom Right and Table 1) Thus, GLP-1-FS7 was deemed to be sufficiently stable and potent to be taken forward to further cellular experiments as well as in vivo studies.
We were also interested in the idea that thioamide modification could alter the signaling bias of an agonist like GLP-1. Signaling bias refers to the way in which subtle changes in the agonists of G-protein coupled receptors like GLP-1R and GIPR can differentially affect activation of various intracellular signaling pathways.13 As a preliminary investigation of bias in our agonists, we performed activity assays using engineered CHO-K1 cells similar to those performed for cAMP production. We found that GLP-1-AS8 has reduced potency for both β-arrestin 1 and 2 activation, with a roughly 30-fold increase in EC50 and a 16–42% decrease in the maximum response. This is only slightly greater than the decrease in potency (11-fold, identical maximum response) seen for Ala8 substitution in GLP-1R cAMP activation (Figure 2). Thus, it seems that thioamide substitution at Ala8 does not significantly affect GLP-1R signaling bias in terms of affinity, but does decrease β-arrestin activation.
GLP-1-FS7 has a ~12-fold higher EC50 and a >75% lower maximum response for both β-arrestin 1 and 2 activation, when compared to GLP-1. However, since GLP-1-F7 has a ~10-fold EC50 and a ~50% lower maximum response in these assays, it seems that much of this effect is the result of His7-to-Phe modification. Comparing GLP-1-FS7 to GLP-1-F7, we find that the thioamide substitution itself only alters the β-arrestin 1 and 2 EC50s by 1.2-fold, albeit with a roughly 50% decrease in the maximum responses. The small change in EC50 is comparable to the 2-fold increase in EC50 seen in comparing GLP-1-FS7 and GLP-1-F7 cAMP activation. Thus, we conclude that thioamide substitution at position 1 also does not significantly affect GLP-1R signaling bias in terms of affinity, but significantly reduces β-arrestin activation. Since this effect is convoluted with the larger effect from Phe substitution, more investigation is warranted. Unfortunately, difficulties in synthesizing GLP-1-HS7 make us unable to directly compare to GLP-1.
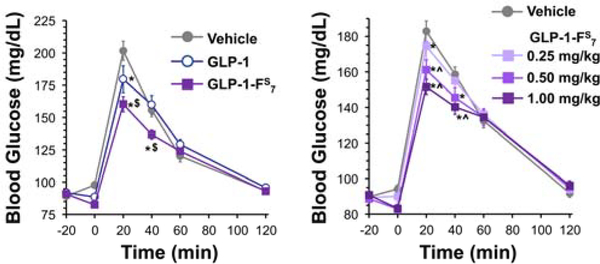
To confirm the in vivo bioactivity of GLP-1-FS7, and as a preliminary step towards evaluating its potential pharmacotherapeutic relevance, the effects of GLP-1-FS7 on glycemic control were compared to the effects of native GLP-1 in adult male Sprague-Dawley rats. Rats were utilized as the animal model for two reasons: 1) their slower metabolism relative to mice provides a more translatable test of pharmacokinetics, and 2) unlike mice, rats recapitulate many of the behavioral and physiological profiles of GLP-1 signaling found in humans.64–67 The glycemic profile of GLP-1-FS7 was compared to that of native GLP-1 in an oral glucose tolerance test (OGTT). Rats were fasted overnight, and OGTT was initiated shortly after the onset of the dark phase. After measuring fasting blood glucose levels using a standard glucometer (time =−20 min), rats (n=16) received intraperitoneal (IP) injection of vehicle (1 mL/kg sterile 0.9% NaCl, pH 7.0–7.4), GLP-1 (0.5 mg/kg), or GLP-1-FS7 (0.5 mg/kg). Twenty min later (time = 0 min), blood glucose was measured again, and each rat received an oral glucose load (2 g/kg) by gavage. Blood glucose was measured again at 20, 40, 60, and 120 min post-glucose load to evaluate the effects of each compound on glycemic control. As expected, native GLP-1 produced a significant reduction in blood glucose levels at 20 min post-glucose load (Fig. 3, Left). Intriguingly, GLP-1-FS7 suppressed blood glucose at 20 and 40 min post-glucose load, and was significantly more effective at lowering blood glucose levels at these times than native GLP-1. These data suggest that the glycemic benefits of GLP-1-FS7 are more potent and more durable than that of native GLP-1 when administered at doses near the GLP-1 EC50 in rats (0.5 mg/kg). This suggests that the glycemic benefits of GLP-1 are retained and even enhanced with thioamide modification, and furthermore, demonstrates that the equipotency seen in cellular assays (Fig. 2, Bottom Right) is maintained in vivo.
Figure 3.
Thioamide-Modified GLP-1 Improves Glycemic Control in Rats. Left: In an oral glucose tolerance test (OGTT), intraperitoneal injection of native GLP-1 (0.5 mg/kg) or GLP-1-FS7 (0.5 mg/kg) suppresses blood glucose levels in rats (n=16) after an oral glucose load (main effect of drug, F2,30=8.11, p<0.01; drug x time interaction, F10,150=5.93, p<0.0001). GLP-1-FS7 more potently and durably reduces blood glucose compared to GLP-1. Right: In a dose-response study of the effects of GLP-1-FS7 on glycemic control (n=16), intraperitoneal injection of compound doses ranging from 0.25–1.0 mg/kg are effective to suppress blood glucose levels (main effect of dose, F3,45=3.61, p<0.03; dose x time interaction, F15,225=4.77, p<0.0001). Higher doses (0.5–1.0 mg/kg) produced greater improvement in glycemic control compared to the lowest dose (0.25 mg/kg). *, p<0.05 compared to vehicle; $, p<0.05 compared to GLP-1; ^, p<0.05 compared to 0.25 mg/kg GLP-1-FS7. Bars represent standard error.
A limited dose-response experiment in the same rats (n=16) was also performed to evaluate the effects of different doses of GLP-1-FS7 on glycemic control. The methods were the same as the first OGTT, except only GLP-1-FS7 was administered at doses ranging from 0.25–1.0 mg/kg (vehicle = 1 mL/kg sterile 0.9% NaCl, pH 7.0–7.4). The results showed that all doses of GLP-1-FS7 tested significantly suppressed blood glucose levels at 20 min after glucose gavage compared to vehicle, while the two higher doses (0.5 and 1.0 mg/kg) continued to reduce blood glucose at 40 min post-gavage (Fig. 3, Right). The two higher doses of GLP-1-FS7 tested also suppressed blood glucose levels significantly more than did the 0.25 mg/kg dose, indicating a dose-responsive effect of the compound on glycemic control.
In preliminary investigations of the pharmacokinetics of GLP-1-FS7, we have performed stability tests in mouse serum and found that its half-life is ~2.5 h, five-fold longer than the half-life of GLP-1-F7 (~35 min). While this shows that protection against DPP-4 proteolysis is sufficient to significantly stabilize the functional form of the peptide, it also implies that other proteases play a role in the degradation of GLP-1-FS7. (Supporting Information, Fig. S10) The longer half-life of GLP-1-F7 itself in serum may be due to binding to serum album proteins. However, addition of albumin to a cleavage assay with purified DPP-4 did not prolong the half-life of GLP-1-F7, so we hypothesize instead that the concentration of active DPP-4 is lower in serum than in our purified DPP-4 assays. Degradation by other proteases in vivo may explain why the GLP-1-FS7 effects in the OGTT assays were not as long-lived as one might have anticipated from the in vitro data. Further stabilization could potentially be achieved with thioamide modification at other protease sites or with attachment of hydrophobic sidechains (e.g., X and Z in Fig. 1) for serum albumin binding.
In order to show that the effects of thioamide modification seen in GLP-1 extend to other DPP-4 substrates, we synthesized versions of GIP with thioamide modifications at the P2 or P1 position. For these peptides, we observed no significant auto-degradation, further confirming that the phenomenon observed with GLP-1-AS8 is unique to a peptide with an N-terminal His and a thioamide at the 2 position. DPP-4 proteolytic stability was tested for each thiopeptide and its parent oxopeptide using in vitro HPLC and MALDI MS assays identical to those performed for GLP-1.
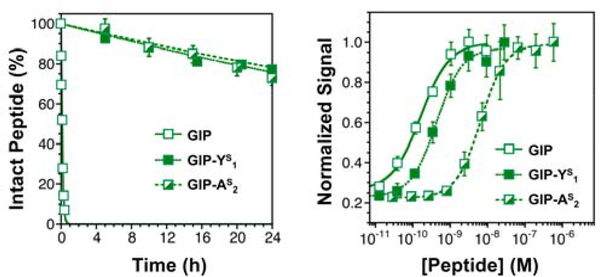
For GIP, modification at the P2 (GIP-YS1) or the P1 (GIP-AS2) position increased peptide half-life in the presence of 2.5 ng/mL DPP-4 to 68 h and 60 h, respectively. (Fig. 4, Left and Table 1) The approximately 750-fold increase in half-life (GIP: 5 min) for these GIP thiopeptides is significantly greater than for GLP-1-F7 thiopeptides. Like GLP-1-FS7 (P2) and GLP-1-AS8 (P1), GIP-YS1 is not a competitive inhibitor of DPP-4 (measured using Ala-Pro-pNA, see Supporting Information, Fig. S12). To measure the potency of GIP-YS1 and GIP-AS2, we again turned to cAMP reporter cell lines. The results of dose-response experiments were similar to those for GLP-1. P1-substituted GIP-AS2 was 40-fold less potent than GIP, but P2-substituted GIP-YS1 was nearly equipotent (Fig. 4, Right and Table 1). This is perhaps not surprising, since the mode of GIP binding to GIPR is believed to be similar GLP-1 binding to GLP-1R.68 However, since this region is not observable at high resolution in the available structures of GLP-1R or other glucagon-like receptors, the precise reasons for P1 being disruptive and P2 being tolerated are not clear.69–72
Figure 4.
Thioamide Substitution Stabilizes GIP Without Disrupting Activity. Left: Proteolysis data demonstrating that GIP-YS1 and GIP-AS2 are cleaved by 2.5 ng/mL DPP-4 more slowly than GIP. Right: Dose response curves for GIP, GIP-YS1, and GIP-AS2, obtained using DiscoveRx GIPR reporter cells, which detect cAMP production following GIPR activation in an enzyme-coupled assay. See Supporting Information for details of both assays, see a list of half-lives and EC50 values in Table 1. Bars represent standard error.
DISCUSSION
These studies show that the single atom modification of thioamide substitution in GLP-1 and GIP can disrupt interactions with a protease without significantly disrupting interactions with that peptide’s receptor. Substitution at P2 or P1 led to a very large decrease in proteolysis rate. For both peptides, we saw that thioamide substitution at P2 had little effect on receptor recognition, but that substitution at P1 decreased the EC50 by 11–40 fold. In the case of GLP-1, we have shown that the increased proteolytic stability of GLP-1-FS7 translated into increased potency in vivo. While there have been a small number of previous animal experiments with peptidyl thioamides, our study demonstrates the clearest evidence for their utility in improving the pharmacotherapeutic utility of bioactive peptides. A full understanding of the pharmacokinetics of thiopeptides will require further study, but we can analyze the in vitro stability and activity of the peptides here in terms of the physical properties of the thioamide and existing structural information.
The intrinsic reactivity of thioamides with nucleophiles is greater than that of oxoamides.73 This is clearly illustrated by the autodegradation of GLP-1-AS8. Thus, the physical basis for thiopeptide proteolytic stability lies not in a difference in reactivity, but in an inability for the thiopeptide to bind to the protease in a productive conformation. It has been shown that peptide backbone thioamides can have highly variable effects on protein structure and stability, depending on local interactions. We have recently performed a comprehensive study in several full-length proteins, and other research groups have previously studied thioamide effects in small proteins and model peptides.74–80 Taken together, these studies demonstrate that the longer C=S bond length (1.71 vs. 1.23 Å) and larger van der Waals radius of sulfur (1.85 vs. 1.40 Å) can lead thioamide substitutions to be disruptive, in some cases weakening interactions by more than 2 kcal/mol. In other cases, the stronger hydrogen bond donor of the thioamide N-H group can compensate for this effect or even lead to net stabilization by thioamide substitution (up to 0.6 kcal/mol). Indeed, Raines showed that thioamides undergo favorable n-to-π* interactions, a stabilizing force in polyproline-type folds.81 This knowledge base can be applied to analyze substrate recognition interactions in an existing DPP-4 structure. Unfortunately, in spite of exciting recent advances in X-ray crystal and cryo-electron microscopy structures of GLP-1R, no structures with sufficient resolution in the GLP-1 N-terminal region exist to perform a similar analysis of receptor activation.72, 82
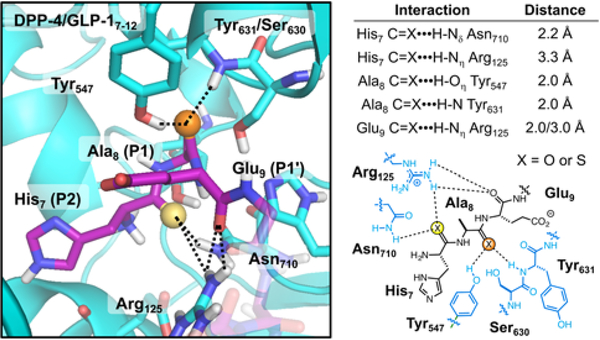
Although many high resolution structures of the DPP-4 enzyme are available with small molecule inhibitors bound, there is only one structure with a peptide substrate bound, a fragment of neuropeptide Y.83 One can use this structure to attempt to understand the effects of thioamide substitution on DPP-4 recognition of GLP-1 through computational modeling. (Fig. 5) A structure of the GLP-1 N-terminus bound to DPP-4 was generated using “FlexPepDock” in the Rosetta computational suite.84–85 The interactions seen in this modeled structure are similar to those seen in the experimental neuropeptide Y-bound structure (see Supporting Information, Fig. S14). Hydrogen bonds of the P2 carbonyl (His7) can be observed with the sidechains of Arg125 and Asn710. The P1 carbonyl (Ala8) accepts hydrogen bonds from the Tyr547 sidechain phenol group and the Ser630/Tyr631 backbone amide. It is not surprising that these relatively short, bifurcated hydrogen bonds would be disrupted by the ~0.5 Å larger thiocarbonyl bond. Since there are no interactions that take advantage of the stabilizing nature of the stronger thioamide N-H donor, it is understandable that we observe a >500-fold weaker interaction with DPP-4 in the P1 and P2 thiopeptides. This would be consistent with a net decrease in DPP-4 binding energy of ~4 kcal/mol, among the largest destabilization effects observed in thioamide peptides and proteins.74, 78, 86 In contrast, the P1’ backbone carbonyl (Glu9) only has significant hydrogen bonding interactions with Arg125 in DPP-4, and these interactions with a single, flexible sidechain may be more tolerant of small changes in geometry. Thus, we believe that we can rationalize the thioamide substitution effects observed for DPP-4 proteolysis.
Figure 5.
Structural Analysis of the Impact of Thioamide Substitution on DPP-4 Substrate Recognition. Left: An image of the DPP-4 (cyan) active site with a GLP-1 N-terminal fragment (purple) bound, modeled based on the neuropeptide Y bound DPP-4 structure in PDB entry 1R9N.83 The P2 and P1 carbonyl oxygens are highlighted as yellow and orange spheres, respectively. Key interactions with DPP-4 are shown as dashed lines. Right: Distances for the interactions shown at left with a schematic representation of the P2 and P1 binding site. See Supporting Information for a detailed discussion of DPP-4 active site modeling results.
The above noted previous DPP-4 studies by Fischer showed that cis/trans isomerization played a prominent role in suppressing proteolysis and that deacylation was the rate-limiting step for both thiopeptide and oxopeptide DPP-4 substrates.50 The relative import of these effects may be different for our full length peptide substrates and their Ala-Pro-pNA substrates, as it has been previously shown that mutational effects have much more dramatic effects in DPP-4 dipeptide fluorescent reporter substrates than in full length peptides.87 The fact that GLP-1-FS7, GLP-1-AS8, and GIP-YS1 do not act as competitive inhibitors supports the idea the thioamides disrupt an initial DPP-4 binding step rather than a later catalytic step. Further mechanistic study may find that multiple thioamide properties are significant. We are systematically studying the positional effects of thioamides in substrates of DPP-4 and several canonical proteases in order to arrive at a structure-based analysis that will eventually allow users to predict useful thioamide locations for preventing proteolysis.
CONCLUSIONS
Thioamide substitution shows potential as a simple and effective way of stabilizing injectable peptides toward proteolysis while maintaining receptor binding. This stability derives from local disruptions that allow thioamides to selectively perturb interactions of a peptide with the protease. In fact, since DPP-4 recognition and receptor activation take place over similar regions of GLP-1 and GIP, these peptides serve as a very stringent test of the ability of thioamides to selectively perturb certain protein interactions. Such interactions need not be only with proteases; our preliminary studies of β-arrestin activation show that we can also subtly affect receptor interactions to alter signaling bias. Although the effect of the O-to-S substitution on GLP-1-F7 signaling bias is convoluted with the effect of the His-to-Phe mutation, in all cases tested, the thioamide significantly reduced β-arrestin 1 and 2 activation without altering affinity. Further investigations in other G-protein coupled receptor ligands are warranted. Regardless of the precise effect of the thioamide, GLP-1-F7 represents a potentially valuable tool for understanding the physiological effects of changes in GLP-1R signaling bias. A peptide with a similar signaling bias profile was reported by Lerner and coworkers, which interestingly lacked the appetite suppressive effects of GLP-1 and many other GLP-1 analogs.88 The stability and unusual signaling bias of GLP-1-F7 may help us to understand the basis for different aspects of native GLP-1 signaling.
In considering the value of thioamides to stabilizing peptides, it must be noted that there are other factors that affect in vivo peptide lifetimes, such as renal clearance. Thioamide modifications can easily be combined with strategies for combatting renal clearance, such as attachment of hydrophobic sidechains (e.g., X and Z in Fig. 1) for serum albumin binding. Thioamide modifications are ideal for some important synthetic peptide applications, such as imaging receptor populations, where one wishes to prevent proteolysis without increasing serum binding. For optimal imaging, the injected peptide should be either intact and bound to its target receptor or eliminated from the blood stream to reduce background. We also anticipate that thioamides will be highly useful for stabilizing newly discovered signaling peptides toward key proteolytic events for early in vivo investigations of their signaling activity.
Supplementary Material
Table 2.
β-Arrestin Activation by GLP-1 and Its Analogs
| β-Arrestin 1 |
β-Arrestin 2 |
|||
|---|---|---|---|---|
| Peptide | EC50 (nM)a | % Max.b | EC50 (nM)a | % Max.b |
| GLP-1 | 07.9 ± 0.7 | - | 005.9 ± 0.4 | - |
| GLP-1-AS8 | 220 ± 14 | 58 | 188 ± 7 | 84 |
| GLP-1-F7 | 72 ± 2 | 35 | 068 ± 2 | 48 |
| GLP-1-FS7 | 088 ± 16 | 16 | 0083 ± 13 | 25 |
Values represent mean ± standard error.
Maximum response computed by normalizing to response to saturating GLP-1 concentrations.
ACKNOWLEDGMENT
This work was supported by funding from the University of Pennsylvania, as well as a pilot grant to EJP and MRH (National Institutes of Health [NIH], National Center for Research Resources, Grant UL1RR024134, and now at the National Center for Advancing Translational Sciences, Grant UL1TR000003), NIH-DK096139 (MRH). EJP acknowledges additional support for the general development of thioamides from the National Science Foundation (NSF CHE-1150351). TMB thanks the NIH for funding through the Chemistry Biology Interface Training Program (T32 GM071399). Instruments supported by the NIH and the National Science Foundation include: MALDI MS (NSF MRI-0820996) and CD (NSF DMR05-20020). We thank Alan Saghatelian and Amanda McFedries for valuable discussions and early thioamide GLP-1 studies in mice.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Descriptions of materials and methods, including peptide synthesis, characterization, additional details on degradation assays, and animal experiments. The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- 1.Mullard A, Nat. Rev. Drug Discov. 2013, 12 (5), 329–332. [DOI] [PubMed] [Google Scholar]
- 2.Projan SJ; Gill D; Lu Z; Herrmann SH, Expert Opin. Biol. Ther. 2004, 4 (8), 1345–1350. [DOI] [PubMed] [Google Scholar]
- 3.Verdine GL; Walensky LD, Clin. Cancer Res. 2007, 13 (24), 7264–7270. [DOI] [PubMed] [Google Scholar]
- 4.Buse JB; Rosenstock J; Sesti G; Schmidt WE; Montanya E; Brett JH; Zychma M; Blonde L, Lancet 2009, 374 (9683), 39–47. [DOI] [PubMed] [Google Scholar]
- 5.Kreymann B; Ghatei M; Williams G; Bloom S, Lancet 1987, 330 (8571), 1300–1304. [DOI] [PubMed] [Google Scholar]
- 6.Weber AE, J. Med. Chem. 2004, 47 (17), 4135–4141. [DOI] [PubMed] [Google Scholar]
- 7.DeFronzo RA; Ratner RE; Han J; Kim DD; Fineman MS; Baron AD, Diabetes Care 2005, 28 (5), 1092–1100. [DOI] [PubMed] [Google Scholar]
- 8.Kratz F, Control J. Release 2008, 132 (3), 171–183. [DOI] [PubMed] [Google Scholar]
- 9.Kratz F, Control J. Release 2014, 190, 331–336. [DOI] [PubMed] [Google Scholar]
- 10.Bird GH; Madani N; Perry AF; Princiotto AM; Supko JG; He X; Gavathiotis E; Sodroski JG; Walensky LD, Proc. Natl. Acad. Sci. USA 2010, 107 (32), 14093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato AK; Viswanathan M; Kent RB; Wood CR, Curr. Opin. Biotechnol. 2006, 17 (6), 638–642. [DOI] [PubMed] [Google Scholar]
- 12.We use the term thiopeptides to describe thioamide-containing peptides while this term is often also used to describe thiazole- and thiazolene-containing natural products such as thiostrepton.
- 13.Koole C; Savage EE; Christopoulos A; Miller LJ; Sexton PM; Wootten D, Mol. Endocrinol. 2013, 27 (8), 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier JJ, Nat. Rev. Endocrinol. 2012, 8 (12), 728–742. [DOI] [PubMed] [Google Scholar]
- 15.Drucker DJ; Nauck MA, Lancet 2006, 368 (9548), 1696–1705. [DOI] [PubMed] [Google Scholar]
- 16.Kim W; Egan JM, Pharmacol. Rev. 2008, 60 (4), 470–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mentlein R; Gallwitz B; Schmidt WE, Eur. J. Biochem. 1993, 214 (3), 829–835. [DOI] [PubMed] [Google Scholar]
- 18.Manandhar B; Ahn JM, J. Med. Chem. 2015, 58 (3), 1020–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadini GP; Simioni N; Frison V; Dal Pos M; Bettio M; Rocchini P; Avogaro A, Acta Diabetol. 2013, 50 (6), 943–949. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen LL; Young AA; Parkes DG, Regul. Pept. 2004, 117 (2), 77–88. [DOI] [PubMed] [Google Scholar]
- 21.Deacon CF; Nauck MA; Meier J; Hucking K; Holst JJ, J. Clin. Endocrinol. Metab. 2000, 85 (10), 3575–3581. [DOI] [PubMed] [Google Scholar]
- 22.Ussher JR; Drucker DJ, Endocr. Rev. 2012, 33 (2), 187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holzer P; Reichmann F; Farzi A, Neuropeptides 2012, 46 (6), 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schjoldager BTG; Baldissera FGA; Mortensen PE; Holst JJ; Christiansen J, Eur. J. Clin. Invest. 1988, 18 (5), 499–503. [DOI] [PubMed] [Google Scholar]
- 25.Clerico A; Iervasi G; Mariani G, Horm. Metab. Res. 1999, 31 (9), 487–498. [DOI] [PubMed] [Google Scholar]
- 26.Bouras M; Huneau JF; Luengo C; Erlansonalbertsson C; Tome D, Peptides 1995, 16 (3), 399–405. [DOI] [PubMed] [Google Scholar]
- 27.Faria ACS; Veldhuis JD; Thorner MO; Vance ML, J. Clin. Endocrinol. Metab. 1989, 68 (3), 535–541. [DOI] [PubMed] [Google Scholar]
- 28.Hokfelt T; Pernow B; Wahren J, J. Intern. Med. 2001, 249 (1), 27–40. [DOI] [PubMed] [Google Scholar]
- 29.Mistrova E; Kruzliak P; Dvorakova MC, Neuropeptides 2016, 58, 41–51. [DOI] [PubMed] [Google Scholar]
- 30.O’Connor TM; O’Connell J; O’Brien DI; Goode T; Bredin CP; Shanahan F, J. Cell. Physiol. 2004, 201 (2), 167–180. [DOI] [PubMed] [Google Scholar]
- 31.Mentlein R, Regul. Pept. 1999, 85 (1), 9–24. [DOI] [PubMed] [Google Scholar]
- 32.Ohnuma K; Takahashi N; Yamochi T; Hosono O; Dang NH; Morimoto C, Front. Biosci. 2008, 13, 2299–2310. [DOI] [PubMed] [Google Scholar]
- 33.Day JW; Ottaway N; Patterson JT; Gelfanov V; Smiley D; Gidda J; Findeisen H; Bruemmer D; Drucker DJ; Chaudhary N; Holland J; Hembree J; Abplanalp W; Grant E; Ruehl J; Wilson H; Kirchner H; Lockie SH; Hofmann S; Woods SC; Nogueiras R; Pfluger PT; Perez-Tilve D; DiMarchi R; Tschop MH, Nat. Chem. Biol. 2009, 5 (10), 749–757. [DOI] [PubMed] [Google Scholar]
- 34.Hojberg PV; Vilsboll T; Rabol R; Knop FK; Bache M; Krarup T; Holst JJ; Madsbad S, Diabetologia 2009, 52 (2), 199–207. [DOI] [PubMed] [Google Scholar]
- 35.Meier JJ; Gallwitz B; Askenas M; Vollmer K; Deacon CF; Holst JJ; Schmidt WE; Nauck MA, Diabetologia 2005, 48 (9), 1872–1881. [DOI] [PubMed] [Google Scholar]
- 36.Skow MA; Bergmann NC; Knop FK, Diabetes Obes. Metab. 2016, 18 (9), 847–854. [DOI] [PubMed] [Google Scholar]
- 37.Ward BP; Ottaway NL; Ma D; Gelfanov VM; Perez-Tilve DP; Giedroc DP; Tschop MH; DiMarchi RD Structural Changes Associated with Peptide Lipidation Broaden Biological Function PDB ID: 2M5P. URL: http://www.rcsb.org/pdb/explore.do?structureId=2m5p.
- 38.Blair HA; Keating GM, Drugs 2015, 75 (6), 651–663. [DOI] [PubMed] [Google Scholar]
- 39.Trujillo JM; Nuffer W, Ann. Pharmacother. 2014, 48 (11), 1494–1501. [DOI] [PubMed] [Google Scholar]
- 40.Deacon CF; Knudsen LB; Madsen K; Wiberg FC; Jacobsen O; Holst JJ, Diabetologia 1998, 41 (3), 271–278. [DOI] [PubMed] [Google Scholar]
- 41.Idris I, Diabetes Obes. Metab. 2016, 18 (3), 312–313. [Google Scholar]
- 42.Lau J; Bloch P; Schaffer L; Pettersson I; Spetzler J; Kofoed J; Madsen K; Knudsen LB; McGuire J; Steensgaard DB; Strauss HM; Gram DX; Knudsen SM; Nielsen FS; Thygesen P; Reedtz-Runge S; Kruse T, J. Med. Chem. 2015, 58 (18), 7370–7380. [DOI] [PubMed] [Google Scholar]
- 43.Johnson LM; Barrick S; Hager MV; McFedries A; Homan EA; Rabaglia ME; Keller MP; Attie AD; Saghatelian A; Bisello A; Gellman SH, J. Am. Chem. Soc. 2014, 136 (37), 12848–12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charron CL; Hickey JL; Nsiama TK; Cruickshank DR; Turnbull WL; Luyt LG, Nat. Prod. Rep. 2016, 33 (6), 761–800. [DOI] [PubMed] [Google Scholar]
- 45.Bartlett PA; Spear KL; Jacobsen NE, Biochemistry 1982, 21 (7), 1608–1611. [DOI] [PubMed] [Google Scholar]
- 46.Bond MD; Holmquist B; Vallee BL, J. Inorg. Biochem. 1986, 28 (2–3), 97–105. [DOI] [PubMed] [Google Scholar]
- 47.Maziak L; Lajoie G; Belleau B, J. Am. Chem. Soc. 1986, 108 (1), 182–183. [Google Scholar]
- 48.Beattie RE; Elmore DT; Williams CH; Guthrie DJS, Biochem. J. 1987, 245 (1), 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foje KL; Hanzlik RP, Biochim. Biophys. Acta 1994, 1201 (3), 447–453. [DOI] [PubMed] [Google Scholar]
- 50.Schutkowski M; Neubert K; Fischer G, Eur. J. Biochem. 1994, 221 (1), 455–461. [DOI] [PubMed] [Google Scholar]
- 51.McElroy J; Guthrie DJS; Hooper NM; Williams CH, Biochem. Soc. Trans. 1998, 26 (1), S31–S31. [DOI] [PubMed] [Google Scholar]
- 52.Yao S; Zutshi R; Chmielewski J, Bioorg. Med. Chem. Lett. 1998, 8 (6), 699–704. [DOI] [PubMed] [Google Scholar]
- 53.Stöckel-Maschek A; Mrestani-Klaus C; Stiebitz B; Demuth H-U; Neubert K, Biochim. Biophys. Acta 2000, 1479 (1–2), 15–31. [DOI] [PubMed] [Google Scholar]
- 54.Zacharie B; Lagraoui M; Dimarco M; Penney CL; Gagnon L, J. Med. Chem. 1999, 42 (11), 2046–2052. [DOI] [PubMed] [Google Scholar]
- 55.Lankiewicz L; Bowers CY; Reynolds GA; Labroo V; Cohen LA; Vonhof S; Siren AL; Spatola AF, Biochem. Biophys. Res. Commun. 1992, 184 (1), 359–366. [DOI] [PubMed] [Google Scholar]
- 56.Zacharie B; Martel R; Sauve G; Belleau B, Bioorg. Med. Chem. Lett. 1993, 3 (4), 619–624. [Google Scholar]
- 57.Jeschke P; Harder A; Etzel W; Gau W; Thielking G; Bonse G; Iinuma K, Pest Manag. Sci. 2001, 57 (11), 1000–1006. [DOI] [PubMed] [Google Scholar]
- 58.Zhang W; Li J; Liu LW; Wang KR; Song JJ; Yan JX; Li ZY; Zhang BZ; Wang R, Peptides 2010, 31 (10), 1832–1838. [DOI] [PubMed] [Google Scholar]
- 59.Wang YJ; Szantai-Kis DM; Petersson EJ, Org. Biomol. Chem. 2015, 13 (18), 5074–81. [DOI] [PubMed] [Google Scholar]
- 60.Szantai-Kis DM; Walters CR; Barrett TM; Hoang EM; Petersson EJ, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goldberg JM; Batjargal S; Petersson EJ, J. Am. Chem. Soc. 2010, 132 (42), 14718–14720. [DOI] [PubMed] [Google Scholar]
- 62.Batjargal S; Wang YJ; Goldberg JM; Wissner RF; Petersson EJ, J. Am. Chem. Soc. 2012, 134 (22), 9172–9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarrauste de Menthière C; Chavanieu A; Grassy G; Dalle S; Salazar G; Kervran A; Pfeiffer B; Renard P; Delagrange P; Manechez D; Bakes D; Ktorza A; Calas B, Eur. J. Med. Chem. 2004, 39 (6), 473–480. [DOI] [PubMed] [Google Scholar]
- 64.Hayes MR; Mietlicki-Baase EG; Kanoski SE; De Jonghe BC, Annu. Rev. Nutr. 2014, 34, 237–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayes MR, Physiol. Behav. 2012, 106 (3), 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanoski SE; Rupprecht LE; Fortin SM; De Jonghe BC; Hayes MR, Neuropharmacology 2012, 62 (5–6), 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayes MR; De Jonghe BC; Kanoski SE, Physiol. Behav. 2010, 100 (5), 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seino Y; Fukushima M; Yabe D, Diabetes Investig J. 2010, 1 (1–2), 8–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Underwood CR; Garibay P; Knudsen LB; Hastrup S; Peters GH; Rudolph R; Reedtz-Runge S, J. Biol. Chem. 2010, 285 (1), 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siu FY; He M; de Graaf C; Han GW; Yang D; Zhang Z; Zhou C; Xu Q; Wacker D; Joseph JS; Liu W; Lau J; Cherezov V; Katritch V; Wang M-W; Stevens RC, Nature 2013, 499 (7459), 444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang D; de Graaf C; Yang L; Song G; Dai A; Cai X; Feng Y; Reedtz-Runge S; Hanson MA; Yang H; Jiang H; Stevens RC; Wang M-W, J. Biol. Chem. 2016, 291 (25), 12991–13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y; Sun B; Feng D; Hu H; Chu M; Qu Q; Tarrasch JT; Li S; Sun Kobilka T; Kobilka BK; Skiniotis G, Nature 2017, 546 (7657), 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jagodziński TS, Chem. Rev. 2003, 103 (1), 197–228. [DOI] [PubMed] [Google Scholar]
- 74.Walters CR; Szantai-Kis DM; Zhang Y; Reinert ZE; Horne WS; Chenoweth DM; Petersson EJ, Chem. Sci. 2017, 8 (4), 2868–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miwa JH; Patel AK; Vivatrat N; Popek SM; Meyer AM, Org. Lett. 2001, 3 (21), 3373–5. [DOI] [PubMed] [Google Scholar]
- 76.Miwa JH; Pallivathucal L; Gowda S; Lee KE, Org. Lett. 2002, 4 (26), 4655–4657. [DOI] [PubMed] [Google Scholar]
- 77.Wildemann D; Schiene-Fischer C; Aumuller T; Bachmann A; Kiefhaber T; Lucke C; Fischer G, J. Am. Chem. Soc. 2007, 129 (16), 4910–4918. [DOI] [PubMed] [Google Scholar]
- 78.Newberry RW; VanVeller B; Raines RT, Chem. Commun. 2015, 51 (47), 9624–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Culik RM; Jo H; DeGrado WF; Gai F, J. Am. Chem. Soc. 2012, 134 (19), 8026–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bachmann A; Wildemann D; Praetorius F; Fischer G; Kiefhaber T, Proc. Natl. Acad. Sci. USA 2011, 108 (10), 3952–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newberry RW; VanVeller B; Guzei IA; Raines RT, J. Am. Chem. Soc. 2013, 135 (21), 7843–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song G; Yang D; Wang Y; de Graaf C; Zhou Q; Jiang S; Liu K; Cai X; Dai A; Lin G; Liu D; Wu F; Wu Y; Zhao S; Ye L; Han GW; Lau J; Wu B; Hanson MA; Liu Z-J; Wang M-W; Stevens RC, Nature 2017, 546 (7657), 312–315. [DOI] [PubMed] [Google Scholar]
- 83.Aertgeerts K; Ye S; Tennant MG; Kraus ML; Rogers J; Sang BC; Skene RJ; Webb DR; Prasad GS, Protein Sci. 2004, 13 (2), 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaufmann KW; Lemmon GH; DeLuca SL; Sheehan JH; Meiler J, Biochemistry 2010, 49 (14), 2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raveh B; London N; Schueler-Furman O, Proteins 2010, 78 (9), 2029–2040. [DOI] [PubMed] [Google Scholar]
- 86.Reiner A; Wildemann D; Fischer G; Kiefhaber T, J. Am. Chem. Soc. 2008, 130 (25), 8079–8084. [DOI] [PubMed] [Google Scholar]
- 87.Kuhn-Wache K; Bar JW; Hoffmann T; Wolf R; Rahfeld JU; Demuth HU, Biol. Chem. 2011, 392 (3), 223–31. [DOI] [PubMed] [Google Scholar]
- 88.Zhang H; Sturchler E; Zhu J; Nieto A; Cistrone PA; Xie J; He L; Yea K; Jones T; Turn R; Di Stefano PS; Griffin PR; Dawson PE; McDonald PH; Lerner RA, Nat. Commun. 2015, 6, 8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.