Short abstract
Many labs have been developing cellular magnetic resonance imaging (MRI), using both superparamagnetic iron oxide nanoparticles (SPIONs) and fluorine-19 (19F)-based cell labels, to track immune and stem cells used for cellular therapies. Although SPION-based MRI cell tracking has very high sensitivity for cell detection, SPIONs are indirectly detected owing to relaxation effects on protons, producing negative magnetic resonance contrast with low signal specificity. Therefore, it is not possible to reliably quantify the local tissue concentration of SPION particles, and cell number cannot be determined. 19F-based cell tracking has high specificity for perfluorocarbon-labeled cells, and 19F signal is directly related to cell number. However, 19F MRI has low sensitivity. Magnetic particle imaging (MPI) is a new imaging modality that directly detects SPIONs. SPION-based cell tracking using MPI displays great potential for overcoming the challenges of MRI-based cell tracking, allowing for both high cellular sensitivity and specificity, and quantification of SPION-labeled cell number. Here we describe nanoparticle and MPI system factors that influence MPI sensitivity and resolution, quantification methods, and give our perspective on testing and applying MPI for cell tracking.
Keywords: magnetic particle imaging, magnetic resonance imaging, cell tracking, superparamagnetic iron oxide, quantification
Introduction
This paper presents our insights on magnetic particle imaging (MPI) as an emerging cell tracking modality. Interest in the use of MPI for cell tracking is growing. Our lab has a unique perspective, as we have been developing and applying magnetic resonance imaging (MRI) cell tracking techniques for the past 15 years. Here we describe the motivation for using MPI for cell tracking, the importance of nanoparticle sensitivity and resolution, and the methods for quantitation of iron mass and cell number from MPI. We also include discussion related to key issues facing the advancement of MPI and areas for future research.
In Vivo Cell Tracking
Cellular therapies are now recognized as effective medicines for treatment of a specific subset of cancers as well as a growing list of autoimmune, degenerative, genetic, and infectious diseases. Cell therapy refers to the administration of immune cells (such as T cells or dendritic cells), which are used to treat cancer, and stem cells (such as mesenchymal stem cells or pluripotent stem cells), which have numerous potential applications including in the treatment of amyotrophic lateral sclerosis, diabetes, graft-vs-host disease, kidney disease, liver disease, multiple sclerosis, myocardial disease, osteoarthritis, Parkinson disease, spinal cord injury, and stroke. Numerous cell therapy clinical trials are underway.
Despite the immense promise of cell therapies, clinical results have been inconsistent and discordant owing to variations in cell source, preparation, and route of administration/implantation methodology. Misinjections also contribute to failed cellular therapies (1). Many fundamental questions about the presence, numbers, persistence, and delivery of cells remain unanswered. In cancer immunotherapy, the magnitude of an antitumor response is proportional to the quantity of antigen-presenting cells that reach a target lymph node (2–4), and therefore, it is crucial to know whether the injected cells have migrated to the target and how many. In stem cell therapy, the survival and persistence of cells at the implant site can be used to inform whether a patient may need repeat dosing or other interventions. In vivo cellular imaging tools have the potential to answer these questions and improve the safety and success of cell therapies. The ideal imaging modality for monitoring cell therapies would be non-invasive, non-ionizing, sensitive enough to allow detection of a few hundred cells, specific, and importantly, quantitative - providing a measure of cell number.
MRI has been widely used for in vivo cell tracking. Cellular MRI uses contrast agents for labeling specific cells, thereby enhancing their detectability (5, 6). The most commonly used agents for cell tracking with MRI are magnetite (Fe2O3)-based superparamagnetic iron oxide nanoparticles (SPIONs). The presence of SPIONs in cells causes a distortion in the magnetic field and leads to abnormal signal hypointensities in iron-sensitive images (5). Areas containing SPION-labeled cells therefore appear as regions of low signal intensity on MRI images, creating negative contrast. Therapeutic cells, including mesenchymal stem cells (7, 8), progenitor cells (9), dendritic cells (10, 11), and pancreatic islets (12), have been labeled with SPIONs and tracked with MRI. The iron label has minimal impact on cell function or phenotype at a wide range of iron loading (13). This technique is highly sensitive, permitting the imaging of single cells in vivo, under ideal conditions (14). There are, however, several limitations of iron-based MRI cell tracking. The first is low specificity owing to other low-signal regions in MR images, such as the air-filled lungs or a region of hemorrhage. Although ultrashort echo time imaging methods have been developed for producing positive contrast from iron-labeled cells, these too have similar problems with specificity. Second, quantification of iron-induced signal loss is complicated, as the measure of the signal void volume is not linear with the number of cells.
Fluorine-19 (19F) MRI with perfluorocarbon (PFC) nanoemulsions to label cells has been also used for cell tracking (15, 16). 19F cell tracking addresses some of the limitations associated with iron-based cell tracking. First, the 19F signal is specific, as endogenous 19F is so low that there is no appreciable tissue background signal. Second, in contrast to the indirect visualization of SPIONs by observed proton signal loss, the spins of 19F nuclei are directly detected and image contrast is proportional to the number of 19F nuclei per voxel. Cell number can be determined if a measurement of 19F nuclei/cell is obtained by NMR spectroscopy; the 19F signal intensity for the cells of interest is compared to the 19F signal intensity of a reference tube containing a known 19F concentration and the NMR calibration value (17). The main limitation of 19F cell tracking is low sensitivity; thousands of PFC-labeled cells per voxel are required. Fluorine sensitivity improves with higher field strengths, and preclinical studies have reported in vivo detection of as few as 1000 cells per voxel (18). However, the first human clinical trial at 3 T showed a cellular detection limit between 1 and 10 million cells (19).
MPI is an emerging imaging modality that directly detects SPIONs (20, 21). MPI is built around a gradient magnet system. Two opposing electromagnets form strong gradient magnetic fields (in the order of T), and a field free region (FFR) is created in the position where these gradient fields cancel out. The gradient field (also known as the selection field) saturates the magnetization of all SPIONs except for SPIONs at the FFR, which experience no magnetic field. The FFR is shifted over an imaging volume, by changing the current through the electromagnets, to produce an image. When the FFR traverses a location containing SPIONs, the SPIONs’ magnetization changes nonlinearly in response to a secondary sinusoidal excitation magnetic field (in the order of mT). This change in SPION magnetization induces a voltage that is detected via a receiver coil, and the resulting signal can be assigned to the instantaneous FFR location to reconstruct the final image (22). The voltages induced are linearly proportional to the number of SPIONs at the FFR location, enabling quantification of SPIONs.
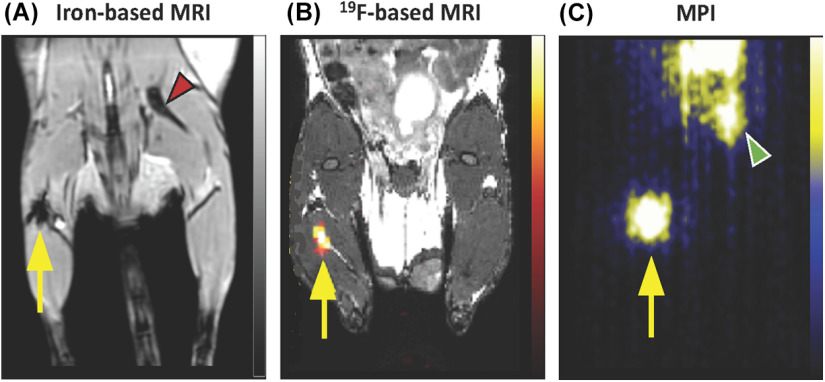
MPI cell tracking has the potential to address many of the limitations presented by SPION- and 19F-based cell tracking (23). First, as the MPI signal is generated only when the magnetic moments of the SPIONs rotate in response to the applied fields, there is no signal from tissue. This imbues MPI with a positive “hot-spot” contrast that provides spatial localization without ambiguity. Second, MPI has high sensitivity, as the signal derives from the direct detection of the electronic magnetization of SPION, which is 108 times larger than the nuclear magnetization of protons seen in MRI (24). Third, the MPI signal is linearly quantitative with SPION concentration, and therefore, the number of SPION-labeled cells can be calculated (25). The shortcomings of MPI include a relatively low spatial resolution, compared to MRI, and the requirement that anatomical images must be acquired separately with a different imaging modality. A comparison of mesenchymal stem cell (MSC) tracking with iron- and 19F-based MRI and MPI is shown in Figure 1. For a detailed description of the physics of MPI readers are referred to review articles by Saritas et al. (2013) (24), Zheng et al. (2017) (21), Yu et al. (2017) (26), Wu et al. (2019) (25), and Talebloo et al. (2020) (27).
Figure 1.
Comparison of iron-based magnetic resonance imaging (MRI), 19F -based MRI, and magnetic particle imaging (MPI) for cell tracking. Mesenchymal stem cells (MSCs) labeled with ferumoxytol (A, C) or perfluorocarbon (B) were implanted intramuscularly and imaged using 1H MRI (A), 19F MRI with overlay to anatomical 1H (B), or MPI (C). The signal associated with MSC is denoted by the yellow arrows in each image. The red arrow (A) shows dark signal coming from the bone, and this shows the lack of specificity of iron-based MRI. Likewise, the green arrow (C) points to MPI signal generated from iron contamination in the mouse digestive system. Images reprinted from Sehl et al. (36). Tomography 2019 (A, C), and Gaudet et al. (76). Magnetic Resonance in Medicine 2017 (B).
MPI Sensitivity and Resolution
Particle sensitivity in MPI refers to the lowest mass of SPION detected per imaging unit (eg, µg/mm3). For MPI cell tracking, the aim is to optimize cellular sensitivity, which refers to the lowest number of SPION-labeled cells detected per imaging unit (i.e., number of cells/mm3). The sensitivity of MPI depends on both nanoparticle and scanner specific factors. Nanoparticle factors include the strength of the nanoparticle magnetization (stronger magnetization improves MPI signal), the rate of SPION relaxation at the FFR (faster change in magnetization leads to higher MPI signal), and the efficiency of the nanoparticle cell labeling (more iron per cell leads to higher sensitivity). Scanner-specific factors include increasing the amplitude of the excitation field (28), decreasing the gradient strength (at the cost of resolution), and signal averaging. For example, increasing the overlap fraction or combining multiple projections (ie, 3D imaging) can improve cell detection. An important consideration is that cellular sensitivity in vitro may be enhanced compared with that in vivo. This outcome is because of the dispersion of cells from the site of administration and additional factors such as breathing motion.
The in vitro detection limit has been reported at 200 cells labeled with ferucarbotran; however, this was an estimate based on the detection of 1000 cells in a 100 µL in vitro suspension with SNR ∼5 (29). Song et al. (30) reported that as few as 250 cells, labeled with a custom-made MPI-tailored SPION (30 pg Fe/cell), could be detected in vivo if a background subtraction method was used. There is no doubt that cellular sensitivity will improve with further advances in acquisition strategies and SPION design. There is still considerable work to be done to show and evaluate MPI cellular detection limits more closely.
The MPI resolution is driven primarily by SPION relaxation. Neel relaxation refers to the reversal of the SPION magnetic moment, whereas Brownian relaxation refers to the physical rotation of the SPION, in response to the MPI excitation magnetic field (31–33). The dominant relaxation mechanism primarily depends on the SPION size, and the MPI resolution worsens with increasing Brownian relaxation. Theoretical modeling based upon the Langevin theory of SPIONs predicts that resolution improves with increasing core size. However, Tay et al. (32) found that improved resolution with increasing magnetic core size follows the steady-state prediction up to approximately 25 nm when the effects of SPION rotational times become significant owing to increasing Brownian relaxation. SPIONs above this size range experience increased drag, slowing their magnetization response and limiting resolution. Importantly, this work looked at single core mono-dispersed nanoparticles. Many of the SPIONs used for cell tracking are multidomain, clustering nanoparticles with more complicated physics.
Resolution is also influenced by the interaction of the nanoparticle and the magnetic field gradient. Stronger gradients increase resolution but at the cost of sensitivity. The spatial resolution of MPI using currently available SPIONs is approximately 1 mm (25), and by using optimized iron oxide nanoparticles, an MPI resolution of 200 µm has been shown (34). With further development, sensitivity and resolution should increase and significant improvements to cell detection limits will be possible.
SPIONs for MPI
As described above, both MPI sensitivity and resolution are closely related to the type of SPION; however, the ideal SPIONs for MPI are still not known. In early MPI cell tracking, commercially available SPIONs used for MRI were evaluated, including ferucarbotran and ferumoxtyol. Ferucarbotran improved MPI characteristics (35–37), and it has been used in MPI studies of mice to detect mesenchymal stem cells (35, 36, 38–40), neural stem cells (40), neural progenitor cells (29), pancreatic islets (41), T-cells (42), and macrophages (25, 37, 43). Although widely used, ferucarbotran is no longer considered optimal for MPI because it has a bimodal size distribution, predominantly containing small cores ∼5 nm in diameter (70%) with a small fraction (30%) of multicore aggregates with an effective size of 24 nm (44). The individual cores are too small to magnetize significantly and so the MPI signal predominately originates from the clustered multicore structures. Approaches to improve MPI sensitivity include increasing the fraction of these larger aggregates (44) or by fractionation of ferucarbotran (45). Lastly, the synthesis of homogeneously distributed single-core SPIONs with optimized core diameters is being investigated (46). Optimizing SPIONs expressly for MPI is emerging as a powerful area of research and will be critical for improving sensitivity and spatial resolution (32).
MPI relaxometry is commonly used as a first step to characterize SPIONs. MPI relaxometry measures the net magnetization and MPI relaxation rate of SPIONs, by turning off the selection field and applying a negative magnetic field and then a positive field, and back. SPIONs in a sample are driven from a negative magnetic saturation to positive, and vice versa. The output is the derivative of the Langevin function, also called the point spread function (PSF). The signal intensity, or height, of the PSF reflects the sensitivity of the SPION. The full-width half maximum (FWHM) relates to the spatial resolution of the SPION (47). A narrower tracer response indicates superior spatial resolution, and a greater signal intensity per mass of iron indicates superior sensitivity. In our experience, relaxometry has significant value for testing SPIONs before the use for MPI as certain SPIONs that may not, in theory, seem suitable for MPI show surprisingly good sensitivity (ie, micron-sized superparamagnetic polystrene beads, MPIO). In Figure 2 we have evaluated 4 commercially available SPIONs using MP relaxometry and MPI: ferucarbotran (Vivotrax, Magnetic Insight Inc., CA) with bimodal core size consisting of 70% 5 nm and 30% 24 nm, synomag-D (Micromod GmbH, Germany) with an average core size of ∼30 nm, MPIO (Bang’s beads, Bangs Laboratories, IN) with a core size of ∼1 µm, and ferumoxytol (Feraheme, AMAG pharmaceuticals, MA) with an average core size of 3.25 nm. Using the same iron mass, relaxometry indicated that the peak sensitivity of synomag-D is superior to MPIO, ferucarbotran, and ferumoxytol (in order). The PSFs also showed that the FWHM of ferucarbotran was superior to that of synomag-D, MPIO, and ferumoxytol (in order). Corresponding MPI images using a standard (5.7 T/m gradient) showed the same trends.
Figure 2.
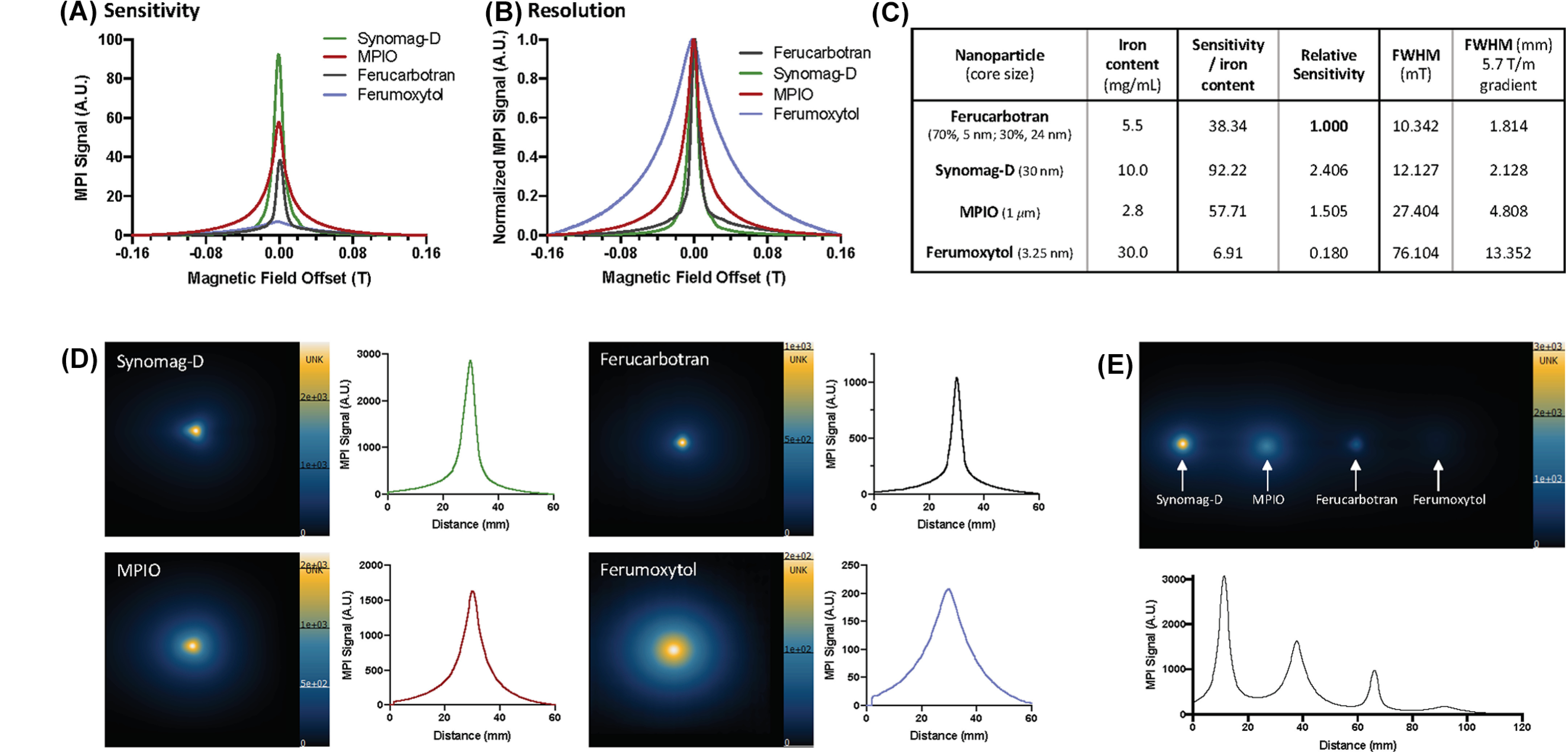
Comparison of commercially available superparamagnetic iron oxide nanoparticles (SPIONs) using MPI relaxometry. Point spread functions (PSFs) show that the relative sensitivity of synomag-D is superior to that of micron-sized superparamagnetic polystyrene beads (MPIO), ferucarbotran, and ferumoxytol (in order) per gram of iron content (A). Normalized PSFs show that the resolution (full-width half maximum; FWHM) of ferucarbotran is superior to that of synomag-D, MPIO, and ferumoxytol (in order) (B). Measurements of sensitivity and resolution for each nanoparticle are outlined in (C). MPI of each individual SPION (30 µg) and the corresponding intensity profile is shown in (D). A single MPI image and intensity profile of all 4 SPIONs (30 µg) show similar results (E). MPI images are displayed in full-dynamic range.
For cell tracking with MPI, it is also critical that the SPION of choice is taken up by cells effectively; not all SPIONs are. Effectively labeled cells have high intracellular iron load, to improve cellular sensitivity, and low extracellular iron. Extracellular iron in cell samples can lead to overestimation of iron content measured by MPI, and in vivo extracellular iron can be taken up by host cells leading to lower specificity. The coating of SPIONs influences their interaction with the cell membrane and the mechanism of uptake. Most SPIONs have a carbohydrate coat, typically dextran or carboxydextran. The carboxyl groups associated with ferucarbotrans lead to a high affinity to the cell membrane (48). Several other surface coating modification strategies have been shown to enhance internalization of SPIONs (49–51). Many cell types can be labeled in vitro simply by overnight coincubation with SPIONs, however, for certain cell types (ie, immune cells) and certain SPIONs [ie, ferumoxytols (52, 53)] coincubation alone is not effective. Transfection agents (TAs) are commonly used to facilitate or enhance cellular incorporation of SPIONs into cells, including poly-l-lysine, lipofectamine, and protamine sulfate (often with heparin) (54–56). TAs coat the surface of SPIONs and improve the rate and/or amount of iron taken up by cells. As shown in Figure 3, A–C, the addition of protamine sulfate and heparin significantly improves uptake of ferucarbotran by mesenchymal stem cells. However, SPION-TA complexes may clump cells together and can lead to higher amounts of free iron in cell preparations (56). It is also necessary to test whether TAs alter cell functionality or phenotype. For example, certain TAs reduce the immunopotency of dendritic cells (57). Importantly, we have recently observed that the MPI characteristics of SPIONs are altered by TAs. Figure 3, C–G, compares the MPI sensitivity and resolution of samples of ferucarbotran and ferucarbotran mixed with protamine sulfate/heparin. The addition of the TA leads to lower MPI signal and lower resolution images. This requires more study but may result from altered relaxation characteristics owing to clustering of iron cores. Therefore, while TAs may be used to achieve higher intracellular iron with the goal of improving MPI sensitivity, this gain may be partially counteracted by this effect.
Figure 3.
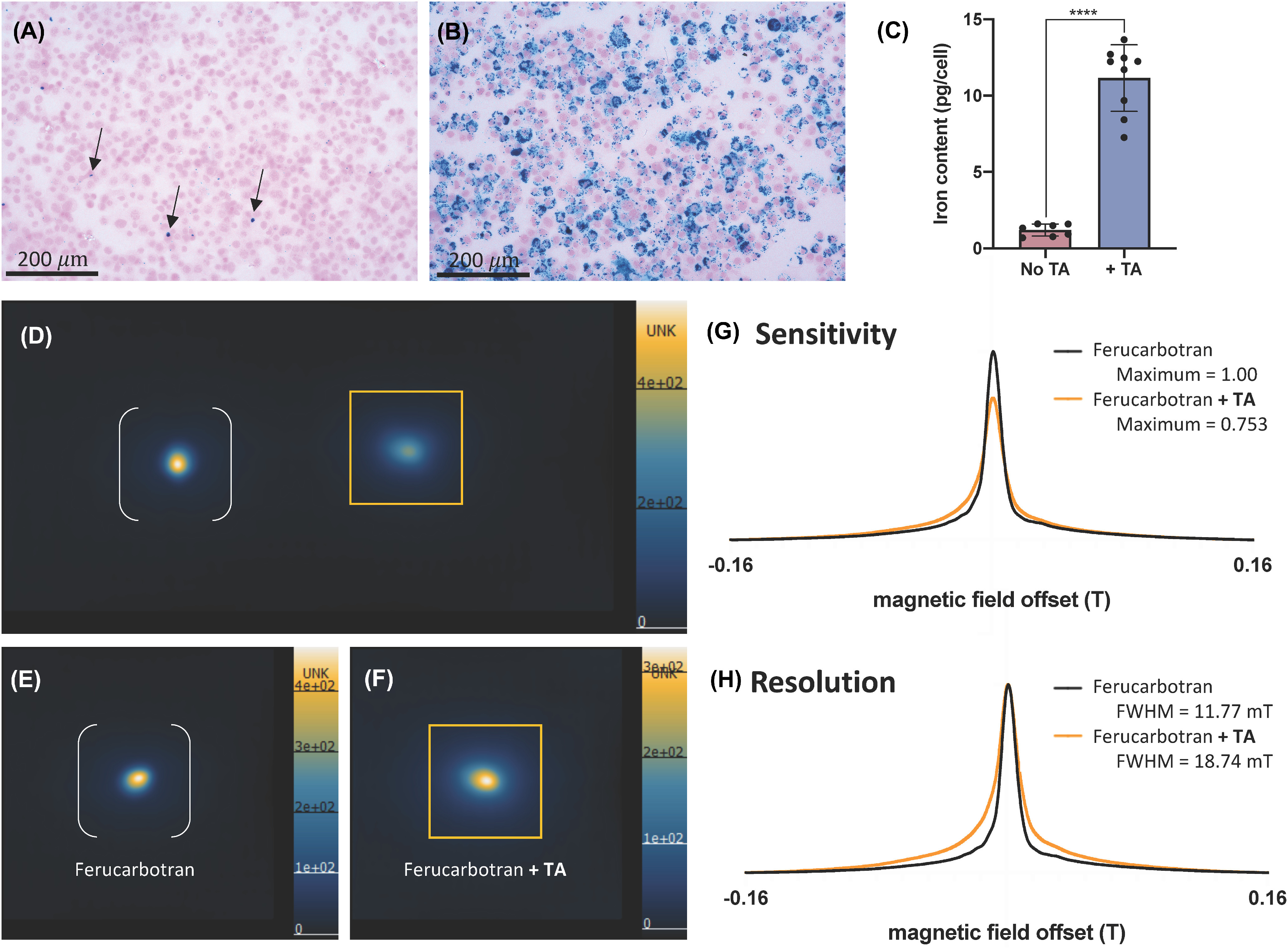
The effect of transfection agents on cell labeling and MPI signal. MSCs labeled with ferucarbotran shows sparse and nonuniform labeling (denoted by black arrows) (A), whereas transfection agents (TAs) protamine sulfate and heparin improve this uptake of ferucarbotran as shown with Perl’s Prussian blue (PPB) stain (B). Inductively coupled plasma mass spectrometry (ICP-MS) measurement of iron mass (pg) per cell quantifies improved uptake of ferucarbotran in MSC using TA (**** P < .0001) (C). MPI of 27.5 µg ferucarbotran alone (white brackets in D and E) and 27.5 µg ferucarbotran with TAs (orange box in D and F) show different MPI features. MPI images are displayed in full-dynamic range. As shown in PSFs, the addition of TAs to ferucarbotran decreases MPI sensitivity (G) and worsens resolution (H) by changing SPION relaxation characteristics.
Quantification of Cell Number
The ability to quantify cell number from MPI images represents a significant improvement for in vivo cell tracking with SPIONs. MPI signal (and the associated iron content) scales linearly with the number of cells present (Figure 4A). We, and others, have previously described techniques to measure MPI signal from images and determine the iron content in a region of interest (ROI) (36, 37, 41, 58, 59). In brief, MPI images are displayed in full-dynamic range and the signal is delineated at the half-maximum by referencing a color lookup table. Total MPI signal for an ROI can be calculated by:
Figure 4.
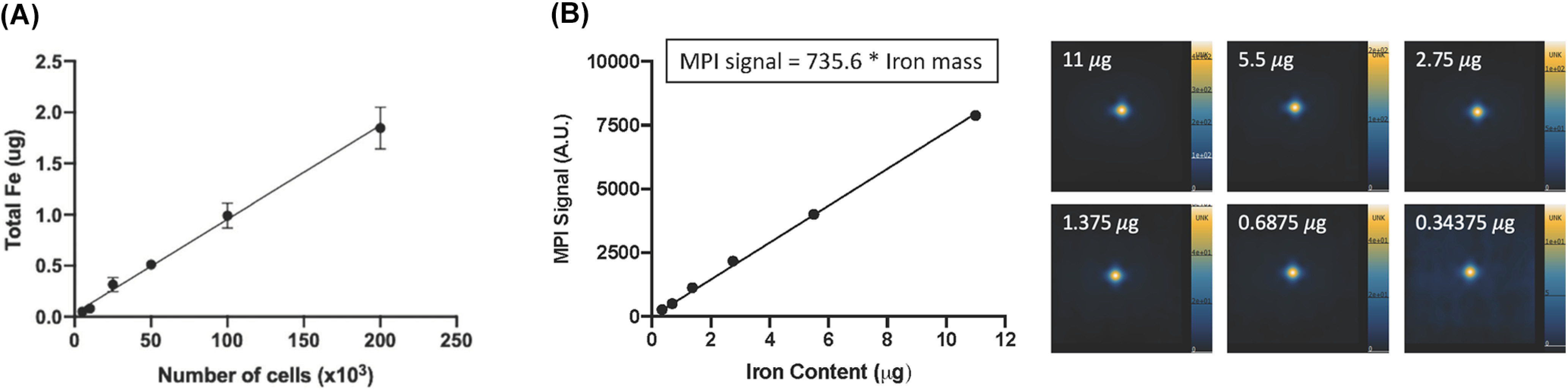
MPI quantification. The iron mass measured from MPI images is directly linear with the number of ferucarbotran-labeled RAW 264.7 macrophages (R2 = 0.9789) (A). Reproduced from Makela et al. (37). MPI signal is directly linear to the amount of iron, as displayed in a calibration line (R2 = 0.9969) (B). This line was produced by imaging multiple samples of ferucarbotran, which varied in iron mass, and the MPI signal from these images was quantified. MPI images are displayed in full-dynamic range in arbitrary units (AU).
A calibration is required to convert measured MPI signal to iron content. The relationship between MPI signal and iron content can be determined by measuring MPI signal from sample(s) of known iron mass. A single reference phantom may be included in MPI images for calibration (37, 41). Alternatively, the construction of a calibration line (MPI signal versus iron mass) using multiple iron samples (5–10) increases the accuracy of this measurement. Subsequently, the equation of this line can be used to determine the iron content associated with MPI signal measured from an ROI. The SPIONs used for calibration must be identical to the SPIONs used in the image that is being calibrated, with similar iron content and volume. In addition, the MPI images of these calibration samples must be acquired using the same imaging parameters. Figure 4B shows MPI images and an example calibration line for ferucarbotran.
The estimation of cell number from MPI images requires a measurement of cellular iron loading. For a sample prepared with a known number of cells (ex vivo), the average iron mass per cell can be measured using inductively coupled plasma mass spectrometry (ICP-MS), spectrophotometry, or MPI. These measurements must be acquired using the same cells used for the in vivo imaging experiment. Subsequently, an estimation of cell number from in vivo MPI images can be calculated:
Caution/Challenges in the Quantification and Interpretation of MPI Data
The calibration method (described above) can be used to relate MPI signal to iron mass, but requires samples with known iron content. Commercially available SPIONs typically report iron concentration in milligram per milliliter. This value is used to properly measure the amount of SPION to be imaged for the calibration and subsequently to calculate the iron mass measured by MPI in vivo. Results in this paper rely on the manufacturer’s report of iron concentration. However we have recently learned it is prudent to independently validate the iron mass in the supplied SPION using spectrophotometry or ICP-MS, as there is often deviation from the reported value.
When interpreting measurements of cell number from MPI images, it is important to consider factors related to cell labeling which introduce uncertainty. First, the measurement of iron mass per cell is an average value; some cells will contain more iron, some less. Second, the amount of iron per cell achieved with each cell labeling experiment has a range of values, even for the same labeling protocol, depending on precise timing and culture conditions. Uniform SPION labeling is optimal for cell tracking but is difficult to achieve. Magnetic sorting of cells is one strategy to improve the uniformity of labeled cells, but this only removes cells with low amounts of iron.
There are also factors which may impact the quantification of cell number over time. When SPION-labeled cells undergo apoptosis in vivo, they are taken up by phagocytes for clearance by the liver (5, 38). This leads to diminished MPI signal at transplant sites (35, 36), and the measurement of MPI signal in the liver can be used to provide a measure of labeled dead cells (17, 41). In the interim, bystander labeling of phagocytes may contribute to false MPI signal at the site of apoptosis. Lastly, intracellular SPIONs that exist within endosomal compartments may be susceptible to degradation by lysosomes (60, 61). This degradation process can reduce the paramagnetism of iron oxide nanoparticles, thereby altering their cellular MPI detectability (62, 63). This is a largely understudied phenomenon; however, it may be reasonable to assume that this effect depends on the biological inertness of the SPION and cellular lysosomal capacity. If cells proliferate in vivo, the MPI signal should not change as long as the cells and progeny remain in the same region. The same iron content would now be retained in more cells; thus, the measurement of cell number would be underestimated. This is different from MRI, where dilution of SPIONs between progeny reduces the ability to detect cells. However, if cell division leads to dispersion of iron further than 1 mm from the original ROI edge, this could lead to a reduction in MPI signal, leading to further underestimation of the number of cells present.
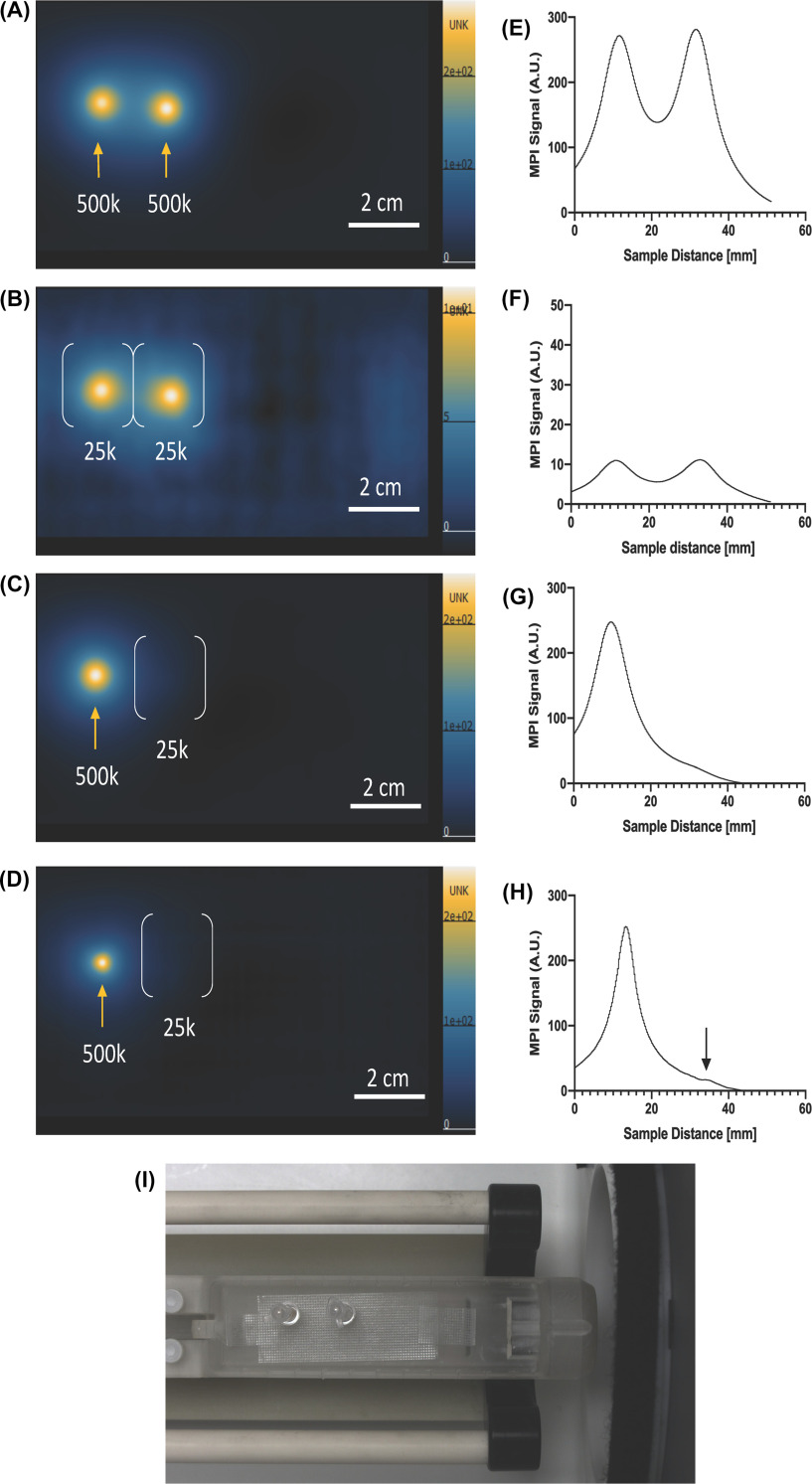
When 2 sources of MPI signal are in close proximity, and are generated from tissues or samples that contain different iron levels, the detection and quantification of the lower-intensity MPI signal can be challenging. This is illustrated in Figure 5. Here, cell pellets of bone marrow dendritic cells (BMDCs) labeled with ferucarbotran containing 2.5 × 104 or 5.0 × 105 cells were imaged at a distance of 2 cm apart. Samples with equal numbers of SPION-labeled cells could be discerned, regardless of the number of cells. However, when 5.0 × 105 cells were imaged alongside 2.5 × 104 cells, the MPI signal from the sample with fewer cells (or, less iron) was hidden by the strong signal from the sample with more cells. This presents a major challenge for studies where the location of MPI signal is unknown, particularly for in vivo cell tracking. Proposed solutions for overcoming this challenge include increasing gradient strengths for improved resolution (as we attempted in Figure 5, D and I), or using postprocessing imaging techniques such as deconvolution filters. This requires further investigation, but is an important consideration for improving in vivo cell detection limits.
Figure 5.
MPI signal from cell samples of low iron content can be concealed by nearby samples with higher iron content. Bone marrow dendritic cells (BMDC) were labeled with ferucarbotran. Two identical samples of 500 × 103 cells (orange arrows) (A) and 25 × 103 cells (white brackets) (B) with 2-cm separation are visible and can be resolved in MPI images using low gradient strengths (3.0 T/m). However, the placement of 500 × 103 cells adjacent to 25 × 103 cells limits the detection of 25 × 103 cells using 3.0 T/m gradients (C) and 5.7 T/m gradients (D). MPI images are displayed in full-dynamic range (in AU). The corresponding signal intensity profiles are shown (E–H). MPI signal cannot be resolved when samples were not identical (G, H); however, increased gradient strength (5.7 T/m) helps to improve the detection of the 25 × 103 cell sample (black arrow, H). A photograph of the imaging bed is shown with 2 cell sample holders (I).
This issue can also arise from unwanted sources of MPI signal. For example, an MPI signal is sometimes detected in regions of the digestive system of mice, owing to the presence of iron in mouse feed. This is shown in Figure 1C and in the supplementary materials of Sehl et al. 2019 (36). This signal is typically low intensity but problematic when attempting to locate and quantify MPI signal which is of similar intensity; such is the case when imaging low cell numbers in vivo and/or when using less-sensitive SPIONs such as ferumoxytol. In addition to the solutions mentioned in the previous paragraph, it may be beneficial to use “clean” iron-free feed and improved SPIONs that produce enhanced MPI signal to dominate over-contamination-related signals. We have also seen MPI signal originating from injection sites in mice, resulting from dried blood in the form of hemosiderin (unpublished results). It is important to clean and monitor injection sites. Both of these MPI signal artifacts can be present in mice even if no SPION-labeled cells are present. If they are close to, or colocalize with, SPION-labeled cells, it can be challenging to detect the source of interest.
Future Opportunities
Anatomical Coregistration
MPI images are paired with computed tomography, optical photographs, or MRI for anatomical reference. MRI provides superior localization owing to soft tissue contrast that provides detailed anatomy, as well as the ability to validate the position of MPI signal as a region of signal void in MRI. However, there are currently no dual MRI/MPI systems. This poses a challenge because the exact alignment of MRI/MPI is difficult to achieve, especially if the position of the mouse has changed between scans. This error may be minimized though the use of a body-conforming animal mold (BCAM) with alignment fiducials for coregistration. There is a need to develop a pipeline and easy-to-use software for MRI/MPI coregistration.
Clinical MPI Hardware and SPIONs
The production of clinical MPI systems is underway. Graeser et al. (64) have shown a human-scale head-only MPI system for cerebral imaging. In addition, the group Mason et al. (65) have created a hand-held MP detector for intraoperative assessment of tumor margins during breast-conserving surgery.
SPIONs are biocompatible and nonionizing, allowing for their safe use for longitudinal imaging in humans. A recent review by JWM Bulte and HE Daldrup-Link highlights 9 clinical trials involving SPION-based MRI cell tracking (66). Clinically approved tracers include Magtrace® (ferucarbotran; Endomagnetics) and Feraheme® (ferumoxytol; AMAG Pharmaceuticals) and there is an investigational new drug application for PrecisionMRX® (Imagion, anti-Her2-labeled SPION; FDA 2019). Although these SPIONs are focused on MRI applications, translation to MPI will require optimization and testing.
Multispectral “Color” MPI for Cell Tracking
A new area of research has been focused on discerning multiple sources of iron based on core size, or viscosity and temperature of the medium (67, 68), owing to differences in Brownian relaxation rates. It has been shown that 2 different SPIONs can be discerned in the same image (69), and this has promising implications to track and distinguish multiple cell types in cell tracking experiments (62). There is also the opportunity to distinguish SPION relaxation in different environments. For example, we have shown in Figure 4 that aggregation of ferucarbotran by TAs changed the MPI relaxation properties. There is potential to monitor viability of cells based on intracellular versus extracellular SPION viscosity changes (33, 70). Similarly this could contribute as a solution to separate contamination-based MPI signal as we have mentioned previously.
Cell Tracking and Hyperthermia
Magnetic hyperthermia is achieved by applying high-frequency alternating magnetic fields to stimulate motion of SPIONs, thus transferring heat to adjacent tissues. MP gradients can be used to target an ROI by aligning the FFR with a heating target and magnetically restrict SPIONs in surrounding tissue (71). In addition, MPI serves as a platform to localize and quantify SPIONs that can be used as a prescription for magnetic hyperthermia. MPI can also be used to monitor the outcome of hyperthermia, in applications of heat-aided drug release or inducing tumor cytotoxicity. A review by Chandrasekharan et al. (72) discusses MP hyperthermia tracers, hardware, and future directions.
There have been some studies that have used cells as a delivery vehicles, such as stem cells, to traffic SPIONs to an ROI for magnetic hyperthermia (73–75). This approach has the potential to deliver SPIONs to previously inaccessible tissue and/or aid in the retention of SPIONs at this tissue. This method combines cell tracking with magnetic hyperthermia. Previously this type of study was inhibited by the lack of precision imaging, and now MPI can serve as a platform to accelerate this area of research.
Conclusions
We are excited by the prospects of cell tracking with MPI. By combining the advantages of particle (SPION) sensitivity and signal specificity, prominent image contrast, and direct quantification, MPI has unique advantages over existing cell tracking modalities. Improvements in cellular sensitivity are expected from SPION development and cellular uptake strategies that may help to advance toward the detection of low cell numbers. Yet there are still many aspects related to the interpretation of cell fate from MPI images that require improvements. These may provide exciting avenues for further investigation. Ultimately, we anticipate that cell tracking with MPI will contribute to our understanding of cellular therapies and guide therapeutic optimization.
Acknowledgment
We would like to acknowledge the Natural Science and Engineering Research Council of Canada (RGPIN-2020 - 06671) and the Canadian Foundation for Innovation.
Footnotes
- MRI
- magnetic resonance imaging
- SPIONs
- superparamagnetic iron oxide nanoparticles
- MPI
- magnetic particle imaging
- PFC
- perfluorocarbon
- FFR
- field free region
- MSC
- mesenchymal stem cell
- PSF
- point spread function
- FWHM
- full-width half maximum
- MPIO
- micron-sized superparamagnetic polystyrene beads
- TAs
- transfection agents
- ROI
- region of interest
- AU
- arbitrary units
- ICP-MS
- inductively coupled plasma mass spectrometry
- BMDCs
- bone marrow dendritic cells
References
- 1.de Vries IJM, Lesterhuis WJ, Barentsz JO, Verdijk P, van Krieken JH, Boerman OC, Oyen WJG, Bonenkamp JJ, Boezeman JB, Adema GJ, Bulte JWM, Scheenen TWJ, Punt CJA, Heerschap A, Figdor CG. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat Biotechnol. 2005;23:1407–1413. [DOI] [PubMed] [Google Scholar]
- 2.Förster R, Braun A, Worbs T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends Immunol. 2012;33:271–280. [DOI] [PubMed] [Google Scholar]
- 3.Martín-Fontecha A, Sebastiani S, HöPken UE, Uguccioni M, Lipp M, Lanzavecchia A, Sallusto F. Regulation of dendritic cell migration to the draining lymph node: impact on T lymphocyte traffic and priming. J Exp Med. 2003;198:615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Sun C, Wang S, Shang N, Shangguan J, Figini M, Pan L, Zhou K, Ma Q, Procissi D, Velichko Y, Yaghmai V, Li G, Zhang Z. Mouse dendritic cell migration in abdominal lymph nodes by intraperitoneal administration. Am J Transl Res. 2018;10:2859–2867. [PMC free article] [PubMed] [Google Scholar]
- 5.Makela AV, Murrell DH, Parkins KM, Kara J, Gaudet JM, Foster PJ. Cellular imaging with MRI. Top Magn Reson Imaging. 2016;25:177–186. [DOI] [PubMed] [Google Scholar]
- 6.Bulte JWM, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–499. [DOI] [PubMed] [Google Scholar]
- 7.Sykova E, Jendelova P. In vivo tracking of stem cells in brain and spinal cord injury. Prog Brain Res. 2007;161:367–383. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Lara LE, Xu X, Hofstetrova K, Pniak A, Chen Y, McFadden CD, Martinez-Santiesteban FM, Rutt BK, Brown A, Foster PJ. The use of cellular magnetic resonance imaging to track the fate of iron-labeled multipotent stromal cells after direct transplantation in a mouse model of spinal cord injury. Mol Imaging Biol. 2011;13:702–711. [DOI] [PubMed] [Google Scholar]
- 9.Hinds KA, Hill JM, Shapiro EM, Laukkanen MO, Silva AC, Combs CA, Varney TR, Balaban RS, Koretsky AP, Dunbar CE. Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allows magnetic resonance imaging of single cells. Blood. 2003;102:867–872. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, de Chickera SN, Willert C, Economopoulos V, Noad J, Rohani R, Wang AY, Levings MK, Scheid E, Foley R, Foster PJ, Dekaban GA. Cellular magnetic resonance imaging of monocyte-derived dendritic cell migration from healthy donors and cancer patients as assessed in a SCID mouse model. Cytotherapy. 2011;13:1234–1248. [DOI] [PubMed] [Google Scholar]
- 11.de Chickera S, Willert C, Mallet C, Foley R, Foster P, Dekaban GA. Cellular MRI as a suitable, sensitive non-invasive modality for correlating in vivo migratory efficiencies of different dendritic cell populations with subsequent immunological outcomes. Int Immunol. 2012;24:29–41. [DOI] [PubMed] [Google Scholar]
- 12.Jirak D, Kriz J, Strzelecki M, Yang J, Hasilo C, White DJ, Foster PJ. Monitoring the survival of islet transplants by MRI using a novel technique for their automated detection and quantification. Magn Reson Mater Phy. 2009;22:257–265. [DOI] [PubMed] [Google Scholar]
- 13.Heyn C, Ronald JA, Ramadan SS, Snir JA, Barry AM, MacKenzie LT, Mikulis DJ, Palmieri D, Bronder JL, Steeg PS, Yoneda T, MacDonald IC, Chambers AF, Rutt BK, Foster PJ. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med Off Med. 2006;56:1001–1010. [DOI] [PubMed] [Google Scholar]
- 14.Heyn C, Ronald JA, Mackenzie LT, MacDonald IC, Chambers AF, Rutt BK, Foster PJ. In vivo magnetic resonance imaging of single cells in mouse brain with optical validation. Magn Reson Med. 2006;55:23–29. [DOI] [PubMed] [Google Scholar]
- 15.Fink C, Gaudet JM, Fox MS, Bhatt S, Viswanathan S, Smith M, Chin J, Foster PJ, Dekaban GA. 19F-perfluorocarbon-labeled human peripheral blood mononuclear cells can be detected in vivo using clinical MRI parameters in a therapeutic cell setting. Sci Rep. 2018;8:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudet JM, Ribot EJ, Chen Y, Gilbert KM, Foster PJ. Tracking the Fate of Stem Cell Implants with Fluorine-19 MRI. PLOS One. 2015;10:e0118544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapelin F, Capitini CM, Ahrens ET. Fluorine-19 MRI for detection and quantification of immune cell therapy for cancer. J Immunother Cancer. 2018;6:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadayakkara DKK, Janjic JM, Pusateri LK, Young W-B, Ahrens ET. In vivo observation of intracellular oximetry in perfluorocarbon-labeled glioma cells and chemotherapeutic response in the CNS using fluorine-19 MRI. Magn Reson Med. 2010;64:1252–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahrens ET, Helfer BM, O'Hanlon CF, Schirda C. Clinical cell therapy imaging using a perfluorocarbon tracer and fluorine-19 MRI. Magn Reson Med. 2014;72:1696–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulte JWM. Superparamagnetic iron oxides as MPI tracers: a primer and review of early applications. Adv Drug Deliv Rev. 2019;138:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng B, Yu E, Orendorff R, Lu K, Konkle JJ, Tay ZW, Hensley D, Zhou XY, Chandrasekharan P, Saritas EU, Goodwill PW, Hazle JD, Conolly SM. Seeing SPIOs directly in vivo with magnetic particle imaging. Mol Imaging Biol. 2017;19:385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu K, Goodwill PW, Saritas EU, Zheng B, Conolly SM. Linearity and shift invariance for quantitative magnetic particle imaging. IEEE Trans Med Imaging. 2013;32:1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulte JWM, Walczak P, Gleich B, Weizenecker J, Markov DE, Aerts HCJ. MPI cell tracking: what can we learn from MRI? Proc SPIE Int Soc Opt Eng. 2011;7965:79650z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saritas EU, Goodwill PW, Croft LR, Konkle JJ, Lu K, Zheng B, Conolly SM. Magnetic particle imaging (MPI) for NMR and MRI researchers. J Magn Reson. 2013;229:116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu LC, Zhang Y, Steinberg G, Qu H, Huang S, Cheng M, Bliss T, Du F, Rao J, Song G, Pisani L, Doyle T, Conolly S, Krishnan K, Grant G, Wintermark M. A review of magnetic particle imaging and perspectives on neuroimaging. AJNR Am J Neuroradiol. 2019;40:206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu EY, Bishop M, Zheng B, Ferguson RM, Khandhar AP, Kemp SJ, Krishnan KM, Goodwill PW, Conolly SM. Magnetic particle imaging: a novel in vivo imaging platform for cancer detection. Nano Lett. 2017;17:1648–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talebloo N, Gudi M, Robertson N, Wang P. Magnetic particle imaging: current applications in biomedical research. J Magn Reson Imaging. 2020;51:1659–1668. [DOI] [PubMed] [Google Scholar]
- 28.Tay ZW, Hensley DW, Chandrasekharan P, Zheng B, Conolly SM. Optimization of drive parameters for resolution, sensitivity and safety in magnetic particle imaging. IEEE Trans Med Imaging. 2020;39:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng B, Vazin T, Goodwill PW, Conway A, Verma A, Ulku Saritas E, Schaffer D, Conolly SM. Magnetic Particle Imaging tracks the long-term fate of in vivo neural cell implants with high image contrast. Sci Rep. 2015;5:14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song G, Chen M, Zhang Y, Cui L, Qu H, Zheng X, Wintermark M, Liu Z, Rao J. Janus iron oxides @ semiconducting polymer nanoparticle tracer for cell tracking by magnetic particle imaging. Nano Lett. 2018;18:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arami H, Ferguson RM, Khandhar AP, Krishnan KM. Size-dependent ferrohydrodynamic relaxometry of magnetic particle imaging tracers in different environments. Med Phys. 2013;40:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay ZW, Hensley DW, Vreeland EC, Zheng B, Conolly SM. The relaxation wall: experimental limits to improving MPI spatial resolution by increasing nanoparticle core size. Biomed Phys End Express. 2017;3:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandrasekharan P, Tay ZW, Zhou Xy YE, Orendorff R, Hensley D, Huynh Q, Barry Fung KL, VanHook CC, Goodwill P, Zheng B, Conolly S. A perspective on a rapid and radiation-free tracer imaging modality, magnetic particle imaging, with promise for clinical translation. Br J Radiol. 2018;91:20180326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay ZW, Conolly SM. Order-of-magnitude resolution and SNR improvement using positive feedback MNP chains in magnetic particle imaging. World Mol Imaging Congr. 2019. [Google Scholar]
- 35.Nejadnik H, Pandit P, Lenkov O, Lahiji AP, Yerneni K, Daldrup-Link HE. Ferumoxytol can be used for quantitative magnetic particle imaging of transplanted stem cells. Mol Imaging Biol. 2019;21:465–472.30194566 [Google Scholar]
- 36.Sehl OC, Makela AV, Hamilton AM, Foster PJ. Trimodal cell tracking in vivo: combining iron- and fluorine-based magnetic resonance imaging with magnetic particle imaging to monitor the delivery of mesenchymal stem cells and the ensuing inflammation. Tomogr Ann Arbor Mich. 2019;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makela AV, Gaudet JM, Schott MA, Sehl OC, Contag CH, Foster PJ. Magnetic particle imaging of macrophages associated with cancer: filling the voids left by iron-based magnetic resonance imaging. Mol Imaging Biol. 2020;22:958–968. [DOI] [PubMed] [Google Scholar]
- 38.Zheng B, von See MP, Yu E, Gunel B, Lu K, Vazin T, Schaffer DV, Goodwill PW, Conolly SM. Quantitative magnetic particle imaging monitors the transplantation, biodistribution, and clearance of stem cells in vivo. Theranostics. 2016;6:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Ma X, Liao H, Liang Z, Li F, Tian J, Ling D. Artificially engineered cubic iron oxide nanoparticle as a high-performance magnetic particle imaging tracer for stem cell tracking. ACS Nano. 2020;14:2053–2062. [DOI] [PubMed] [Google Scholar]
- 40.Bulte JWM, Walczak P, Janowski M, Krishnan KM, Arami H, Halkola A, Gleich B, Rahmer J. et al. Quantitative “hot spot” imaging of transplanted stem cells using superparamagnetic tracers and magnetic particle imaging (MPI). Tomography. 2015;1:91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, Goodwill PW, Pandit P, Gaudet J, Ross A, Wang J, Yu E, Hensley DW, Doyle TC, Contag CH, Conolly S, Moore A. Magnetic particle imaging of islet transplantation in the liver and under the kidney capsule in mouse models. Quant Imaging Med Surg. 2018;8:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivera-Rodriguez A, Hoang-Minh LB, Chiu-Lam A, Sarna N, Marrero-Morales L, Mitchell DA, Rinaldi C. et al. Tracking adoptive T cell therapy using magnetic particle imaging. bioRxiv. 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaudet J, Mansfield J, Goodwill P. Imaging cancer immunology: tracking immune cells in vivo with magnetic particle imaging. J Immunol. 2019;202:130.7–130.7. [Google Scholar]
- 44.Eberbeck D, Dennis CL, Huls NF, Krycka KL, Gruttner C, Westphal F. Multicore magnetic nanoparticles for magnetic particle imaging. IEEE Trans Magn. 2013;49:269–274. [Google Scholar]
- 45.Yoshida T, Othman NB, Enpuku K. Characterization of magnetically fractionated magnetic nanoparticles for magnetic particle imaging. J Appl Phys. 2013;114:173908. [Google Scholar]
- 46.Ferguson RM, Minard KR, Krishnan KM. Optimization of nanoparticle core size for magnetic particle imaging. J Magn Magn Mater. 2009;321:1548–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houston WV. A compound interferometer for fine structure work. Phys Rev. 1927;29:478–484. [Google Scholar]
- 48.Mailänder V, Lorenz MR, Holzapfel V, Musyanovych A, Fuchs K, Wiesneth M, Walther P, Landfester K, Schrezenmeier H. Carboxylated superparamagnetic iron oxide particles label cells intracellularly without transfection Agents. Mol Imaging Biol. 2008;10:138–146. [DOI] [PubMed] [Google Scholar]
- 49.Arbab AS, Bashaw LA, Miller BR, Jordan EK, Lewis BK, Kalish H, Frank JA. Characterization of biophysical and metabolic properties of cells labeled with superparamagnetic iron oxide nanoparticles and transfection agent for cellular MR imaging. Radiology. 2003;229:838–846. [DOI] [PubMed] [Google Scholar]
- 50.Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, Lewis BK, Bryant LH, Bulte JWM. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228:480–487. [DOI] [PubMed] [Google Scholar]
- 51.Tang KS, Shapiro EM. Enhanced magnetic cell labeling efficiency using –NH2 coated MPIOs. Magn Reson Med. 2011;65:1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant LH, Kim SJ, Hobson M, Milo B, Kovacs ZI, Jikaria N, Lewis BK, Aronova MA, Sousa AA, Zhang G, Leapman RD, Frank JA. Physicochemical characterization of ferumoxytol, heparin and protamine nanocomplexes for improved magnetic labeling of stem cells. Nanomedicine Nanotechnol Biol Med. 2017;13:503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang X, Loc WS, Dong C, Matters GL, Butler PJ, Kester M, Meyers C, Jiang Y, Adair JH. The use of nanoparticulates to treat breast cancer. Nanomed. 2017;12:2367–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arbab AS, Bashaw LA, Miller BR, Jordan EK, Bulte JWM, Frank JA. Intracytoplasmic tagging of cells with ferumoxides and transfection agent for cellular magnetic resonance imaging after cell transplantation: methods and techniques. Transplantation. 2003;76:1123–1130. [DOI] [PubMed] [Google Scholar]
- 55.Thu MS, Bryant LH, Coppola T, Jordan EK, Budde MD, Lewis BK, Chaudhry A, Ren J, Varma NRS, Arbab AS, Frank JA. Self-assembling nanocomplexes by combining ferumoxytol, heparin and protamine for cell tracking by magnetic resonance imaging. Nat Med. 2012;18:463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Buul GM, Farrell E, Kops N, van Tiel ST, Bos PK, Weinans H, Krestin GP, van Osch GJVM, Bernsen MR. Ferumoxides–protamine sulfate is more effective than ferucarbotran for cell labeling: implications for clinically applicable cell tracking using MRI. Contrast Media Mol Imaging. 2009;4:230–236. [DOI] [PubMed] [Google Scholar]
- 57.Rughetti A, Biffoni M, Sabbatucci M, Rahimi H, Pellicciotta I, Fattorossi A, Pierelli L, Scambia G, Lavitrano M, Frati L, Nuti M. Transfected human dendritic cells to induce antitumor immunity. Gene Ther. 2000;7:1458–1466. [DOI] [PubMed] [Google Scholar]
- 58.Parkins KM, Melo KP, Ronald JA, Foster PJ. Visualizing tumour self-homing with magnetic particle imaging. bioRxiv. 2020. [DOI] [PubMed] [Google Scholar]
- 59.Melo KP, Makela AV, Hamilton AM, Foster PJ. Development of magnetic particle imaging (MPI) for cell tracking and detection. bioRxiv. 2020 [Google Scholar]
- 60.Golovko DM, Henning T, Bauer JS, Settles M, Frenzel T, Mayerhofer A, Rummeny EJ, Daldrup-Link HE. Accelerated stem cell labeling with ferucarbotran and protamine. Eur Radiol. 2010;20:640–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toth GB, Varallyay CG, Horvath A, Bashir MR, Choyke PL, Daldrup-Link HE, Dosa E, Finn JP, Gahramanov S, Harisinghani M, Macdougall I, Neuwelt A, Vasanawala SS, Ambady P, Barajas R, Cetas JS, Ciporen J, DeLoughery TJ, Doolittle ND, Fu R, Grinstead J, Guimaraes AR, Hamilton BE, Li X, McConnell HL, Muldoon LL, Nesbit G, Netto JP, Petterson D, Rooney WD, Schwartz D, Szidonya L, Neuwelt EA. Current and potential imaging applications of ferumoxytol for magnetic resonance imaging. Kidney Int. 2017;92:47–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuka H, Mimura A, Inaoka Y, Murase K. Magnetic nanoparticles in macrophages and cancer cells exhibit different signal behavior on magnetic particle imaging. J Nanosci Nanotechnol. 2019;19:6857–6865. [DOI] [PubMed] [Google Scholar]
- 63.Guzy J, Chakravarty S, Buchanan FJ, Chen H, Gaudet JM, Hix JML, Mallett CL, Shapiro EM. Complex relationship between iron oxide nanoparticle degradation and the signal intensity in magnetic particle imaging. ACS Appl Nano Mater. 2020;3:3991–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Graeser M, Thieben F, Szwargulski P, Werner F, Gdaniec N, Boberg M, Griese F, Möddel M, Ludewig P, van de Ven D, Weber OM, Woywode O, Gleich B, Knopp T. Human-sized magnetic particle imaging for brain applications. Nat Commun. 2019;10:1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mason E, Mattingly E, Herb K, Franconi S, Sliwiak M, Cooley C, et al. Magnetic particle imaging for intraoperative margin analysis in breast-conserving surgery. Int J Magn Part Imaging. 2020;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bulte JWM, Daldrup-Link HE. Clinical tracking of cell transfer and cell transplantation: trials and tribulations. Radiology. 2018;289:604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu K, Su D, Saha R, Liu J, Chugh VK, Wang J-P. Magnetic particle spectroscopy: a short review of applications using magnetic nanoparticles. ACS Appl Nano Mater. 2020;3:4972–4989. [Google Scholar]
- 68.Utkur M, Muslu Y, Saritas EU. Relaxation-based color magnetic particle imaging for viscosity mapping. Appl Phys Lett. 2019;115:152403. [Google Scholar]
- 69.Shasha C, Teeman E, Krishnan KM, Szwargulski P, Knopp T, Möddel M. Discriminating nanoparticle core size using multi-contrast MPI. Phys Med Biol. 2019;64:074001. [DOI] [PubMed] [Google Scholar]
- 70.Teeman E, Shasha C, Evans JE, Krishnan KM. Intracellular dynamics of superparamagnetic iron oxide nanoparticles for magnetic particle imaging. Nanoscale. 2019;11:7771–7780. [DOI] [PubMed] [Google Scholar]
- 71.Tay ZW, Chandrasekharan P, Chiu-Lam A, Hensley DW, Dhavalikar R, Zhou XY, Yu EY, Goodwill PW, Zheng B, Rinaldi C, Conolly SM. Magnetic particle imaging-guided heating in vivo using gradient fields for arbitrary localization of magnetic hyperthermia therapy. ACS Nano. 2018;12:3699–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandrasekharan P, Tay ZW, Hensley D, Zhou XY, Fung BK, Colson C, Lu Y, Fellows BD, Huynh Q, Saayujya C, Yu E, Orendorff R, Zheng B, Goodwill P, Rinaldi C, Conolly S. Using magnetic particle imaging systems to localize and guide magnetic hyperthermia treatment: tracers, hardware, and future medical applications. Theranostics. 2020;10:2965–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalber T, Ordidge K, Southern P, Loebinger M, Kyrtatos P, Pankhurst Q, Lythgoe MF, Janes SM. Hyperthermia treatment of tumors by mesenchymal stem cell-delivered superparamagnetic iron oxide nanoparticles. Int J Nanomedicine. 2016;11:1973.– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Altanerova U, Babincova M, Babinec P, Benejova K, Jakubechova J, Altanerova V, Zduriencikova M, Repiska V, Altaner C. Human mesenchymal stem cell-derived iron oxide exosomes allow targeted ablation of tumor cells via magnetic hyperthermia. Int J Nanomedicine. 2017;12:7923–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basel MT, Balivada S, Wang H, Shrestha TB, Moon Seo G, Pyle M, Abayaweera G, Dani R, Koper OB, Tamura M, Chikan V, Bossmann SH, Troyer DL. et al. Cell-delivered magnetic nanoparticles caused hyperthermia-mediated increased survival in a murine pancreatic cancer model. Int J Nanomedicine. 2012;7:297– Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gaudet JM, Hamilton Am, Chen Y, Fox MS, Foster PJ. Application of dual 19F and iron cellular MRI agents to track the infiltration of immune cells to the site of a rejected stem cell transplant. Magn Reson Med. 2017;78:713–720. [DOI] [PubMed] [Google Scholar]