Abstract
Radiology reports contain important clinical information about patients which are often tied through spatial expressions. Spatial expressions (or triggers) are mainly used to describe the positioning of radiographic findings or medical devices with respect to some anatomical structures. As the expressions result from the mental visualization of the radiologist’s interpretations, they are varied and complex. The focus of this work is to automatically identify the spatial expression terms from three different radiology sub-domains. We propose a hybrid deep learning-based NLP method that includes – 1) generating a set of candidate spatial triggers by exact match with the known trigger terms from the training data, 2) applying domain-specific constraints to filter the candidate triggers, and 3) utilizing a BERT-based classifier to predict whether a candidate trigger is a true spatial trigger or not. The results are promising, with an improvement of 24 points in the average F1 measure compared to a standard BERT-based sequence labeler.
1. Introduction
Radiology reports contain a radiologist’s interpretations of an imaging study of a patient. The mental interpretations often get expressed through descriptions of important radiological entities with reference to a particular anatomical structure (Datta et al., 2020a). The radiological entities whose positions are described mainly include radiographic findings (e.g., clinical findings like interstitial emphysema and imaging observations like ground-glass opacity) and medical devices (e.g., endotracheal tube and central venous catheter). There exists a wide variation in the spatial language used by radiologists in expressing the exact positioning of the radiological entities. Limited research has focused on effectively identifying the spatial expressions from multiple imaging modalities. Therefore, the focus of this work is to investigate different automatic approaches to extract the spatial expressions from the report sentences along with highlighting the various challenges involved in this task. These extracted spatial expressions, if predicted accurately, can also facilitate clinical applications such as automatic labeling of radiographic images for training image classifiers (Wang et al., 2017).
Identification of spatial expressions in a sentence forms the foundation for other downstream spatial information extraction tasks. Much of the clinically relevant information appears in the context of a spatial expression. Consider the following sentence:
A lytic lesion at the left vertex extending into the epidural region, scalp and soft tissues is grossly unchanged in appearance.
Here, we note that it is very crucial to accurately identify the spatial expressions such as at and extending into. Firstly, these trigger terms denote the specific positioning of the lesion by associating the lesion term with anatomical entities such as left vertex and epidural region. Secondly, this indirectly helps in identifying all the modifier information about the lesion including its density (i.e., lytic) and status (i.e., unchanged in appearance).
Spatial expressions are also used to describe the positioning of the medical devices that are inserted into specific body locations. Radiologists often document the current position status of the devices (e.g., malpositioned, satisfactory position) and often times indicate their changes in positioning. The following is an example of the radiologist’s interpretation about a device position:
PICC line enters from left arm, descends to the lower inferior vena cava, then turns and extends peripherally to left subclavian vein.
This captures the mental visualization of the radiologist as they interpret the specific position of the PICC line from the corresponding image. We note that there are diverse expressions belonging to multiple part-of-speech categories (e.g., verbs, prepositions, verbs followed by prepositions) that the radiologists use in documenting the spatial position of both findings and devices.
In this work, our aim is to identify all the spatial expressions given a radiology report sentence. We experiment with a pre-trained language model, BERT (Devlin et al., 2019), used as a sequence labeler to extract the spatial expressions (or triggers). We further propose a hybrid deep learning method where we use BERT as a classifier in combination with domain-dependent heuristics. Specifically, in this hybrid approach, we first extract the candidate trigger terms from the sentences with high recall leveraging the terms from the training corpus. We then filter the candidates by applying a set of radiology-specific constraints. Finally, we utilize BERT as a classification model to identify if each of the filtered candidate terms is a trigger expression or not.
2. Related Work
Some previous studies have focused on extracting spatial relations from radiology reports (Roberts et al., 2012; Rink et al., 2013). However, both these studies are specific to appendicitis-related reports. Our previous work has also aimed at identifying spatial expressions (mainly prepositional) from chest X-ray reports (Datta et al., 2020a; Datta and Roberts, 2020). Moreover, all these studies have focused on identifying spatial relations associated only with radiographic findings. We aim to identify more complex and varied spatial expressions associated with descriptions of both findings and medical devices. Importantly, descriptions of devices often utilize far richer spatial language, as shown in the PICC line example above.
Both in the general and medical domains, hybrid deep learning approaches have been used lately for various natural language processing (NLP) tasks such as document classification (Asim et al., 2019) and named entity recognition (Li et al., 2019). A recent work has also demonstrated the promising results of applying a hybrid approach for extracting clinical information from CT scan reports (Gupta et al., 2019). Moreover, many NLP tasks have leveraged the contextualized representations of pre-trained language models such as BERT. However, not much effort has been directed toward building hybrid methods based on BERT. Extracting spatial expressions from text often requires domain knowledge of language characteristics. Thus, we investigate the impact of combining radiology-specific constraints with a BERT-based model to extract spatial expressions from radiology reports.
3. Dataset
We use a dataset of 400 radiology reports containing annotated spatial expressions (Datta et al., 2020b). These reports are taken from the MIMIC III clinical corpus (Johnson et al., 2016). Our dataset consists of an equal distribution of three different imaging modalities, namely, chest X-rays, brain MRIs, and babygrams. Some basic statistics related to the spatial expressions in this dataset are shown in Table 1. Note that this dataset includes multi-word spatial triggers and triggers with varied part-of-speech categories. This makes the task more challenging compared to using single word triggers, mostly prepositions as in Rad-SpRL (Datta et al., 2020a).
Table 1:
Corpus statistics of spatial expressions.
| Item | Frequency |
|---|---|
| Spatial triggers | 1372 |
| POS sequence categories | 33 |
| Item | Frequency | Examples |
|---|---|---|
| Prepositions (IN) | 1093 | within, throughout |
| Verbs (VBG|VBP|VBZ|VB|VBD) |
163 | demonstrate, shows |
| Verb followed by preposition | 52 | extending into, projected at |
| Noun followed by preposition | 32 | projects over, grows into |
| Longest triggers (TO DT NN IN, PDT DT NN TO) | 7 | to the left of, all the way to |
4. Methods
4.1. Sequence Labeling Method (Baseline)
We take a BERTBASE model pre-trained on MIMIC (Si et al., 2019) and fine-tune on our annotated corpus to identify the spatial triggers. We treat this as a sequence labeling task where each sentence is WordPiece-tokenized and represented as [[CLS] sentence [SEP]] to construct an input sequence to the BERT encoder as in Devlin et al. (2019). The encoder output is fed into a linear classification layer to predict labels per token. We use the BIO scheme for tagging the spatial triggers.
4.2. Proposed Hybrid Method
The proposed pipeline is shown in Figure 1.
Figure 1:
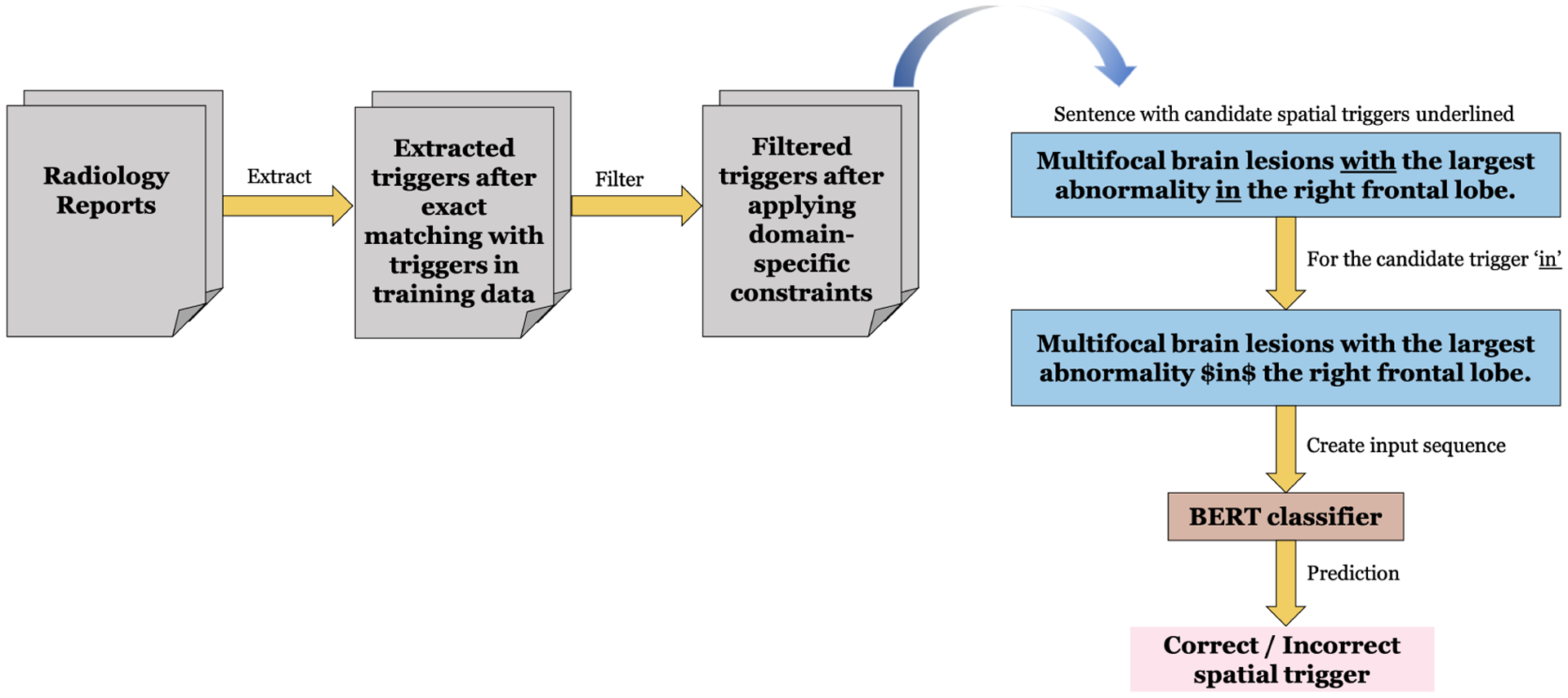
Pipeline of the proposed hybrid approach.
Generating candidate spatial triggers
This focuses on extracting spatial triggers in a sentence with high recall. First, we construct a set of all the unique spatial triggers encountered in the training set. Then, we identify the triggers in an unseen radiology report using case-insensitive exact matching against the triggers in the constructed set. In case of the triggers having overlapping spans, we use the longest span as the final candidate trigger. For example, when both the triggers - extends in and in are identified in a sentence and have overlapping spans, extends in is selected as the candidate trigger for the next step.
Applying radiology-specific constraints
As the candidate generation phase aims to improve recall, this results in the common problem of low precision. Thus we introduce a set of radiology language-specific rules to reduce the number of false positives. We develop the constraints such that they are generalizable across different types of radiology reports. For example, the left or the right are frequent phrases which are usually followed by the spatial trigger on. However, left and right do not indicate any specific anatomical location. So we exclude on from our final candidate trigger list if it occurs in scenarios like this. Thus, for each of the common spatial triggers such as with, in, and at, we develop a set of frequent terms or phrases such that when any of the phrase is seen surrounding a trigger, that particular trigger will be excluded from our final candidate trigger list. Note that we construct separate list of such surrounding phrases for left and right side of a spatial trigger. A few examples of the developed phrases are shown in Table 2. The complete list is in appendix Table 4.
Table 2:
Example terms for 3 common spatial triggers.
| Spatial trigger | Terms to left | Terms to right |
|---|---|---|
| of | evidence, | time, |
| suggestive, | position, | |
| possibility,… | uncertain, … | |
| with | correlation, | previous, |
| compatible, | prior, | |
| consistent, … | known, … | |
| to | appear, | suggest, |
| compared, | assess, | |
| rotated,… | be,… |
BERT Classifier
A BERT-based classification model is used determine whether each candidate trigger identified in the previous step is correct. We construct the input data as follows:
Identify the triggers with distinct spans combining both gold triggers and the candidate triggers in a sentence.
Create a separate sentence instance for each of the triggers obtained from the above step.
Assign a positive (correct) label to an instance if the associated trigger is a gold trigger and a negative (incorrect) label otherwise.
We construct an input sequence to BERT by converting each of the above instances to the standard BERT input format [[CLS] sentence [SEP]], similar to Section 4.1. Note that the classification of being correct/incorrect is based on a specific spatial trigger in a sentence. In order to inform the model about the positional information of the spatial trigger, we insert a special character sequence ‘$’ both to the left and right of the trigger. The other aspects of the model architecture is similar to the original BERT paper’s implementation (Devlin et al., 2019).
5. Evaluation and Experimental settings
We perform 10-fold cross validation (CV) to evaluate the performance of both the BERT-based methods. For each of the 10 iterations, reports in 8 folds are used for training and 1 fold each are used for validation and testing. Average precision, recall, and F1 measures are reported for these methods. We also report these performance metric values for the rule-based methods (both for exact matching and exact matching + constraints) by evaluating on the same test folds. Note that we train the BERT classifier using the candidates directly obtained after the exact matching step. While evaluating, we apply the additional domain constraints over the candidate triggers generated from exact matching. We make this decision based on the results of our preliminary experiments.
We use the BERTBASE variant for both sequence labeling and classification models. The models are pre-trained on MIMIC-III clinical notes (Si et al., 2019) for 320K steps. The maximum sequence length for both the tasks is set at 128 and learning rate at 2e–5. Based on the validation set performance, we select the number of training epochs as 4. We use the cased version of the models.
6. Results
The results of spatial trigger extraction are shown in Table 3. The average accuracy of the BERT-based classifier model over 10-fold CV is 88.7%.
Table 3:
Spatial trigger extraction results. Average Precision (P), Recall (R), and F1 across 10 test folds. 10-fold cross validation is performed for the BERT-based models (first and last rows).
| Method | P(%) | R(%) | F1 |
|---|---|---|---|
| BERT-Based Sequence Labeling | 92.20 | 43.04 | 57.52 |
| Exact matching | 18.71 | 96.77 | 31.32 |
| Exact matching + Constraints | 34.56 | 93.02 | 50.21 |
| Exact matching + Constraints + BERT-Based Classification | 84.43 | 79.19 | 81.10 |
We notice that the sequence labeling method obtained high precision and low recall. The exact matching achieved a much improved recall (96.77%) compared to the sequence labeling system. However, it resulted in too many false positive spatial triggers, mainly because of common prepositional and verb terms such as of, with, and are (as indicated by a very low precision). We achieved slightly better precision by applying constraints over the exact matched triggers (shown in the third row of Table 3). Our proposed method which utilizes a set of domain-inspired constraints on top of a BERT-based classifier helps in obtaining a balanced precision and recall, improving the F1 by almost 24 points compared to standard BERT sequence labeling.
7. Discussion
We focus on extracting varied spatial expressions from radiology reports using a sequence labeling method as well as a hybrid approach that first applies domain-specific rules to extract the candidate triggers and later employs a deep learning-based classifier to judge every candidate. Our proposed method (Exact matching + Constraints + BERT-based classification) achieves much improved average F1 measure in CV.
Error Analysis
We observe that, after applying constraints, most of the triggers that are missed by the rule-based approach are uncommon phrases that are not seen in the training data, e.g., verbs followed by prepositions such as grows into and verbs such as filling. Whereas, for the proposed hybrid approach, missed triggers are usually verbs such as demonstrates and appears.
Challenges
Many of the spatial expressions which describe the presence of an abnormality in a specific anatomical location are common English language terms such as of, with, and are. Some other challenges include identifying whether a ‘verb followed by prepositional/adverb’ phrase always indicates a spatial expression or not, since in a few cases they imply intermediate change in position (e.g., kinks back) rather than the position where a radiological entity is actually located.
Future Directions
Our next steps include examining the generalizability of our proposed approach when applied to other types of radiology reports (e.g., ultrasound, computed tomography, etc.). We also aim to incorporate additional rules that can extract spatial expressions beyond the ones seen in the training set. One of the potential rules may be to automatically generate more variations of triggers with the form ‘verb followed by preposition’. Using part-of-speech (POS) information to automatically extract triggers holds potential but this may introduce errors from the POS taggers.
8. Conclusion
This work proposes a BERT-based hybrid method to extract spatial expressions from radiology reports. This method achieves satisfactory performance with an average F1 measure of 81.10 over 10-fold CV. We also extract spatial expressions by formulating the problem as a sequence labeling task (used as baseline). We find that the BERT-based sequence labeling model suffers from low recall. Our proposed hybrid approach combining radiology-specific constraints with a BERT-based classifier helps to improve the recall by around 36%. We also address some of the challenges involved in the task of spatial trigger extraction in the radiology domain. We plan to further improve the performance of the system by adding more granular domain constraints as well as evaluate the generalizability of the method across multi-institutional datasets.
Acknowledgments
This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering (NIBIB: R21EB029575) and the Patient-Centered Outcomes Research Institute (PCORI: ME-2018C1-10963).
A Appendix
Table 4:
A more exhaustive list of the terms used for building the constraints for a set of spatial triggers in the dataset.
| Spatial trigger | Terms to the left | Terms to the right |
|---|---|---|
| of | mri, mra, mrv, xray, projection, babygram, head, view, history, evidence, suggestive, possibility, evaluation, repositioning, ct, loop, free, level, floor, top, area, circle, suggestion, remainder, shortness, position, component, foci, focus, hour, examination, date, study, reposition, positioning | time, position, normal, certain, uncertain |
| with | made, correlation, brain, xray, ct, mri, mra, mrv, projection, infant, patient, baby, compatible, consistent, catheter, associated | previous, prior, known, exam, lead, port, sideport, wire, its, no, tip, distal |
| to | appear, similar, due, compared, rotated, made, secondary, attributed, related | suggest, assess, be |
| in | increase, decrease, normal, result | size, position, good, satisfactory, appearance, (+ limit as the second or third term to the right) |
| show/demonstrate | mri, mra, mrv, xray, projection, babygram, head, view, history, ct, which | - |
| from | mri, mra, mrv, xray, projection, ct, lead, port, sideport, wire | - |
| on | as, appearance | mri, mra, mrv, xray, projection, ct, chest, brain, head |
| for | evidence, assess | - |
| is/are | there, finding, findings, noted, demonstrated, size, this, nonspecific | normal, stable, clear, maintained, preserved, age-appropriate, unchanged, identified, made, within, recommended, removed, repositioned, replaced, low, high, seen, located, present, absent, unremarkable, remarkable, marked, approximately, noted, no, in, large, larger, small, smaller, otherwise, intact, slightly |
| has been | - | removed, repositioned, replaced, reviewed, resolution |
| within | - | normal |
| has/have | - | increased, decreased, improved |
| above | described | - |
| by | edited, recommended, suggested, reviewed, signed, read | - |
| over | - | to |
| at | - | time mentions (example format-8:18 AM) |
| on/to | - | left, right (second term to the right) |
| all | image, imaging, radiograph, radiography, film, series, comparison is made, time, map (any of these terms to the left of the trigger with window length 3) | image, imaging, radiograph, radiography, film, series, comparison is made, time, map (any of these terms to the right of the trigger with window length 3) |
References
- Asim Muhammad Nabeel, Khan Muhammad Usman Ghani, Malik Muhammad Imran, Dengel Andreas, and Ahmed Sheraz. 2019. A Robust Hybrid Approach for Textual Document Classification.
- Datta Surabhi and Roberts Kirk. 2020. A dataset of chest X-ray reports annotated with Spatial Role Labeling annotations. Data in Brief, 32:106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta Surabhi, Si Yuqi, Rodriguez Laritza, Shooshan Sonya E, Demner-Fushman Dina, and Roberts Kirk. 2020a. Understanding spatial language in radiology: Representation framework, annotation, and spatial relation extraction from chest X-ray reports using deep learning. Journal of Biomedical Informatics, 108:103473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta Surabhi, Ulinski Morgan, Godfrey-Stovall Jordan, Khanpara Shekhar, Riascos-Castaneda Roy F., and Roberts Kirk. 2020b. Rad-SpatialNet: A Frame-based Resource for Fine-Grained Spatial Relations in Radiology Reports. In Proceedings of The 12th Language Resources and Evaluation Conference, pages 2251–2260. [PMC free article] [PubMed] [Google Scholar]
- Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pages 4171–4186. Association for Computational Linguistics. [Google Scholar]
- Gupta Khushbu Er., Thammasudjarit Ratchainant, and Thakkinstian Ammarin. 2019. A Hybrid Engine for Clinical Information Extraction from Radiology Reports. In 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), pages 293–297. [Google Scholar]
- Alistair E.W. Johnson, Pollard Tom J., Shen Lu, Lehman Li Wei H., Feng Mengling, Ghassemi Mohammad, Moody Benjamin, Szolovits Peter, Celi Leo Anthony, and Mark Roger G.. 2016. MIMIC-III, a freely accessible critical care database. Scientific Data, 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Xusheng, Fu Chengcheng, Zhong Ran, Zhong Duo, He Tingting, and Jiang Xingpeng. 2019. A hybrid deep learning framework for bacterial named entity recognition with domain features. BMC Bioinformatics, 20(16):583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink Bryan, Roberts Kirk, Harabagiu Sanda, Scheuermann Richard H, Toomay Seth, Browning Travis, Bosler Teresa, and Peshock Ronald. 2013. Extracting actionable findings of appendicitis from radiology reports using natural language processing. In AMIA Joint Summits on Translational Science Proceedings, volume 2013, page 221. [PMC free article] [PubMed] [Google Scholar]
- Roberts Kirk, Rink Bryan, Harabagiu Sanda M, Scheuermann Richard H, Toomay Seth, Browning Travis, Bosler Teresa, and Peshock Ronald. 2012. A machine learning approach for identifying anatomical locations of actionable findings in radiology reports. In AMIA Annual Symposium Proceedings, volume 2012, pages 779–788. [PMC free article] [PubMed] [Google Scholar]
- Si Yuqi, Wang Jingqi, Xu Hua, and Roberts Kirk. 2019. Enhancing clinical concept extraction with contextual embeddings. Journal of the American Medical Informatics Association, 26(11):1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xiaosong, Peng Yifan, Lu Le, Lu Zhiyong, Bagheri Mohammadhadi, and Summers Ronald M.. 2017. ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 3462–3471. [Google Scholar]


