Abstract
The currently emerging pathogen SARS-CoV-2 has produced the global pandemic crisis by causing COVID-19. The unique and novel genetic makeup of SARS-CoV-2 has created hurdles in biological research, due to which the potential drug/vaccine candidates have not yet been discovered by the scientific community. Meanwhile, the advantages of bioinformatics in viral research had created a milestone since last few decades. The exploitation of bioinformatics tools and techniques has successfully interpreted this viral genomics architecture. Some major in silico studies involving next-generation sequencing, genome-wide association studies, computer-aided drug design etc. have been effectively applied in COVID-19 research methodologies and discovered novel information on SARS-CoV-2 in several ways. Nowadays the implementation of in silico studies in COVID-19 research has not only sequenced the SARS-CoV-2 genome but also properly analyzed the sequencing errors, evolutionary relationship, genetic variations, putative drug candidates against SARS-CoV-2 viral genes etc. within a very short time period. These would be very needful towards further research on COVID-19 pandemic and essential for vaccine development against SARS-CoV-2 which will save public health.
Keywords: SARS-CoV-2, COVID-19, Bioinformatics, Next generation sequencing, Genome wide association study, Drug design
1. Introduction
Due to the small genome size, viruses have complex methods to maximize the coding potential of genomes and evaluation (Gautam et al., 2019). Meanwhile, the introduction of genomics and bioinformatics have contributed enormously to understand the infectious disease from disease pathogenesis, mechanisms and the spread of antimicrobial resistance to host immune responses (Bah et al., 2018).
SARS-CoV-2, which has created world pandemic scenario by affecting not only public health but also the socio-economic status of the entire humankind. The genome of the novel severe acute respiratory syndrome 2 (SARS-CoV-2) has been observed to be between 29.8 kb to 29.9 kb in size, and its sequence differs substantially from some of the previously identified human corona viruses including SARS and the Middle East respiratory syndrome (MERS) (Khailany et al., 2020; Chaw et al., 2020). However, the proper investigation of epidemiological, virological and pathogenic characteristics of SARS-CoV-2 is crucial to introduce novel treatment approaches and to develop effective prevention strategies (Messina et al., 2020). For the above bioinformatics tools and techniques have been implemented.
2. Next-generation sequencing
Advances in Next-Generation sequencing (NGS) innovations have brought about a remarkable multiplication of genomic sequence data (Suwinski et al., 2019). NGS has revolutionized the scale and deepness of biomedical sciences. During an outbreak condition in a health care system, the fast and effective identification of causative pathogen with epidemiological surveys are needed to permit a focused on disease control reaction. The accuracy of NGS in viral variants has productively analyzed and quantify the extremely high diversity within viral quasi-species. Many low frequency discovered drug or vaccine resistant mutations of therapeutic importance (Lu et al., 2020). High throughput sequencing technologies, including whole-genome sequencing (WGS) metagenomics technique, are providing the possibility to rapidly obtain the full sequence of pathogen genomes.
2.1. Metagenomics
The in silico virus sequencing is often based on alignments mapping of reads against a reference sequence (Maurier et al., 2019). Whereas a simple, cost-effective approach metagenomics is the only approach, which does not require reference sequence for analysis. It represents a powerful application for pathogen identification from the environmental samples and directly accessing the genetic content of the organism during emerging pandemics situations (Peddu et al., 2020; Thomas et al., 2012). Metagenomics applications have also introduced in recent COVID-19 pandemics to reveal some critical novel information regarding SARS-CoV-2. The metagenomics has been used for rapid identification and quick characterization of the first few cases of COVID19 (Chen et al., 2020; Manning et al., 2020), for examining the SARS-CoV-2 with other co-infections in nasopharyngeal throat swabs of patients (Vardhan and Sahoo, 2020), identification of the intermediate host in transferring the infection to human body (Lam et al., 2020), screening of the homologous sequence of SARS-CoV-2 in other organisms (Wahba et al., 2020), the effect of SARS-CoV-2 in human faecal microbiome alterations (Zuo et al., 2020), clinical SARS-CoV-2 infection with bacterial co-infections (Peddu et al., 2020) etc. These findings have helped and are helping, the clinicians for better isolation of COVID-19 patients with different symptoms (Table 1 ). There are certain software and databases have reportedly used for interpretation of metagenomics applications (Table 2 ).
Table 1.
Application of metagenomics in different experimental studies on SARS-CoV-2.
| Author and publication year | Objectives of the study | Sequencing platform | Findings |
|---|---|---|---|
| Peddu et al., 2020 | Studied on SARS-CoV-2 epidemic, laboratory-confirmed positive and negative samples from Seattle, Washington | Illumina MiSeq |
|
| Chen et al., 2020 | Investigated two pneumonia patients who developed acute respiratory syndromes after independent contact history with Wuhan sea food market | Illumina Miseq |
|
| Manning et al., 2020 | Quick characterization of Cambodia's first case of COVID-2019 | iSeq100 Illumina |
|
| Van Tan et al., 2020 | Isolation of other pathogen co-infections in people with COVID-19 | Illumina MiSeq |
|
| Tsan-Yuk-Lam et al., 2020 | Identification of any intermediate host for SARS-CoV-2 infection transmission to human | Illumina HiSeq |
|
| Wahba et al., 2020 | Examined close matches to the severe acute respiratory syndrome coronavirus 2 | NA |
|
| Zuo et al., 2020 | Investigated temporal transcriptional activity of SARS-CoV-2 and its association with longitudinal faecal microbiome alterations in patients with COVID-19 | Illumina NextSeq 550 |
|
Table 2.
Basic Bioinformatics Databases/Tools useful in COVID19 Next Generation Sequencing Data Analysis (Meta Genomics and Whole Genome Sequencing).
| Databases/Tools | Applications | References |
|---|---|---|
| Sequence Read Archive (SRA) Database (https://www.ncbi.nlm.nih.gov/sra) | It is the largest publicly available repository of high throughput sequencing data, stores raw sequencing data and alignment information. | Leinonen et al., 2011a |
| European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/browser/) | Provides a comprehensive record on DNA and RNA raw sequencing and assembly data. | Leinonen et al., 2011a, Leinonen et al., 2011b |
| Metagenomics | ||
| FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) | Used to check quality control on raw sequences generated from high throughput sequencing pipelines. | Brown et al., 2017 |
| Cutadapt (https://cutadapt.readthedocs.io/en/stable/) | Used to clean the sequences. It finds and removes adapter sequences, primers, poly-A tails and other types of unwanted sequence from the high-throughput sequencing reads. | Martin, 2011 |
| Qiime (http://qiime.org/) | An open-source bioinformatics pipeline for performing microbiome analysis from raw DNA sequencing data. It interprets demultiplexing and quality filtering, OTU picking, taxonomic assignment, and phylogenetic reconstruction, and diversity analyses and visualizations through command lines. | Kuczynski et al., 2011 |
| Whole genome sequencing | ||
| FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) | Used to check quality control on raw sequences generated from high throughput sequencing pipelines. | Brown et al., 2017 |
| Cutadapt (https://cutadapt.readthedocs.io/en/stable/) | Used to clean the sequences. It finds and removes adapter sequences, primers, poly-A tails and other types of unwanted sequence from the high-throughput sequencing reads. | Martin, 2011 |
| MaSuRCA (https://github.com/alekseyzimin/masurca) | Genome Assembler | Zimin et al., 2013 |
| Ragout (https://github.com/fenderglass/Ragout) | A reference assisted assembly tool. Records contigs to create high quality scaffolds by using a genome rearrangement approach and multiple closely related genome references as a guide. | Kolmogorov et al., 2014 |
| Prokka (https://kbase.us/applist/apps/ProkkaAnnotation/annotate_contigs/release?gclid=Cj0KCQiAzZL-BRDnARIsAPCJs729c42yhrdcRV0tbPIaJ5NVefVzYHwx5kDILF1ndoV-P5_Ue1qstiYaAgWrEALw_wcB) | Rapid annotation of prokaryotic genomes. | Seemann, 2014 |
| AUGUSTUS (http://augustus.gobics.de/) | A tool to predict genes in eukaryote genome sequences. | Stanke and Morgenstern, 2005 |
2.2. Whole genome sequencing
Obtaining virus genome sequence directly from clinical samples is still a challenging task due to the low load of virus genetic material compared to the host DNA and the difficulty to get an accurate genome assembly (Maurier et al., 2019). By the time genome sequencing procedure of virus has become a convenient method for better understanding of virus pathogenicity and epidemiological surveillance. Whole-genome sequencing (WGS) is a potent implement for studying virus evolution and genetic association to diseases or for tracking outbreaks. The depth of the sequencing data and the quality of the obtained sequences make this approach particularly efficient in this context (Kremer et al., 2017).
For the early understanding and diagnosis of COVID-19, the whole genome sequencing of SARS-CoV-2 was done for the samples collected from different countries throughout the world by using NGS platforms like Illumina miseq, Roche etc. (Sah et al., 2020; Yadav et al., 2020; Sekizuka et al., 2020; Chong et al., 2020; Caly et al., 2020) (Table 2). The use of nanopore sequencing is used for genome sequencing of SARS-CoV-2 (Caly et al., 2020) (Table 3 ). The available whole genome sequences of SARS-CoV-2 in various online databases, and data analysis software provides insights into the further genomic data analysis to offer better medications to the patients (Table 2).
Table 3.
Whole genome sequencing (WGS) of SARS-CoV-2 strains in different COVID19 research studies.
| Author and Publication Year | Objectives of the Study | Platform | Findings |
|---|---|---|---|
| Sah et al., 2020 | Whole genome sequencing of SARS-CoV-2 specimen isolated from COVID-19 patients of Nepal | Illumina miSeq |
|
| Yadav et al., 2020 | Characterization of SARS-CoV-2 sequences isolated from India with travel history of China | Illumina miniseq |
|
| Sekizuka et al., 2020 | Characterization of SARS-CoV-2 genome, isolated from Japan with travel history of Egypt | Illumina |
|
| Chong et al., 2020 | Whole genome sequencing and analysis of SARS-CoV-2 isolated from Malaysia | Illumina iseq |
|
| Caly et al., 2020 | To describe the first isolation and sequencing of SARS-CoV-2 in Australia and rapid sharing of the isolate | Oxford Nanopore Technologies and Illumina short-read |
|
3. Genome-wide association study
GWAS has rehabilitated the complex disease genetics in to modest by providing various convincing links between complex characteristics of human and disease. Comprehensive and accurate detection of variants from whole-genome sequencing is a definite prerequisite for translational genomic research (Hwang et al., 2019). GWAS has involved in the screening of genetic variants across the genomes of many individuals to identify genotype-phenotype associations. Genetic variants discovered by GWAS are used to identify individuals at high risk of deadly diseases, which influences the early detection and prevention of diseases (Tam et al., 2019).
A genome wide association study (GWAS) is an extensive genetic analysis of the disease-associated observable alleles in the host/pathogen in the form of single nucleotide polymorphisms (SNP) (Patron et al., 2019). The use of GWAS applications including sequence analysis, alignment, genetic/nucleotide variations in the form of SNPs, genomic structure and alterations, primer design etc. have represented novel insights in case of SARS-CoV-2 experiments by accurately detect and quantify rare viral variants within the species (Khailany et al., 2020; Ellinghaus et al., 2020; Aiewsakun et al., 2020; Ray et al., 2020a) (Table 4 ). In addition to the SNP analysis, the incorporation of haplotype diversity analysis with phylogenetic analysis has been frequently used in the SARS-CoV-2 research analyses to study the evolution and population demography of SARS-CoV-2 globally (Ramírez et al., 2020; Fang et al., 2020). The molecular and evolutionary relationship with other coronavirus species, closely related species identification etc. have been efficaciously analyzed through phylogenetic study. This provides additional data for proper genomic assessment of SARS-CoV-2 (Ray et al., 2020b; Tabibzadeh et al., 2020; Satpathy, 2020; Joshi and Paul, 2020; Zhou et al., 2020; Lopes et al., 2020) (Table 4).
Table 4.
Interpretation of genome wide association studies (GWAS) for characterization of SARS-CoV-2 genomes.
| Author and Publication Year | Objective | Findings |
|---|---|---|
| Khailany et al., 2020 | Understand the genomic structure and variations in SARS-CoV-2 complete genome sequences |
|
| Ellinghaus et al., 2020 | Identification of potential genetic factors involved in the development of Covid-19 |
|
| Aiewsakun et al., 2020 | Identification of Genetic variation associated with COVID-19 severity |
|
| Ray et al., 2020b | Elucidation of Nucleotide polymorphisms in whole genome sequences of SARS-CoV-2 |
|
| Tabibzadeh et al., 2020 | Investigate and track SARS-CoV-2 in Iranian COVID-19 patients |
|
| Satpathy, 2020 | Investigation on source of origin of this novel coronavirus |
|
| Joshi and Paul, 2020 | Highlight the similarities and changes observed in the submitted Indian viral strains |
|
| Zhou et al., 2020 | Analyse the evolution and variation of SARS-CoV-2 during the epidemic starting at the end of 2019 |
|
| Lopes et al., 2020 | Investigate bats and pangolin as hosts in SARS-CoV-2 cross-species transmission |
|
Also to prevent the false positive results during testing of COVID-19 through real-time polymerase chain reaction (rtPCR) and decreasing the need for standardization across different PCR protocols, some primers have been designed through in silico algorithms by targeting conserved segments in viral genome (Lanza et al., 2020; Lopez-Rincon et al., 2020; Toms et al., 2020). This generated novel information on SARS-CoV-2 infectious genes are helping the researchers in the vaccine development against SARS-CoV-2, according to the identified viral genes coding regions, genetic sequence variations and molecular differentiations between the isolated species throughout the world. All the reported genomic experiments and analyses including SNP study, phylogenetic analysis, primer designing etc. have been carried out through high throughput bioinformatics tools and techniques which provide an appropriate pipeline for data analyses and annotations (Table 5 ).
Table 5.
List of researches reported on in silico drug design (CADD) against viral proteins of SARS-CoV-2.
| Author and Publication Year | Objective of the Study | Target Protein | Findings |
|---|---|---|---|
| Prasanth et al., 2020 | identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2 | Mpro and Spike |
|
| Hall Jr and Ji, 2020 | Identification of effective inhibors against Spike glycoprotein and 3CL protease of SARS-CoV-2 | Spike and 3CL Pro |
|
| Wei et al., 2020 | Selection of potential molecules that can target viral spike proteins | Spike protein |
|
| Fantini et al., 2020 | Studied the effects of Chloroquine and Hydroxychloroquine for treating Covid-19 | Spike Protein |
|
| BR et al., 2020 | Screening of small molecules to bind ACE2 specific RBD on Spike glycoprotein of SARS-CoV-2 | Spike protein |
|
| Cavasotto and Di Filippo, 2020 | Docking-based screening from approved drugs and compounds undergoing clinical trials, against three SARS-CoV-2 target proteins | Spike, M pro, Papain like protease |
|
| Vardhan and Sahoo, 2020 | Virtual screening of phytochemicals against viral proteins of SARS-CoV-2 | Spike, Mpro, 3CL pro, PL pro, ACE2, RdRp |
|
| Panda et al., 2020 | Structure-based drug designing and immunoinformatics approach for SARS-CoV-2 |
Spike glycoprotein, M pro, ACE2 |
|
| Sarma et al., 2020 | Homology assisted identification of inhibitor against RNA binding domain of N protein | Nucleocapsid protein |
|
| Ray et al., 2020a | Potential drug compound identification against Covid-19 | Nucleocapsid protein |
|
| Bhowmik et al., 2020 | Identify potential drug candidates against SARS-CoV-2 structural proteins | Membrane, Envelope and Nucleocapsid protein |
|
| Lavecchia and Fernandez, 2020 | Stabilization of non-native Protein-Protein Interactions (PPIs) of the nucleocapsid protein for inhibit viral replication in SARS-CoV-2 | Nucleocapsid Protein |
|
| Gupta et al., 2020 | Detection of inhibitors of SARS-CoV-2 ion channel to control covid-19 | Envelope protein |
|
| Jo et al., 2020 | Screening of flavonoinds against 3CL pro of SARS-CoV-2 | 3CL pro |
|
| Kumar et al., 2020 | Inhibitors screening and drug discovery against main protease (Mpro) of SARS-CoV-2 | Mpro |
|
4. Computer aided drug design
Drug design is very challenging, expensive, time consuming and an integrated rising discipline (Bisht and Singh, 2019). In the interim, the field of bioinformatics has become a crucial part of the drug design that plays a vital role for the validation of drug targets. It can help in the understanding of complex biological processes to improve drug discovery (Choudhury and Saikia, 2018). The in silico screening or computer-aided drug design (CADD) has signified as a dominant practice because of its proper algorithms including the development of digital repositories for the study of chemical interaction relationships, computer programs for designing compounds with unusual physicochemical characteristics as well as tools for systematic assessment of potential lead candidates etc. in drug discovery and development (Song et al., 2009). Also, the additional benefits like cost-saving, time to market, in-sight knowledge of drug-receptor interaction, speed up in drug discovery and development increases its popularity in scientific researches (Ramírez et al., 2020).
The potentiality of CADD has been exploited to the fullest in finding a solution for this COVID-19 outbreak. Researchers have taken the privilege of CADD including structure-based drug design, network-based drug design towards the identification of potential drug candidates against the identified viral proteins including Spike (S) protein (Prasanth et al., 2020; Hall Jr and Ji, 2020; Wei et al., 2020; Fantini et al., 2020; BR et al., 2020; Cavasotto and Di Filippo, 2020; Vardhan and Sahoo, 2020; Panda et al., 2020), Nucleocapsid (N) protein (Sarma et al., 2020; Ray et al., 2020a; Bhowmik et al., 2020; Lavecchia and Fernandez, 2020), Envelop protein (Bhowmik et al., 2020; Lavecchia and Fernandez, 2020; Gupta et al., 2020), Membrane (M) Protein (Bhowmik et al., 2020), Main protease (M pro) (Prasanth et al., 2020; Cavasotto and Di Filippo, 2020; Vardhan and Sahoo, 2020; Panda et al., 2020; Kumar et al., 2020), 3CL protease (Hall Jr and Ji, 2020; Vardhan and Sahoo, 2020; Jo et al., 2020) of SARS-CoV-2 by using the bioinformatics tools and software (Table 6 ). This immediate and effective action has not only predicted novel putative natural inhibitors but also re-experimented some previously used ancient synthetic drugs with antiviral activities like chloroquine (malaria), hydroxylchloroquine (maalaria), zanamivir (influenza A & B virus), indinavir (HIV), saquinavir (HIV), remdesivir (SARS-CoV), ralterravin (HIV), streptomycine, ciprofloxacin, zanamivir (influenza virus), glycyrrhizic acid (anti inflammation) etc. against SARS-CoV-2 (Hall Jr and Ji, 2020; Fantini et al., 2020; BR et al., 2020; Panda et al., 2020; Ray et al., 2020b) (Table 6). For the successful completion of CADD, various bioinformatics tools and databases have been used since last decades and would be used in further research (Table 7 ).
Table 6.
Basic Bioinformatics Databases/Tools useful for COVID19 genomics research.
| Databases/ Tools | Application | References |
|---|---|---|
| GEO (Gene Expression Omnibus) database (https://www.ncbi.nlm.nih.gov/geo/) | It is a repository of functional genomics data generated from experiments and stores curate gene expression profiles. | Clough and Barrett, 2016 |
| NCBI Gene database (https://www.ncbi.nlm.nih.gov/gene/) | Repository of gene related information from a wide range of species. | Brown et al., 2015 |
| UCSC genome Browser (https://genome.ucsc.edu/) | Broad collection of vertebrate and model organism assemblies and annotations, along with a large suite of tools for viewing, analyzing and downloading genomic data. | Karolchik et al., 2009 |
| UniProt (https://www.uniprot.org/) | Resource of protein sequence and functional information | UniProt Consortium, 2008 |
| CD (Conserved Domain) Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) | Conserved domain search through multiple and pair wise sequence alignments. | Ray et al., 2020a |
| DAVID (Database for Annotation, Visualization and Integrated Discovery) | Functional annotation of genes (Biological process, Molecular function, Cellular component) | Huang et al., 2007 |
| KEGG (Kyoto Encyclopaedia of Genes and Genome) | Metabolic pathway analysis | Kanehisa and Goto, 2000 |
| Discovery of Single Nucleotide Polymorphisms | ||
| dbSNP (https://www.ncbi.nlm.nih.gov/snp/) | A crucial repository for each single base nucleotide substitutions and quick deletion and insertion polymorphisms | Sherry et al., 2001 |
| SIFT (https://sift.bii.a-star.edu.sg/) | Predicts effects of an amino acid substitution on protein function based on sequence homology and the physical properties of amino acids. | Sim et al., 2012 |
| PredictSNP1 (https://loschmidt.chemi.muni.cz/predictsnp1/) | Consensus classifier for prediction of disease related amino acid mutations. | Rath et al., 2020 |
| PredictSNP2 (https://loschmidt.chemi.muni.cz/predictsnp2/) | Platform for prediction of effects of SNPs in genomic region. | Bendl et al., 2016 |
| PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/) | Predicts possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations. | Ray et al., 2019 |
| PROVEAN (http://provean.jcvi.org/index.php) | Predicts impact of an amino acid substitution or indel on the biological function of a protein. | Ray et al., 2019 |
| SNAP2 (https://rostlab.org/services/snap/) | Predicts functional effects of sequence variants. | Ray et al., 2019 |
| Phylogenetic Analysis | ||
| MEGA (Molecular Evolutionary Genetics Analysis) (https://www.megasoftware.net/) | Multiple sequence alignment, phylogenetic tree generation and statistical analyses. | Kumar et al., 2008 |
| Phylogeny.fr (https://www.phylogeny.fr/) | Reconstruct and analyse phylogenetic relationships between molecular sequences. | Dereeper et al., 2008 |
| PAUP (https://paup.phylosolutions.com/) | Reconstruct and analyse phylogenetic relationships between molecular sequences using parsimony method. | Wilgenbusch and Swofford, 2003 |
| DnaSP (http://www.ub.edu/dnasp/) | Analyse DNA polymorphisms using data from a single locus, and also generate haplotype diversity between the sequences. | Rozas et al., 2017 |
| PopArt (http://popart.otago.ac.nz/index.shtml) | Population genetic software which visualizes haplotype diversity network. | Leigh and Bryant, 2015 |
| Primer Design | ||
| Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/) | Primer design, often in high-throughput genomics applications. | Untergasser et al., 2012 |
| NCBI Primer-Blast (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) | Design new target-specific primers in one step as well as to check the specificity of pre-existing primers and also placing primers based on exon/intron locations and excluding single nucleotide polymorphism (SNP) sites in primers. | Ye et al., 2012 |
Table 7.
Basic Bioinformatics Databases/Tools useful for COVID19 In silico drug design.
| Databases/ Tools | Application | References |
|---|---|---|
| BLAST (Basic local alignment search tool) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) | Used for local similarity between sequences by comparing nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. | Boratyn et al., 2013 |
| PDB (Protein databank) (https://www.rcsb.org/) | Protein three dimensional structure database, it conation information about the 3D shapes of proteins, nucleic acids, and complex assemblies. | Berman et al., 2000 |
| PubChem (https://pubchem.ncbi.nlm.nih.gov/) | Chemical structure database, contains information on chemical compounds including name, molecular formula, chemical and physical properties, biological activities, toxic effects, literatures etc. | Kim et al., 2016 |
| Drug Bank (https://www.drugbank.ca/) | Drugbank contains information on FDA approved drugs and drug targets. It is a both bioinformatics and chemoinformatics resource. | Wishart et al., 2018 |
| Modeller (https://salilab.org/modeller/) | Used for homology or comparative modeling of protein three-dimensional structures by aligning query sequence with known structure. | Eswar et al., 2006 |
| AutoDock (http://autodock.scripps.edu/) | Molecular docking between protein and ligand (small compounds) molecules. | Forli et al., 2016 |
| Autodockvina (http://vina.scripps.edu/) | An open source for molecular docking and it significantly improves the average accuracy of the binding mode predictions compared to AutoDock 4. | Trott and Olson, 2010 |
| Zdock (http://zdock.umassmed.edu/) | An automatic protein docking online server, which simply interprets the protein structures. | Pierce et al., 2011 |
| SwissDock (http://www.swissdock.ch/) | A web service to predict the molecular interactions between a target protein and a small molecule. | Grosdidier et al., 2011 |
| PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/) | A simple molecular docking algorithm based on shape complementarity principles. | Schneidman-Duhovny et al., 2005 |
| Glide (https://www.schrodinger.com/glide) | It offers the full range of speed vs. accuracy options, from the high-throughput virtual screening mode for efficiently enriching million compound libraries for reliably docking tens to hundreds of thousands of ligand with high accuracy, advanced scoring, and higher enrichment of results. | Richard et al., 2004 |
| PyMol (https://pymol.org/2/) | Molecular structure visualization and editing tool. | Seeliger and de Groot, 2010 |
| Discovery Studio Visualizer (https://discover.3ds.com/discovery-studio-visualizer-download) | Structure visualization, and analysis of 3D molecules. | Ray et al., 2020a |
| UCSF Chimera (https://www.cgl.ucsf.edu/chimera/) | Visualization and analysis of molecular structures and related data, including density maps, trajectories, and sequence alignments. Also used for energy minimization of molecules. | Pettersen et al., 2004 |
| Open Babel (http://openbabel.org/wiki/Main_Page) | A chemical toolbox designed to search, convert file format, analyse, or store data from molecular modeling, chemistry, solid-state materials, biochemistry, or related areas. | O'Boyle et al., 2011 |
| Gromacs (http://www.gromacs.org/About_Gromacs) | Molecular dynamics simulation tool | Abraham et al., 2015 |
| NAMD (https://www.ks.uiuc.edu/Research/namd/) | Parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. | Phillips et al., 2005 |
| VMD (https://www.ks.uiuc.edu/Research/vmd/) | Molecular visualization program for displaying, animating, and analyzing large biomolecular systems using 3-D graphics and built-in scripting. | Hsin et al., 2008 |
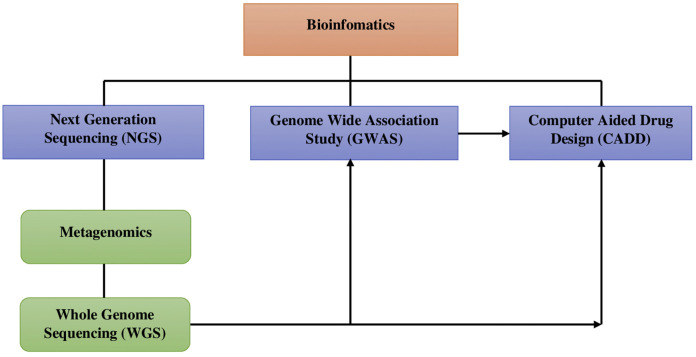
The overall in silico processes are established in an order to perform a task in a sequential manner. From the beginning metagenomics to the end CADD have interconnected and represented the applications of bioinformatics in a single flow diagram (Fig. 1 ).
Fig. 1.
The graphical representation of interconnected bioinformatics applications implemented in COVID-19 research.
5. Limitations
Wide application of robust algorithm based tools and information perceived from several public repository have enriched the knowledge spheres of modern life science research. The available bioinformatics tools and techniques are simple, accurate, cost effective, economical and freely available on internet, enabling their universal use for different research purposes. The above mentioned online repositories including PDB, PubChem, DrugBank, NCBI gene/genome databases, UCSC genome database, Uniprot, dbSNP, GEO, SRA, ENA (Table 2), (Table 5), (Table 7) etc. have updated frequently with huge novel datasets, which provides much authenticated and useful information to the users to carry out their research purposes. However, there is some limitations in use of certain tools particularly used for drug design such as Modeller (Table 7) or any other software generated 3D structure of proteins is approximate, which needs to be properly validated through crystallographic method for further study. The analyzed docking parameters based on predefined algorithms of autodock (Table 7) should be simulate further to analyse the proper stability between target and drug candidate interactions. Likewise, some softwares including Schrodinger, Discovery studio (Table 7), PAUP (Table 5) etc. are creating limitations for researchers during data analysis and accession, as they are customized or paid software. Apart from the above major drawbacks/limitations some minor flaws are associated with the using of tools and software i.e. error during software installation, software dependencies particularity the type of operating systems, high speed internet network connection, high core computer facility etc. The designed tools and software are meant for respective analyses, the user cannot modify the algorithms and outputs according to own interest, the user need to use different respective software for different purposes to get the authenticate results. The knowledge about different programming languages like Perl, R, Python and Linux operating system is necessary to work with different bioinformatics software as well as to rewrite the codes needed to solve particular biological problem computationally, in particular for software used for next generation sequencing analyses.
6. Future aspects
The observations on SARS-CoV-2 will be explored extensively through bioinformatics and its applications variously. The researchers can also elucidate the SNPs in host body after affected with COVID-19. According to the modified nucleotides/genes novel primers can be designed for polymerase chain reaction through computational primer design algorithms. Apart from the drug design, putative inhibitory peptide can be created against SARS-CoV-2 viral genes. These further ideas would exploit many more denovo information of SARS-CoV-2, which will help the clinicians to add novel medication insights in the diagnosis procedures.
7. Conclusion
The outbreak of COVID-19 throughout the world is a big challenge for people to overcome this. Advances in bioinformatics techniques have been proved as the most advanced and effective technique in biomedical research. The high throughput screening and accuracy of data analysis have made this possible. The vast utilization of computational approaches in the current pandemic situation has effectively used from the preliminary stage of viral sample identification to the end stage of drug design by discovering novel information on SARS-CoV-2 genomic contents, variations, diversity within the species and predicted potential drug/ vaccine candidates against the viral genes within a very short period. In the present economically down condition, the successfully implementation of bioinformatics approaches against SARS-CoV-2 is a great achievement for scientific community.
Funding
No funding has been received for this work.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. 2015. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers 1–2, 19–25. [Google Scholar]
- Aiewsakun P., Wongtrokoongate P., Thawornwattana Y., Hongeng S., Thitithanyanont A. SARS-CoV-2 genetic variations associated with COVID-19 severity. MedRxiv [Preprint] 2020 doi: 10.1101/2020.05.27.20114546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah S.Y., Morang’a C.M., Kengne-Ouafo J.A., Amenga-Etego L., Awandare G.A. Highlights on the application of genomics and bioinformatics in the fight against infectious diseases: challenges and opportunities in Africa. Front. Genet. 2018;9 doi: 10.3389/fgene.2018.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendl J., Musil M., Štourač J., Zendulka J., Damborský J., Brezovský J. PredictSNP2: a unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput. Biol. 2016;12(5) doi: 10.1371/journal.pcbi.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik D., Nandi R., Jagadeesan R., Kumar N., Prakash A., Kumar D. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect. Genet. Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht N., Singh B.K. Role of computer aided drug design in drug development and drug discovery. IJPSR. 2019;9(4):1405–1415. [Google Scholar]
- Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Br B., Damle H., Ganju S., Damle L. In silico screening of known small molecules to bind ACE2 specific RBD on Spike glycoprotein of SARS-CoV-2 for repurposing against COVID-19. F1000Research. 2020;9:663. doi: 10.12688/f1000research.24143.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015;43:D36–D42. doi: 10.1093/nar/gku1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J., Pirrung M., McCue L.A. FQC dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017;33(19):3137–3139. doi: 10.1093/bioinformatics/btx373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Druce J., Roberts J., Bond K., Tran T., et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020;212(10):459–462. doi: 10.5694/mja2.50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasotto C., Di Filippo J. In silico drug repurposing for COVID-19: targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol. Inform. 2020 doi: 10.1002/minf.202000115. [DOI] [PubMed] [Google Scholar]
- Chaw S.M., Tai J.H., Chen S.L., Hsieh C.H., Chang S.Y., Yeh S.H., et al. The origin and underlying driving forces of the SARS-CoV-2 outbreak. J. Biomed. Sci. 2020;27(1) doi: 10.1186/s12929-020-00665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y.M., Sam I.C., Ponnampalavanar S., Syed Omar S.F., Kamarulzaman A., Munusamy V. Complete genome sequences of SARS-CoV-2 strains detected in Malaysia. Microbiol Resour Announc. 2020;9(20) doi: 10.1128/MRA.00383-20. e00383-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury M.D., Saikia R. Essential basic protocol in computer aided drug designing: efficiency and challenges. Int J Biotech Bioeng. 2018;4(4):77–80. [Google Scholar]
- Clough E., Barrett T. The gene expression omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., Buti M., et al. Genomewide association study of severe Covid-19 with respiratory failure. NEJM. 2020 doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.Y., Pieper U., Sali A. Comparative protein structure modeling using Modeller. Current Protocols Bioinformatics. 2006;5(5.6) doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B., Liu L., Yu X., Li X., Ye G., Xu J., et al. Genome-wide data inferring the evolution and population demography of the novel pneumonia coronavirus (SARS-CoV-2) bioRxiv [Preprint] 2020 doi: 10.1101/2020.03.04.976662. [DOI] [Google Scholar]
- Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55(5):105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli S., Huey R., Pique M.E., Sanner M.F., Goodsell D.S., Olson A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016;11(5):905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam A., Tiwari A., Malik Y.S. Bioinformatics applications in advancing animal virus research. Recent Adv. Anim. Virol. 2019;6:447–471. [Google Scholar]
- Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1751300. 1538–0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.C., Jr., Ji H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis. 2020;35:101646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J., Arkhipov A., Yin Y., Stone J.E., Schulten K. Using VMD: an introductory tutorial. Curr. Protoc. Bioinformatics. 2008 doi: 10.1002/0471250953.bi0507s24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K.B., Lee I.H., Li H., Won D.G., Hernandez-Ferrer C., Negron J.A., Kong S.W. Comparative analysis of whole-genome sequencing pipelines to minimize false negative findings. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-39108-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Kim D.Y., Kim M.S., Shin D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhibition Medicinal Chemistry. 2020;35(1):1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., Paul S. Phylogenetic analysis of the novel coronavirus reveals important variants in Indian strains. bioRxiv [Preprint] 2020 doi: 10.1101/2020.04.14.041301. [DOI] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Kent W.J. The UCSC genome browser. Curr. Protoc. Bioinformatics. 2009;1:4. doi: 10.1002/0471250953.bi0104s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Thiessen P.A., Cheng T., Yu B., Shoemaker B.A., Wang J., Bolton E.E., Wang Y., Bryant S.H. Literature information in PubChem: associations between PubChem records and scientific articles. J. Cheminformatics. 2016;8:32. doi: 10.1186/s13321-016-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M., Raney B., Paten B., Pham S. Ragout—a reference-assisted assembly tool for bacterial genomes. Bioinformatics. 2014;30(12):i302–i309. doi: 10.1093/bioinformatics/btu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer F.S., McBride A.J.A., Pinto L.S. Approaches for in silico finishing of microbial genome sequences. Genet. Mol. Biol. 2017;40(3):553–576. doi: 10.1590/1678-4685-GMB-2016-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J., Stombaugh J., Walters W.A., González A., Caporaso J.G., Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics. 2011;10:10.7. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y., Singh H., Patel C.N. In silico prediction of potential inhibitors for the Main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J. Infect. Public Health. 2020 doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.Y., Shum M.H.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lanza D.C.F., Lima J.P.M.S., Jeronima S.M.B. Research Square [Preprint] 2020. Design and in silico validation of polymerase chain reaction primers to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavecchia, M., and Fernandez, J., 2020. In silico study of SARS-CoV-2 Nucleocapsid protein-protein interactions and potential candidates for their stabilization. [preprint] 2020070558.
- Leigh J.W., Bryant D. Popart: full-feature software for haplotype network construction. Methods Ecol. Evolut. 2015;6:1110–1116. [Google Scholar]
- Leinonen R., Sugawara H., Shumway M., et al. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R., Akhtar R., Birney E., Bower L., Cerdeno-Tárraga A., Cheng Y., Cleland I., et al. The European nucleotide archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes L.R., de Mattos Cardillo G., Paiva P.B. Molecular evolution and phylogenetic analysis of SARS-CoV-2 and hosts ACE2 protein suggest Malayan pangolin as intermediary host. Braz. J. Microbiol. 2020:1–7. doi: 10.1007/s42770-020-00321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rincon A., Tonda A., Mendoza-Maldonado L., Mulders D.G.J.C., Molenkamp R., Claassen E., et al. Specific primer Design for Accurate Detection of SARS-CoV-2 using deep learning. [preprint] 2020. [DOI] [PMC free article] [PubMed]
- Lu I.N., Muller C.P., He F.Q. Applying next-generation sequencing to unravel the mutational landscape in viral quasispecies. Virus Res. 2020;283:197963. doi: 10.1016/j.virusres.2020.197963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning J.E., Bohl J.A., Lay S., Chea S., Sovann L., Sengdoeurn Y., et al. 2020. Rapid metagenomic characterization of a case of imported COVID-19 in Cambodia. bioRxiv [Preprint] [DOI] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- Maurier F., Beury D., Fléchon L., Varré J.S., Touzet H., Goffard A., Hot D., Caboche S. A complete protocol for whole-genome sequencing of virus from clinical samples: application to coronavirus OC43. Virology. 2019;531:141–148. doi: 10.1016/j.virol.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina F., Giombini E., Agrati C., Vairo F., Bartoli T.A., Aoghazi Sal, et al. COVID-19: viral–host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. J. Transl. Med. 2020;233 doi: 10.1186/s12967-020-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N.M., Banck M., James C.A., et al. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.G. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020;6(28):eabb8097. doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron J., Serra-Cayuela A., Han B., Li C., Wishart D.S. Assessing the performance of genome-wide association studies for predicting disease risk. PLoS One. 2019;14(12) doi: 10.1371/journal.pone.0220215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddu V., Shean R.C., Xie H., Shrestha L., Perchetti G.A., Minot S.S., Roychoudhury P., Huang M.L., et al. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin. Chem. 2020;66(7):966–972. doi: 10.1093/clinchem/hvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen T.D., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce B.G., Hourai Y., Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth D.S.N.B.K., Murahari M., Chandramohan V., Panda S.P., Atmakuri L.R., Guntupalli C. In silico identification of potential inhibitors from cinnamon against main protease and spike glycoprotein of SARS CoV-2. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J.D., Muñoz M., Hernández C., Flórez C., Gomez S., Rico A., Pardo L., Barros E.C., Paniz-Mondolfi A.E. Genetic diversity among SARS-CoV2 strains in South America may impact performance of molecular detection. Pathogens. 2020;9(7):580. doi: 10.3390/pathogens9070580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S.N., Ray M., Patri M. Computational discovery and assessment of non-synonymous single nucleotide polymorphisms from target gene pool associated with Parkinson's disease. Gene Reports. 2020 doi: 10.1016/j.genrep.2020.100947. [DOI] [Google Scholar]
- Ray M., Mishra J., Priyadarshini A., Sahoo S. In silico identification of potential drug target and analysis of effective single nucleotide polymorphisms for autism spectrum disorder. Gene Reports. 2019;16 doi: 10.1016/j.genrep.2019.100420. [DOI] [Google Scholar]
- Ray M., Sarkar S., Rath S.N., Sable M.N. 2020. Elucidation of genome polymorphisms in emerging SARS-CoV-2. bioRxiv [preprint] [DOI] [Google Scholar]
- Ray M., Sarkar S., Rath S.N. Druggability for COVID19 – in silico discovery of potential drug compounds against Nucleocapsid (N) protein of SARS-CoV-2. ChemRxiv [preprint] 2020 doi: 10.26434/chemrxiv.12387290.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard A., Friesner Jay L., Banks Robert B., Murphy Thomas A., Halgren Jasna J., Klicic Daniel T., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Sah R., Rodriguez-Morales A.J., Jha R., Chu D.K.W., Gu H., Peiris M., et al. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol. Resourc. Announc. 2020;9(11) doi: 10.1128/MRA.00169-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Shekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D.P., Medhi B. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain) J. Biomol. Struct. Dyn. 2020;18:1–9. doi: 10.1080/07391102.2020.1753580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy R. In silico based whole genome phylogenetic analysis of novel coronavirus (SARS-CoV-2) Int. J. Emerging Technol. 2020;11(3):1157–1163. [Google Scholar]
- Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24(5):417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. Jul 15, Epub 2014 Mar 18. [DOI] [PubMed] [Google Scholar]
- Sekizuka T., Kuramoto S., Nariai E., Taira M., Hachisu Y., et al. SARS-CoV-2 genome analysis of Japanese travelers in Nile River cruise. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.M., Lim S.J., Tong J.C. Recent advances in computer-aided drug design. Brief. Bioinform. 2009;10(5):579–591. doi: 10.1093/bib/bbp023. [DOI] [PubMed] [Google Scholar]
- Stanke M., Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwinski P., Ong C., Ling M.H.T., Poh Y.M., Khan A.M., Ong H.S. Advancing Personalized Medicine Through the Application of Whole Exome Sequencing and Big Data Analytics. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabibzadeh A., Zamani F., Laali A., Esghaei M., Tameshkel F.S., Keyvani H., et al. SARS-CoV-2 molecular and phylogenetic analysis in COVID-19 patients: a preliminary report from Iran. Infect. Genet. Evol. 2020;104387 doi: 10.1016/j.meegid.2020.104387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019 doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- Thomas T., Gilbert J., Meyer F. Metagenomics - a guide from sampling to data analysis. Microbial Inform. Exp. 2012;2(1):3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toms D., Li J., Cai H.Y. Evaluation of WHO listed COVID-19 qPCR primers and probe in silico with 375 SERS-CoV-2 full genome sequences. MedRxiv [Preprint] 2020 doi: 10.1101/2020.04.22.20075697. [DOI] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40(15) doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tan L., Thi Thu Hong N., My Ngoc N., Tan Thanh T., Thanh Lam V., et al. SARS-CoV-2 and co-infections detection in nasopharyngeal throat swabs of COVID-19 patients by metagenomics. J. Inf. Secur. 2020;81(2):e175–e177. doi: 10.1016/j.jinf.2020.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhan S., Sahoo S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020;124:103936. doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Jain N., Fire A.Z., Shoura M.J., Artiles K.L., McCoy M.J., Jeong D.E. An extensive Meta-metagenomic search identifies SARS-CoV-2-homologous sequences in pangolin lung viromes. mSphere. 2020;5(3) doi: 10.1128/mSphere.00160-20. e00160-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T.Z., Wang H., Wu X.Q., Lu Y., Guan S.H., Dong F.Q., Dong C.L., et al. In silico screening of potential spike glycoprotein inhibitors of SARS-CoV-2 with drug repurposing strategy. Chin J Integr Med. 2020;1:1–7. doi: 10.1007/s11655-020-3427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilgenbusch J.C., Swofford D. Inferring evolutionary trees with PAUP. Curr Protoc Bioinformatics. 2003;6(6.4) doi: 10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Potdar V.A., Choudhary M.L., Nyayanit D.A., Agrawal M., Jadhav S.M., et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J. Med. Res. 2020;151(2 & 3):200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhang S., Chen J., Wan C., Zhao W., Zhang B. Analysis of variation and evolution of SARS-CoV-2 genome. Nan Fang Yi Ke Da Xue Xue Bao. 2020;40(2):152–158. doi: 10.12122/j.issn.1673-4254.2020.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V., Marçais G., Puiu D., Roberts M., Salzberg S.L., Yorke J.A. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T., Liu Q., Zhang F., Lui G.C., Tso E.Y., Yeoh Y.K., et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2020:2020–322294. doi: 10.1136/gutjnl-2020-322294. [DOI] [PMC free article] [PubMed] [Google Scholar]