Abstract
The regenerating islet-derived family member 4 (Reg4) in the gastrointestinal tract is up-regulated during intestinal inflammation. However, the physiological function of Reg4 in the inflammation is largely unknown. In the current study, the functional roles and involved mechanisms of intestinal epithelial Reg4 in intestinal inflammation were studied in healthy and inflamed states using human intestinal specimens, an intestinal conditional Reg4 knockout mouse (Reg4ΔIEC) model and dextran sulfate sodium (DSS)-induced colitis model. We showed that the elevated serum Reg4 in pediatric intestinal failure (IF) patients were positively correlated with the serum concentrations of proinflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α). In inflamed intestine of IF patients, the crypt base Reg4 protein was increased and highly expressed towards the luminal face. The Reg4 was indicated as a novel target of activating transcription factor 2 (ATF2) that enhanced Reg4 expression during the intestinal inflammation. In vivo, the DSS-induced colitis was significantly ameliorated in Reg4ΔIEC mice. Reg4ΔIEC mice altered the colonic bacterial composition and reduced the bacteria adhere to the colonic epithelium. In vitro, Reg4 was showed to promote the growth of colonic organoids, and that this occurs through a mechanism involving activation of signal transducer and activator of transcription 3 (STAT3). In conclusion, our findings demonstrated intestinal-epithelial Reg4 deficiency protects against experimental colitis and mucosal injury via a mechanism involving alteration of bacterial homeostasis and STAT3 activation.
Introduction
The regenerating islet-derived family member 4 (Reg4) is the most recently discovered member of the Reg gene family.1–3 In human, the Reg gene family members Reg1A, Reg1B, and Reg3A are found encoded in tandem on chromosome 2p12, whereas Reg4 is located on chromosome 1p12.4,5 Unlike Reg1A, Reg1B, and Reg3A that are derived from Paneth cells,6–9 the Reg4 is physiologically expressed in enteroendocrine cells and expands to epithelial cells during intestinal inflammation.10–14 Recent studies reported that Reg4 is overexpressed in several types of gastrointestinal tract (GI tract) malignancies, indicating Reg4 might have a prognostic or a predictive value in cancers of the GI tract.15–20 Although the biological function of Reg4 in cancer of GI tract is still unclear, Reg4 protein seems to act as growth factor in malignant cells.21,22 Additionally, Reg4 is also strongly up-regulated during intestinal inflammation,23,24 but the functional roles of Reg4 as well as the regulation of Reg4 expression in intestinal inflammation remained elusive. In intestine, the accumulation of several Paneth and epithelial cell-derived antimicrobial peptides and proteins (AMP) is vital to maintain the immune homeostasis via avoiding colonization of the epithelial cell surface and invasion by opportunistic pathogens.25 One key human antimicrobial protein is Reg3α, which has been reported to be bactericidal via binding to peptidoglycan of Gram-positive bacteria.6,26 It is reminded that Reg4 may be involved in intestinal inflammation via alteration of interacted relationships between microbiota and the intestinal surface.
In the present study, to unravel the roles and mechanisms of Reg4 in the intestinal inflammation, we initially performed population based cross-sectional study on Reg4 in relation to inflammatory response in pediatric intestinal failure (IF) patients, who usually have severe intestinal inflammation.27 In addition, we generated mice lacking Reg4 specifically in intestinal epithelial cells (Reg4ΔIEC) to study the exact roles of Reg4 in intestinal inflammation.
Results
Reg4 is selectively expressed in the mucosa of uninflammed intestine
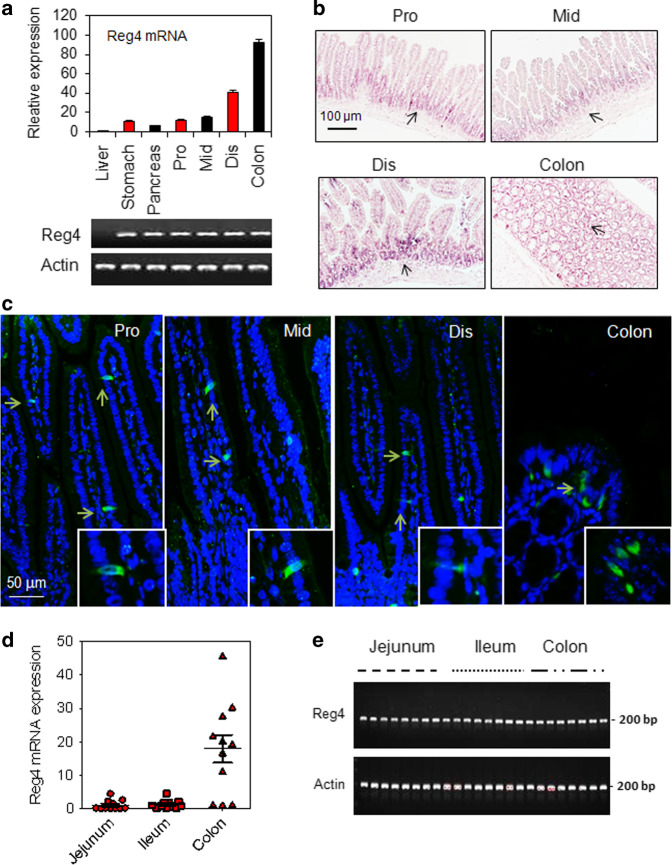
Under normal states, Reg4 expressed at the mucosa of mouse proximal (pro), middle (mid), distal (dis) small bowel and colon (Fig. 1a–c). The expression of Reg4 mRNA was higher in the colon then in the small intestine (Fig. 1a). The colorimetric in situ hybridization (CISH) assay and immunofluorescence (IF) staining showed that Reg4 protein exclusively expressed in intestinal mucosa under normal condition, mainly presenting at the basement of crypts (Fig. 1b, c). Consistent with this finding, the real-time PCR (RT-PCR) showed that Reg4 mRNA was also strongly expressed in mucosa of human jejunum, Ileum, and colon (Fig. 1d, e).
Fig. 1.
Reg4 predominantly expressed in intestine of mice and human. a Quantification of Reg4 mRNA in the mouse liver, stomach, pancreas, proximal (pro), middle (mid), distal (dis) small bowel and colon. b Colorimetric in situ hybridization (CISH) analysis for Reg4 in mouse proximal (pro), middle (mid), distal (dis) small bowel and colon. c Immunofluorescence staining for Reg4 in mouse proximal (pro), middle (mid), distal (dis) small bowel and colon. d Real time-PCR (RT-PCR) analysis for Reg4 in jejunum, ileum and colon of intestinal failure (IF) patients. e Representative images of the DNA agarose gels of panel d
Reg4 is increasingly expressed in intestinal epithelial cells during inflammation
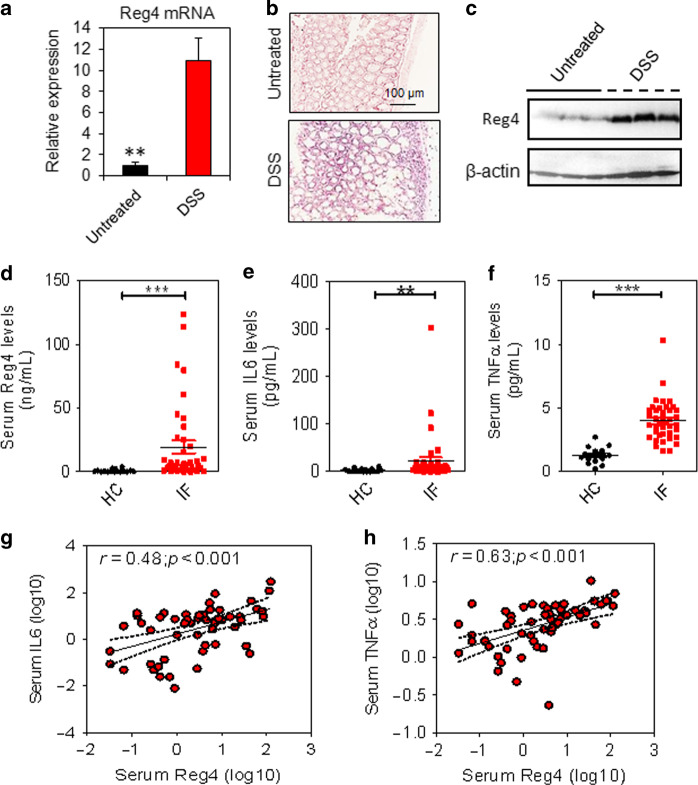
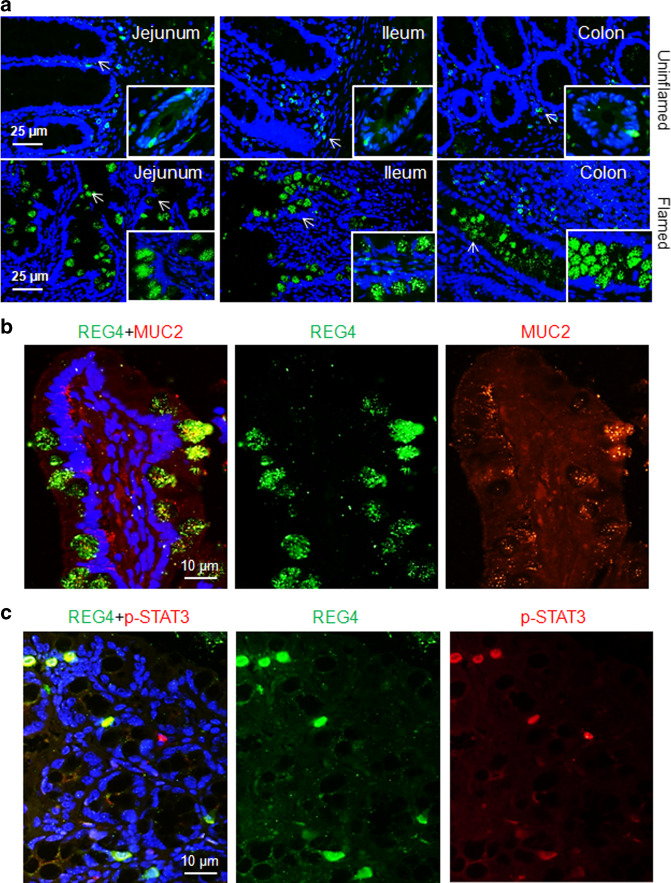
When mice challenged with 2% dextran sulfate sodium (DSS), the Reg4 mRNA increased significantly in the mucosa of colon (Fig. 2a). In mice given DSS, Reg4 expanded from crypt base to the epithelial cells (Fig. 2b). Consistent with the findings in Reg4 mRNA, the Reg4 protein was evidently increased in the mucosa of mice with DSS treatment (Fig. 2c). In pediatric IF patients, serum Reg4 levels were significantly higher [n = 40, 5.441 ng/ml (1.626–24.06), p < 0.001] than in age-matched healthy controls [n = 16, 0.284 ng/ml (0.082–.910)] using ELISA analysis (Fig. 2d). The proinflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) were also increased significantly in serum of pediatric IF patients when compared to controls (Fig. 2e, f). In addition, serum Reg4 protein levels were positively correlated with the concentrations of serum IL-6 (r = 0.48, p < 0.001) and serum TNF-α (r = 0.63, p < 0.001) (Fig. 2g, f). In uninflammed areas of patients’ intestine, the Reg4 protein was selectively expressed at crypts or villus of jejunum, ileum, and colon (Fig. 3a). In contrast, Reg4 protein expression markedly increased in epithelial surface of inflamed mucosa (Fig. 3a). Furthermore, Reg4 co-localized with the goblet cell marker MUC2 and the phosphorylated STAT3 (p-STAT3, Tyr705) in the inflamed mucosa of patients (Fig. 3b, c).
Fig. 2.
Reg4 levels are correlated with intestinal inflammation. a Quantification of Reg4 mRNA in mouse colon with or without DSS treatment using RT-PCR assay. b Colorimetric in situ hybridization (CISH) analysis for Reg4 in mouse colon. c The Reg4 protein levels were determined in mouse colon using western blot. d–f The levels of serum Reg4, IL-6 and TNF-α were determined in healthy controls (HC, n = 16) and children with intestinal failure (IF, n = 40) using the enzyme linked immunosorbent assay (ELISA). (g, h) The serum Reg4 levels were correlated with serum IL-6 and TNF-α. **p < 0.01, ***p <0.001
Fig. 3.
Intestinal Reg4 increased in inflamed mucosa of IF patients. a Immunofluorescence staining for Reg4 in the uninflammed and inflamed intestine of IF patients (n = 8 each group) (b, c). Reg4 and Muc2 co-staining, and Reg4 and p-STAT3 co-staining were analyzed in mucosa of patients (n = 6 each group)
Activating transcription factor 2 (ATF2) enhances Reg4 expression during intestinal inflammation
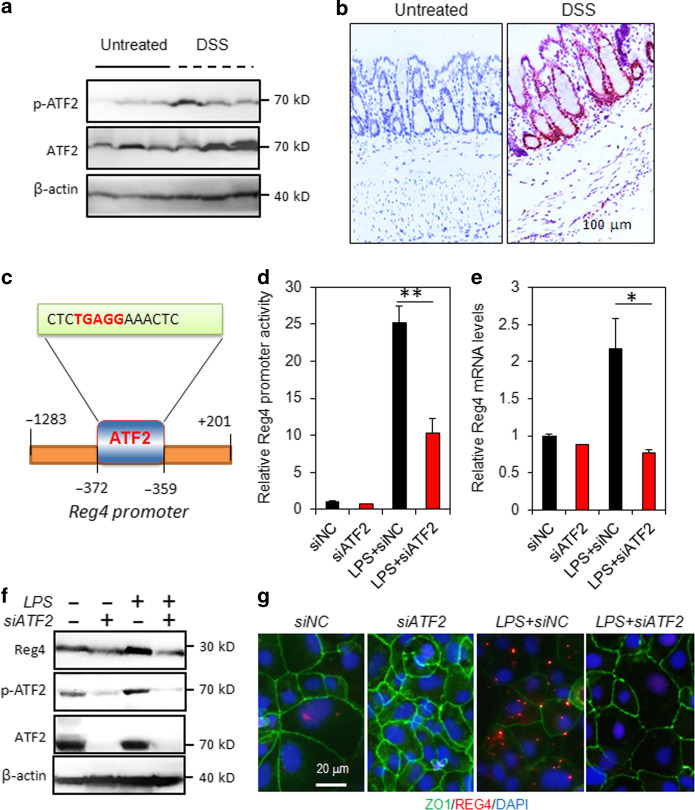
During the DSS induced intestinal inflammation, Western blot analysis showed that the levels of activated ATF2 (phosphorylated-ATF2, p-ATF2, Thr71) increased evidently in colonic mucosa (Fig. 4a). Consistently, immunohistochemistry (IHC) staining indicated that the p-ATF2-positive cells were markedly increased after DSS administration (Fig. 4b). Using bioinformative software, it predicted that there was a binding site for ATF2 (CTCTGAGGAAACTC) located at 1 kb region upstream of Reg4 transcription start site (TSS) (Fig. 4c). Intestinal cells Caco2 transfected with Reg4 promoter-luciferase vectors and ATF2 siRNA for 72 h were treated with lipopolysaccharide (LPS, 100 μg/mL) for 30 min. As shown Fig. 4d, Caco2 cells treatment with LPS resulted in a 25-fold induction of Reg4 promoter activity (Fig. 4d). The ATF2 knockdown significantly counteracted the LPS-induced Reg4 promoter activity (Fig. 4d). Consistently, Reg4 mRNA and protein were reduced after ATF2 depletion (Fig. 4e–g).
Fig. 4.
ATF2 promotes Reg4 transcription in during the intestinal inflammation. a Western blot analysis for p-ATF2 and ATF2 in the colonic mucosa with or without DSS treatment. b Representative images of the immunohistochemistry (IHC) staining of p-ATF2. c The binding motif of ATF2 on the promoter of Reg4. a The binding motif of ATF2 on the promoter of Reg4. c–f The promoter activity, mRNA and protein of Reg4 were reduced by ATF2 knockdown. C, The promoter activity was analyzed by luciferase reporter system. D, The mRNA levels were determined by RT-PCR. E, Western blot analysis for Reg4 and ATF2. F, Immunofluorescence staining for Reg4 and ZO1. *p < 0.05, **p < 0.01
Intestinal epithelial Reg4 deficiency protects against DSS-induced colitis
To study the potential roles of Reg4 in intestinal inflammation, we generated mice lacking Reg4 specifically in intestinal epithelial cells (Reg4ΔIEC) by crossing mice carrying loxP-flanked (floxed, fl) Reg4 alleles (Reg4fl/fl) mice with villin-Cre transgenics (Supplementary Fig. 1). As shown in Supplementary Fig. 2, the average body weight, villus height and crypt number in Reg4ΔIEC mice did not obviously differ from the Reg4fl/fl littermates (Supplementary Fig. 2A–C). IHC staining indicated Reg4ΔIEC mice had fewer cells that stained positive with Muc2, Ki67 or p-STAT3 than Reg4fl/fl mice (Supplementary Fig. 2C and 2D). In colonic epithelium, transmission electron microscopy (TEM) showed Reg4ΔIEC mice had immature enterocyte cells with having numerous brush borders (Supplementary Fig. 2E). The RT-PCR analysis showed the mRNA levels of Muc2, Ki67, Lgr5 and especially Alpi were decreased in colon of Reg4ΔIEC mice (Supplementary Fig. 2f).
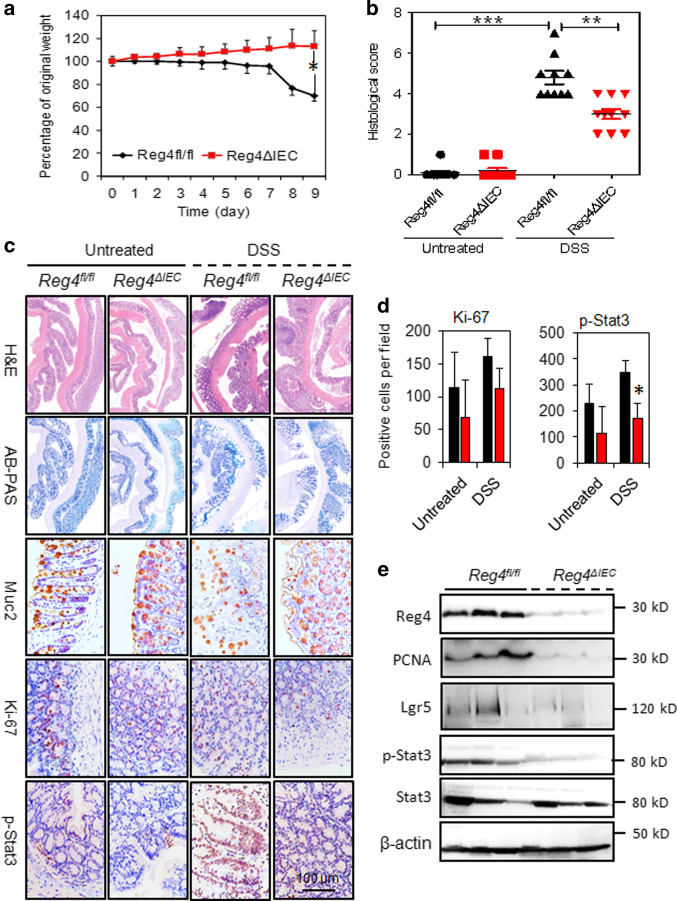
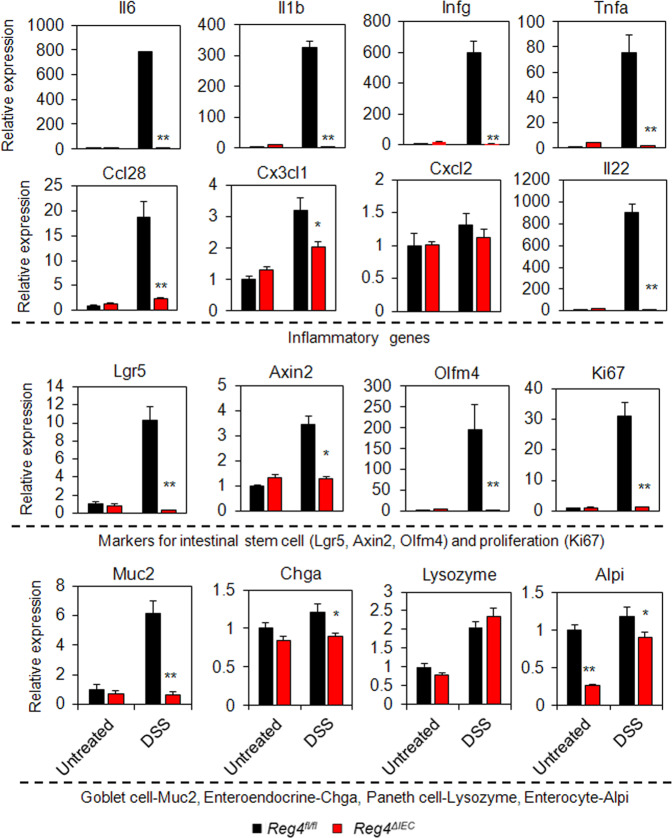
During DSS treatment, Reg4ΔIEC mice exhibited less body weight loss than Reg4fl/fl mice (Fig. 5a). Histologically, Reg4ΔIEC mice had less colonic mucosal damage and decreased inflammatory infiltrates compared to DSS-treated Reg4fl/fl mice (Fig. 5b, c). IHC analysis indicated Reg4ΔIEC mice exhibited had lower expression of Muc2, Ki67, and p-STAT3 in colonic mucosa (Fig. 5c, d). Indeed, western-blots analysis revealed the levels of PCNA, Lgr5, and p-STAT3 reduced evidently in colon of Reg4ΔIEC mice with DSS treatment (Fig. 5e). In agreement with histological findings, the inflammatory genes, including Il6, Il1b, Infg, Tnfa, Ccl28, Cx3cl1, and Il22, were significantly reduced in colonic mucosa of Reg4ΔIEC mice following DSS-treatment compared to DSS-treated Reg4fl/fl mice (Fig. 6). Moreover, the ISC genes, including Lgr5, Axin2, Olfm4, and also proliferative gene Ki67 decreased significantly in colon of DSS-treated Reg4ΔIEC mice (Fig. 6). The DSS-treated Reg4ΔIEC mice also had a reduction in levels of goblet cell maker, Muc2, enteroendocrine gene, Chga and enterocyte marker, Alpi (Fig. 6).
Fig. 5.
Intestinal-epithelial Reg4 deficiency ameliorated DSS-mediated histological changes. a The body weight was monitored in DSS-treated Reg4ΔIEC mice and DSS-treated Reg4fl/fl mice. b Quantification of histological scores. c Hematoxylin and eosin (H&E) staining, Alcian blue/periodic acid Schiff base (AB-PAS) staining and Immunohistochemical analysis with Muc2, Ki67 and p-STAT3 antibodies in Reg4ΔIEC mice and Reg4fl/fl mice (n = 8 per group). (d) Quantification of Immunohistochemical analysis for Ki67 and p-STAT3. (G) The colonic proteins Reg4, p-STAT3, STAT3, Lgr5 and PCNA were analysed in DSS-treated Reg4ΔIEC mice and DSS-treated Reg4fl/fl mice. *p < 0.05, **p < 0.01
Fig. 6.
Reg4 deficiency inhibited DSS-induced colon inflammation. The genes for Il6, Il1b, Infg, Tnfa, Ccl28, Cx3cl1, Cxcl2, Il22, Lgr5, Axin2, Olfm4, Ki67, Muc2, Lysozyme, Chga, and Alpi were determined in colon of Reg4ΔIEC mice and Reg4fl/fl mice with or without DSS-treatment using RT-PCR analysis (n = 8 per group). *p < 0.05, **p < 0.01
Intestinal epithelial Reg4 deficiency alters the colonic bacterial composition
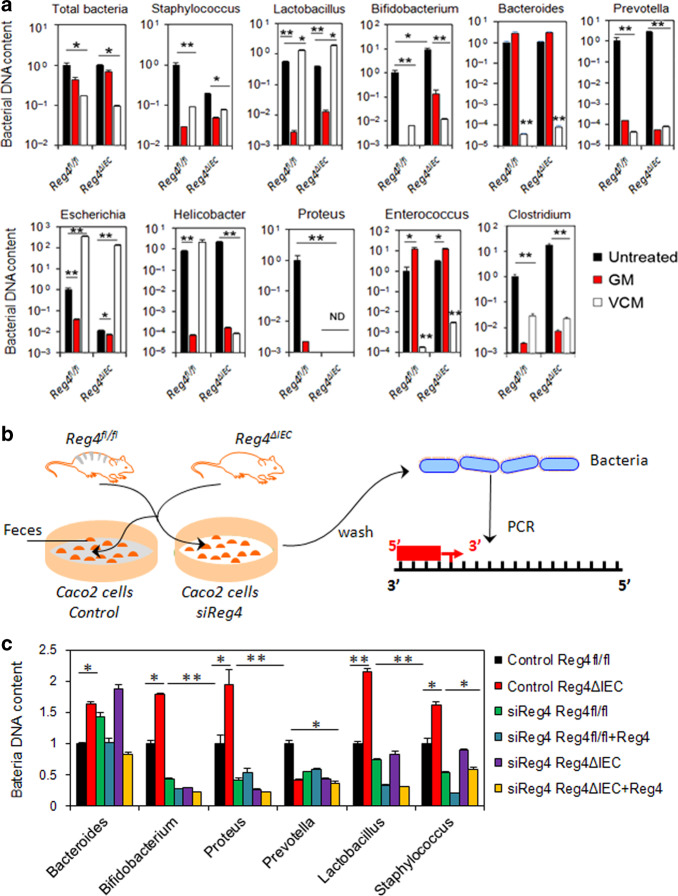
Given that Reg4 is a member of the calcium-dependent (C-type) lectin superfamily, we further study its roles in the commensal microbiota homeostasis. We initially homogenized colonic tissues and feces from Reg4fl/fl and Reg4ΔIEC mice, and cultured on agar plates for 16 h. As shown in Supplementary Fig. 3A, the number of colony-forming units (CFU) in colon tissues and in feces of Reg4ΔIEC mice did not evidently differ from Reg4fl/fl mice (Supplementary Fig. 3A). The fluorescence in situ hybridization (FISH) using a universal bacterial 16S ribosomal RNA (rRNA) probe and co-stained with Reg4 showed that presence of intestinal bacteria overlapped or adjacent to Reg4 protein in inflamed intestine of Reg4fl/fl mice (Supplementary Fig. 3B). In Reg4ΔIEC mice, bacteria were not observed adjacent to epithelial cell layers (Supplementary Fig. 3B). We further isolated DNA from the colonic tissues and fecal content and analyzed for the presence of bacteria by PCR using bacterial genus-specific primers. It showed that the numbers of several bacterial genera were decreased in colonic tissues of Reg4ΔIEC mice, particularly the Prevotella, Escherichia and Lactobacillus (Supplementary Fig. 3C). Mice were then orally treated with GM, which is active against Gram-negative bacteria. Reg4ΔIEC mice treated with GM had decreased numbers of Prevotella, Escherichia, Helicobacter, and Proteus in feces (Fig. 7a). In addition, GM treatment ameliorated the colitis in DSS-treated Reg4ΔIEC mice (Supplementary Figs. 4 and 5). Mice treated with vancomycin (VCM), which is active against Gram-positive bacteria, resulted in increased numbers of fecal Lactobacillus and Escherichia (Fig. 7a). VCM treatment did not ameliorated DSS-induced colitis in Reg4ΔIEC mice (Supplementary Figs. 4 and 5).
Fig. 7.
Intestinal Reg4 potentially affects the intestinal bacteria homeostasis. a Quantitative PCR of bacterial DNA isolated from 1 mg of feces from untreated, gentamicin (GM)-treated and vancomycin (VCM)-treated mice. Data show the bacterial DNA amounts compared to untreated Reg4fl/fl mice group (n = 8 per group). b Schema of binding assay of fecal bacteria. c Bound analysis for fecal bacteria attached to the Caco2 cells with or without Reg4 expression. *p <0.05, **p < 0.01
Reg4 knockdown reduces bacterial adhering to the colonic epithelial cells
In this study, we further address the mechanisms by which Reg4 affected bacterial homeostasis. The fecal suspensions from Reg4ΔIEC mice or Reg4fl/fl mice were incubated with or without recombinant Reg4 protein. After removing larger particles, the bacterial suspensions were added to cultures of polarized Caco2 cells with or without Reg4 siRNA transfection, and after 8 h the bacteria that had attached to Caco2 cells were analyzed (Fig. 7b). As shown in Fig. 7c, the fecal suspensions from Reg4ΔIEC mice had more Proteus, Lactobacillus, Bifidobacterium, Staphylococcus, and Bacteroides adherence to Caco2 cells (Fig. 7c). Reg4 knockdown in Caco2 cells or pretreatment with recombinant Reg4 protein reduced the Proteus, Lactobacillus, Bifidobacterium, Staphylococcus, and Bacteroides bound to Caco2 cells (Fig. 7c).
Reg4 promotes growth of colonic organoids via activation of STAT3
Since the above data show the Reg4 is critical to colitis, we next investigate its roles and mechanisms in colonic regeneration. Consistent with findings in vivo (Fig. 5), we found that the number of colonic organoids from Reg4ΔIEC mice evidently less than the ones from Reg4fl/fl mice (Fig. 8a). After 3 days of recombinant Reg4 protein treatment, the colonic organoids increased significantly compared to untreated ones (Fig. 8a). In line with increased number of organoids, Reg4 treatment enhanced EdU incorporation in colonic organoids (Fig. 8b). Given found that Reg4 increased phosphorylation of STAT3 (Y705), we further evaluated STAT3 in Reg4-induced growth of colonic organoids. As shown in Fig. 8a, b, the treatment with the STAT3 inhibitor Stattic successfully abrogated Reg4-mediated the growth of colonic organoids (Fig. 8a, b). We also showed that recombinant Reg4 protein increased the expression of Lgr5 and Ki67, and this effect was blocked by Stattic treatment (Fig. 8c, d).
Fig. 8.
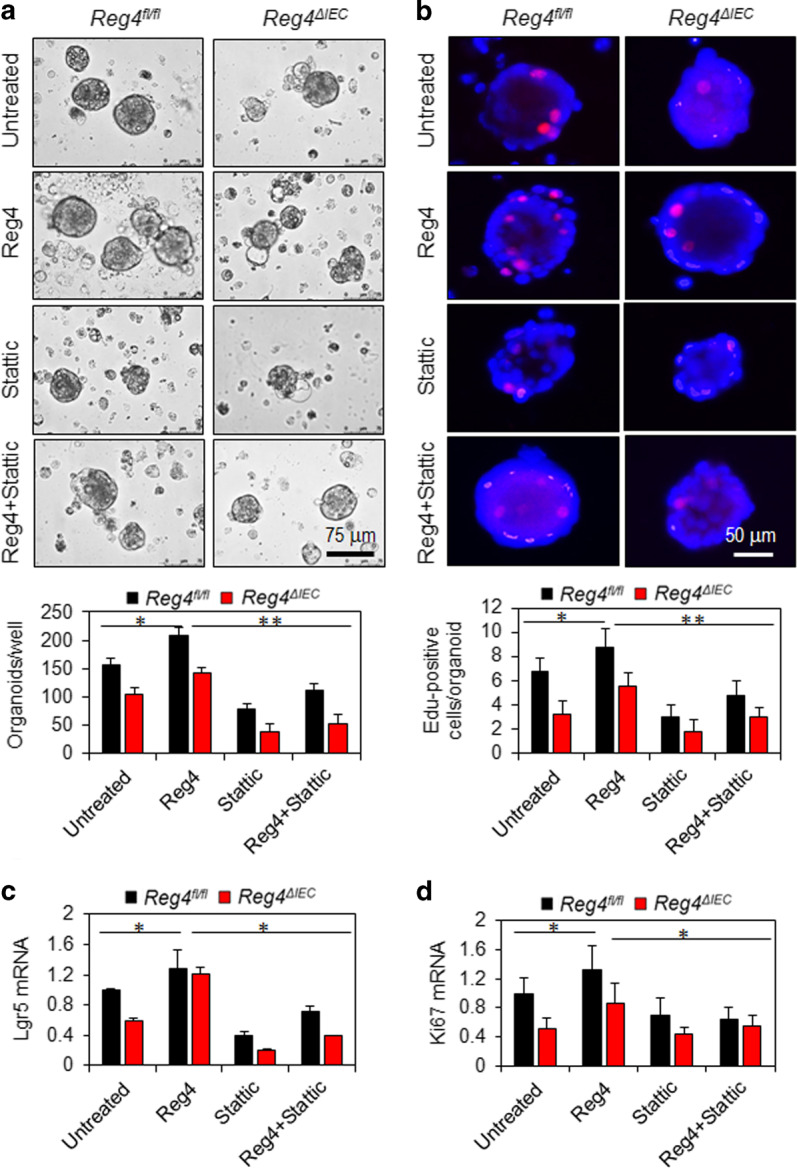
Reg4 promotes growth of colonic organoids dependent on STAT3 activation. a The number of colonic organoids was calculated following incubation with or without Reg4 protein for 3 days. b EdU staining and its quantification. c, d Levels of Lgr5 mRNA and Ki67 mRNA were determined in colonic organoids with RT-PCR. *p < 0.05, **p < 0.01
Discussion
Reg4, a newly discovered member of the Reg gene family, was firstly isolated from a cDNA library of ulcerative colitis (UC) tissues,3 implying that the Reg4 may play some roles in the intestinal inflammation. In this study, we demonstrated that in the normal crypt base Reg4 protein expands to epithelial cells may due to ATF2 activation during intestinal inflammation. The population-based cross-sectional study indicated that the levels of serum Reg4 reflect the presence and the degrees of intestinal inflammation in pediatrics with IF. In DSS-induced murine colitis, intestinal-specific Reg4 deficiency protects intestinal inflammation and alters the bacterial composition.
In normal intestine, other investigators and we showed that Reg4 protein was selectively expressed at crypts or villus at low levels, which was consistent with it’s a marker of enteroendocrine cells.14,23 In inflamed mucosa of intestine, Reg4 protein markedly increased in outer parts of crypts and in a surface pattern. In addition, serum Reg4 protein levels were significantly higher in IF patients and positively correlated with cytokines IL-6 and TNF-α, indicating that serum Reg4 could reflect the degrees of intestinal inflammation. Up to date, the mechanisms involved in regulating Reg4 expression during the intestinal inflammation remains elusive. It reported that other Reg members Reg1 and Reg3 expression could be enhanced by several cytokines, such as IL-6.28–30 In contrast to Reg1 or Reg3 expression, proinflammatory cytokines had none stimulatory effect on Reg4 gene expression in the human colon cancer cell line.24 It thus suggests that the Reg4 gene expression is cannot be directly stimulated by cytokines. As reported previously, Reg4 is a transcriptional target of GATA6 in colon cancer cells.31 It has also been reported that Reg4 is a direct target of the intestinal transcriptional factor CDX2.32 In the present study, we identified Reg4 as a novel target gene of ATF2, a member of the leucine zipper family of DNA-binding proteins,33,34 which has been shown to play an important role in many of the inflammatory responses.35,36 It reported that GATA6 acts in combination with other transcriptional factors, including TCF437 and CDX2,38 to stimulate or repress Reg4 gene expression. We thus proposed that the Reg4 might be regulated by ATF2 and/or with other co-factors in the inflamed intestine.
To study the exact roles of Reg4 in intestinal inflammation, we conditionally knockout intestinal Reg4 in mice (Reg4ΔIEC) and challenged with a DSS for 10 days. We demonstrated that Reg4fl/fl mice exhibited profound weight loss, increased disease severity, increased proinflammatory genes expression, and extensive intestinal ulceration, loss of crypt architecture, while Reg4ΔIEC mice were minimally affected, indicating that Reg4 deficiency is critical for limiting DSS-induced intestinal damage and inflammation. Dysregulation of intestinal homeostasis and susceptibility to intestinal inflammation are often associated with alterations in commensal bacterial populations.39–41 Recent studies have shown that other members of the Reg family, such as Reg3, have protective effects against experimental colitis via its antimicrobial effects. The murine Reg3γ has antibacterial activities against Gram-positive bacteria by interacting with peptidoglycan carbohydrate.6 Reg3β exerts bactericidal activity against Gram-negative bacteria by binding to lipopolysaccharide.42,43 Unlike Reg3 showed an apparently paneth-cell-dependent expansion from the colonic crypts during inflammation,44 whereas we showed that Reg4 was expressed in enteroendocrine cells and expanded to epithelial cells of the upper colonic crypts during inflammation, suggests that Reg4 may have different effects on microbiota composition. Indeed, we demonstrated that several bacteria, including Prevotella, Escherichia, and Lactobacillus, decreased in colonic tissues of Reg4ΔIEC mice, and Reg4ΔIEC mice had less bacteria presenting adjacent to the epithelial cell layers. Mice were then orally treated with gentamicin (GM), which is active against Gram-negative bacteria, decreased numbers of Prevotella, Escherichia, Helicobacter, and Proteus, and, accordingly, no bacteria were present just above the intestinal epithelial layers. Additionally, GM treatment ameliorated the intestinal inflammation in DSS-treated Reg4ΔIEC mice, suggests that Reg4 deficiency decreased mucus penetration by Gram-negative bacteria such as Proteus, Escherichia, and Helicobacter and reduced sensitivity to intestinal inflammation. Furthermore, Reg4 knockdown in Caco2 cells significantly reduced the bacteria, including Proteus, Bacteroides, Lactobacillus, Bifidobacterium, and Staphylococcus, attaching to the cells. Given that Reg4 had a carbohydrate recognition domain (CRD),3,23 which suggests that CRD might be essential to Reg4 bounding the bacteria.
In response to inflammation-induced mucosal injury, epithelial regeneration is critical for barrier maintenance and organ function. The intestinal stem cell (ISC) niche provides Wnt, Notch, and epidermal growth factor (EGF) signals supporting Lgr5-positive crypt base columnar ISCs for epithelial maintenance.45,46 Sasaki et al.47 recently reported that Reg4 could promote organoids formation of single Lgr5-positive colon stem cells, but involved mechanisms remained not fully understood. The signal transducer and activator of transcription 3 (STAT3) has been reported to promote ISCs survival and ISCs-mediated epithelial regeneration.48,49 In vivo, we showed that Reg4 deficiency significantly inhibited DSS-induced activation of STAT3 in colonic muscoa. In vitro, the recombinant Reg4 protein treatment promoted the growth of colonic organoids, but this effect was successfully abrogated by treatment of STAT3 inhibitor. Two key cytokines are IL6 and IL22, which activate their respective receptors, followed by phosphorylation and activation of STAT3 in intestine to promote their regeneration or growth.50,51 We here demonstrated that several proinflammatory cytokines, including IL6 and IL22, reduced significantly in colon of DSS-treated Reg4ΔIEC mice. Thus, it suggests that inflammation-induced Reg4 may promote the colon regeneration via IL6/22-STAT3 signaling.
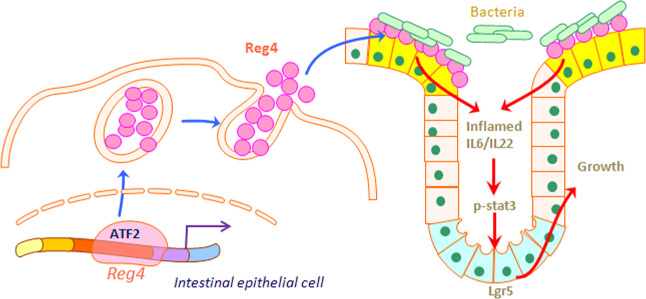
Taken together, our studies demonstrate that Reg4 is highly expressed towards the intestinal luminal face during inflammation, and ATF2 plays transcriptional role in the inflammation-mediated Reg4 expression. Intestinal-specific depletion of Reg4 ameliorates DSS-induced colonic inflammation and injury might through decreasing bacterial aggregating onto the intestinal epithelial cells, thus reducing the risk of inflammation. During the colonic mucosal regeneration, Reg4 enhances the growth of organoids via activating the STAT3 signaling (Fig. 9).
Fig. 9.
Schema of that Reg4 was involved in intestinal inflammation and regeneration. The activated ATF2 enhanced the Reg4 expression during the intestinal inflammation. The intestinal epithelial Reg4 bounded bacteria and induced the colitis. During the inflammation inducing-colonic injuries, Reg4 increased the regeneration via activating the IL6/22-STAT3 signaling
Materials and methods
Animal experiments
To generate intestinal-specific Reg4 deficiency mice, the Reg4-floxed alleles (Reg4fl/fl) were bred with Villin-cre+ mice to generated Reg4 conditional knockout mice (Reg4ΔIEC). Six-week-old Reg4ΔIEC mice and their littermate Reg4fl/fl mice were used for DSS-induced colitis experiments. Acute colitis was induced by administration of 2% DSS (36–50 kDa) in the drinking water for 10 days. The changes in weight were monitored each day. All animal experiments were performed according to guidelines of the Institutional Animal Care and Use Committee of the Xin hua hospital, School of Medicine, Shanghai Jiao Tong University.
Statistical analysis
The statistics are presented as mean ± SD. In human population, the Kolmogorov-Smirnov test was used to assess distributions. Abnormally distributed data were logarithmically transformed before analysis. Correlations between serum Reg4 and serum IL6 or TNFα were tested by Spearman rank correlation test. Mann Whitney U-test, Fisher exact test or one-way ANOVA were used to compare differences between groups. The level of statistical significance was set at 0.05.
Detailed protocols are provided in the Supplementary Materials and Methods.
Supplementary information
Acknowledgements
We thank Yinuo Xu, Ping Wu, Chuanqi Zhao and Beilin Gu for the administrative, technical, or material support.
Author contributions
Y.X. and W.C. accomplished the study concept and design, acquisition of data, analysis and interpretation of data, obtained funding and drafting of the manuscript; Y.X., Y.W., and Y.L. performed most of the experiments. Y.L. and W.Y. gave the administrative, technical, or material support. This study was supported by National Natural Science Foundation of China (81770517 and 81630039), Shanghai Key Laboratory of Pediatric Gastroenterology and Nutrition (17DZ2272000) and Research Funding of Shanghai Health and Family Planning Commission (201640153).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://www.nature.com/articles/s41385-020-00367-2"
Change history
12/16/2020
A Correction to this paper has been published: 10.1038/s41385-020-00367-2
Contributor Information
Yongtao Xiao, Phone: +86 21 25076438, Email: xiaoyongtao@xinhuamed.com.cn.
Wei Cai, Phone: +86 21 25076441, Email: caiw204@sjtu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41385-019-0161-5) contains supplementary material, which is available to authorized users.
References
- 1.Dusetti NJ, et al. Molecular cloning, genomic organization, and chromosomal localization of the human pancreatitis-associated protein (PAP) gene. Genomics. 1994;19:108–114. doi: 10.1006/geno.1994.1019. [DOI] [PubMed] [Google Scholar]
- 2.Itoh T, Teraoka H. Cloning and tissue-specific expression of cDNAs for the human and mouse homologues of rat pancreatitis-associated protein (PAP) Biochim. Et. Biophys. Acta. 1993;1172:184–186. doi: 10.1016/0167-4781(93)90290-t. [DOI] [PubMed] [Google Scholar]
- 3.Hartupee JC, Zhang H, Bonaldo MF, Soares MB, Dieckgraefe BK. Isolation and characterization of a cDNA encoding a novel member of the human regenerating protein family: Reg IV. Biochim. Et. Biophys. Acta. 2001;1518:287–293. doi: 10.1016/s0167-4781(00)00284-0. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto H, Takasawa S. Recent advances in the Okamoto model: the CD38-cyclic ADP-ribose signal system and the regenerating gene protein (Reg)-Reg receptor system in beta-cells. Diabetes. 2002;51(Suppl 3):S462–S473. doi: 10.2337/diabetes.51.2007.s462. [DOI] [PubMed] [Google Scholar]
- 5.Takasawa S. Regenerating gene (REG) product and its potential clinical usage. Expert Opin. Ther. Targets. 2016;20:541–550. doi: 10.1517/14728222.2016.1123691. [DOI] [PubMed] [Google Scholar]
- 6.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayabe T, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 9.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl Acad. Sci. USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroder N, et al. Identification of mouse genes with highly specific expression patterns in differentiated intestinal epithelium. Gastroenterology. 2006;130:902–907. doi: 10.1053/j.gastro.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Heiskala K, Arola J, Heiskala M, Andersson LC. Expression of Reg IV and Hath1 in neuroendocrine neoplasms. Histol. Histopathol. 2010;25:63–72. doi: 10.14670/HH-25.63. [DOI] [PubMed] [Google Scholar]
- 12.Oue N, et al. Expression and localization of Reg IV in human neoplastic and non-neoplastic tissues: Reg IV expression is associated with intestinal and neuroendocrine differentiation in gastric adenocarcinoma. J. Pathol. 2005;207:185–198. doi: 10.1002/path.1827. [DOI] [PubMed] [Google Scholar]
- 13.van Beelen Granlund A, et al. REG gene expression in inflamed and healthy colon mucosa explored by in situ hybridisation. Cell Tissue Res. 2013;352:639–646. doi: 10.1007/s00441-013-1592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grun D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi T, et al. Immunohistochemical analysis of Reg IV in urogenital organs: Frequent expression of Reg IV in prostate cancer and potential utility as serum tumor marker. Oncol. Rep. 2009;21:95–100. [PubMed] [Google Scholar]
- 16.Heiskala K, Giles-Komar J, Heiskala M, Andersson LC. High expression of RELP (Reg IV) in neoplastic goblet cells of appendiceal mucinous cystadenoma and pseudomyxoma peritonei. Virchows Arch. : Int. J. Pathol. 2006;448:295–300. doi: 10.1007/s00428-005-0105-1. [DOI] [PubMed] [Google Scholar]
- 17.Mitani Y, et al. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene. 2007;26:4383–4393. doi: 10.1038/sj.onc.1210215. [DOI] [PubMed] [Google Scholar]
- 18.Ohara S, et al. Reg IV is an independent prognostic factor for relapse in patients with clinically localized prostate cancer. Cancer Sci. 2008;99:1570–1577. doi: 10.1111/j.1349-7006.2008.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eguchi H, et al. Serum REG4 level is a predictive biomarker for the response to preoperative chemoradiotherapy in patients with pancreatic cancer. Pancreas. 2009;38:791–798. doi: 10.1097/MPA.0b013e3181ac5337. [DOI] [PubMed] [Google Scholar]
- 20.Oue N, et al. Serum concentration of Reg IV in patients with colorectal cancer: overexpression and high serum levels of Reg IV are associated with liver metastasis. Oncology. 2007;72:371–380. doi: 10.1159/000113147. [DOI] [PubMed] [Google Scholar]
- 21.Bishnupuri KS, et al. Reg IV activates the epidermal growth factor receptor/Akt/AP-1 signaling pathway in colon adenocarcinomas. Gastroenterology. 2006;130:137–149. doi: 10.1053/j.gastro.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Rafa L, et al. REG4 acts as a mitogenic, motility and pro-invasive factor for colon cancer cells. Int. J. Oncol. 2010;36:689–698. doi: 10.3892/ijo_00000544. [DOI] [PubMed] [Google Scholar]
- 23.Kamarainen M, et al. RELP, a novel human REG-like protein with up-regulated expression in inflammatory and metaplastic gastrointestinal mucosa. Am. J. Pathol. 2003;163:11–20. doi: 10.1016/S0002-9440(10)63625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanakin A, et al. Expression of the REG IV gene in ulcerative colitis. Lab. Investig. 2007;87:304–314. doi: 10.1038/labinvest.3700507. [DOI] [PubMed] [Google Scholar]
- 25.Ouellette AJ. Paneth cells and innate mucosal immunity. Curr. Opin. Gastroenterol. 2010;26:547–553. doi: 10.1097/MOG.0b013e32833dccde. [DOI] [PubMed] [Google Scholar]
- 26.Loonen LM, et al. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 27.Schalamon J, Mayr JM, Hollwarth ME. Mortality and economics in short bowel syndrome. Best. Pract. Res. Clin. Gastroenterol. 2003;17:931–942. doi: 10.1016/s1521-6918(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 28.Sekikawa A, et al. Possible role of REG Ialpha protein in ulcerative colitis and colitic cancer. Gut. 2005;54:1437–1444. doi: 10.1136/gut.2004.053587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akiyama T, et al. Activation of Reg gene, a gene for insulin-producing beta -cell regeneration: poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc. Natl Acad. Sci. USA. 2001;98:48–53. doi: 10.1073/pnas.240458597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechreche H, et al. pap, reg Ialpha and reg Ibeta mRNAs are concomitantly up-regulated during human colorectal carcinogenesis. Int. J. Cancer. 1999;81:688–694. doi: 10.1002/(sici)1097-0215(19990531)81:5<688::aid-ijc3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki Y, et al. REG4 is a transcriptional target of GATA6 and is essential for colorectal tumorigenesis. Sci. Rep. 2015;5:14291. doi: 10.1038/srep14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito Y, et al. Reg IV is a direct target of intestinal transcriptional factor CDX2 in gastric cancer. PLoS ONE. 2012;7:e47545. doi: 10.1371/journal.pone.0047545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Campbell D, Derijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 34.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Sadi R, et al. Mechanism of IL-1beta modulation of intestinal epithelial barrier involves p38 kinase and activating transcription factor-2 activation. J. Immunol. 2013;190:6596–6606. doi: 10.4049/jimmunol.1201876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu T, et al. The regulatory role of activating transcription factor 2 in inflammation. Mediat. Inflamm. 2014;2014:950472. doi: 10.1155/2014/950472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whissell G, et al. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat. Cell Biol. 2014;16:695–707. doi: 10.1038/ncb2992. [DOI] [PubMed] [Google Scholar]
- 38.Verzi MP, et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raetz M, et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nat. Immunol. 2013;14:136–142. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Ampting MT, et al. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect. Immun. 2012;80:1115–1120. doi: 10.1128/IAI.06165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miki T, Holst O, Hardt WD. The bactericidal activity of the C-type lectin RegIIIbeta against Gram-negative bacteria involves binding to lipid A. J. Biol. Chem. 2012;287:34844–34855. doi: 10.1074/jbc.M112.399998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Granlund A, et al. Activation of REG family proteins in colitis. Scand. J. Gastroenterol. 2011;46:1316–1323. doi: 10.3109/00365521.2011.605463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 46.Langlands AJ, et al. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLoS Biol. 2016;14:e1002491. doi: 10.1371/journal.pbio.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasaki N, et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl Acad. Sci. USA. 2016;113:E5399–E5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindemans CA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matthews JR, Sansom OJ, Clarke AR. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death Differ. 2011;18:1934–1943. doi: 10.1038/cdd.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou, Q. et al. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ.25, 1657–1670 (2018). [DOI] [PMC free article] [PubMed]
- 51.Li Y, et al. Disease-related expression of the IL6/STAT3/SOCS3 signalling pathway in ulcerative colitis and ulcerative colitis-related carcinogenesis. Gut. 2010;59:227–235. doi: 10.1136/gut.2009.184176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.