Abstract
Engineering microbial cell factories has been widely applied to produce compounds spanning from intricate natural products to bulk commodities. In each case, host robustness is essential to ensure the reliable and sustainable production of targeted metabolites. However, it can be negatively affected by metabolic burden, pathway toxicity, and harsh environment, resulting in a decreased titer and productivity. Enhanced robustness enables host to have better production performance under complicated growth circumstances. Here, we review current strategies for boosting host robustness, including metabolic balancing, genetic and phenotype stability enhancement, and tolerance engineering. In addition, we discuss the challenges and future perspectives on microbial host engineering for increased robustness.
Keywords: robustness, dynamic control, plasmids stability, tolerance engineering
Graphical abstract
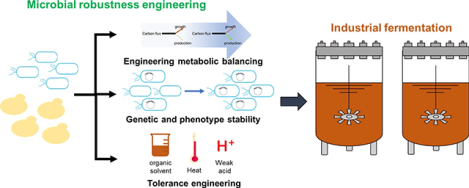
1. Introduction
Engineering microbial cell factories is a sustainable and promising approach for the synthesis of various compounds, such as natural products [1], biofuels [2], and bulk chemicals [3]. It can be achieved by combining heterologous or non-natural biosynthetic pathways into genetically advantageous microbial cell factories, such as Escherichia coli [4], Saccharomyces cerevisiae [5], and Bacillus sublitis [6]. To construct efficient microbial cell factories with improved titers, a number of metabolic engineering strategies have been developed, including enzyme selection/screening and engineering to overcome rate-limiting steps [7], pathway engineering for increased supply of precursors and cofactors [8], fine-tuning gene expression for optimal pathway performance [9], and host engineering to block competing pathways [10].
Despite the great progress achieved by these strategies, engineering industrial microbial cell factories remains a challenge. Engineered microbial cell factories constantly encounter disturbances caused by metabolic imbalance, genetic and phenotype instability, and harsh external environment in the process of large-scale fermentation [11]. The fluctuations of intracellular environment and concentrations of intermediates will affect the metabolic balance [12]. Besides, engineered strains usually contain recombinant pathways and tend to be more sensitive to stress factors than the wild type because of genetic and phenotype instability [13]. Moreover, harsh extracellular circumstances, such as high temperature, low pH, and metabolite toxicity, in laboratory- and commercial-scales require the host to be extremely robust to accomplish desired performance [14]. Therefore, increasing the host robustness against these perturbations becomes one of the major considerations in design and construction of microbial cell factories. We review the current research to increase robustness for high titer and productivity, focusing on engineering metabolic balancing, improving genetic and phenotype stability, and enhancing tolerance towards harsh environmental conditions. In addition, we discuss challenges and future perspectives on microbial host engineering for increased robustness.
2. Metabolic balancing
2.1. Engineering intermediates and cofactors balancing
The performance and robustness of industrial strains are often hindered by the accumulation of toxic metabolites and byproducts, imbalance of cofactors, and competition between production and cell growth. Traditionally, the accumulation of toxic intermediates can be addressed through modularized optimization to fine-tune the carbon fluxes (Fig. 1a) [15]. In a case of pyrogallol overproduction where the amino acid biosynthesis pathway were boosted, the relative expression levels of aroL, ppsA, tktA and aroGfbr (APTA), entCBA and nahGopt were fine adjusted to balance the carbon flux and successfully avoided the accumulation of harmful 2,3-dihydroxybenzoic acid in E. coli, resulting in a 2.44-fold improvement in pyrogallol production (893 mg/L) [16]. The balance of cofactors or redox homeostasis are usually achieved by the removal of cofactor competitive pathways or introduction of cofactor generating reactions [17]. However, rigid pathway engineering and control sometimes can exhibit undesired side effects and cause metabolic burdens when strains are exposed to fluctuated environmental perturbations [18]. Recent decades have witnessed the rapid development of a more responsive method, namely the dynamic pathway regulation. Dynamic regulation is using biosensors to autonomously control the metabolic fluxes based on intracellular chemical signals or extracellular environmental status to avoid unbalanced co-factors or reduce the amounts of toxic metabolites [19–21]. The well-known example is the dynamic regulation of toxic intermediate farnesyl pyrophosphate (FPP) in isoprenoid production, resulting in a 2-fold increase in final titer of amorphadiene (1.6 g/L) [22]. Similar strategy has also been demonstrated in biosynthesis of fatty acid [23]. Bi-functional dynamic regulation was applied in cis,cis-muconic acid synthesis pathway. Through up-regulation of salicylic acid synthesis and down-regulation of competing pathway for the cosubstrate malonyl-CoA, a 4.72-fold increase in titer (1861.9 mg/L) was achieved compared with strains with only static control (394.5 mg/L) [12].
Figure 1.
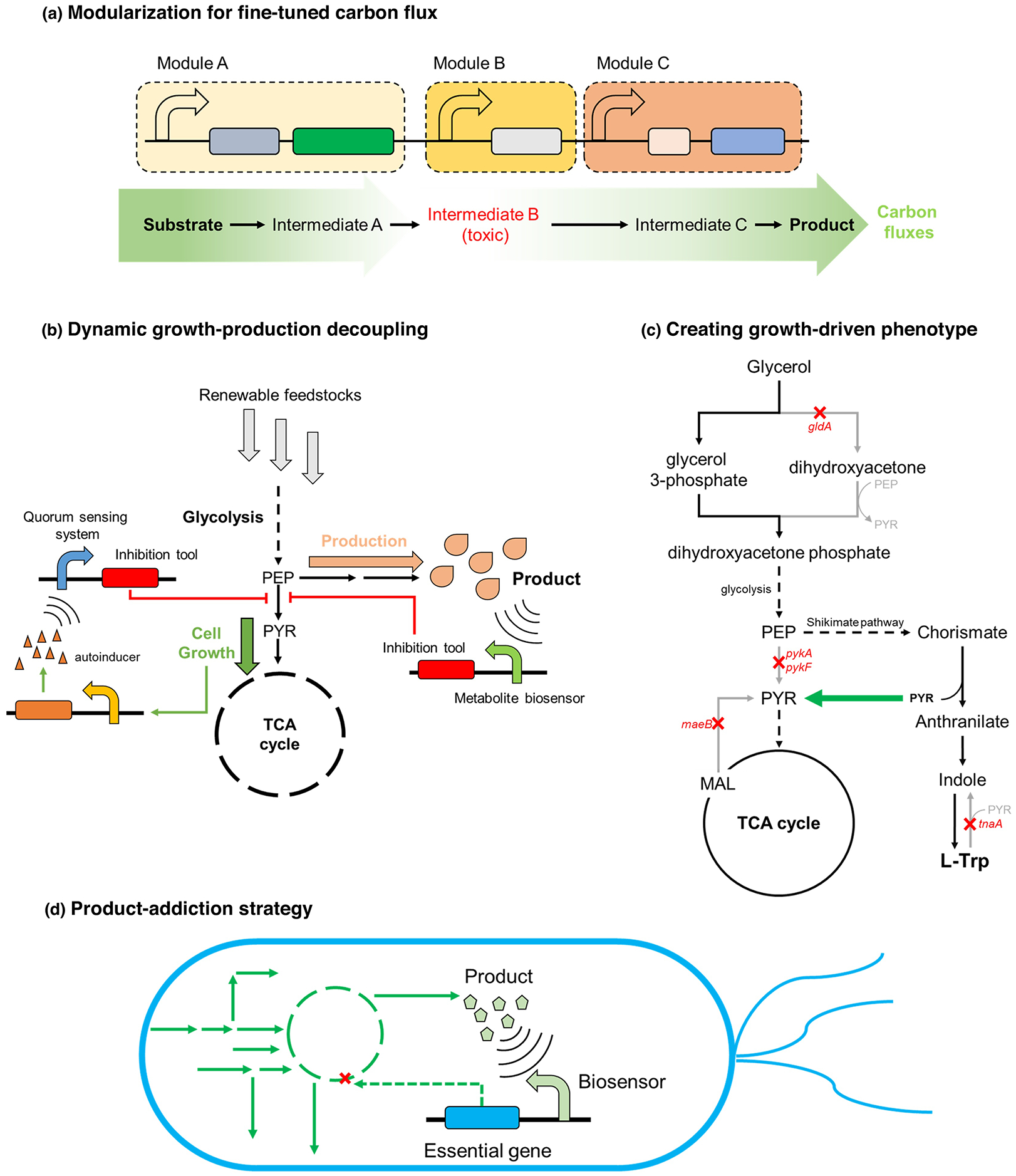
(a) Fine-tuning the carbon flux through modularization of gene expression. Module A, B and C are responsible for the production of intermediate A, B, and C, respectively. The accumulation of toxic intermediate B can be minimized by tuning the expression level of every module. (b) Dynamic regulation for decoupling cell growth and production. At initial stage, the cell can utilize most of resources to grow, with only a limited concentration of product accumulating. As fermentation proceeds, the accumulation of product will activate the inhibition on competing pathways and cell growth, redirecting carbon fluxes and other sources to synthesis of target compounds. (c) Growth-driven production. For example, all major source of pyruvate is removed except the one in L-Trp synthesis. Thus, the L-Trp production will become obligatory for cell growth and biomass generation based on pyruvate. (d) Product-addiction strategy. An essential gene was removed in genome and another copy is placed under the control of a product-inducible biosensor, so only the cells that are able to synthesize the target product will be able express the essential gene and survive.
2.2. Decoupling cell growth and production
In addition to the burdens from toxic intermediates and unbalanced cofactors, some biosynthetic pathways also compete with cell growth. Conventionally, this can be resolved by blocking the competing pathway [10], but it may hamper the cell viability and thus decrease the productivity. Another option is to perform two-stage fermentation (growth stage and production stage), but this kind of fermentation requires manual stage change by adding inducers to activate the production pathway. Although researchers were given full control of stage change, it is often hard to determine the optimal switch time point. Autonomous dynamical pathway control, aided by metabolite biosensors or quorum sensing systems, becomes a more viable option for decoupling cell growth and production to avoid competition between product synthesis and cell growth (Fig. 1b) [24,25]. Applying a “nutrition” sensor responding to the concentration of glucose delayed the synthesis of vanillic acid and thus enabled decoupled cell growth and production in E. coli. The metabolic stress test revealed that the introduction of this nutrient-sensing module lowered the metabolic burden by 2.4-fold and achieved a robust growth rate during bioconversion to vanillic acid [26]. A layered dynamic pathway control strategy, combining a myo-inositol biosensor ispA and an AHL-responsive quorum sensing system, was employed in biosynthesis of glucaric acid. This strategy simultaneously balanced the production and consumption rates of the key intermediate myo-inositol and decoupled cell growth and production of glucaric acid to avoid competing with essential glycolysis, resulting in a 5-fold increase in glucaric acid titer (2 g/L) [27]. Biosensor-based dynamic pathway regulation allowed real-time monitoring of intracellular metabolism and environmental signals, and thus could provide more precise control and reduce the metabolic stress on microbial cell factories.
2.3. Growth-driven/product-addiction strategy
Instead of decoupling cell growth and production, another promising strategy to increase cell robustness is coupling the production of target compounds with cell growth. By manipulating and rewriting the microbial metabolism, the synthesis of target compounds become obligatory for cell growth, enabling a “growth-driven” phenotype and increased cell robustness (Fig.1c) [28]. A pyruvate-driven L-tryptophan strain was constructed by removing all major pyruvate generating steps but the one in the synthesis of L-tryptophan in E. coli. The strain with pyruvate-driven phenotype achieved 2.37-fold and 2.04-fold increase of titer in L-tryptophan (1.73 g/L) and cis-cis muconic acid production (1.82 g/L), respectively [3]. A similar strategy was applied in butanone production driven by acetyl-CoA. By removing native acetate fermentation pathway (encoded by poxB, ackA and pta) and acetyl-CoA formation from acetate (encoded by acs), the only source of acetyl-CoA for E. coli was through the transfer of CoA moiety from 3-hydroxyvaleryl-CoA to acetate. The strain with acetyl-CoA driven phenotype produced 855 mg/L butanone, with complete deprivation of all supplied acetate [29]. However, establishing growth-driven phenotypes often requires that there are one or more essential components for cell growth can be generated from the pathway, which subsequently limits the generality of this method. A more general strategy is placing the essential genes folP and glmM under the control of a biosensor which only responds to the specific product, and thus forming a “product-addiction” phenotype (Fig. 1d). A synthetic product addiction system was constructed and enabled a stable mevalonate-overproducing strain, which can retain the performance over 95 generations [30]. The product addiction strategy holds greater generality than creating growth-driven phenotypes, but the application of product addiction could also suffer from limited biosensor availability.
3. Improving genetic and phenotype stability in engineered hosts
While genomic engineering is believed to be a stable approach for host engineering, plasmids-based systems are widely used due to its convenient modification and tunable expression levels. Selection and stable existence of plasmids are conventionally ensured by the resistance markers rescuing cells from antibiotic supplemented culture. However, cost, environmental and health concerns of antibiotic use have induced global discourage of such in food and biotech segments. To this end, strategies have been developed to maintain plasmids and strain phenotypes while minimizing antibiotics involvement. So far, developed plasmid maintenance approaches mainly include applying the toxin/antitoxin (TA) system, RNA-based interactions, operator-repressor titration (ORT), and auxotrophy complementation [13].
The TA system consists of a stable toxin protein interacting with a less stable antidote molecule (Fig. 2a). Such system has been adapted for use in plasmid-based production. For instance, a yefM/yoeB pair was introduced in Streptomyces by genome integration of the toxin gene and plasmid expression of the antitoxin gene. The resultant strain achieved stable production of proteins over 8-day incubation [31]. Meanwhile, RNA-based approaches and ORT were less explored recently. RNA-based approaches utilized the cis-acting of such oligonucleotides to interfere certain target genes (Fig.2b). ORT has been devised as a plasmid maintenance approach based on competitive binding of repressor on DNA-binding operator copies located between plasmids and the chromosome (Fig.2c) [32].
Figure 2.
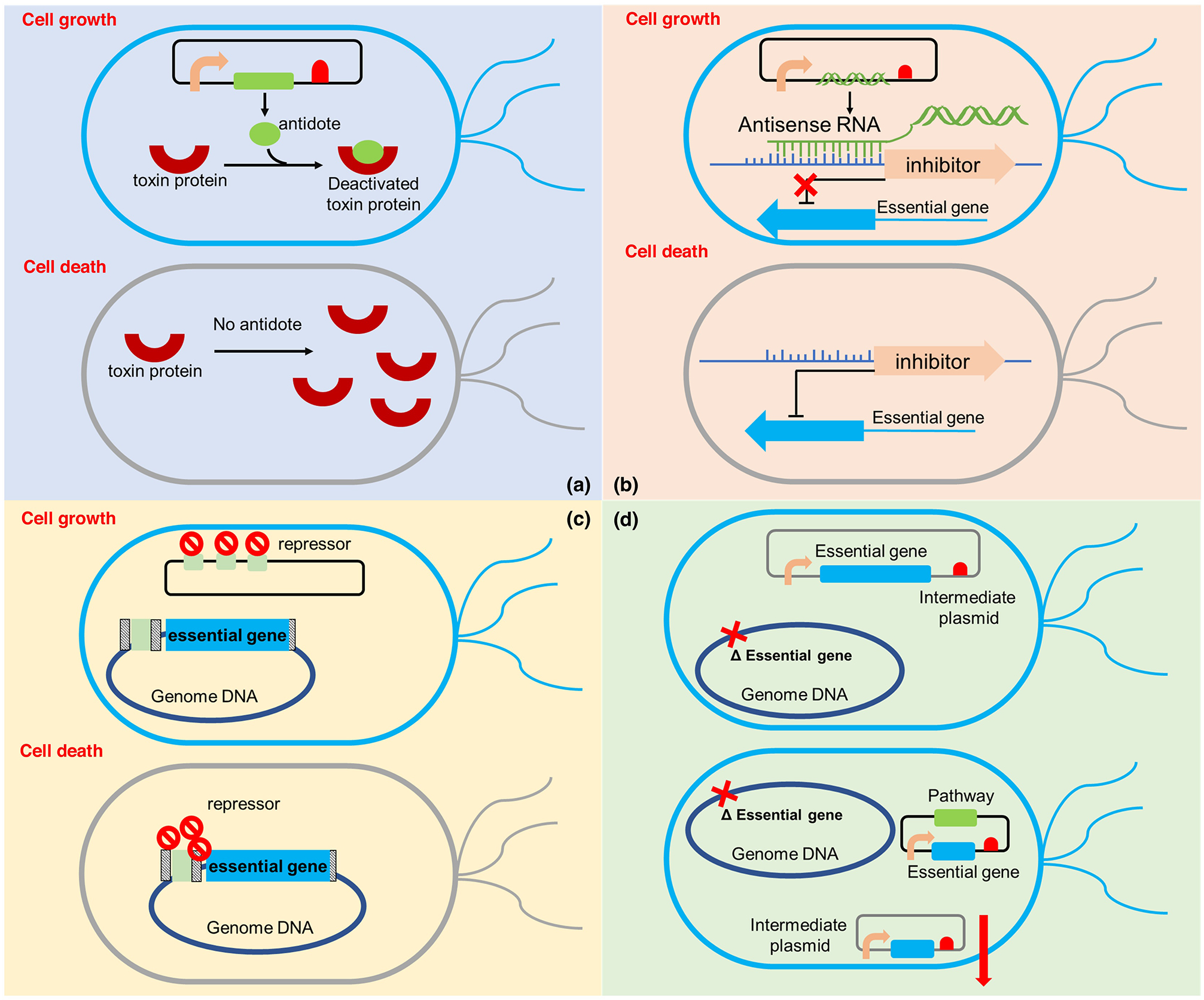
Strategies have been developed to maintain plasmids and strain phenotypes stability. (a) TA system; it consists of a stable toxin protein in genome interacting with a less stable antidote molecule in plasmid. If losing the antidote-containing plasmid, the toxin protein cannot be repressed by antidote and the host will be killed by the toxin. (b) RNA-based interactions; the essential gene is under the control of a regulatory promoter, which can be repressed by a corresponding inhibitor. A plasmid-transcribed antisense RNA (asRNA) can interact with the promoter sequence of this inhibitor. When the plasmid is present, the transcribed asRNA can bind with the promoter sequence of inhibitor and repress the expression of the inhibitor. Then the inhibition on the essential gene will be relieved. With the successfully expressed essential gene, the cells can grow well. However, when the plasmid is absent, the inhibitor cannot the repressed by asRNA and the expression of this inhibitor will repress the essential gene, which will cause growth defects on cells. (c) operator-repressor titration (ORT); it has been devised as a plasmid maintenance approach based on competitive binding of repressor on DNA-binding operator copies between plasmids and the chromosome. In the existence of a plasmid containing the same operator, most of the repressors will bind to the operators in the plasmid and only a small fraction of repressors will bind to the operators in the genome, because of the competitive binding between repressors and operators. Thus, the inhibition efficiency on the essential gene will decrease and it can be expressed to help the cell growth. However, if no plasmid is present, all the repressors will bind to the operator located in the genome and inhibit the expression of the essential gene. Without the essential gene, the cells will be unbale to grow. (d) auxotrophy complementation; this system is to optimize the process of traditional auxotrophic strain construction. In the optimized process, first, the essential gene was expressed in a temperature-sensitive intermediate plasmid and the essential gene in genome was deleted. Then, the production plasmid containing pathway gene and the deleted essential gene were transferred into host with the intermediate plasmid. When increasing the incubating temperature to 42 °C, the intermediate plasmid will be degraded and only the production plasmid was kept in the cell, resulting an auxotrophic strain. In this strain, if the plasmid is lost or mutated during fermentation, the cells will be unbale to grow due to the loss of essential gene.
In addition to the above strategies, auxotrophy complementation appears to be a straightforward and effective way to confer symbiosis between the plasmids and cells, and thus has attracted great attention recently (Fig. 2d). Auxotrophy complementation sequesters cell growth-related genes to the plasmids so that cells with plasmids obtain a fitness advantage. Non-essential genes have been targeted due to ease of manipulation, however, in defined nutrient conditions. Knockout of triosephosphate isomerase (tpiA) was found to significantly impair E. coli viability, while its complementation on the plasmid led to growth rescue and stable expression of β-glucanase [33]. On the other hand, sequestration of essential genes was also explored, but with strategies to bypass the strain fitness issue. Nonetheless, all the strategies have been focused on protein and plasmid expression. Not till recently, has the improved plasmid stability approaches been applied in cases of biochemical production. Kang et al. constructed a synthetic auxotrophic system based on the essential gene infA and further demonstrated the plasmid copy number can be controlled by varying its expression levels [34]. Although effective, deleting essential genes from chromosome requiring either complemented copy from plasmid or supplemented nutrient at all times to ensure cell viability, and thus extends the strain development timeframe. Zhang et al. introduced an intermediate plasmid to expedite the process, which harbors essential genes and can be easily cured and displaced by incoming production plasmids [13]. Sequestration of the essential genes folP from genome was found to largely improve plasmid maintenance and phenotype stability on strains producing salicylic acid for 80 generations [13].
While tremendous efforts have been put for engineering at individual cell level, cell-to-cell variation and heterogeneity pose great challenges for industrial applications [35]. Population quality control and heterogeneity mitigations were often used to tackle these challenges. For instance, by controlling a tetracycline efflux pump (TetA) through product-responsive biosensors, Xiao and colleagues linked cell fitness with production and demonstrated 3-fold enhanced production of both free fatty acid (FFA) and tyrosine [36].
4. Tolerance engineering
4.1. Thermal stability
Cooling system is required to maintain the optimal temperature (usually ≤ 37 °C) for fermentation since cell growth is an exothermic process releasing up to half of the energy stored in substrates as heat [37]. Tolerance engineering to maintain host performance at high temperature saves the expensive cooling cost [37]. In addition, fermentation at higher temperature also helps prevent bacteria contamination and reduces the cost of cooling the fermenter or heating for product separation [37]. Heat shock causes multiple cell damages including intracellular protein aggregation, cell structure destruction, and nuclear processing interference (Fig. 3a). Heat shock proteins (HSPs) can protect cells against these damages. Heterogenous expression of HSPs in a riboflavin-producing strain Bacillus subtilis 446 improved cell survival by up to 5 folds after the 10h heat shock treatment at 50 °C, and increased the riboflavin titers by 38–59% under 39 °C fermentation [38].
Figure 3.
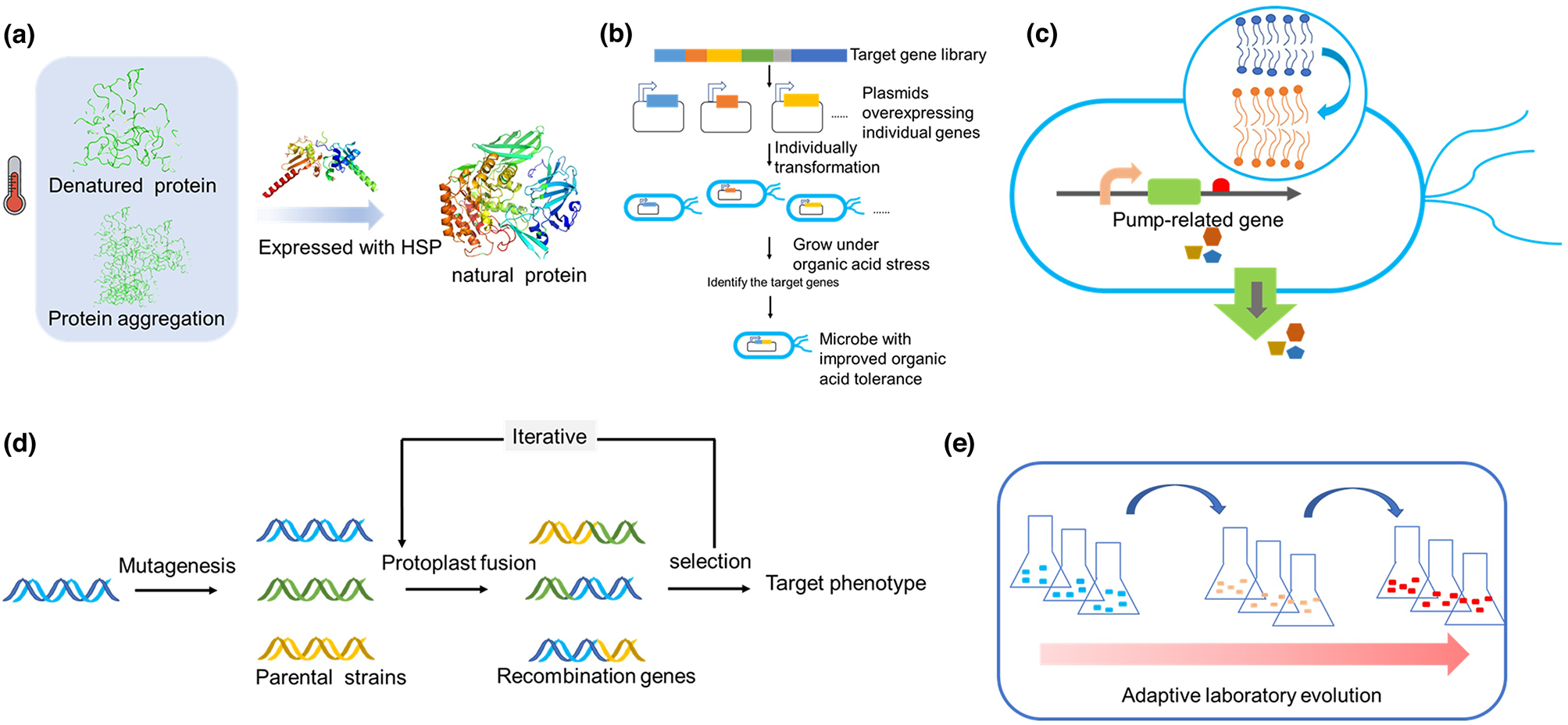
(a) Thermal stability; HSP reduces denatured protein and protein aggregation by promoting the re-folding or degradation of unfolded proteins. HSP, heat shock protein. (b) Acid tolerance engineering. (c) organic solvent tolerance; Increasing acyl chain tail length of membrane lipids and overexpressing efflux pumps can improve microbial tolerance against organic solvents. (d) Genome shuffling; phenotype can be rapidly improved by recursive genomic recombination and screening of generated strains. (e) Adaptive laboratory evolution; strain tolerance can be improved after adaptive evolution under severe selection pressure.
Adaptive laboratory evolution (ALE) is another approach to improve microbial heat tolerance (Fig. 3e). Heat shock response has complicated mechanism, and ALE has the advantage of letting many nonintuitive beneficial mutations occur in parallel. In previous research, thermotolerant yeast strains were obtained through ALE, with ~1.5 times [39] faster growth at 40°C. Correspondingly, ethanol productivity of the evolved strains was 1.6 times faster [40]. The Tmax of C. glutamicum strain GLY3 was also improved from 37 to 40.5 °C in the minimal medium [41].
4.2. Weak acid
The accumulation of acidic intermediates and by-products often occurs during the microbial fermentation. In these processes, the excessive proton (H+) disrupts transmembrane components, negatively affects internal pH homeostasis, and damages the conformation of structural proteins, leading to defected host growth and decreased final titer and productivity. The maintenance of intracellular pH requires the exportation of protons, which will consume a large amount of ATP. The addition of neutralizing reagents, such as NH3•H2O and CaCO3, has been used in industrial fermentation, but this can increase the cost and introduce unnecessary elements [42].
Thus, it is necessary to equip the microbial cells with increased robustness in acidic environment. Researchers have already characterized some genes effective for acid resistance (Fig. 3b). Overexpression of WHI2 gene, encoding a cytoplasmatic globular scaffold protein, in S. cerevisiae can increase the acetic acid tolerance and the pH. The final titer of ethanol was 5-times higher than the control in glucose fermentation with acetic acid [43]. Overexpression of HAA1, encoding a transcriptional activator involved in weak acid stress response, resulted in a 10% increase of final lactic acid titer without neutralization [44]. Some other genes related to acid resistance are also identified, such as EUG1, SAC6, ASG1, ADH3, SKS1, GIS4 [45]. In addition to rational engineering of specific genes, some random strategies were also applied for screening of acid tolerance strains. Genome shuffling (Fig. 3d) was used in Actinobacillus succinogenes for improved acid tolerance leading to a 2.6-fold succinic acid titer increase [46].
4.3. Organic solvents
Many biochemicals, such as biofuels and biopolymers, are competitive replacements for gasoline and diesel. With high industrial values, however, they are solvent-like hydrocarbons and thus toxic to microorganisms [47]. Therefore, engineering strains to improve their tolerance on organic solvents is necessary. Altering membrane lipid composition can be an effective approach (Fig. 3c). It is previously suggested that increasing acyl chain tail length of membrane lipids is beneficial to host tolerance [48]. E. coli evolved under octanoic acid pressure was proved to have increased average lipid length and demonstrated around 15% higher n-butanol and isobutanol tolerance [49].
Another more straightforward strategy is introducing pumps to export the toxic solvents from cells. Recently, Baslet et al. reported that controlled expression of the native resistance-nodulation-cell division (RND)-type efflux pump TtgABC effectively improved the survival rate of Pseudomonas putida exposing to multiple short-chain alcohols by at least 10 folds [50]. Zhang et al. evaluated native efflux pumps of E. coli and recognized four major transporters (MdtJ, Bcr, MdtH and YdeA) able to improve short and medium carbon chain alcohol tolerance by 35% – 50% [51]. Considering the similarities between damaging mechanisms of heat shock and organic solvents, HSPs are also capable of improving solvent tolerance. Co-overexpressing GrpE, GroESL, and ClpB increased cell growth by 200%, 390%, and 78% respectively in 7% ethanol, 1% n-butanol, or 25% 1,2,4-butanetriol [52].
5. Conclusion and future perspectives
Engineering host robustness is essential to maintain production performance of microbial cell factories. A stable host is more economically feasible to be scaled-up to industrial fermentation. In this review, we summarized the current approaches to improve host robustness, including engineering metabolic balancing, enhancing genetic and phenotype stability, and tolerance engineering. In addition to the strategies we reviewed, there are other methods to enhance the host robustness, such as organelle engineering, omics technology, global transcription machinery engineering, functional redundancy, or mathematical model [53–55]. The strategies summarized above effectively increased the microbial hosts robustness and expanded their applications in industrial biotechnology. However, there are still some challenges in engineering microbial hosts robustness. First, dynamic regulation application could be hindered by the limitations of biosensors, such as low sensitivity, narrow dynamic range, and limited spectrum of effectors. The sensitivity of the biosensor determines when to switch to production and the dynamic range affects the productivity of the biosynthesis pathway. Systematic optimization is required to modify the sensitivity and alter the dynamic range of biosensors to facilitate their application in dynamic regulation. In terms of sensor engineering, computational strategies can be effective to engineer robust biosensor systems [56,57]. Meanwhile, developing new biosensors is an effective approach to broaden the spectrum of detectable metabolites. Novel biosensors can be possibly mined and characterized based on sequence similarity with existing ones. Besides, existing biosensors can also be modified to sense various compounds by either directed evolution or rational protein engineering [58].
Second, conventional microbial hosts are usually mesophilic and have limited capabilities to overcome harsh environmental stresses even with certain tolerance enhancement. Some non-conventional hosts, such as thermophilic and acidophilic strains, can possibly remedy the limitations. As for tolerance engineering, elucidating the key resistance mechanism is challenging, but it is important to better guide the engineering direction for enhanced microbial tolerance. A series of potential tools can be used to reveal the mechanism, including global metabolite profiling, adaptive lab evolution combined with sequence analysis and omics analysis [59]. Besides, CRISPR-Cas9 as an unprecedentedly flexible tool for gene knockout or knockdown has also been adopted for microbial tolerance mechanism elucidation [60]. Further engineering and development of non-conventional hosts may be a future direction for building robust microbial cell factories.
Highlights.
Robustness engineering is essential to increase the production performance of microbial cell factories.
Typical strategies for enhancing the microbial host robustness were reviewed and summarized, including engineering metabolic balancing, enhancing plasmid and phenotype stability, and tolerance engineering.
The challenges and future perspectives of robustness engineering were discussed.
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM128620. We also acknowledge the support from the College of Engineering, The University of Georgia, Athens
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no competing interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Yang Y, Lin Y, Li L, Linhardt RJ, Yan Y: Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab Eng 2015, 29:217–226. [DOI] [PubMed] [Google Scholar]
- 2.Humphreys CM, Minton NP: Advances in metabolic engineering in the microbial production of fuels and chemicals from C1 gas. Curr Opin Biotechnol 2018, 50:174–181. [DOI] [PubMed] [Google Scholar]
- 3.•.Wang J, Zhang R, Zhang Y, Yang Y, Lin Y, Yan Y: Developing a pyruvate-driven metabolic scenario for growth-coupled microbial production. Metab Eng 2019, 55:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]; This research demonstrated the great potential of creating growth-driven production phenotype for constructing robust microbial cell factories.
- 4.•.Fujiwara R, Noda S, Tanaka T, Kondo A: Metabolic engineering of Escherichia coli for shikimate pathway derivative production from glucose-xylose co-substrate. Nat Commun 2020, 11:279. [DOI] [PMC free article] [PubMed] [Google Scholar]; This research used parallel metabolic pathway engineering (PMPE) for the shikimate pathway derivative production. Using glucose for product synthesis and xylose for cell growth, the authors separated the glycolysis and the pentose phosphate pathway from the tricarboxylic acid (TCA) cycle.
- 5.Luo X, Reiter MA, d’Espaux L, Wong J, Denby CM, Lechner A, Zhang Y, Grzybowski AT, Harth S, Lin W, et al. : Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567:123–126. [DOI] [PubMed] [Google Scholar]
- 6.Tian R, Liu Y, Chen J, Li J, Liu L, Du G, Chen J: Synthetic N-terminal coding sequences for fine-tuning gene expression and metabolic engineering in Bacillus subtilis. Metab Eng 2019, 55:131–141. [DOI] [PubMed] [Google Scholar]
- 7.Longwell CK, Labanieh L, Cochran JR: High-throughput screening technologies for enzyme engineering. Curr Opin Biotechnol 2017, 48:196–202. [DOI] [PubMed] [Google Scholar]
- 8.Wu W, Zhang Y, Liu D, Chen Z: Efficient mining of natural NADH-utilizing dehydrogenases enables systematic cofactor engineering of lysine synthesis pathway of Corynebacterium glutamicum. Metab Eng 2019, 52:77–86. [DOI] [PubMed] [Google Scholar]
- 9.Noh M, Yoo SM, Kim WJ, Lee SY: Gene Expression Knockdown by Modulating Synthetic Small RNA Expression in Escherichia coli. Cell Syst 2017, 5:418–426 e414. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Shen X, Jain R, Wang J, Yuan Q, Yan Y: Establishing a novel biosynthetic pathway for the production of 3,4-dihydroxybutyric acid from xylose in Escherichia coli. Metab Eng 2017, 41:39–45. [DOI] [PubMed] [Google Scholar]
- 11.Gong Z, Nielsen J, Zhou YJ: Engineering Robustness of Microbial Cell Factories. Biotechnol J 2017, 12. [DOI] [PubMed] [Google Scholar]
- 12.•.Yang Y, Lin Y, Wang J, Wu Y, Zhang R, Cheng M, Shen X, Wang J, Chen Z, Li C, et al. : Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis. Nat Commun 2018, 9:3043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Synergetic bi-functional dynamical control combining up-regulation of rate-limiting biosynthesis steps and down-regulation of competing pathway was applied to balance the carbon flux for glycolysis and cis, cis-muconic acid synthesis.
- 13.•.Zhang R, Yang Y, Wang J, Lin Y, Yan Y: Synthetic symbiosis combining plasmid displacement enables rapid construction of phenotype-stable strains. Metab Eng 2019, 55:85–91. [DOI] [PubMed] [Google Scholar]; This research established a standardized protocol for creating synthetic auxotrophic strains by introducing an intermediate plasmid to facilitate the process, which simplified and accelerated the process for construction of phenotype-stable strains.
- 14.Choi KR, Jang WD, Yang D, Cho JS, Park D, Lee SY: Systems Metabolic Engineering Strategies: Integrating Systems and Synthetic Biology with Metabolic Engineering. Trends Biotechnol 2019, 37:817–837. [DOI] [PubMed] [Google Scholar]
- 15.Ma SM, Garcia DE, Redding-Johanson AM, Friedland GD, Chan R, Batth TS, Haliburton JR, Chivian D, Keasling JD, Petzold CJ, et al. : Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab Eng 2011, 13:588–597. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Shen X, Yuan Q, Yan Y: Microbial synthesis of pyrogallol using genetically engineered Escherichia coli. Metab Eng 2018, 45:134–141. [DOI] [PubMed] [Google Scholar]
- 17.Wang P, Yang X, Lin B, Huang J, Tao Y: Cofactor self-sufficient whole-cell biocatalysts for the production of 2-phenylethanol. Metab Eng 2017, 44:143–149. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, Mannan AA, Han Y, Oyarzun DA, Zhang F: Dynamic metabolic control: towards precision engineering of metabolism. J Ind Microbiol Biotechnol 2018, 45:535–543. [DOI] [PubMed] [Google Scholar]
- 19.Shen X, Wang J, Li C, Yuan Q, Yan Y: Dynamic gene expression engineering as a tool in pathway engineering. Curr Opin Biotechnol 2019, 59:122–129. [DOI] [PubMed] [Google Scholar]
- 20.Lv Y, Gu Y, Xu J, Zhou J, Xu P: Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab Eng 2020, 61:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Carothers JM, Keasling JD: Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 2012, 30:354–359. [DOI] [PubMed] [Google Scholar]
- 22.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, et al. : Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol 2013, 31:1039–1046. [DOI] [PubMed] [Google Scholar]
- 23.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M: Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U S A 2014, 111:11299–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A, Reizman IM, Reisch CR, Prather KL: Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 2017, 35:273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soma Y, Hanai TJMe: Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng 2015, 30:7–15. [DOI] [PubMed] [Google Scholar]
- 26.Lo TM, Chng SH, Teo WS, Cho HS, Chang MW: A Two-Layer Gene Circuit for Decoupling Cell Growth from Metabolite Production. Cell Syst 2016, 3:133–143. [DOI] [PubMed] [Google Scholar]
- 27.••.Doong SJ, Gupta A, Prather KLJ: Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc Natl Acad Sci U S A 2018, 115:2964–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]; This research applied dynamic regulation in both pathway-specific and pathway-independent manner to simultaneously address the low stability of myo-inositol and decouple cell growth and production of glucaric acid.
- 28.Lan EI, Liao JC: ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc Natl Acad Sci U S A 2012, 109:6018–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehrer CR, Rand JM, Incha MR, Cook TB, Demir B, Motagamwala AH, Kim D, Dumesic JA, Pfleger BF: Growth-coupled bioconversion of levulinic acid to butanone. Metab Eng 2019, 55:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.•.Rugbjerg P, Sarup-Lytzen K, Nagy M, Sommer MOA: Synthetic addiction extends the productive life time of engineered Escherichia coli populations. Proc Natl Acad Sci U S A 2018, 115:2347–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]; The metabolite biosensor-based product-addiction strategy enabled a high-yield mevalonate producing strain that can maintain the phenotype over 95 generations in industrial scale fermentation.
- 31.Sevillano L, Díaz M, Santamaría RI: Stable expression plasmids for Streptomyces based on a toxin-antitoxin system. Microbial cell factories 2013, 12:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams SG, Cranenburgh RM, Weiss AM, Wrighton CJ, Hanak JA, Sherratt DJ: Repressor titration: a novel system for selection and stable maintenance of recombinant plasmids. Nucleic acids research 1998, 26:2120–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selvamani RSV, Telaar M, Friehs K, Flaschel E: Antibiotic-free segregational plasmid stabilization in Escherichia coli owing to the knockout of triosephosphate isomerase (tpiA). Microbial cell factories 2014, 13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang CW, Lim HG, Yang J, Noh MH, Seo SW, Jung GY: Synthetic auxotrophs for stable and tunable maintenance of plasmid copy number. Metabolic engineering 2018, 48:121–128. [DOI] [PubMed] [Google Scholar]
- 35.Tonn MK, Thomas P, Barahona M, Oyarzun DA: Stochastic modelling reveals mechanisms of metabolic heterogeneity. Commun Biol 2019, 2:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao Y, Bowen CH, Liu D, Zhang F: Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nature Chemical Biology 2016, 12:339–344. [DOI] [PubMed] [Google Scholar]
- 37.Thorwall S, Schwartz C, Chartron JW, Wheeldon I: Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. Nat Chem Biol 2020, 16:113–121. [DOI] [PubMed] [Google Scholar]; The authors insightfully analyzed current core problems limiting the industrialization of bioprocess and provided interesting perspectives.
- 38.••.Wang J, Wang W, Wang H, Yuan F, Xu Z, Yang K, Li Z, Chen Y, Fan K: Improvement of stress tolerance and riboflavin production of Bacillus subtilis by introduction of heat shock proteins from thermophilic bacillus strains. Appl Microbiol Biotechnol 2019, 103:4455–4465. [DOI] [PubMed] [Google Scholar]; This research demonstrates a typical strategy for tolerance engineering. The authors identified heterogenous HSPs able to improve heat tolerance of B. subtilis and fully evaluated other notable effects of these HSPs. The industrial B. subtilis stain gained not only higher heat tolerance, but also osmic tolerance and thus riboflavin productivity by overexpressing the heterogenous HSPs.
- 39.Caspeta L, Coronel J, Montes de Oca A, Abarca E, González L, Martínez AJB, bioengineering: Engineering high‐ gravity fermentations for ethanol production at elevated temperature with Saccharomyces cerevisiae. 2019, 116:2587–2597. [DOI] [PubMed] [Google Scholar]
- 40.Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallström BM, Petranovic D, Nielsen J: Altered sterol composition renders yeast thermotolerant. Science 2014, 346:75–78. [DOI] [PubMed] [Google Scholar]
- 41.Oide S, Gunji W, Moteki Y, Yamamoto S, Suda M, Jojima T, Yukawa H, Inui M: Thermal and solvent stress cross-tolerance conferred to Corynebacterium glutamicum by adaptive laboratory evolution. Appl Environ Microbiol 2015, 81:2284–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baek SH, Kwon EY, Bae SJ, Cho BR, Kim SY, Hahn JS: Improvement of d-Lactic Acid Production in Saccharomyces cerevisiae Under Acidic Conditions by Evolutionary and Rational Metabolic Engineering. Biotechnol J 2017, 12. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Stabryla L, Wei N: Improved Acetic Acid Resistance in Saccharomyces cerevisiae by Overexpression of the WHI2 Gene Identified through Inverse Metabolic Engineering. Appl Environ Microbiol 2016, 82:2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baek SH, Kwon EY, Kim YH, Hahn JS: Metabolic engineering and adaptive evolution for efficient production of D-lactic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 2016, 100:2737–2748. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Ramos D, Gorter de Vries AR, Grijseels SS, van Berkum MC, Swinnen S, van den Broek M, Nevoigt E, Daran JM, Pronk JT, van Maris AJ: A new laboratory evolution approach to select for constitutive acetic acid tolerance in Saccharomyces cerevisiae and identification of causal mutations. Biotechnol Biofuels 2016, 9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu S, You Y, Xia F, Liu J, Dai W, Liu J, Wang Y: Genome shuffling improved acid-tolerance and succinic acid production of Actinobacillus succinogenes. Food Sci Biotechnol 2019, 28:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Nielsen J: Recent trends in metabolic engineering of microbial chemical factories. Curr Opin Biotechnol 2019, 60:188–197. [DOI] [PubMed] [Google Scholar]
- 48.Sandoval NR, Papoutsakis ET: Engineering membrane and cell-wall programs for tolerance to toxic chemicals: Beyond solo genes. Curr Opin Microbiol 2016, 33:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Royce LA, Yoon JM, Chen Y, Rickenbach E, Shanks JV, Jarboe LR: Evolution for exogenous octanoic acid tolerance improves carboxylic acid production and membrane integrity. Metab Eng 2015, 29:180–188. [DOI] [PubMed] [Google Scholar]
- 50.•.Basler G, Thompson M, Tullman-Ercek D, Keasling J: A Pseudomonas putida efflux pump acts on short-chain alcohols. Biotechnol Biofuels 2018, 11:136. [DOI] [PMC free article] [PubMed] [Google Scholar]; For the first time, a native RND-type efflux system was applied to export various short-chain alcohols. In addition, the results also suggested that the extensively adopted growth assay might have some limitations on identifying the function of efflux pumps.
- 51.Zhang Y, Dong R, Zhang M, Gao H: Native efflux pumps of Escherichia coli responsible for short and medium chain alcohol. Biochemical Engineering Journal 2018, 133:149–156 [Google Scholar]
- 52.Zingaro KA, Papoutsakis ETJM: Toward a semisynthetic stress response system to engineer microbial solvent tolerance. mBio 2012, 3:e00308–00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammer SK, Avalos JLJNcb: Harnessing yeast organelles for metabolic engineering. Nat Chem Biol 2017, 13:823. [DOI] [PubMed] [Google Scholar]
- 54.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A: Engineering microbial biofuel tolerance and export using efflux pumps. J Molecular systems biology 2011, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, et al. : Function and functional redundancy in microbial systems. Nature Ecology & Evolution 2018, 2:936–943. [DOI] [PubMed] [Google Scholar]
- 56.Xu P: Branch point control at malonyl-CoA node: A computational framework to uncover the design principles of an ideal genetic-metabolic switch. Metab Eng Commun 2020, 10:e00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mannan AA, Liu D, Zhang F, Oyarzun DA: Fundamental Design Principles for Transcription-Factor-Based Metabolite Biosensors. ACS Synth Biol 2017, 6:1851–1859. [DOI] [PubMed] [Google Scholar]
- 58.Li C, Zhang R, Wang J, Wilson LM, Yan YJTiB: Protein Engineering for Improving and Diversifying Natural Product Biosynthesis. Trends in Biotechnology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu B, Yang YM, Beck DA, Wang QW, Chen WJ, Yang J, Lidstrom ME, Yang S: Comprehensive molecular characterization of Methylobacterium extorquens AM1 adapted for 1-butanol tolerance. Biotechnol Biofuels 2016, 9:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otoupal PB, Chatterjee A: CRISPR Gene Perturbations Provide Insights for Improving Bacterial Biofuel Tolerance. Front Bioeng Biotechnol 2018, 6:122. [DOI] [PMC free article] [PubMed] [Google Scholar]


