Abstract
Nicotinamide cofactors enable oxidoreductases to catalyze a myriad of important reactions in biomanufacturing. Decades of research has focused on optimizing enzymes which utilize natural nicotinamide cofactors, namely nicotinamide adenine dinucleotide (phosphate) (NAD(P)+). Recent findings reignite the interest in engineering enzymes to utilize noncanonical cofactors, the mimetics of NAD+ (mNADs), which exhibit superior industrial properties in vitro and enable specific electron delivery in vivo. We compare recent advances in engineering natural versus noncanonical cofactor-utilizing enzymes, discuss design principles discovered, and survey emerging high-throughput platforms beyond the traditional 96-well plate-based methods. Obtaining mNAD-dependent enzymes remains challenging with a limited toolkit. To this end, we highlight design principles and technologies which can potentially be translated from engineering natural to noncanonical cofactor-dependent enzymes.
Keywords: Nicotinamide redox cofactor, Oxidoreductases, Noncanonical cofactor, NAD mimetics, protein engineering, High-throughput screening, directed evolution
Graphical Abstract
INTRODUCTION
Nicotinamide cofactor-utilizing enzymes are versatile catalysts for both in vitro chemical synthesis and in vivo metabolic engineering. Although more than 15,000 sequences have been confirmed or predicted to encode NAD(P)+ or NAD(P)H utilizing enzymes [1], natural enzymes frequently do not meet the catalytic needs of compatibility with the metabolism of a chassis host in vivo and viability at large scales in vitro, and often require engineering of the enzyme’s natural cofactor specificity, substrate scope, and robustness.
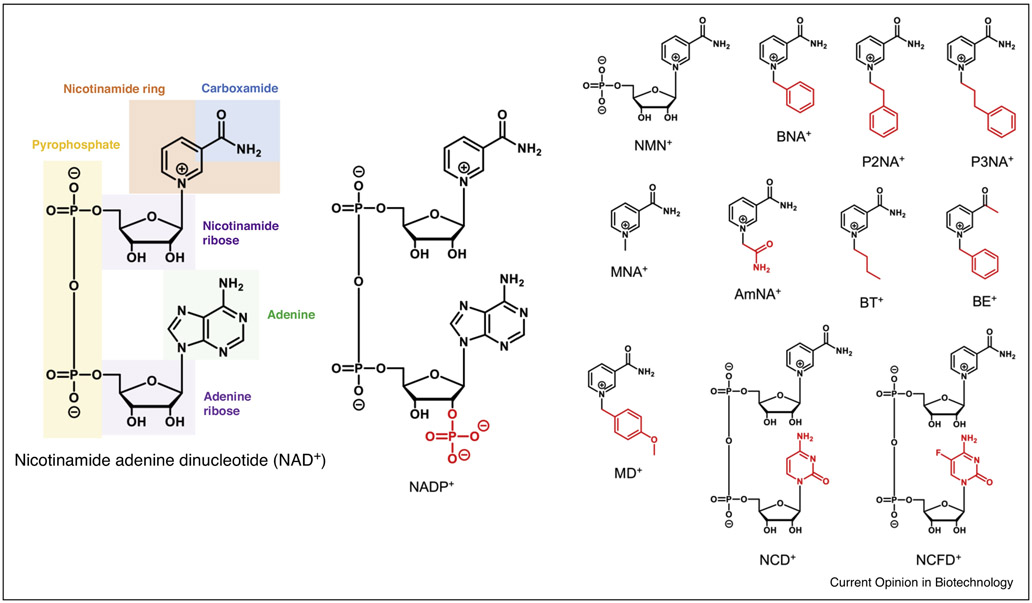
Recent studies highlight the value of engineering these enzymes to use noncanonical nicotinamide cofactors, which are mimetics of NAD+ (mNADs). The nicotinamide ring is the only fragment required for a small molecule to function as a redox cofactor [2-4]. Molecules with alternative functional groups replacing the NAD(P)/H carboxamide [4], the adenine base [5-7], the nicotinamide ribose [8], and mimics truncated at different atoms have been explored as artificial redox cofactors [9,10] (Figure 1). These mimics have industrial value for lowering feedstock costs as mNADs are often simpler to synthesize [11] and have greater stability than native cofactors [12], permit access to new chemistries with altered redox potential [13], reduce oxygenase decoupling [14,15], and importantly enable specific delivery of electrons in both whole cells and crude cell lysates [9,11,16]. Metabolic pathways engineered to specifically utilize mNADs are orthogonal from the host’s metabolism, as they do not cross-talk with native pathways which only use natural cofactors. This allows precise control of chemical reactions in the cells without interference and has been demonstrated in vivo for the production of malate from the carboxylation of pyruvate via nicotinamide cytosine dinucleotide (NCD+) [6, 16], and selective generation of the pharmaceutical intermediate levodione by nicotinamide mononucleotide (NMN+) mediated reduction [9].
Figure 1. Chemical structures of natural nicotinamide redox cofactors and mNADs.
The natural nicotinamide redox cofactor NAD+ is composed of the catalytic nicotinamide ring attached to a ribose, pyrophosphate, a second ribose, and adenine base. NADP+ differs in that a phosphate group replaces a hydroxyl on the 2’ carbon of the adenine ribose. mNADs maintain the central nicotinamide ring, but are truncated or incorporate alternative functional groups compared to NAD+. The cofactors are illustrated in their oxidized from, complete cofactor names are listed under Tables 1 and 2.
Design principles in engineering natural cofactor-dependent enzymes
Many general design principles are derived from decades of research in engineering NAD(P)/H-dependent enzymes. For example, altering specificity between natural cofactors commonly relies on the mutagenesis of binding pocket amino acids interacting with the signature 2’-phosphate or 2’-hydroxyl groups that differentiate NADP(H) and NAD(H), respectively. Fundamental semi-rational design rules have been captured by an easy-to-use web tool Cofactor Specificity Reversal-Structural Analysis and Library Design (CSR-SALAD) [17]. This computational method incorporates structural, activity, and genetic information to automatically design focused libraries [18]; however, it has met with limited success for enzymes that utilize cofactors in complex reaction mechanisms [17, 19]. Flexible loop grafting has emerged as another design principle in engineering the cofactor preference of TIM barrel oxidoreductases. Swapping of cofactor binding loops between homologous ene reductases (ERs) with NADPH and NADH preference [20] shows promise as a means of generating flexibility in cofactor preferences. Additional studies on aldo-keto reductase (AKR) inverted cofactor preference from NAD+ to NADP+ by inserting either additional residues [21] or a calcium controllable repeats-in-toxin (RTX) domain [22] into substrate binding loops.
Beyond cofactor specificity, complex traits such as stability and conformational dynamics are a challenging task for rational design. Recent reports revealed the effects of modulating the microenvironment surrounding oxidoreductases, which can potentially be a universal design principle in engineering both NAD(P)/H and mNAD-dependent enzymes. For example, fusing a variant of superfolding green fluorescent protein (sfGFP) with extreme surface charges enhanced the activity of AKR, possibly by influencing the apparent ionic strength of the active site [23]. Furthermore, increased cofactor availability has been explored with DNA-enzyme nanostructures [24] acting as local reservoirs of cofactors, fusing of redox cycling partners by co-expression [25-29], and directly tethering NAD(H) to proteins with polyethylene glycol chains [30].
Design principles in engineering noncanonical cofactor-dependent enzymes
We summarize efforts in enhancing mNAD catalysis and evaluate the extent of success through the following metrics [31] (Tables 1 and 2):
Table 1.
Performance of wild type enzymes with noncanonical cofactors
| Enzyme | Uniprot | Native Cofactor |
Noncanonical Cofactora |
CSRb | Source |
|---|---|---|---|---|---|
| D-lactate dehydrogenase (L. helveticus) | P30901 | NAD+ | NCD+ | 4.8×10−2 | Liu et al, 2020 [5] |
| Formate dehydrogenase (Pseudomonas sp. 101) | P33160 | NAD+ | NCD+ | 4.7×l0−2 | Guo et al., 2020 [6] |
| Glucose Dehydrogenase (B. subtilis) | A0A1B2ATD9 | NAD+ | NMN+ | 2.6×l0−6 | Black et al., 2019 [9] |
| NADP+ | NMN+ | 1.7×l0−6 | |||
| 6-phospho gluconate dehydrogenase (T. maritima) | A0A2N5RL69 | NADP+ | NMN+ | 3.1×l0−6 | Huang et al., 2019 [10] |
| Phosphite dehydrogenase (Ralstonia sp. 4506) | G4XDR8 | NAD+ | NCD+ | 5.8×10−2 | Liu et al., 2019 [7] |
| 3-hydroxy benzoate 6-hydroxylase (R. jostii) | Q0SFK6 | NADH | BNAH | 7.5×10−4 | Guarneri et al., 2019 [14] |
| AmNAH | 4.9×10−4 | ||||
| NADPH | BNAH | 4.8×10−3 | |||
| AmNAH | 3.2×10−3 | ||||
| para-Hydroxybenzoate hydroxylase (P. fluorescens) | P20586 | NADH | AmNAH | 2.1 | |
| NADPH | AmNAH | 2.4×10−3 | |||
| Salicylate hydroxylase (P. putida) | - | NADH | BNAH | 8.3×10−3 | |
| NADPH | BNAH | 8.8×10−1 | |||
| Glucose dehydrogenase (S. solfataricus) | O93715 | NAD+ | BNA+ | 1.0×10−2 | Nowak et al., 2017 [8] |
| P2NA+ | 1.5×10−2 | ||||
| P3NA+ | 5.6×10−3 | ||||
| NADH oxidase (L. pentosus) | F6IXY6 | NADH | MNAH | - | Nowak et al., 2015, 2017 [12, 15] |
| BNAH | - | ||||
| P2NAH | - | ||||
| P3NAH | - | ||||
| Xenobiotic reductase (P. putida) | Q3ZDM6 | NADH | BNAH | 3.1×102 | Knaus et al., 2016 [32] |
| BTH | 1.8×103 | ||||
| BEH | 2.7×102 | ||||
| NADPH | BNAH | 2.7 | |||
| BTH | 1.5×10 | ||||
| BEH | 2.7×102 | ||||
| Pentaerythritol tetranitrate reductase (E. cloacae) | P71278 | NADPH | BNAH | 1.4 | |
| BTH | 5.2 | ||||
| BEH | 1.0Δ10−1 | ||||
| Thennophilic old yellow enzyme (T. pseudethanolicus) | B0KAH1 | NADH | BNAH | 8.2×10−1 | |
| BTH | 1.2 | ||||
| BEH | 3.3×10−1 | ||||
| NADPH | BNAH | 2.2×10−2 | |||
| BTH | 3.2×10−2 | ||||
| BEH | 8.7×10−3 | ||||
| Styrene monooxygenase (R. opacus) | C7ACG0 | NADH | BNAH | - | Paul et al.. 2015 [54] |
| Alcohol dehydrogenase (P. furiosus) | - | NAD+ | NMN+ | 8.6×10−6 | Campbell et al.. 2012 [35] |
| Malic enzyme (E. coli) | P26616 | NAD+ | NFCD+ | 9.3×10−3 | Ji et al.. 2011 [53] |
| NCD+ | 1.3×10−2 |
Full names of the noncanonical cofactors: AmNA+, 1-(2-carbamoylmethyl)-1,4-dihydronicotinamide; BNA+, 1-benzyl-1,4-dihydronicotinamide; BT+, 1-butyl-1,4-dihydronicotinamide; BE+, 1-(1-benzyl-1,4-dihydro-3-yl) ethanone; MD+, N-4-methoxybenzyl-1,4-dihydronicotinamide; MNA+, 1-methyl-1,4-dihydropyridine-3-carboxamide; NCD+, Nicotinamide cytosine dinucleotide; NCFD+, Nicotinamide flucytosine dinucleotide; NMN+, Nicotinamide mononucleotide; P2NA+, 1-phenethyl-1,4-dihydropyridine-3-carboxamide; P3NA+, 1-(3-phenylpropyl)-1,4-dihydropyridine-3-carboxamide. Reduced cofactor ends with “H”
CSR, Cofactor Specificity Ratio (Equation 1)
Table 2.
Performance of engineered enzymes with noncanonical cofactors
| Enzyme | Uniprot | Strategy | Native Cofactor |
Noncanonical Cofactora |
Mutations | CSRb | RCEC | RSd | Source |
|---|---|---|---|---|---|---|---|---|---|
| D-lactate dehydrogenase (L. helveticus) | P30901 | Bibliography, Saturation, Structure | NAD+ | NCD+ | V152R-N213E | 4.3×10 | 2.1×10−1 | 9.0×102 | Liu et al., 2020 [5] |
| V152R-I177K-N213I | 4.2×10 | 3.1×10−1 | 8.8×102 | ||||||
| Formate dehydrogenase (Pseudomonas sp. 101) | P33160 | Bibliography, Saturation, Structure | NAD+ | NCD+ | V198I-C256I-P260S-E261P-S381N-S383F | 1.7×102 | 3.7×10−2 | 3.5×103 | Guo et al., 2020 [6] |
| Glucose Dehydrogenase (B. subtilis) | A0A1B2ATD9 | Computational, Structure | NADP+ | NMN+ | Y34Q-A93K-I195R | 6.8×10−2 | 1.8×10−3 | 4.1×104 | Black et al., 2019 [9] |
| S17E-Y34Q-A93K-I195R | 1.9×10 | 7.5×10−4 | 1.1×107 | ||||||
| NAD+ | NMN+ | Y34Q-A93K-I195R | 4.6 | 2.8×10−3 | 1.8×106 | ||||
| S17E-Y34Q-A93K-I195R | 5.5×10 | 1.2×10−3 | 2.1×107 | ||||||
| 6-phospho gluconate dehydrogenase (T. maritima) | A0A2N5RL69 | Computational, Random, Saturation, Structure | NADP+ | NMN+ | Mut 5-1e | 1.4×10−2 | 1.2×10−4 | 4.6×103 | Huang et al., 2019 [10] |
| Mut 6-1f | 1.4×10−2 | 1.5×10−4 | 4.4×103 | ||||||
| Glucose-6-phosphate dehydrogenase (T. maritima) | A0A2N5RPI0 | Computational, Random, Saturation, Structure | NADP+ | NMN+ | A64S-R65I-T66I | - | - | - | |
| Phosphite dehydrogenase (Ralstonia sp. 4506) | G4XDR8 | Bibliography, Computational, Saturation, Structure | NAD+ | NCD+ | I151R-P176R | 2.0×10 | 1.3×10−1 | 3.4×102 | Liu et al., 2019 [7] |
| I151R-P176R-M207A | 4.5×10 | 9.4×10−3 | 7.8×102 | ||||||
| Glucose dehydrogenase (S. solfataricus) | O93715 | Bibliography, Saturation, Structure | NAD+ | BNA+ | I192T-V306G | 5.5×10 | 6.5×10−2 | 5.5×102 | Nowak et al., 2017 [8] |
| I192T-V306I | 2.9×10−1 | 1.0×10−1 | 2.9×10 | ||||||
| P2NA+ | I192T-V306G | 3.1 | 3.6×10−2 | 2.0×102 | |||||
| I192T-V306I | 9.2×10−1 | 3.2×10−1 | 6.1×10 | ||||||
| P3NA+ | I192T-V306G | 3.8×10 | 2.6×10−1 | 6.8×102 | |||||
| I192T-V306I | 3.0×10−1 | 1.1× 10−1 | 5.3×10 | ||||||
| Phosphite dehydrogenase (Ralstonia sp. 4506) | G4XDR8 | Bibliography | NAD+ | NCD+ | I151R | 4.3×10 | 1.4×10−2 | 5.4×102 | Wang et al., 2017 [16] |
| P450-BM3 (B. megaterium) | P14779 | Bibliography, Structure | NADH | BNAH | R966D-W1046S | 1.0×10−1 | 9.6×10 | - | Lo et al., 2017 [33] |
| MDH | R966D-W1046S | 6.4×10−2 | 6.0×10 | - | |||||
| Alcohol dehydrogenase (P. furiosus) | - | MSAg, Structure | NAD+ | NMN+ | K249G-H255R | 1.6×10−3 | 4.4×10−4 | 1.8×102 | Campbell et al., 2012 [35] |
| Malic enzyme (E. coli) | P26616 | Computational, MSA, Saturation, Structure | NAD+ | NFCD+ | L310R | 9.8×10 | 4.1×10−1 | 1.1×104 | Ji et al., 2011 [53] |
| L310R-Q401C | 2.7×102 | 4.5×10−1 | 2.9×104 | ||||||
| NCD+ | L310R | 1.9×102 | 7.9×10−1 | 1.4×104 | |||||
| L310R-Q401C | 4.3×102 | 7.2×10−1 | 3.3×104 | ||||||
| D-lactate dehydrogenase (L. helveticus) | P30901 | MSA, Structure | NAD+ | NCFD+ | V152R | - | - | - | |
| Malate dehydrogenase (E. coli) | P61889 | MSA, Structure | NAD+ | NCFD+ | L6R | - | - | - | |
| Lactate dehydrogenase (B. stearothermophilus) | P00344 | MSA, Saturation, Structure | NAD+ | NMN+ | C81S-N85R-F16Q | - | - | - | Flores et al., 2005 [34] |
Full names of the noncanonical cofactors: AmNA+, 1-(2-carbamoylmethyl)-1,4-dihydronicotinamide; BNA+, 1-benzyl-1,4-dihydronicotinamide; BT+, 1-butyl-1,4-dihydronicotinamide; BE+, 1-(1-benzyl-1,4-dihydro-3-yl) ethanone; MD+, N-4-methoxybenzyl-1,4-dihydronicotinamide; MNA+, 1-methyl-1,4-dihydropyridine-3-carboxamide; NCD+, Nicotinamide cytosine dinucleotide; NCFD+, Nicotinamide flucytosine dinucleotide; NMN+, Nicotinamide mononucleotide; P2NA+, 1-phenethyl-1,4-dihydropyridine-3-carboxamide; P3NA+, 1-(3-phenylpropyl)-1,4-dihydropyridine-3-carboxamide. Reduced cofactor ends with “H”
CSR, Cofactor Specificity Ratio (Equation 1)
RCE, Relative Catalytic Efficiency (Equation 2)
RS, Relative Specificity (Equation 3)
Mut 5-1 contains A11G-K27R-R33I-T34I-F60Y-D82L-T83L-Q86L-K118N-I120F-D294V-F326S-Y383C-N387S-A447V
Mut 6-1 contains A11G-K27R-R33I-T34I-F60Y-D82L-T83L-Q86L-K118N-I120F-D251E-D294V-F326S-F329Y-Y383C-N387S-V390G-A447V
MSA, Multiple Sequence Alignment
(1) Coenzyme Specificity Ratio (CSR), a measure of preference for the mNAD over natural cofactors. While most wild type enzymes use mNADs very poorly, as reflected by near zero CSR, many flavoenzymes including enoate reductases, nitroreductases, and para-hydroxybenzoate hydroxylase exhibit promising activities [9,14,32] (Table 1). In particular, the xenobiotic reductase from Pseudomonas putida (P. putida XenA) utilizes a range of mNADs more efficiently than natural cofactors [32] (Table 1). High CSR is desirable for creating orthogonal redox circuitry [6,9,16]; however, while most studies only consider NAD+, it is important to measure CSR for both NAD+ and NADP+ [9] when determining orthogonality in vivo.
| (1) |
(2) Relative Catalytic Efficiency (RCE) is the ratio of the mutant’s catalytic efficiency with mNAD compared to wild type with native cofactor. Since wild type enzymes have been optimized by Nature with its native cofactor, RCE essentially indicates how effective the engineering approaches are compared to natural evolution. RCEs for mNADs are extremely low for most engineered enzymes, indicating that catalytic activities are a small fraction of the wild type with native cofactor. We note that P450-BM3 R966D-W1046S reported by Lo et al. [33] had an exceptional RCE of ~96 for 1-benzyl-1,4-dihydronicotinamide (BNAH) and ~60 for N-4-methoxybenzyl-1,4-dihydronicotinamide (MDH) (Table 2). In comparison, RCEs of >1 are frequently achieved in switching NAD+ and NADP+ specificity [31].
| (2) |
(3) Relative Specificity (RS), the CSR of a variant compared to that of the wild type, which is often referred to as the fold of cofactor specificity switch toward noncanonical cofactors. This parameter is useful for comparing the effectiveness of different engineering approaches in general, independently of the specific enzymes targeted (Table 2).
| (3) |
Because the number of successful cases is still relatively small, core design principles for switching cofactor specificity toward noncanonical cofactors have yet to clearly emerge. The field still largely relies on semi-rational and random engineering which often yields beneficial mutations with unknown mechanisms. Gaining fundamental understanding on enzyme-mNAD interaction through structural and kinetic studies is crucial to deriving design principles to streamline engineering. Nevertheless, the following trends are notable:
First, relaxation of cofactor specificity is linked to enhanced activity with mNADs. Bacillus stearothermophilus lactate dehydrogenase F16Q-C81S-N85R with specificity switched from NAD+ to NADP+ was found to reduce NMN+ with trace activity [34]. The K249G-H255R variant of Pyrococcus furiosus alcohol dehydrogenase designed to increase the volume of the active site for NADP+ binding unexpectedly gained the ability to utilize NMN+ (Table 2), and showed a 40% increase in maximum current density when used in a biofuel cell, postulated to be due to improved mass transfer of NMN+ compared to NAD+ [35]. The P450-BM3 mutant R966D-W1046S (Table 2) capable of using both NADPH and NADH was also able to utilize BNAH for the reduction of cytochrome c with a catalytic efficiency of 41.3 min−1 μM−1, while the wild type had no detectable activity [33,36]. A similar variant P450-BM3 W1046S also gained activity for utilizing both natural cofactors and reduced NMN+ (NMNH) [9].
Second, size reduction of the cofactor binding pocket to improve packing often affords increased activity toward mNADs. For example, the phosphite dehydrogenase from Ralstonia sp. 4506 harboring I151R-P176R-M207A mutations had significantly enhanced activity toward NCD+. Crystallography suggested activity was achieved through compression of the binding pocket around the smaller cytosine [7]. Interestingly, natural flavoenzymes that efficiently utilize mNADs also employ this strategy. The bulky Trp302 residue in P. putida XenA active site adopts a different conformation when smaller mNADs are bound to pack more tightly against the cofactors [32].
Third, design to install polar interactions, which in principle contribute more strongly to binding affinity than hydrophobic packing, is effective for achieving stringent binding of mNADs. We recently engineered a highly orthogonal Bacillus subtilis glucose dehydrogenase S17E-Y34Q-A93K-I195R to use NMN+ [9] which showed the highest RS (Equation 3) reported to date of 1.1 × 107 for NADP+ and 2.1 × 107 for NAD+. We first utilized Rosetta modeling to identify the positively charged I195R mutation which is predicted to form a salt bridge with the highly negative NMN+ phosphate. Next, we achieved exclusive specificity for NMN+ by introducing S17E which is modeled to repel the phosphate in the adenosine monophosphate (AMP) moiety that is only present in the natural cofactors but not in NMN+. Due to the high conservation of residues lining the cofactor binding pocket, we hypothesize that these mutations should be readily transferable and support NMN+ binding in homologs.
Technology development for engineering natural cofactor-dependent enzymes
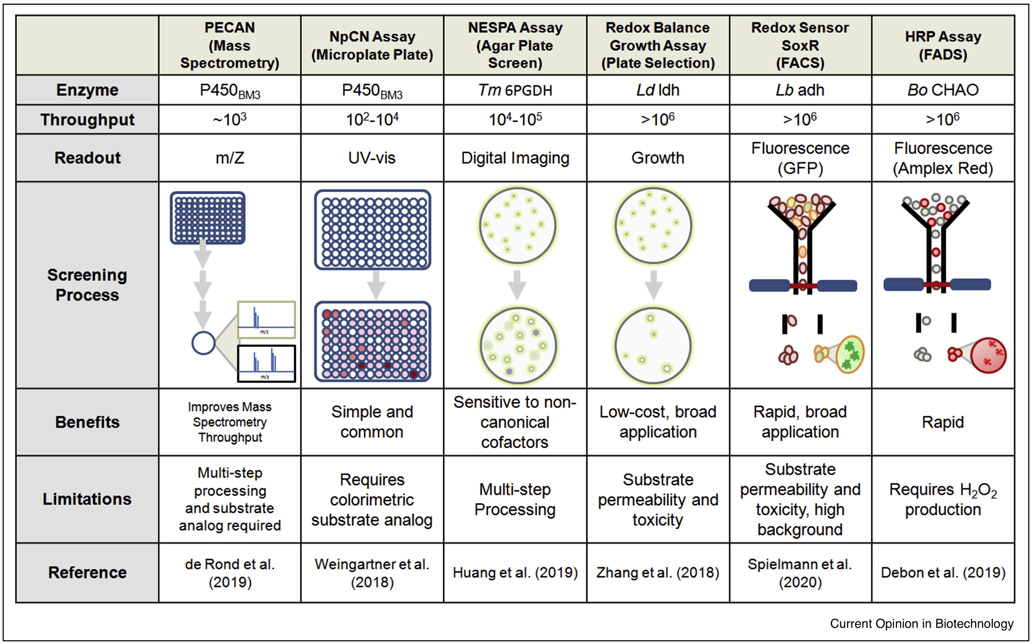
Limited throughput has driven the use of semi-rational strategies to minimize the number of variants screened and to maximize the likelihood of isolating promising candidates. Many of these focused libraries have been screened based on readouts that can be determined by a microplate reader [37,38] or visualized on an agar plate [39,40]. Application of a 4-nitrophenylacetonitrile microplate assay provided a colorimetric screen to isolate cytochrome P450-BM3 variants for hydroquinone production with 70-fold improvement over wild type activity [37] (Figure 2). In another example, an agar screen leveraged the solubility difference of the substrate and product to evolve the substrate scope of a cyclohexanone monooxygenase (CHMO) for pilot-scale applications [40,41]. For enzymes that do not produce color or absorbance change during catalysis, a mass spectrometry-based screening platform was developed (Figure 2) to use “click” chemistry to enhance throughput [42]. This mass spectrometry-based platform may be readily applicable to engineering mNAD-dependent enzymes. Despite success, throughput remains limiting (103-105); furthermore, reduced library sizes may miss potential cooperative effects critical for dramatic improvements [9].
Figure 2. Representative screening methods used to facilitate the directed evolution of oxidoreductases.
PECAN (probing enzymes with click-assisted NIMS), NpCN (4-nitrophenylacetonitrile), NESPA (NAD(P)-eliminated solid-phase assay), soxR (Redox-sensitive transcriptional activator), FACS (Fluorescence Activated Cell Sorting), HRP (Horse radish peroxidase), and FADS (Fluorescence Activated Droplet Sorting). Targeted Oxidoreductases: P450BM3 (NADPH-dependent Cytochrome P450 BM3), Tm 6PGDH (NADP+-dependent Thermotoga maritima 6-phosphogluconate dehydrogenase), Ld ldh (NADH-dependent Lactobacillus delbrueckii d-lactate dehydrogenase), Lb adh (NADPH-dependent Lactobacillus brevi alcohol dehydrogenase), Bo CHAO (FADH2-dependent Brevibacterium oxydans cyclohexamine oxidase)
Recent campaigns apply “ultra-throughput” (>106) methods using reactions that can be detected by fluorescence sorting [43-45]. For example, Brevibacterium oxydcms cyclohexylamine oxidase (Bo CHAO) variants were compartmentalized in droplets and screened for their activity towards a non-natural substrate using fluorescent activated droplet sorting (FADS) (Figure 2), which yielded a mutant with 960-fold increased catalytic efficiency [45]. However, this method is only applicable to enzymes that produce H2O2 which is detected by a fluorescent dye, Amplex-Ultra Red. To overcome this limitation, an Escherichia coli strain harboring SoxR-regulated GFP cassette to report the intracellular NADPH/NADP+ ratio was developed to screen NADPH-dependent enzymes via fluorescent activated cell sorting (FACS) [43,44]. This system enabled screening of a random library and isolated a Lactobacillus brevi alcohol dehydrogenase variant with improved activity for the reduction of 2,5-hexanedione to (2R,5R)-hexanediol [44] (Figure 2). Advanced sorting techniques offer rapid screening of the library being explored, but are often hindered by narrow dynamic ranges and high background signal. Selections, as opposed to screens, do not rely on special instrumentation and automatically eliminate undesirable candidates.
In vivo selection platforms modulate cell growth by disrupting intracellular cofactor cycling within engineered E. coli strains. These platforms were pioneered in early work aiming to accumulate NADH in anaerobic condition by disrupting the host’s native fermentative pathways, for example in strain JCL166 (ΔadhE ΔldhA Δfrd). In this strain, anaerobic growth is only restored when an NADH-recycling enzyme is present. This system has identified endogenous E. coli enzymes which form a 2,3-butanediol production pathway [46]. The same principle of cofactor recycling is the foundation for a variety of ultra-high throughput (>106) growth-based selections of nicotinamide-dependent oxidoreductases in directed evolution [47-51]. A recent growth-based selection strain has an engineered NADPH-dependent glycolysis, and therefore required a NADPH-consuming “fermentative” reaction to grow anaerobically. This platform enabled the selection of ~6.2 × 107 variants in one round, and produced a Lactobacillus delbrueckii D-lactate dehydrogenase with a 470-fold increase in activity with NADPH [49] (Figure 2).
This selection strategy has since been expanded to include both NADPH and NADH-dependent selections in aerobic conditions, to be compatible with engineering oxygenases such as p-hydroxybenzoate hydroxylase [47] and cyclohexanone monooxygenase [48]. These results highlight the usefulness of in vivo growth platforms for oxidoreductase selections.
Growth selection has not been applied in engineering noncanonical cofactor-dependent enzymes. However, our recent work where E. coli growth was obligately linked to the cycling of the noncanonical cofactor NMN+ presents a platform for future studies. This was achieved by disrupting standard glycolysis networks and directing glucose entry into the life-essential carbon metabolism through our NMN+-specific glucose dehydrogenase (GDH) [9]. Cell growth was only restored when the NMN+-cycling partner of GDH was present to complete the NMN+ based redox cycle and prevent cofactor depletion. The specific function of the partner is not linked to cell survival and we anticipate that the complementary partner can be exchanged.
Technology development for engineering noncanonical cofactor-dependent enzymes
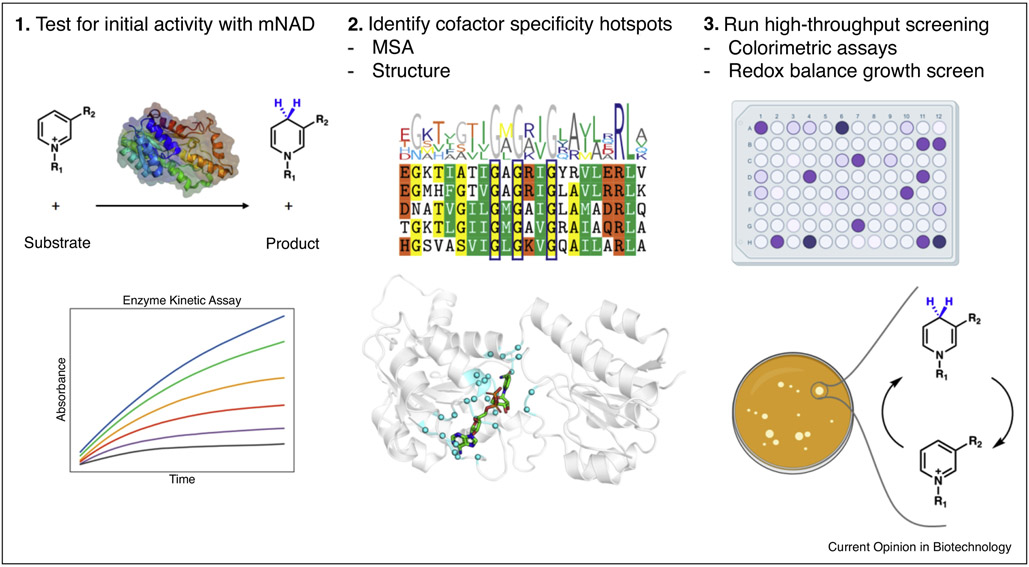
In general, efforts in engineering mNAD-dependent enzymes follow three steps (Figure 3): First, wild type enzymes from different organisms are screened with the mNAD of interest to identify a starting template. Second, sequence alignment or computational models predict positions surrounding the cofactor binding pocket. Third, identified positions are targeted by mutagenesis, often in combinatorial fashion. To achieve high diversity, site-saturation mutagenesis is typically performed with degenerate primers, and variants are screened with 96-well plate-based absorbance assays detecting reduced cofactor [5,8] or colorimetric assays detecting reactions of reduced cofactor with nitroblue tetrazolium and phenazine methosulfate producing the purple dye formazan [7,52]. Future tool developments will include growth-based selection where the ability of the cell to cycle the target mNAD is linked to life-essential functions such as carbon metabolism. Variants with more active mNAD cycling will readily outcompete those with lower fitness resulting in facile, high-throughput selection of mNAD-dependent enzymes through readout of cell growth.
Figure 3. Outline of protocol to engineer enzymes for mNAD activity.
An initial screen is performed with the mNAD of interest and wild type enzyme to determine the baseline performance. Positions surrounding the cofactor binding pocket and those that contribute to cofactor specificity found through sequence alignment are chosen for mutagenesis. Variants are screened through colorimetric assay measuring activity through color development reflecting production of reduced cofactor. Future tool developments to improve throughput will include growth-based selection assays where the ability of the cell to regenerate mNAD is linked to survival.
We highlight two recent reports departing from the standard saturation mutagenesis and 96-well plate based screening approach. Huang et al. [10] developed the NAD(P)-eliminated solid phase assay (NESPA) (Figure 2), a colorimetric screen performed with colonies grown on agar plate advancing throughput to over 105 samples per round while managing low background noise. A heat treatment step is performed to permeabilize cells, followed by washing to remove endogenous NAD(P)+. Rounds of saturation mutagenesis at the cofactor binding site, followed by error prone PCR to raise diversity in more distal regions, resulted in a Thermotoga maritima 6-phosphogluconate dehydrogenase variant with 50-fold enhanced NMN+-dependent activity. However, heat treatment limits the assay to screening thermostable enzymes, and manual washing steps may lead to high variance. In our recent study [9], in silico screening was performed in lieu of experimental screening. Bioinformatic analysis was used to identify positions with high plasticity to tolerate mutations. Then, by simulating the effects of mutations on the mNAD binding pose using Rosetta, we greatly narrowed down candidates that warranted experimental testing and eliminated the need to broadly sample with site-saturation mutagenesis. The best mutant B. subtilis glucose dehydrogenase S17E-Y34Q-A93K-I195R was obtained from just experimentally testing <20 candidates.
CONCLUSION
When engineering enzymes to utilize noncanonical cofactors, even the most developed variants often show low relative catalytic efficiencies with mNADs. The sampling cap from utilizing 96-well plate-based screens greatly restricts our ability to identify rare, highly functional variants. Future directions to expand the mNAD evolution toolbox will involve adapting principles and methods currently used for natural cofactors.
In addition, computational methods will be highly instrumental in engineering mNAD-dependent enzymes. Without crystal structures of the target enzymes with noncanonical cofactors bound, molecular modeling tools are essential for visualizing enzyme-cofactor interaction. Furthermore, homology modeling tools and sequence alignment facilitate the translation of successful mutations between different enzymes. For example, E. coli malic enzyme L310R gained the ability to utilize nicotinamide flucytosine dinucleotide (NCFD+) and NCD+ [53]. High sequence conservation at L310 inspired the rational design of E. coli malate dehydrogenase L6R for NCFD+ binding [53], Lactobacillus helveticus D-lactate dehydrogenase V152R [5,53], and Ralstonia sp. 4506 phosphite dehydrogenase I151R for NCD+ binding [16].
ACKNOWLEDGEMENT
H.L. acknowledges support from University of California, Irvine, the National Science Foundation (NSF) (award no. 1847705), and the National Institutes of Health (NIH) (award no. DP2 GM137427). S.M. acknowledges support from the NSF Graduate Research Fellowship Program (grant no. DGE-1839285).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
* of special interest
** of outstanding interest
- 1.Sellés Vidal L, Kelly CL, Mordaka PM, Heap JT: Review of NAD(P)H-dependent oxidoreductases: Properties, engineering and application. Biochim Biophys Acta: Proteins Proteomics 2018, 1866:327–347. [DOI] [PubMed] [Google Scholar]
- 2.Sicsic S, Durand P, Langrené S, Le Goffic F: Activity of NMN+, nicotinamide ribose and analogs in alcohol oxidation promoted by horse-liver alcohol dehydrogenase. Improvement of this activity and structural requirements of the pyridine nucleotide part of the NAD+ coenzyme. Ear J Biochem 1986, 155:403–407. [DOI] [PubMed] [Google Scholar]
- 3.Sicsic S, Durand P, Langrene S, le Goffic F: A new approach for using cofactor dependent enzymes: example of alcohol dehydrogenase. FEBS Lett 1984, 176:321–330. [DOI] [PubMed] [Google Scholar]
- 4.Anderson BM, Kaplan NO: Enzymatic studies with analogues of diphosphopyridine nucleotide. J Biol Chem 1959, 234:1226–1232. [PubMed] [Google Scholar]
- 5.Liu Y, Li Q, Wang L, Guo X, Wang J, Wang Q, Zhao ZK: Engineering d-Lactate Dehydrogenase to Favor an Non-natural Cofactor Nicotinamide Cytosine Dinucleotide. Chembiochem 2020, doi: 10.1002/cbic.201900766. [DOI] [PubMed] [Google Scholar]
- 6.Guo X, Liu Y, Wang Q, Wang X, Li Q, Liu W, Zhao ZK: Non-natural Cofactor and Formate-Driven Reductive Carboxylation of Pyruvate. Angew Chem Int Ed Engl 2020, 59:3143–3146. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Feng Y, Wang L, Guo X, Liu W, Li Q, Wang X, Xue S, Zhao ZK: Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor. ACS Catal 2019,* Example of performing saturation mutagenesis on residues surrounding the adenosine binding pocket and screening through colorimetric assay. Final mutants were crystallized and the effects of the mutations in enabling NCD binding were determined to be through improved packing at the cytosine.
- 8.Nowak C, Pick A, Lommes P, Sieber V: Enzymatic Reduction of Nicotinamide Biomimetic Cofactors Using an Engineered Glucose Dehydrogenase: Providing a Regeneration System for Artificial Cofactors. ACS Catal 2017, 7:5202–5208.* First report of a non-flavin enzyme reducing mNADs without a ribonucleotide
- 9.Black WB, Zhang L, Mak WS, Maxel S, Cui Y, King E, Fong B, Sanchez Martinez A, Siegel JB, Li H: Engineering a nicotinamide mononucleotide redox cofactor system for biocatalysis. Nat Chem Biol 2020, 16:87–94.** Application of molecular modeling for rational design resulted in the most orthogonal design to date. The enzyme was further shown to function in vivo for NMN+ regeneration to support central carbon metabolism and specific electron delivery to NMN+ dependent reactions.
- 10.Huang R, Chen H, Upp DM, Lewis JC, Zhang Y-HPJ: A High-throughput Method for Directed Evolution of NAD(P)+-dependent Dehydrogenases for the Reduction of Biomimetic Nicotinamide Analogues. ACS Catal 2019, doi: 10.1021/acscatal.9b03840.** Agar plate based high-throughput screen of colonies for NMN+ activity coupled with rounds of saturation and random mutagenesis. Reduced background noise through heat treatment and washing.
- 11.Paul CE, Gargiulo S, Opperman DJ, Lavandera I, Gotor-Femández V, Gotor V, Taglieber A, Arends IWCE, Hollmann F: Mimicking nature: synthetic nicotinamide cofactors for C─C bioreduction using enoate reductases. Org Lett 2013, 15:180–183. [DOI] [PubMed] [Google Scholar]
- 12.Nowak C, Pick A, Csepei L-I, Sieber V: Characterization of Biomimetic Cofactors According to Stability, Redox Potentials, and Enzymatic Conversion by NADH Oxidase from Lactobacillus pentosus. Chembiochem 2017, 18:1944–1949. [DOI] [PubMed] [Google Scholar]
- 13.Löw SA, Löw IM, Weissenbom MJ, Hauer B: Enhanced Ene-Reductase Activity through Alteration of Artificial Nicotinamide Cofactor Substituents. ChemCatChem 2016, 8:911–915. [Google Scholar]
- 14.Guarneri A, Westphal AH, Leertouwer J, Lunsonga J, Franssen MCR, Opperman DJ, Hollmann F, van Berkel WJH, Paul CE: Flavoenzyme-mediated regioselective aromatic hydroxylation with coenzyme biomimetics. ChemCatChem 2019, doi: 10.1002/cctc.201902044. [DOI] [Google Scholar]
- 15.Nowak C, Beer B, Pick A, Roth T, Lommes P, Sieber V: A water-forming NADH oxidase from Lactobacillus pentosus suitable for the regeneration of synthetic biomimetic cofactors. Front Microbiol 2015, 6:957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Ji D, Liu Y, Wang Q, Wang X, Zhou YJ, Zhang Y, Liu W, Zhao ZK: Synthetic Cofactor-Linked Metabolic Circuits for Selective Energy Transfer. ACS Catal 2017, 7:1977–1983. [Google Scholar]
- 17.Cahn JKB, Brinkmann-Chen S, Arnold FH: Enzyme Nicotinamide Cofactor Specificity Reversal Guided by Automated Structural Analysis and Library Design In Synthetic Metabolic Pathways: Methods and Protocols. Edited by Jensen MK, Keasling JD. Springer; New York; 2018:15–26. [DOI] [PubMed] [Google Scholar]
- 18.Gmelch TJ, Sperl JM, Sieber V: Molecular dynamics analysis of a rationally designed aldehyde dehydrogenase gives insights into improved activity for the non-native cofactor NAD. ACS Synth Biol 2020, doi: 10.1021/acssynbio.9b00527. [DOI] [PubMed] [Google Scholar]
- 19.Beier A, Bordewick S, Genz M, Schmidt S, van den Bergh T, Peters C, Joosten H-J, Bornscheuer UT: Switch in Cofactor Specificity of a Baeyer-Villiger Monooxygenase. Chembiochem 2016, 17:2312–2315. [DOI] [PubMed] [Google Scholar]
- 20.Mähler C, Kratzl F, Vogel M, Vinnenberg S, Weuster- Botz D, Castiglione K: Loop Swapping as a Potent Approach to Increase Ene Reductase Activity with Nicotinamide Adenine Dinucleotide (NADH). Adv Synth Catal 2019, 106:946. [Google Scholar]
- 21.Solanki K, Abdallah W, Banta S: Engineering the cofactor specificity of an alcohol dehydrogenase via single mutations or insertions distal to the 2’-phosphate group of NADP(H). Protein Eng Des Sel 2017, 30:373–380.* The article shows that modification of substrate binding loops outside of regions conventionally targeted for cofactor specificity engineering can enable inverted cofactor preference for TIM barrel aldo-keto reductases
- 22.Abdallah W, Solanki K, Banta S: Insertion of a Calcium-Responsive Beta Roll Domain into a Thermostable Alcohol Dehydrogenase Enables Tunable Control over Cofactor Selectivity. ACS Catal 2018, doi: 10.1021/acscatal.7b03809. [DOI] [Google Scholar]
- 23.Abdallah W, Chirino V, Wheeldon I, Banta S: Catalysis of Thermostable Alcohol Dehydrogenase Improved by Engineering the Microenvironment through Fusion with Supercharged Proteins. Chembiochem 2019, 20:1827–1837.* The article investigated complexes of superfolding green fluorescent protein (sfGFP) with extreme surface charges and a thermostable alcohol dehydrogenase to demonstrate an accessible method of modulating enzymatic microenvironment to control activity
- 24.Gao Y, Roberts CC, Zhu J, Lin J-L, Chang C-EA, Wheeldon I: Tuning Enzyme Kinetics through Designed Intermolecular Interactions Far from the Active Site. ACS Catal 2015, 5:2149–2153. [Google Scholar]
- 25.Lu C, Shen F, Wang S, Wang Y, Liu J, Bai W-J, Wang X: An Engineered Self-Sufficient Biocatalyst Enables Scalable Production of Linear a-Olefins from Carboxylic Acids. ACS Catal 2018, 8:5794–5798. [Google Scholar]
- 26.Beyer N, Kulig JK, Bartsch A, Hayes MA, Janssen DB, Fraaije MW: P450BM3 fused to phosphite dehydrogenase allows phosphite-driven selective oxidations. Appl Microbiol Biotechnol 2017, 101:2319–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corrado ML, Knaus T, Mutti FG: A Chimeric Styrene Monooxygenase with Increased Efficiency in Asymmetric Biocatalytic Epoxidation. Chembiochem 2018, 19:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L, Aalbers FS, Tang W, Rollig R, Fraaije MW, Kara S: Convergent Cascade Catalyzed by Monooxygenase–Alcohol Dehydrogenase Fusion Applied in Organic Media. Chembiochem 2019, 20:1653–1658. [DOI] [PubMed] [Google Scholar]
- 29.Aalbers FS, Fraaije MW: Design of Artificial Alcohol Oxidases: Alcohol Dehydrogenase-NADPH Oxidase Fusions for Continuous Oxidations. Chembiochem 2019, 20:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozbakir HF, Garcia KE, Banta S: Creation of a formate: malate oxidoreductase by fusion of dehydrogenase enzymes with PEGylated cofactor swing arms. Protein Eng Des Sel 2018, 31:103–108. [DOI] [PubMed] [Google Scholar]
- 31.Chánique AM, Parra LP: Protein Engineering for Nicotinamide Coenzyme Specificity in Oxidoreductases: Attempts and Challenges. Front Microbiol 2018, 9:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knaus T, Paul CE, Levy CW, de Vries S, Mutti FG, Hollmann F, Scrutton NS: Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes. J Am Chem Soc 2016, 138:1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo HC, Ryan JD, Kerr JB, Clark DS, Fish RH: Bioorganometallic chemistry: Co-factor regeneration, enzyme recognition of biomimetic 1,4-NADH analogs, and organic synthesis; tandem catalyzed regioselective formation of N-substituted-1,4-dihydronicotinamide derivatives with [Cp*Rh(bpy)H]+, coupled to chiral S-alcohol formation with HLADH, and engineered cytochrome P450s, for selective C-H oxidation reactions. J Orgcmomet Chem 2017, 839:38–52. [Google Scholar]
- 34.Flores H, Ellington AD: A modified consensus approach to mutagenesis inverts the cofactor specificity of Bacillus stearothermophilus lactate dehydrogenase. Protein Eng Des Sel 2005, 18:369–377. [DOI] [PubMed] [Google Scholar]
- 35.Campbell E, Meredith M, Minteer SD, Banta S: Enzymatic biofuel cells utilizing a biomimetic cofactor. Chem Commun 2012, 48:1898–1900. [DOI] [PubMed] [Google Scholar]
- 36.Ryan JD, Fish RH, Clark DS: Engineering cytochrome P450 enzymes for improved activity towards biomimetic 1,4-NADH cofactors. Chembiochem 2008, 9:2579–2582. [DOI] [PubMed] [Google Scholar]
- 37.Weingartner AM, Sauer DF, Dhoke GV, Davari MD, Ruff AJ, Schwaneberg U: A hydroquinone-specific screening system for directed P450 evolution. Appl Microbiol Biotechnol 2018, 102:9657–9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandenberg OF, Chen K, Arnold FH: Directed Evolution of a Cytochrome P450 Carbene Transferase for Selective Functionalization of Cyclic Compounds. J Am Chem Soc 2019, doi: 10.1021/jacs.9b02931. [DOI] [PubMed] [Google Scholar]
- 39.Časaitė V, Sadauskas M, Vaitekūnas J, Gasparavičiūtė R, Meškienė R, Skikaitė I, Sakalauskas M, Jakubovska J, Tauraitė D, Meškys R: Engineering of a chromogenic enzyme screening system based on an auxiliary indole-3-carboxylic acid monooxygenase. Microbiologyopen 2019, 8:e00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Wu Y-Q, Xu N, Zhao Q, Yu H-L, Xu J-H: Engineering of Cyclohexanone Monooxygenase for the Enantioselective Synthesis of (S)-Omeprazole. ACS Sustainable Chem Eng 2019, 7:7218–7226. [Google Scholar]
- 41.Xu N, Zhu J, Wu Y-Q, Zhang Y, Xia J-Y, Zhao Q, Lin G-Q, Yu H-L, Xu J-H: Enzymatic Preparation of the Chiral (S)-Sulfoxide Drug Esomeprazole at Pilot-Scale Levels. Org Process Res Dev 2020, doi: 10.1021/acs.oprd.0c00115. [DOI] [Google Scholar]
- 42.de Rond T, Gao J, Zargar A, de Raad M, Cunha J, Northen TR, Keasling JD: A High- Throughput Mass Spectrometric Enzyme Activity Assay Enabling the Discovery of Cytochrome P450 Biocatalysts. Angew Chem Int Ed 2019, 58:10114–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siedler S, Schendzielorz G, Binder S, Eggeling L, Bringer S, Bott M: SoxR as a single-cell biosensor for NADPH-consuming enzymes in Escherichia coli. ACS Synth Biol 2014, 3:41–47. [DOI] [PubMed] [Google Scholar]
- 44.Spielmann A, Brack Y, van Beek H, Flachbart L, Sundermeyer L, Baumgart M, Bott M: NADPH biosensor-based identification of an alcohol dehydrogenase variant with improved catalytic properties caused by a single charge reversal at the protein surface. AMB Express 2020, 10:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debon A, Pott M, Obexer R, Green AP, Friedrich L, Griffiths AD, Hilvert D: Ultrahigh-throughput screening enables efficient single-round oxidase remodelling. Nature Catalysis 2019, 2:740–747.* This article describes an effective hydrogen peroxide-coupled fluorescence activated droplet sorting (FASDS) assay that facilitated the rapid screen of a large cyclohexylamine oxidase library (107 members) and isolation of a variant with a 960-fold catalytic efficiency improvement for non-native substrate.
- 46.Liang K, Shen CR: Selection of an endogenous 2,3-butanediol pathway in Escherichia coli by fermentative redox balance. Metab Eng 2017, 39:181–191. [DOI] [PubMed] [Google Scholar]
- 47.Maxel S, Aspacio D, King E, Zhang L, Acosta AP, Li H: A growth-based, high-throughput selection platform enables remodeling of 4-hydroxybenzoate hydroxylase active site. ACS catalysis 2020, doi: 10.1021/acscatal.0c01892** The article demonstrates the practicality of high-throughput redox-based selection for the directed evolution of NADPH-dependent monooxygenases that feature complex reaction mechanisms.
- 48.Maxel S, Zhang L, King E, Aspacio D, Acosta AP, Luo R, Li H: Pairing two growth-based, high-throughput selections to fine tune conformational dynamics in oxygenase engineering. bioRxiv 2020, doi: 10.1101/2020.05.22.111575. [DOI] [Google Scholar]
- 49.Zhang L, King E, Luo R, Li H: Development of a High-Throughput, In Vivo Selection Platform for NADPH-Dependent Reactions Based on Redox Balance Principles. ACS Synth Biol 2018, doi: 10.1021/acssynbio.8b00179.** The article demonstrates how reversing the natural cofactor preference of glycolysis from NADH to NADPH creates redox imbalance and can be utilized as selection pressure to provide an effective growth-based platform for the directed evolution of a D-lactate dehydrogenase
- 50.Lindner SN, Calzad Iacute Az Ramirez L, Krüsemann J, Yishai O, Belkhelfa S, He H, Bouzon M, Döring V, Bar-Even A: NADPH-auxotrophic E. coli: a sensor strain for testing in vivo regeneration of NADPH. ACS Synth Biol 2018, doi: 10.1021/acssynbio.8b00313. [DOI] [PubMed] [Google Scholar]
- 51.Ramírez LC, Calvó-Tusell C, Stoffel GMM, Lindner S, Osuna S, Erb TJ, Garcia-Borràs M, Bar-Even A, Acevedo-Rocha CG: In vivo selection for formate dehydrogenases with high efficiency and specificity towards NADP+. bioRxiv 2020, doi: 10.1101/2020.04.02.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Zhou YJ, Wang L, Liu W, Liu Y, Peng C, Zhao ZK: Engineering Escherichia coli Nicotinic Acid Mononucleotide Adenylyltransferase for Fully Active Amidated NAD Biosynthesis. Appl Environ Microbiol 2017, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji D, Wang L, Hou S, Liu W, Wang J, Wang Q, Zhao ZK: Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide. J Am Chem Soc 2011, 133:20857–20862.* First in a series of works that engineered enzymes for NCD binding. The discovered mutation was transferred successfully to homologs phosphite dehydrogenase, lactate dehydrogenase, and formate dehydrogenase.
- 54.Paul CE, Tischler D, Riedel A, Heine T, Itoh N, Hollmann F: Nonenzymatic Regeneration of Styrene Monooxygenase for Catalysis. ACS Catal 2015, 5:2961–2965. [Google Scholar]