Abstract
The biogeography of the mammalian intestine is remarkable in that a vast microbial consortium exists inside the organism, surrounded by intestinal epithelial cells. The microbiome and the intestinal epithelium have developed a complex network of interactions that maintain intestinal homeostasis. We now recognize that functions of the epithelium are compartmentalized in specific intestinal epithelial cell subtypes. Furthermore, we are beginning to understand the ways in which microbes and their metabolic products impact the specific epithelial subsets. Here, we survey the mechanisms utilized by the microbiome to regulate intestinal epithelial function, and inversely, how different epithelial cell subtypes cooperate in regulating the microbiome.
Introduction
The ability for the microbiome to alter and manipulate physiology and morphology of the host intestine has been appreciated for decades [1]. However, as we enter the age of single-cell biology [2••], it is becoming apparent that treating the intestinal epithelium as a homogeneous layer of cells is grossly inappropriate. Furthermore, as the use of conditional Cre-lox systems continues to be developed, we gain a more nuanced appreciation of how individual cells regulate host–microbiome interactions that was never before possible. Indeed, the intestine is composed of a great variety of intestinal epithelial cell (IEC) sub-types, each with their own specialized reciprocal relationship with the microbiome (Table 1). All IECs originate from the intestinal stem cell which divide into transit amplifying cells (TACs) that serve as intermediates between the stem cell and the terminally differentiated IEC [3]. TACs subsequently populate the intestine with the various IEC subtypes following commitment towards a secretory or absorptive lineage [4]. Paneth cells, goblet cells, tuft cells, and enteroendocrine cells, which release large amounts of antimicrobial peptides (AMPs), mucins, type 2 immune mediators, or hormones, respectively, are derived from the secretory lineage [4]. Meanwhile, enterocytes and colonocytes, which are major nutrient absorbers, and Microfold (M) cells, which act principally in microbial antigen uptake, are derived from the absorptive lineage [4].
Table 1.
Summary of IEC subtype function and localization
| Cell type | Primary canonical functions | Location | Interactions with microbiome |
|---|---|---|---|
| Intestinal stem cells | Replenish the epithelial cell layer every ~4–5 days | Small intestine Colon | Microbiome-derived lactate stimulates ISC proliferation |
| Goblet cells | Secrete mucins and other factors into the gut lumen; goblet cell-associated antigen passages | Small intestine Colon | Provide a carbon source to bacteria in the form of mucus |
| Paneth cells | Secrete antimicrobial peptides into the mucosa; support the stem cell niche of the crypt | Small intestine only | Alteration of microbial colonization niche through antimicrobial peptides, |
| Tuft cells | Trigger type 2 immune responses against parasites | Small intestine Colon | Stimulation by microbiome-derived succinate |
| Enteroendocrine cells | Release hormones in response to a variety of stimuli | Small intestine Colon | 5-HT production in response to SCFA stimulation |
| M cells | Uptake of luminal antigens and delivery to the Peyer’s patches | Small intestine only | Uptake of microbial antigen, regulation of SFB levels |
| Enterocytes | Physical barrier; nutrient absorption; epithelial cell shedding | Small intestine only | Endocytosis of microbial antigen. Microbiome-derived lactate and acetate regulate chylomicron release |
| Colonocytes | Physical barrier; nutrient absorption; epithelial cell shedding | Colon only | Metabolism of microbiome-derived SCFAs, provide glycans as a carbon source to bacteria |
Herein, we lay the foundation for future research working to interrogate cell-specific host-microbiome interactions, by reviewing seminal data that exemplifies prominent ways in which particular epithelial cell functions affect the microbiome and reciprocally, how the microbiome affects particular epithelial cell functions. Although our review is not an exhaustive list of all previously described interactions, we touch upon common themes and conceptual modules of cell-specific host-microbiome interactions.
Inside-out: cell type-specific regulation of the microbiome
The large intestine houses the highest number of bacteria in terms of quantity and diversity, so the existence of a reciprocal relationship between the microbiome and colonocytes becomes immediately apparent. Perhaps the best described interaction involves the microbial production of short-chain fatty acids (SCFAs), which colonocytes use as fuel [5]. SCFA metabolism by colonocytes promotes aerobic respiration, maintaining the hypoxic environment of the large intestine that most commensals require [6]. In the absence of SCFA, colonocytes undergo anaerobic respiration releasing oxygen and nitrates into the lumen that facilitate the expansion of pathogens such as Escherichia coli and Salmonella [7••]. In contrast to mature colonocytes, TACs exhibit low basal oxygen consumption [8], leading to increased availability of oxygen and nitrate, allowing for facultative anaerobic bacteria, such as E. coli, to expand in ulcerative colitis mouse models [9–11]. These phenotypes are also present in ulcerative colitis patients, and may contribute to disease pathology [12].
Along the same lines, host enterocytes can provide carbon sources utilized by host commensals. While mucus had long been conceptualized simply as a lubricant for fecal matter moving through the gastrointestinal tract, it is now clear that mucus acts as a medium for the colonization of commensal organisms to maintain immune homeostasis [13]. The highly glycosylated mucins that constitute a major architectural component of the mucosal layer are primary carbon sources for some commensals, like Akkermansia muciniphila [14], whose abundance is inversely correlated with severity of metabolic and inflammatory diseases [15–17]. Lipid modification, such as sialylation of mucin glycans, have also been recently described as metabolic resources for commensals like Ruminococcus gnavus [18•]. Indeed, mucin remains an important modality whereby goblet cells feed microbes, some requiring the fermentation products of their neighbors [19]. Fucosylation of small intestinal IEC also is utilized as fuel for the microbiome, including Bacteroides, in times of starvation [20]. Feeding commensals, which colonize the mucus layer and hoard potential resources that pathogens could use otherwise [21], may represent a colonization resistance strategy of the host, by which the function of the microbiome is used to prevent infection.
Additionally, initiation of IEC inflammatory cascades can result in an antimicrobial response, modulating the bacterial composition. By virtue of their role as producers of AMPs [22], Paneth cells inherently have a strong influence over microbial composition in the small intestine. Both chemical and genetic depletion of Paneth cells in mice resulted in robust and long-lasting changes in the microbiome, including a significant reduction of Proteobacteria [23]. Furthermore, the most abundant antimicrobial peptides produced by humans, a-Defensin-5, is shown to have direct bactericidal activity towards several members of the human microbiome, and can therefore alter bacterial communities in vivo [24]. More generally, Paneth cells maintain species-specific microbiome communities, as transgenic expression of human defensins in mice results in large shifts in detectable bacteria [25].
Goblet cells sense pathogen-associated molecular patterns (PAMPs) in the gut and maintain the mucosal barrier accordingly. TLR-dependent signaling and microbial metabolites trigger the NLRP6-dependent inflammasome in goblet cells to stimulate mucus production [26–28]. Furthermore, it was recently shown that neuronal-derived IL-18 is responsible for driving goblet cell production of antimicrobial peptides upon infection [29•]. These recent findings, together with the prior literature, demonstrate how crucial microbial responses of goblet cells are to reinforce barrier integrity.
Tuft cells also indirectly regulate the gut microbiome during type 2 immune responses. By triggering ILC2s to release IL-13, tuft cells can engage IL-13-responsive goblet cells to release mucus to brush away not only eukaryotic but also bacterial pathogens. Tuft cell derived IL-25 has also shown protection against cancer-associated dysbiosis [30]. How tuft cells communicate, and with whom they communicate with to maintain barrier integrity is certainly a growing field of interest with much to be explored.
As the major epithelial mediators of antigen uptake, M cells are especially critical for regulating the microbiome. Conditional deletion of RANKL using villin-Cre results in loss of M cells in mice [31]. Although these mice showed similar levels of microbial diversity in the intestine, the levels of IgA-coated bacteria increased [31]. IgA-coating has previously been shown to mark bacterial members of the microbiome with strong inflammatory capacity [32]. Transient depletion of M cells, however, has been shown to increase the levels of segmented filamentous bacteria (SFB), demonstrating that M cells regulate ileal SFB abundance [33••].
Situated at the bottom of intestinal crypts are the intestinal stem cells (ISCs). While undergoing continuous proliferation, new daughter cells migrate upwards along the crypt-villus axis towards the gut lumen and differentiate into the many specialized cell types along the way in a controlled manner [34]. While the majority of gut microbes are situated in the mucosal layer above the villi, a subset of microbes called the crypt-specific core microbiota are maintained within the crypt in close proximity to the stem cell niche [35]. The abundances of various crypt-associated and mucosa-associated microbes have been correlated to colon cancer development [36].
In all, various IEC subtypes exert regulation over the microbiome. Nearly all described microbiome effects from IEC have been studied in the context of a single IEC subtype. However, it would be interesting to understand if and how IEC subtypes communicate with each other to coordinate responses towards the microbiome. Furthermore, given that most of the work only measures microbial abundances, it is likely that deeper levels of microbiome regulation by epithelial cell subsets escape our current knowledge, for example, at the level of commensal transcriptomes.
Outside-in: cell type-specific regulation by the microbiome
The microbiome utilizes diverse mechanisms to influence the intestinal epithelium, which can be organized into a few discrete conceptual modules. A major form of communication is through pattern recognition receptor (PRR) signaling, for example, in Paneth cells. The fact that germ-free (GF) animals have lower levels of AMP production and decreased Paneth cell number is evidence of microbial regulation of Paneth cell differentiation and function [37,38]. One such mechanism involves activation of Paneth cell AMPs via MyD88 signaling, a downstream adaptor of several PRRs [39•]. In fact, loss of MyD88 results in a number of defects in the intestinal epithelium, including decreased mucin production, suggesting PRR signaling in goblet cells also influences their behavior [40].
Beyond PRRs, several IEC subtypes also express receptors that can respond to microbial metabolites, circumventing the need to sense bacteria directly, by instead monitoring a metabolite proxy for the state of intestinal microbial colonization. In addition to the impact of SCFAs on enterocyte metabolism as discussed above, a recent report [41] identified microbial metabolites regulating epithelial lipid metabolism. l-lactate from Lactobacillus paracasei enhances lipid storage by inhibiting beta-oxidation, while Escherichia coli-derived acetate promotes beta-oxidation. Paneth cells can also be affected by similar microbial metabolites. Microbial-derived lactic acid is capable of signaling through GPR81 on Paneth cells to increase Paneth cell number [42••].
Tuft cells are also known to respond to the microbiome, as antibiotic-induced dysbiosis resulted in tuft cell hyperplasia [30]. Using single-cell transcriptomics, intestinal tuft cells were recently shown to specifically express Sucnr1 and Ffar3, receptors for the microbially derived metabolites succinate and SCFAs, respectively [43•]. Parasite-derived succinate has been shown to activate type 2 immune responses by tuft cells [44]; however, recent evidence extends this finding to commensal bacteria-derived succinate [34]. Interestingly, one group identified a tuft cell hyperplasia phenotype in germ-free mice colonized with helminth-free microbiota, while goblet cell hyperplasia was not simultaneously observed [45]. Altogether, these studies implicate tuft cells in responding to bacterial presence in the intestinal mucosa, broadening their roles described in anti-parasitic responses, which have been the major focus of tuft cell research (Figure 1).
Figure 1. The reciprocal interactions of microbiome and IEC.
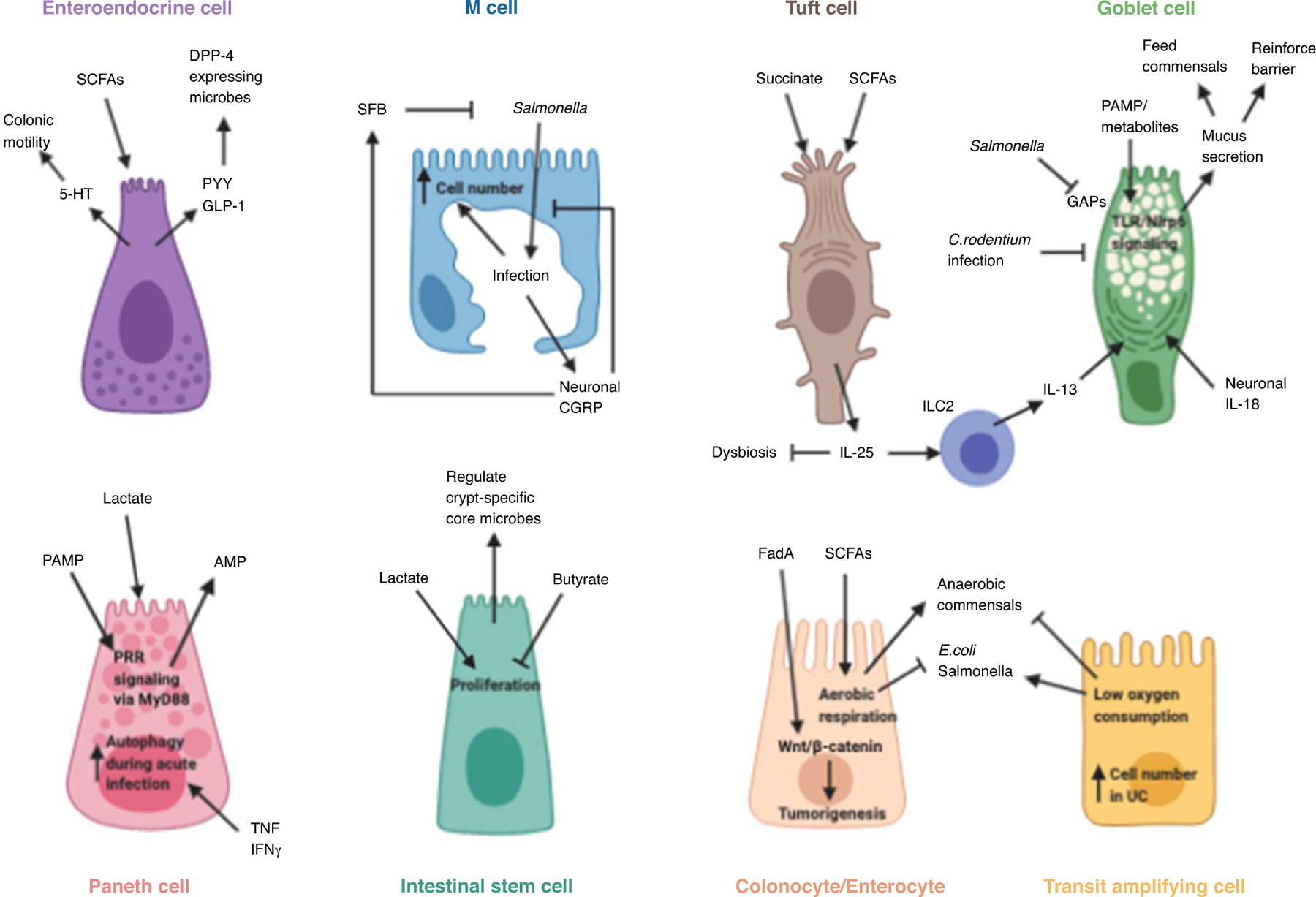
A number of different modules define the relationships between host cell and microbe. Metabolites, for example, can initiate and enhance physiological functions of immune cells, such as EECs, tuft cells, colonocytes, and Paneth cells. Reciprocally, host effector molecules, such as AMPs from Paneth cells, or mucins from Goblet cells, can alter microbiome composition.
Enteroendocrine cells (EECs), the largest endocrine system in the body [46], express receptors that are sensitive to microbial products such as SCFAs [47•], indole [48], secondary bile acids [49], and structural components of the microbial membrane [50], allowing the microbiome to exert control over host metabolism. Moreover, butyrate released by spore-forming bacteria upregulates serotonin (5-HT) synthesis by EECs [51]. Germ-free mice consequently have lower levels of 5-HT [52], leading to abnormal colonic motility. Indeed Clostridia-derived cellular components can also induce 5-HT secretion [53]. Besides 5-HT, EECs also secrete anorectic gut hormones such as Peptide YY (PYY) and glucagon-like peptide (GLP-1) [46]. PYY is a satiety factor that inhibits food intake and gastrointestinal motility, while GLP-1 is an incretin hormone [54]. In primary colonic cell cultures and enteroendocrine model cell lines, PYY production has been found to be strongly stimulated by butyrate and propionate [47•]. However, a separate report finds that GF mice had upregulated functional capacity in their EECs, increasing PYY production and secretion [55], suggesting GLP-1 resistance. This discrepancy could be due to differences in model systems, but could also be attributed to differences in protease activity. For example, a recent study found that genera such as Prevotella or Lactobacillus express enzymes similar to dipeptidyl peptidase IV (DPP-4), the enzyme responsible for GLP-1 and PYY breakdown [54]. This can then serve as a feedback loop, as decreasing DPP-4 leads to altered microbiota composition and microbial metabolite abundance [54].
Models of bacterial infection have uncovered several mechanisms implicating microbial metabolites, reactive oxygen species, and PAMPs as stimuli driving ISC proliferation [56,57]. Interestingly, while microbial-derived butyrate was found to suppress ISC proliferation, lactate stimulated ISC differentiation [42••]. Furthermore, Lactobacillus-derived indole metabolites, which signal through aryl hydrocarbon receptor (AhR) on epithelial cells, can also induce ISC proliferation [58]. Peptostreptococcus-derived tryptophan metabolites, which also can signal through AhR, similarly induce goblet cell proliferation [59]. Additionally, the microbiome can have profound effects on the cellular composition of the intestinal epithelial layer by polarizing the differentiation of TACs [60].
Furthermore, bacteria and bacterial antigen can directly interact with IECs. M cells have long been known to endocytose microbial antigen, and it has been demonstrated that commensals utilize M cells to maintain tolerance [61]. Despite the critical role of M cells in regulating bacterial antigen in the intestine, GF mice have no loss in M cells [62]. This is particularly interesting, as it is known that Peyer’s patches, where M cells typically reside, are underdeveloped in GF animals [63]. Although commensals have no apparent role in M cell development [62], pathogens appear to influence their cellularity, as M cell numbers increase following Salmonella infection [64]. These data would imply that the intestine maintains a pool of M cells poised to respond to antigen, and upon pathogenic challenge, the host responds by increasing the sites of antigenic uptake.
In addition to M cells, enterocytes also seem to endocytose microbial antigens. Recently, it has been shown that SFB antigens are taken up by IECs in a clathrin-dependent manner, which is required for the induction of antigen-specific TH17 cells [65]. Furthermore, in colonocytes, tumorigenesis can be mediated by direct interactions with microbes. Several groups have shown reduced tumor formation in several mouse models of colorectal cancer (CRC) inGFmice[66]. The abundancesofseveral members of the microbiome have been shown to correlate with CRC in humans, the most prevalent being Fusobacterium species [67]. Although mechanistic understanding is incomplete, recent reports demonstrate that Fusobacterium produce a virulence factor, FadA, capable of binding E-cadherin on colonocytes to induce cell proliferation via Wnt/β-catenin signaling [68,69].
These direct bacterial-epithelial interactions also occur in other IEC subtypes. Goblet cells are capable of delivering soluble antigens from the gut lumen via goblet cell-associated antigen passages (GAPs) to tolerogenic CD103+ dendritic cells [70]. The enteric pathogen Salmonella Typhimurium was shown to inhibit GAPs during infection [71]. Interestingly, while goblet cell hyperplasia has been associated with parasitic infections [72], bacterial infections, including Citrobacter rodentium, have been associated with the depletion of goblet cells [73], suggesting their significant role in maintaining barrier integrity.
Overall, IECs can sense their microbial environment to regulate diverse processes. These interactions typically occur via PRR recognition of bacterial ligands, or sensing of microbe-specific metabolites, such as SCFA. In general, the level of IEC response correlates to the severity of signal. For example, PRR signaling indicative of pathogenic crypt invasion results in enhanced AMP expression and mucin production. Metabolite sensing, on the other hand, can communicate to IECs without pathogenic invasion of the microbe, resulting in a wider range of both pro-inflammatory and anti-inflammatory responses. Given the large abundance of potential metabolite sensors in humans [74,75], it is possible we are only scratching the surface of microbiome-mediated IEC regulation.
Conclusions and perspectives
The regulatory nature of the microbiome and specific cell types in the intestine is inherently reciprocal. Direct recognition of bacterial molecules, for example through PRRs, can result in proliferation of epithelial cells, and production of antibacterial proteins. Metabolic byproducts from bacteria can alter epithelial activation states, and metabolic byproducts of the epithelia can alter bacterial composition.
Recent studies have uncovered a profound impact of the microbiome on fundamental cell biological processes of IECs, including protein turnover [76] and circadian rhythms [77–80], suggesting that several additional features of host–microbiome interactions remain to be uncovered.
As research in host-microbiome interactions progresses, more emphasis should be placed on cell-type specific responses. Although the use of germline-transgenic and pan-epithelial Cre-lines has unmistakably enhanced our understanding of host-microbe interactions, utilization of cell-specific Cre-lines that allows for the dissection of exact cellular mechanisms will provide clarity. Such models have been enabled by recent large-scale single cell transcriptome, mass cytometry, and spatial profiling studies that identified more comprehensive lists of cell type-specific genes, and have also revealed novel IEC cell states and subpopulations [81,82]. It is also clear that significant differences in microbiome diversity exist between laboratory-maintained and wild mice [83]. Experimental designs that account for this may help to address concerns of reproducibility in studies of host-microbiome interactions. Taking advantage of all such resources will allow us to refine our understanding of bidirectional host-microbiome interactions and their impacts on human health. This information can be useful in the treatment of several etiologies with underlying epithelial dysfunction, such as inflammatory bowel disease and cancer.
Acknowledgements
We thank the members of the Levy laboratory for their helpful discussions. We apologize to the investigators whose relevant work was not included in this review, owing to space constraints. The Figure was generated using BioRender.com.
A.S. is supported by the National Institute of General Medical Sciences of the National Institutes of Health under (K12GM081259). M.L. is a 2019 Searle Scholar and supported by the NIH Director’s New Innovator Award (DP2AG067511), The University of Pennsylvania Institute for Immunology, Center for Molecular Studies in Digestive and Liver Diseases, Center for Excellence in Environmental Toxicology, and the Institute on Aging and Alzheimer’s Disease.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Donaldson RM: Normal bacterial populations of the intestine and their relation to intestinal function. New Engl J Med 1964, 270:938–945 CONTD. [DOI] [PubMed] [Google Scholar]
- 2. ••.Haber AL et al. : A single-cell survey of the small intestinal epithelium. Nature 2017, 551:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides a comprehensive analysis of mouse small intestines using single-cell expression profiling to provide new organizational principles of gut homeostasis and physiology.
- 3.Hsu YC, Li L, Fuchs E: Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 2014, 157:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gehart H, Clevers H: Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 2019, 16:19–34 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 5.Wong JMW, De Souza R, Kendall CWC, Emam A, Jenkins DJA: Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 2006, 40:235–243. [DOI] [PubMed] [Google Scholar]
- 6.Kelly CJ et al. : Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015, 17:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ••.Byndloss MX et al. : Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science (80-) 2017, 357:570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates a novel function of pathogenic protection via SCFAs whereby host cells consume oxygen that may otherwise be utilized by bacteria such as E. coli.
- 8.Winter SE et al. : Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science (80-) 2013, 339:708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan YY et al. : A bioassay to measure energy metabolism in mouse colonic crypts, organoids, and sorted stem cells. Am J Physiol - Gastrointest Liver Physiol 2015, 309:G1–G9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litvak Y, Byndloss MX, Bäumler AJ: Colonocyte metabolism shapes the gut microbiota. Science (80-) 2018, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes ER et al. : Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe 2017, 21:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudhgaonkar SP, Tandan SK, Kumar D, Raviprakash V, Kataria M: Influence of simultaneous inhibition of cyclooxygenase-2 and inducible nitric oxide synthase in experimental colitis in rats. Inflammopharmacology 2007, 15:188–195. [DOI] [PubMed] [Google Scholar]
- 13.Jakobsson HE et al. : The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 2015, 16:164–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derrien M, Vaughan EE, Plugge CM, de Vos WM: Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004:1469–1476 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 15.Everard A et al. : Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013, 110:9066–9071 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cani PD, de Vos WM: Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol 2017, 8:1765 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bian X et al. : Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front Microbiol 2019, 10:2259 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. •.Bell A et al. : Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol 2019, 4:2393–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revealed a unique sialic acid pathway in bacteria that has important implications for the spatial adaptation of mucin-foraging gut symbionts.
- 19.Garber JM et al. : The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus. Commun Biol 2020, 3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickard JM et al. : Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 2014, 514:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zúñiga M, Monedero V, Yebra MJ: Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front Microbiol 2018, 9:1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clevers HC, Bevins CL: Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol 2013, 75:289–311. [DOI] [PubMed] [Google Scholar]
- 23.Lueschow SR et al. : Loss of murine Paneth cell function alters the immature intestinal microbiome and mimics changes seen in neonatal necrotizing enterocolitis. PLoS One 2018, 13: e0204967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehmann D et al. : Paneth cell a-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proc Natl Acad Sci U S A 2019, 116:3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzman NH et al. : Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 2010, 11:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wlodarska M et al. : NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 2014, 156:1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy M et al. : Microbiota-Modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2015, 163:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birchenough GMH, Nystrom EEL, Johansson MEV, Hansson GC: A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science (80-) 2016, 352:1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. •.Jarret A et al. : Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell 2020, 180:50–63.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that enteric neurons are capable of modifying epithelial cell function in response to pathogen, suggesting yet undiscovered role in normal microbiome maintenance or monitoring capabilities.
- 30.Li Q et al. : Intestinal dysbacteriosis-induced IL-25 promotes development of HCC via alternative activation of macrophages in tumor microenvironment. J Exp Clin Cancer Res 2019, 38:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rios D et al. : Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol 2016, 9:907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palm NW et al. : Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158:1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. ••.Lai NY et al. : Gut-innervating nociceptor neurons regulate Peyer’s patch Microfold cells and SFB levels to mediate salmonella host defense. Cell 2020, 180:33–49.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates how nociceptor sensory neurons, in response to pathogen, can influence both SFB colonization and M cell density.
- 34.Lei W et al. : Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A 2018, 115:5552–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou Q, Ye L, Huang L, Yu Q: The research progress on intestinal stem cells and its relationship with intestinal microbiota. Front Immunol 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saffarian A et al. : Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenborn AA et al. : The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes 2019, 10:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gulati AS et al. : Mouse background strain profoundly influences paneth cell function and intestinal microbial composition. PLoS One 2012, 7:e32403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. •.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV: Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 2008, 105:20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates that homeostatic PRR signaling by commensal PAMPs is necessary for normal functions of IECs, such as Paneth cells.
- 40.Frantz AL et al. : Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol 2012, 5:501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Araújo JR et al. : Fermentation products of commensal bacteria alter enterocyte lipid metabolism. Cell Host Microbe 2020, 27:358–375.e7. [DOI] [PubMed] [Google Scholar]
- 42. ••.Lee YS et al. : Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe 2018, 24:833–846.e6. [DOI] [PubMed] [Google Scholar]; This work reveals how lactic-acid producing bacteria support intestinal epithelial cell regeneration suggesting a critical developmental and oncogenic consequences.
- 43. •.Nadjsombati MS et al. : Detection of succinate by intestinal tuft cells triggers a Type 2 innate immune circuit. Immunity 2018, 49:33–41.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a range of receptors present on Tuft cells capable of sensing microbiome signals, include a receptor which initiates downstream immune processes in response to microbial succinate.
- 44.Schneider C et al. : A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 2018, 174:271–284.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinley ET et al. : Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI insight 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahlman H, Nilsson: The gut as the largest endocrine organ in the body. Ann Oncol Off J Eur Soc Med Oncol 2001, 12(Suppl. 2): S63–68. [DOI] [PubMed] [Google Scholar]
- 47. •.Larraufie P et al. : SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep 2018, 8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates that human epithelial cells, in response to SCFAs, can regulate hormone secretion responsible for neurological behaviors such as food intake.
- 48.Chimerel C et al. : Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep 2014, 9:1202–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brighton CA et al. : Bile acids trigger GLP-1 release predominantly by accessing basolaterally located G protein-coupled bile acid receptors. Endocrinology 2015, 156:3961–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebrun LJ et al. : Enteroendocrine L cells sense LPS after Gut barrier injury to enhance GLP-1 secretion. Cell Rep 2017, 21:1160–1168. [DOI] [PubMed] [Google Scholar]
- 51.Vincent AD, Wang XY, Parsons SP, Khan WI, Huizinga JD: Abnormal absorptive colonic motor activity in germ-free mice is rectified by butyrate, an effect possibly mediated by mucosal serotonin. Am J Physiol - Gastrointest Liver Physiol 2018, 315:G896–G907. [DOI] [PubMed] [Google Scholar]
- 52.Yano JM et al. : Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161:264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandić AD et al. : Clostridium ramosum regulates enterochromaffin cell development and serotonin release. Sci Rep 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olivares M et al. : The potential role of the Dipeptidyl peptidase-4-like activity from the gut microbiota on the Host Health. Front Microbiol 2018, 9:1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arora T et al. : Microbial regulation of the L cell transcriptome. Sci Rep 2018, 8:1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ: The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 2014, 15:792–798. [DOI] [PubMed] [Google Scholar]
- 57.Kaiko GE et al. : The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 2016, 165:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou Q et al. : Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ 2018, 25:1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wlodarska M et al. : Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017, 22:25–37.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abo H et al. : Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration. Nat Commun 2020, 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura Y et al. : Microfold cell-dependent antigen transport alleviates infectious colitis by inducing antigen-specific cellular immunity. Mucosal Immunol 2020:1–12 10.1038/s41385-020-0263-0. [DOI] [PubMed] [Google Scholar]
- 62.Nagashima K et al. : Targeted deletion of RANKL in M cell inducer cells by the Col6a1-Cre driver. Biochem Biophys Res Commun 2017, 493:437–443. [DOI] [PubMed] [Google Scholar]
- 63.Jung C, Hugot J-P, Barreau F: Peyer’s patches: the immune sensors of the intestine. Int J Inflamm 2010:823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tahoun A et al. : Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microbe 2012, 12:645–656. [DOI] [PubMed] [Google Scholar]
- 65.Ladinsky MS et al. : Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science (80-) 2019, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon K, Kim N: The effect of microbiota on colon carcinogenesis. J Cancer Prev 2018, 23:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J et al. : Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer 2016, 139:1318–1326. [DOI] [PubMed] [Google Scholar]
- 68.Rubinstein MR et al. : Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rubinstein MR et al. : Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDole JR et al. : Goblet cells deliver luminal antigen to CD103 + dendritic cells in the small intestine. Nature 2012, 483:345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kulkarni DH et al. : Goblet cell associated antigen passages are inhibited during Salmonella typhimur/cium infection to prevent pathogen dissemination and limit responses to dietary antigens article. Mucosal Immunol 2018, 11:1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim J, Khan W: Goblet cells and mucins: role in innate defense in enteric infections. Pathogens 2013, 2:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan JM et al. : CD4+ T cells drive goblet cell depletion during citrobacter rodentium infection. Infect Immun 2013, 81:4649–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perland E, Fredriksson R: Classification systems of secondary active transporters. Trends Pharmacol Sci 2017, 38:305–315. [DOI] [PubMed] [Google Scholar]
- 75.Lin L, Yee SW, Kim RB, Giacomini KM: SLC transporters as therapeutic targets: Emerging opportunities. Nat Rev Drug Discov 2015, 14:543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arike L et al. : Protein turnover in epithelial cells and mucus along the gastrointestinal tract is coordinated by the spatial location and microbiota. Cell Rep. 2020, 30:1077–1087.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mukherji A, Kobiita A, Ye T, Chambon P: Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013, 153:812–827. [DOI] [PubMed] [Google Scholar]
- 78.Thaiss CA et al. : Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 2016, 167:1495–1510.e12. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y et al. : The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science (80-) 2017, 357:912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuang Z et al. : The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science (80-) 2019, 365:1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ayyaz A et al. : Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019, 569:121–125. [DOI] [PubMed] [Google Scholar]
- 82.Herring CA et al. : Unsupervised trajectory analysis of single-cell RNA-Seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst 2018, 6:37–51.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosshart SP et al. : Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 2017, 171:1015–1028.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]


