Abstract
Hereditary diseases and complex traits often manifest in specific tissues, whereas their causal genes are expressed in many tissues that remain unaffected. Among the mechanisms that have been suggested for this enigmatic phenomenon is dosage-sensitive compensation by paralogs of causal genes. Accordingly, tissue-selectivity stems from dosage imbalance between causal genes and paralogs that occurs particularly in disease-susceptible tissues. Here, we used a large-scale dataset of thousands of tissue transcriptomes and applied a linear mixed model (LMM) framework to assess this and other dosage-sensitive mechanisms. LMM analysis of 382 hereditary diseases consistently showed evidence for dosage-sensitive compensation by paralogs across diseases subsets and susceptible tissues. LMM analysis of 135 candidate genes that are strongly associated with 16 tissue-selective complex traits revealed a similar tendency among half of the trait-associated genes. This suggests that dosage-sensitive compensation by paralogs affects the tissue-selectivity of complex traits, and can be used to illuminate candidate genes' modes of action. Next, we applied LMM to analyze dosage imbalance between causal genes and three classes of genetic modifiers, including regulatory micro-RNAs, pseudogenes, and genetic interactors. Our results propose modifiers as a fundamental axis in tissue-selectivity of diseases and traits, and demonstrates the power of LMM as a statistical framework for discovering treatment avenues.
Keywords: Hereditary diseases, Complex traits, Paralogs, Data integration, Linear mixed models
1. Introduction
Technological advancements in genome mapping and sequencing have largely increased our knowledge of the genetic determinants of monogenic and complex diseases. Disease-causing genes and disease-associated genetic variations have been recorded for hundreds of diseases and complex traits [1], [2]. However, functional understanding of the molecular mechanisms by which these genetic determinants impact health remains limited. Across hundreds of genetic diseases, clinical manifestation is often limited to few tissues, as demonstrated by neurodegenerative disorders, muscular dystrophies, and even cancers [3]. In an effort to illuminate disease mechanisms, several studies have turned to analyze the mechanisms that underlie the tissue-specific manifestation of genetic diseases (e.g., [4], [5], [6], [7], reviewed in [8]). These were made possible by large-scale tissue profiling resources, such as the Genotype-Tissue Expression (GTEx) consortium [9] and the Human Protein Atlas [10], covering tens of tissues. Using these datasets, studies repeatedly showed that tissue-specific diseases manifestation does not stem from tissue-specific expression of disease-causing genes in disease-susceptible tissues. Rather, disease-causing genes and variants are often expressed in many additional tissues that do not show disease symptoms [4], [5], [11]. Among the explanations for tissue-selectivity were expression-based mechanisms, such as over-expression of disease causing genes in disease-susceptible tissues [4], [5]; Regulatory-based mechanisms, such as the presence of tissue-specific eQTLs in disease-susceptible tissues [7]; and network-based mechanisms, such as the occurrence of tissue-specific protein interactions in disease-susceptible tissues [5], [11] (reviewed in [8]).
A general conception for the tissue-selectivity of diseases relates to tissue-selective compensation. Accordingly, tissue-wide robustness to the causal aberration occurs owing to the presence of a compensatory factor, and thus disease phenotypes emerge wherever this factor is limited. Gene duplicates, namely paralogs, are known to have common functionalities and were shown to compensate for each other’s loss (reviewed in [12]). In human, with the exception of a few paralogs that showed mutual dependence [13], large-scale assays of human cell lines revealed mostly compensatory relationships between paralogs [13], [14], [15]. The relationship between gene dosage and paralogs was also studied with respect to Mendelian disease genes with distinct modes of inheritance, leading to somewhat controversial results [16], [17], [18]. For example, autosomal dominant disease genes were enriched for paralogs that arose from whole genome duplication [19], [18], whereas autosomal recessive genes were enriched for paralogs that arose from old small-scale duplication [17].
We recently tested this conception at large-scale by studying the role of paralogs in over 120 hereditary diseases based on hundreds of tissue transcriptomes gathered by GTEx [20]. We showed that paralogs of disease-causing genes tend to be under-expressed relative to the causal gene in disease-susceptible tissues, suggesting that their compensatory function is relatively limited in these tissues and allows for disease phenotypes to emerge (Fig. 1A). The tendency for relative under-expression of paralogs was common across various subsets of causal genes, diseases, and tissues.
Fig. 1.
Quantitative relationships between causal genes and their paralogs. A. A model of the relationship between a causal gene (marked C) and its functionally-redundant paralog (marked P). In the healthy state, the redundant functions of both genes are represented as common interactors. In the aberrant state, the causal gene loses some of its functionality (dashed lines). In the unaffected tissues, the limited functionality of the causal gene is masked by the presence of its paralog. In the diseases-susceptible tissue (right), masking is insufficient due to relatively low expression of the paralog (small circle). B. The distribution of 382 hereditary diseases (yellow) and their 295 causal genes (red) across disease-susceptible tissues. C. The distribution of causal genes according to the number of paralogs they have. D. The distribution of hereditary diseases according to the number of susceptible tissues, and the distribution of causal genes and their paralogs according to number of tissues expressing at 1 TPM or above. E. The distribution of expression ratios (log-transformed) of causal genes and their paralogs in disease-susceptible versus unaffected tissues. The distributions are shown as follows: Left: 158 causal genes with a single paralog. Middle: 137 causal genes that were compared to each of their two most closely related paralogs. Right: 137 causal genes that were compared to the summed expression of all their paralogs. The black line indicates the median value. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Here, we aimed to expand our view of dosage-sensitive molecular mechanisms in several ways. Firstly, we gathered a dataset composing of 382 tissue-selective diseases and 11,215 tissue transcriptomes that was over 3-fold larger than used in Ref. [20]. Secondly, to assess simultaneously the contribution of multiple factors we employed the linear mixed model (LMM) methodology. By using LMM, we were able to account for the clustered nature of the data formed by the donors, and to efficiently adjust the association of the outcome with the main independent factor (tissue type) to multiple covariates potentially confounding the association at question [21]. LMM findings pointed to paralog identity levels and disease mode of inheritance as additional modulators of causal-gene and paralog expression. However, the relative under-expression of paralogs in disease-susceptible tissue was the strongest consistent factor. Thirdly, we analyzed dosage-relationships in complex traits. Specifically, we focused on 16 tissue-selective traits, and analyzed paralogs relationships in 135 candidate trait-associated genes identified via genome-wide association studies (GWAS). We found that tissue-selective under-expression of paralogs is also common among complex traits. Fourthly, we turned to analyze the relevance of additional classes of genetic modifiers, including micro-RNA regulation, transcribed pseudogenes, and genetic interactors of disease-causing genes, to the tissue-selectivity of Mendelian diseases. Altogether, our findings provide positive evidence for the relevance of dosage-sensitive mechanisms in tissue-selective disease manifestation. They also demonstrate the power of LMM as a statistical framework for analysis of quantitative genetic relationships.
2. Results
2.1. Factors affecting expression ratios of causal genes and paralogs
To study the quantitative relationships between causal genes and paralogs across tissues, we gathered a set of 382 hereditary diseases with manually-curated tissue-selective manifestations (Fig. 1B, Table S1). These diseases were caused by mutations in 295 distinct causal genes, each of which having at least one high-confidence paralog (Methods, Fig. 1C). To obtain a quantitative molecular view into these diseases we employed the large-scale transcriptomic dataset of GTEx, consisting of 11,215 transcriptome profiles sampled from 51 tissues and 714 donors [9]. This dataset was over 3.7-fold and 26.7-fold larger in terms of genes and transcriptomic profiles, respectively, than the dataset used in Ref. [20] (Fig. S1). We first examined the tissue-selectivity of these diseases, their causal genes and their paralogs. As observed previously [5], [11], diseases were highly tissue-specific, whereas their causal genes were broadly expressed across the human body (Fig. 1D).
Next, we examined the quantitative relationship between causal genes and paralogs in disease-susceptible versus unaffected tissues. We modeled the quantitative relationships by the expression ratios of causal genes and their paralogs across tissues (Methods). Focusing on the subset of 158 (54%) causal genes with a single paralog, revealed a tendency for higher expression ratios in disease-susceptible tissues (p < E16, Mann-Whitney (MW), Fig. 1E left). The same tendency was shared by the 137 remaining causal genes, where expression ratios were computed for causal genes and each of their top two paralogs (p < E16, MW, Fig. 1E middle). Lastly, to account for causal genes with multiple paralogs that provide collective compensation, we examined the expression ratio between these 137 causal genes and the summed expression levels of their paralogs. The tendency for higher expression ratio in disease-susceptible tissues remained significant (p < E16, MW, Fig. 1E right). Thus, the trend observed previously in a smaller disease dataset was maintained [20].
The above analysis did not consider the potential impact of other factors on the observed expression ratios, including transcriptomic and disease-related factors (Fig. 2A). Candidate transcriptomic factors included donor parameters (e.g., sex, age, cause of death), sample parameters (e.g., autolysis score, ischemic time), and profile parameters (e.g., RNA integrity number), extracted from GTEx. Candidate disease-related factors included disease mode of inheritance, namely autosomal dominant versus recessive, and paralogs’ sequence identities, which have already been pointed out by other studies [17], [18], [22]. Along with the impact of tissue type that we accounted for above, namely disease-susceptible versus unaffected tissues, we considered a total of 12 distinct factors (Methods).
Fig. 2.
LMM analysis of expression ratios between causal genes and their paralogs. A. The different factors that might impact expression ratios (left) and the resulting LMM formulation (right). B. The standardized coefficient values that were determined by the LMM. Left: LMM analysis of 158 causal genes with a single paralog. Middle: LMM analysis of 137 causal genes that were compared to each of their two most closely related paralogs. Right: LMM analysis of 137 causal genes that were compared to the summed expression of all their paralogs. C. The standardized coefficient values that were determined by the LMM for subsets of causal genes with distinct susceptible tissues. Results are shown in the single paralog, top two paralogs, and sum of all paralogs scenarios. In each scenario case, LMM was applied separately to causal genes associated with brain, heart, skeletal muscle and skin diseases.
To test the impact of each of these factors while controlling for confounding factors, and to account for the inherently clustered nature of GTEx (where each subject is represented by a distinct cluster), we turned to a multivariable model that combines mixed and fixed effects. Given the log normal distribution of the expression ratios (Fig. S2A), LMM was the best fit (Methods). In the LMM we regressed expression ratios of causal genes and paralogs on 12 independent variables, each representing a different factor, as specified in the regression equation (Methods, Fig. 2A). We then applied the formulation to model expression ratios in the single paralog and multi paralogs scenarios described above (Fig. 1E). Regression findings included the standardized coefficients determined by the LMM per variable and their statistical significance (Fig. 2B and Table S2). Only five factors were significant in all scenarios, including tissue type, paralogs sequence identity, mode of inheritance, RNA integrity number, and the date of nucleic acid isolation batch. Out of those, tissue type, paralogs’ sequence identities, and disease mode of inheritance had the strongest independent impacts. Notably, paralogs’ sequence identities and disease mode of inheritance coefficients were positive in certain scenarios and negative in others, implying that they contributed to higher or lower expression ratio, respectively. In contrast, tissue type coefficient was consistently positive in all scenarios, supporting the aforementioned relationship between causal genes and their paralogs in the disease-susceptible tissues.
To test the generalizability of the LMM findings, we analyzed separately subsets of diseases sharing the same susceptible tissue. We focused on the four tissues with over 15 causal genes: brain (123 causal genes), heart (32 causal genes), muscle (18 causal genes), and skin (18 causal genes). Per tissue, we applied the LMM separately to causal genes with a single paralog, top two paralogs, and sum of paralogs. Similar to the analysis above, the standardized coefficients of causal gene-paralog similarity and disease mode of inheritance were relatively high, yet inconsistent across tissues and across scenarios (the potential association with disease mode of inheritance was not due to the fraction of genes with dominant inheritance per tissue, Fig. S3). In contrast, across all but one case, the standardized coefficient of tissue type was both relatively high and consistently positive (Fig. 2C). Thus, tissue type positively affected expression ratios between causal genes and their paralogs across scenarios and disease subsets.
2.2. Tilted expression ratios between paralogs in tissue-selective complex traits
Given that the impact of tissue type on expression ratios is common among genes that are causal for tissue-selective phenotypes, we went on to test whether it is also common among genes that are likely causal for tissue-selective complex traits. We concentrated on complex traits that were analyzed via GWAS, a genetic screening methodology that identifies genetic variations that are common across patients versus healthy individuals. The identified genetic variations are typically spread across the genome and often implicate several genes in the same genomic region, challenging the identification of trait-causing genes [23]. We analyzed 16 tissue-selective traits for which candidate genes harboring trait-associated genetic variations were identified and ranked by the statistical significance of their trait association [24]. The tissue-selectivity of each trait was manually curated (Methods). Per trait, we considered only candidate genes with highly significant trait associations (p < E−15), and that had at least one high-confidence paralog (Methods, Fig. 3A, Table S3). Altogether, we analyzed 135 genes, including 78 genes with a single paralog and 57 genes with multiple paralogs. Similar to hereditary diseases, these traits were highly tissue-specific, whereas candidate genes and their paralogs tended to be expressed broadly across the body (Fig. 3B).
Fig. 3.
LMM analysis of expression ratios between candidate trait-causing genes and their paralogs. A. The fractions of complex traits, candidate genes, and their paralogs, per trait-susceptible tissue. B. The distribution of 16 complex traits according to the number of tissues in which they manifest, and the distribution of 135 candidate genes and their 178 paralogs according to number of tissues expressing them at 1 TPM or above. C. The scheme for applying LMM to each candidate gene, resulting in identification of candidate genes with positive and significant impact of tissue type on candidate gene – paralog expression ratio. D. The fraction of candidate genes per trait out of the total set of candidate genes, with LMM positive genes marked in red. E. The fraction of candidate genes per trait-susceptible tissue with LMM positive genes marked in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To test whether tissue type impacts expression ratios of candidate genes for complex traits, we employed the LMM regression separately to each candidate gene and its paralog(s). We then extracted candidate genes where, similarly to disease-causing genes, expression ratio was positively and significantly affected by tissue type, which we denoted as LMM positive (Fig. 3C). LMM regression revealed that 71 out of 135 candidate genes were LMM positive. LMM positive genes were found in 15 out of 16 traits and consisted of 35–56% of the candidate genes per trait (Fig. 3D). They were also similarly frequent per susceptible tissue (Fig. 3E).
It is compelling to suggest that LMM positive genes are more likely to be trait-associated. One such LMM positive gene was TCF7L2, which was significantly associated with fasting proinsulin (adjusted p = 1.01E−176). Although proinsulin is synthesized in pancreas, TCF7L2 was not preferentially expressed in pancreas, implying that TCF7L2 expression alone would not reveal its tissue-specific impact. However, by considering its expression relative to its paralog TCF7L1, its pancreas-specificity is revealed (Fig. 4A). An intriguing example is presented by the fatty acid desaturase (FADS) gene cluster consisting of FADS1, FADS2, and FADS3. FADS1 and FADS2 encode the delta-5-desaturase and delta-6-desaturase, respectively, but the role of FADS3 in fatty acid metabolism is unclear. All FADS genes were detected by GWAS as candidate genes associated with blood measures of total cholesterol, HDL, LDL, fasting glucose, and triglyceride. All of these traits are associated with the liver, while triglycerides are also associated with the small intestine and adipose tissues. Given that the FADS genes are closely related, we used the sum of paralogs model. FADS1 was LMM positive in liver (adjusted p = 7.97E−61), supporting its association with liver-specific traits. Though its liver-susceptibility cannot be foretold by its liver expression, it became clearer upon considering its expression ratio relative to its paralogs, which is maximal in liver (Fig. 4B). FADS2 had similar results. FADS3, in contrast, was LMM positive only for the triglycerides trait (adjusted p = 7.47E−57, Fig. 4C). In support of this, high expression of FADS3 in adipose tissue was previously associated with increased risk for familial combined hyperlipidemia and higher triglyceride levels in Mexican population [25].
Fig. 4.
Examples for candidate trait-associated genes showing tissue type effects on expression ratios. Trait-associated tissues were marked red. A. Top: The expression of TCF7L2 across tissues according to GTEx. Its expression in pancreas, its trait-associated tissue, is smaller than its expression in most other tissues. Bottom: The expression ratio between TCF7L2 and its paralog TCF7L1 across tissues according to GTEx. Median expression ratio was highest in pancreas (p = 1.01E-176). B. Top: The expression of FADS1 across tissues according to GTEx. Its expression in liver, its trait-associated tissue, is not up-regulated relative to other tissues. Bottom: The expression ratio between FADS1 and the sum of its paralogs, FADS2 and FADS3. Median expression ratio was particularly high in liver (p = 7.79E-61). C. Top: The expression of FADS3 across tissues according to GTEx. FADS3 was associated with multiple traits and trait-associated tissues, including liver, small intestine, and adipose tissues. Bottom: The expression ratio between FADS3 and the sum of its paralogs, FADS1 and FADS2. Median expression ratio was high particularly in adipose tissues (p = 7.47E-52). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Beyond paralogs: LMM analysis of additional dosage-compensatory relationships
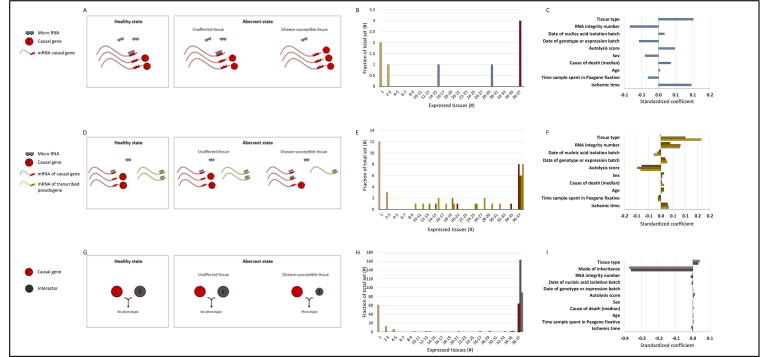
The analyses above focused on causal gene compensation by its paralogs. Next, we turned to investigate additional mechanisms that could modify the impact of widely expressed causal genes on the tissue-selectivity of diseases. The first mechanism we considered was causal gene regulation by micro-RNAs (miRNAs). miRNAs are small non-coding RNA molecules that regulate gene expression post-transcriptionally by inhibiting mRNA translation potentially leading to its degradation (Fig. 5A). Here we explored the possibility that the quantitative relationships between causal genes and their regulating miRNAs were altered in disease-susceptible tissues. Data of miRNAs that were shown experimentally to interact with the transcripts of causal genes were extracted from miRecords [26], of which only three miRNAs were measured in GTEx (Fig. 5B, Table S4). The LMM revealed that tissue type positively affected expression ratio between causal genes and their miRNAs (p = 4.6E−70, Fig. 5C). These results suggest that the inhibition of causal genes by their miRNAs was reduced in the disease-susceptible tissue.
Fig. 5.
Applying LMM to analyze quantitative relationships between causal genes and additional types of modifier genes. A. A regulatory miRNA binds its target causal gene in unaffected tissues, thereby exerting post-transcriptional regulation. In the disease-susceptible tissue (right), a lower level of the miRNA would result in reduced post-transcriptional regulation over the causal gene. B. The tissue-specificity of three diseases, and the distribution of three causal genes and two miRNAs according to number of tissues expressing them at 1 TPM or above. C. The standardized coefficient values that were determined by the LMM. D. A transcribed pseudogene might indirectly regulate the causal gene, potentially by binding a miRNA that is a common regulator of both. A lower level of the transcribed pseudogene in the disease-susceptible tissue (right) would result in reduced pseudogene-exerted effects. E. The tissue-specificity of diseases, and the distribution of causal genes and pseudogenes according to number of tissues expressing them at 1 TPM or above (dark gold; nine diseases and seven causal genes), 0.5 TPM or above (gold; 10 diseases and eight causal genes), and 0.1 TPM or above (light gold; 15 diseases and 10 causal genes). F. The standardized coefficient values that were determined by the LMM. G. Regular levels of causal genes and their genetic interactors across unaffected tissues do not lead to phenotype. However, the combination of causal gene aberration and low expression level of the genetic interactor might lead to phenotype specifically in disease-susceptible tissues (right). H. The tissue-specificity of 79 diseases, and the distribution of 67 causal genes and their genetic interactors (92 positive interactors, light grey; 169 negative interactors, dark grey), according to number of tissues expressing them at a level ≥ 1 TPM. I. The standardized coefficient values that were determined by the LMM for positive (light grey) and negative (dark grey) genetic interactors using the summed expression model.
The second mechanism we tested involved pseudogenes of causal genes. Pseudogenes are DNA sequences that resemble functional genes yet lack the capacity to encode for proteins. Transcribed pseudogenes were previously shown to indirectly regulate their respective coding genes, for example by acting as microRNA sponges [27] (Fig. 5D). We examined whether the quantitative relationships between causal genes and their pseudogenes were altered in disease-susceptible tissues. For that, we extracted pseudogenes that were members of a causal gene homologs family [28], and that were expressed in GTEx. Owing to their low expression levels, we considered three expression thresholds for pseudogenes: 1, 0.5, and 0.1 transcripts per million reads (TPM), which allowed us to analyze 7, 8, and 10 causal genes, respectively (Fig. 5E, Table S5). We applied LMM to all three sets. Tissue type coefficient was insignificant at the 1TPM subset, yet was positive at the lower thresholds (p < E−195, Fig. 5F), and had stronger impact relative to other factors. This suggests that the relative expression of causal genes and their pseudogenes is higher in disease-susceptible tissues, implying less indirect regulation by pseudogenes.
The third mechanism that we examined was epistasis, or genetic interaction, where the phenotypic effect of an aberrant causal gene was dependent on the presence or absence of an aberrant modifier genes. However, instead of the modifier gene being aberrant, we tested whether it is relatively lowly expressed (mimicking aberration) in disease-susceptible tissues (Fig. 5G). We extracted data of positive and negative genetic interactors of 67 causal genes and 79 hereditary diseases from BioGRID. Notably, 38 causal genes corresponding to 45 diseases had both positive and negative genetic interactors (Fig. 5H, Table S6). We applied LMM to each set separately, while considering the ratio between causal genes and the summed expression levels of either their positive or negative genetic interactors. Tissue type coefficient was positive in both cases (p < E−101, Fig. 5I), implying more likelihood for epistatic interactions in those tissues. However, a much larger impact on ratios was due to disease mode of inheritance (p < E−300). Altogether, despite the limited available data, our analysis suggests a role for additional classes of genetic modifiers in tissue-selectivity of diseases.
3. Discussion
We presented a computational analysis of molecular mechanisms that potentially underlie the tissue-selective manifestation of hereditary diseases and complex traits. These diseases and traits tend to manifest in a highly tissue-specific manner, albeit the ubiquitous expression of their causal or associated genes. Knowledge of the molecular mechanisms that govern diseases and traits manifestation is critical for better understanding of human physiology and pathophysiology, but is currently limited [8]. Here we focused on mechanisms involving dosage-sensitive relationships between causal genes and potential modifier genes. To model these relationships, we relied on large-scale human tissue transcriptomes, encompassing 51 tissues and 11,215 transcriptomic profiles, that were mapped in a consistent manner by GTEx [9]. Though these profiles might be challenging for protein-level analyses, they constitute a powerful resource for comparative analyses at the transcriptomic level.
Using this large-scale dataset, we modeled quantitative relationships by calculating, per transcriptomic profile, the ratios between transcript levels of relevant molecules [20]. We then analyzed these relationships across profiles via LMM regression, which was not previously used for analyses of tissue-selectivity. LMM provided a statistical framework for simultaneous analysis of the relationships between multiple factors on expression ratios. Specifically, LMM allowed to point to factors that could affect the relationships between paralogs while controlling for other factors. This is feasible only in a multivariable model and was not possible with a univariable analysis. Additionally, the natural clustering of observations within one subject was controlled for by assigning each subject with a random effect (cluster). The flexibility of the LMM scheme allowed us to analyze sample- and disease-related factors (Fig. 2A), to analyze subsets of genes and individual genes via several models (Figs. 2C and 4), and to assess multiple modifier classes (Fig. 5).
We first applied LMM to study the quantitative relationships between causal genes and their paralogs. The relevance of paralogs was shown previously for a subset of hereditary diseases, supporting insufficient compensation by paralogs in disease-susceptible tissues [20]. Here, by using LMM and larger disease and transcriptomic datasets, we found that expression ratios were tilted in disease-susceptible tissues (Fig. 2B, C), in agreement with previous results [20]. Whereas interpreting regression coefficients could be problematic when crude estimates of association are considered, the usage of a multivariable statistical tool enabled to account for the possibility of confounding factors that could have been associated with the tissue type variable. Consequently, the inclusion of other factors in the model enabled a valid estimation of the relationship between paralog expression ratios and tissue type. Nevertheless, the presence of residual confounding factors, which were not measured and therefore were not part of the analysis and not adjusted for, could not be ruled out.
LMM findings also pointed to the impact of paralog identity levels and disease mode of inheritance on expression ratios (Fig. 2B). Across diseases, expression ratios tended to be higher for causal genes and paralogs with larger sequence identity, potentially due to dosage sharing between closely-related paralogs [29]. Expression ratios also tended to be higher for autosomal recessive disease genes, for which previous studies had conflicting conclusions [17], [18]. However, these results were not consistent across disease subsets (Fig. 2C).
Following the analysis of hereditary diseases, we turned to analyze whether quantitative relationships between causal genes and their paralogs may play a role in the tissue-selectivity of complex traits. Using single-gene LMM regression, we found that tissue type was a significant factor in about half of the trait-associated genes (Fig. 3D, E). This analysis could therefore be used to illuminate disease mechanisms (Fig. 4A), or to prioritize candidate genes for tissue-selective complex traits, as demonstrated by the different FADS genes (Fig. 4B, C).
We used the LMM framework to analyze three additional classes of modifiers. The first two modifier classes that we tested revolved around post-transcriptional regulation of causal genes, either via miRNAs (Fig. 5A) or pseudogenes (Fig. 5D). These modifiers were demonstrated previously to affect disease emergence, though not in a tissue-selective manner [27]. Although relying on small datasets, LMM analysis of both classes revealed that the ratio between a causal gene and its potential modifier was higher in the disease-susceptible tissue, supporting their potential impact on tissue-selective manifestation (Fig. 5C, F). These results should be revisited when more data are available. The last class was related to epistasis (Fig. 5G). LMM findings revealed that regardless of the type of genetic interaction, tissue type had a significant impact, though its effect was smaller compared to disease mode of inheritance (Fig. 5I). LMM regression can readily be used to assess the impact of additional factors and modifier classes across diseases, genes, or tissues. It could be applied to other questions, for example, to model or predict tissue type based on various factors [30], such as the factors that were modeled in the current study.
Altogether, our analyses support the role of genetic modifiers as a fundamental axis in the tissue selective manifestation of hereditary diseases and complex traits. Unraveling functional redundancy in paralogs of causal genes was already shown to set the ground for drug development, as in cases of hereditary spinal muscular atrophy [31], [32], and cancer [33], [34]. Revealing additional genetic modifiers may clarify the pathogenesis and open novel treatment avenues.
4. Methods
Transcriptomic dataset: RNA-sequencing profiles of human tissues were obtained from GTEx portal (version 7) [9], and consisted 11,215 transcriptomic profiles sampled from 51 primary tissues (samples from transformed cells were not included). We united sub-tissues of the same main tissue, including sub-tissues of skin, of heart, and of brain, which resulted in 37 tissues. Henceforth, we analyzed only causal genes that were expressed above 1 TPM in at least half of the samples of a given tissue, and in at least 20% of the tissues, including the disease-susceptible tissue.
Diseases, traits, and genes datasets: We analyzed 382 hereditary diseases with manually-curated tissue-selective manifestation [11]. Disease-causing genes with a known molecular basis were downloaded from OMIM [1] (Table S1). The dataset of complex traits included 16 traits that were previously analyzed via GWAS. We manually-curated the tissue manifestation of each trait. The genes associated with each traits were collected by Marbach et al. [24], which applied the Pascal tool to assemble single-nucleotide polymorphisms (SNPs) summary statistics into gene probability scores [35]. Using these scores, we considered as candidate genes per trait only genes with p-value < E−15, which resulted in 135 trait-associated genes (Table S3).
Genetic modifiers of causal genes: Paralogs of causal genes were extracted from Ensembl-biomart using R package ‘biomart’ (download date: 8/7/2019). We included paralogs with over 40% reciprocal sequence identity that were co-expressed with their respective causal gene above 1 TPM in at least 20% of the tissues. miRNAs that interact with causal genes via experimentally-validated interactions were extracted from miRecords [26] (Table S4). Transcribed pseudogenes of causal genes were collected from a dataset of 3281 pseudogene-gene families, where a pseudogene was associated with a gene family based on its sequence similarity to the family consensus sequence [28] (Table S5). We included in the analysis pseudogenes that were expressed at levels exceeding 0.1, 0.5, and 1 TPM. Positive and negative genetic interactors of causal genes were extracted from BioGRID [36] (Table S6).
Computation of quantitative relationships: To quantify the relationship between a causal gene and its modifier gene we computed the ratio between their expression levels per sample, across all samples that expressed both genes at levels exceeding 1 TPM (different thresholds were applied to pseudogenes). In the top two paralogs analysis, we computed the ratio between the expression level of a causal gene and (i) the expression level of its paralogs with highest reciprocal sequence identity, and (ii) the expression level of its paralogs with the second highest reciprocal sequence identity, resulting in two ratios computed per sample per causal gene. In the sum of paralogs analysis, we computed the ratio between the expression level of a causal gene and the sum of the expression levels of all its paralogs, per sample. In the analysis of genetic interactors, we computed the ratio between the expression level of a causal gene and the sum of the expression levels of all its positive interactors, or all its negative interactors, per sample. In paralogs and genetic interactors analyses, we excluded pairs where the paralog or the genetic interactor were disease-causing in the same tissue as the causal gene.
LMM analysis: To account for the clustered nature of the GTEx dataset (where each cluster represents a distinct subject), it was imperative to use a modeling technique that combines mixed and fixed effects. To identify a suitable modeling technique, we analyzed the distribution of expression ratios between causal genes and their paralogs. The log expression ratio was normally distributed (Fig. S2A). Since our dataset met the assumptions of the log normal model, and since the normal distribution has been profoundly used in statistical modeling, we selected LMM. As part of the sensitivity analysis, we also applied the quasi-Poisson model. The estimates obtained with the quasi-Poisson model followed the trends obtained with the log normal model (Fig. S2B). Next, we used LMM to identify factors that impact quantitative relationships between causal genes and potential modifier genes. For that, we log-transformed the expression ratio, and designated it as the dependent outcome of the LMM. We defined it as normally distributed per sample. The different factors served as independent variables. These factors consisted of donor parameters, sample parameters, and transcriptomic parameters, which were downloaded from GTEx; and disease-related parameters, including tissue type, mode of inheritance, and paralogs’ sequence identities. The formulation appears in Eq. (1) below. We accounted for the clustered nature of the data formed by the same donors by assigning a random intercept to each donor. Each factor was represented as a vector to LMM. We applied LMM to gene sets as described in the Results, and per gene for candidate genes for complex traits. LMM findings in each analysis included a coefficient per factor and its statistical significance. To enable comparison between factors with different ranges of values, each coefficient was standardized. The standardized coefficients expressed the expected change in the log-transformed expression ratios in standard deviation units, per change of a standard deviation in the independent regression terms. The statistical significance of each factor was adjusted for multiple hypothesis testing via Bonferroni correction. LMM was implemented using R (version 3.5.2) and the lme4 package.
| (1) |
A short description of the factors and their values range appears below: 1. Tissue type: unaffected tissues (0), disease-susceptible tissues (1). 2. Sex: male (0), female (1). 3. Age: age of donor, 20 to 79. 4. Cause of death: original values were 0, 1, 2, 3, and 4, which we represented by dummy variables. Cause 0 was kept out of the regression, and a factor was created for each remaining cause; the value of each factor value was set to 1 if matched donor cause of death, and 0 otherwise. 5. Ischemic time: −1226 to 1739. 6. Time sample spent in PAXgene fixative: 240 to 1673. 7. RNA integrity number: 3 to 10. 8. Autolysis score: none (0), mild (1), moderate (2), and severe (3). 9. Date of nucleic acid isolation batch: 17/05/2011 to 21/11/2014. 10. Date of genotype or expression batch: 30/10/2011 to 16/01/2015. 11. Mode of inheritance: autosomal dominant (0), autosomal recessive (1). 12. Paralogs sequence identity: 40 to 100. The factors 2–10 were extracted from GTEx [9], factor 11 was extracted from OMIM [1], and factor 12 was extracted from bioMart.
Acknowledgement
This research was supported by the Israel Science Foundation (ISF) through grant number 317/19 to E.Y.L.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.10.030.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucl Acids Res. 2019;47:D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buniello A. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucl Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haigis K.M., Cichowski K., Elledge S.J. Tissue-specificity in cancer: the rule, not the exception. Science. 2019;363(6432):1150–1151. doi: 10.1126/science:aaw3472. [DOI] [PubMed] [Google Scholar]
- 4.Lage K., Hansen N.T., Karlberg E.O., Eklund A.C., Roque F.S., Donahoe P.K., Szallasi Z., Jensen T.S., Brunak S. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci. 2008;105(52):20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barshir R., Shwartz O., Smoly I.Y., Yeger-Lotem E. Comparative analysis of human tissue interactomes reveals factors leading to tissue-specific manifestation of hereditary diseases. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C., Chasman D.I., FitzGerald G.A., Dolinski K., Grosser T., Troyanskaya O.G. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamazon E.R. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hekselman I., Yeger-Lotem E. Mechanisms of tissue and cell-type specificity in heritable traits and diseases. Nat Rev Genet. 2020;21:137–150. doi: 10.1038/s41576-019-0200-9. [DOI] [PubMed] [Google Scholar]
- 9.GTEx Consortium, et al. Genetic effects on gene expression across human tissues. Nature 550, 204-213, DOI:10.1038/nature24277 (2017). [DOI] [PMC free article] [PubMed]
- 10.Uhlen M. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 11.Basha O. et al. Differential network analysis of multiple human tissue interactomes highlights tissue-selective processes and genetic disorder genes. Bioinformatics (2020), DOI:10.1093/bioinformatics/btaa034. [DOI] [PubMed]
- 12.Diss G., Ascencio D., DeLuna A., Landry C.R. Molecular mechanisms of paralogous compensation and the robustness of cellular networks. J Exp Zool B Mol Dev Evol. 2014;322:488–499. doi: 10.1002/jez.b.22555. [DOI] [PubMed] [Google Scholar]
- 13.Dandage R., Landry C.R. Paralog dependency indirectly affects the robustness of human cells. Mol Syst Biol. 2019;15 doi: 10.15252/msb:20198871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science:aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Kegel B. & Ryan CJ. Paralog buffering contributes to the variable essentiality of genes in cancer cell lines. PLoS Genet 15 (2019), e1008466, DOI:10.1371/journal.pgen.1008466. [DOI] [PMC free article] [PubMed]
- 16.Kondrashov F.A., Koonin E.V. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen W.H., Zhao X.M., van Noort V., Bork P. Comments on “Human dominant disease genes are enriched in paralogs originating from whole genome duplication”. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh P.P., Affeldt S., Malaguti G., Isambert H. Human dominant disease genes are enriched in paralogs originating from whole genome duplication. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makino T., McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barshir R. et al. Role of duplicate genes in determining the tissue-selectivity of hereditary diseases. PLoS Genet 14 (2018), e1007327, DOI:10.1371/journal.pgen.1007327. [DOI] [PMC free article] [PubMed]
- 21.Wu L. Taylor and Francis Group; Boca Raton, FL, USA: 2010. Mixed effects models for complex data. [Google Scholar]
- 22.Chen W.H., Zhao X.M., van Noort V., Bork P. Human monogenic disease genes have frequently functionally redundant paralogs. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 24.Marbach D. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016;13:366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plaisier C.L. A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucl Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson T.S., Li S., Kho J.R., Huang K., Zhang Y. Network analysis of pseudogene-gene relationships: from pseudogene evolution to their functional potentials. Pac Symp Biocomput. 2018;23:536–547. [PMC free article] [PubMed] [Google Scholar]
- 29.Lan X., Pritchard J.K. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science. 2016;352(6288):1009–1013. doi: 10.1126/science:aad8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finucane H.K. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50(4):621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finkel R.S. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 32.Mercuri E. Nusinersen versus sham control in later-onset spinal muscular atrophy. N Engl J Med. 2018;378:625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 33.Jdey W. Drug-driven synthetic lethality: bypassing tumor cell genetics with a combination of AsiDNA and PARP inhibitors. Clin Cancer Res. 2017;23:1001–1011. doi: 10.1158/1078-0432.CCR-16-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.S. Harnessing synthetic lethality to predict the response to cancer treatment. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamparter D., Marbach D., Rueedi R., Kutalik Z., Bergmann S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oughtred R. The BioGRID interaction database: 2019 update. Nucl Acids Res. 2019;47:D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.