Abstract
G-quadruplexes are tetrahelical structures. They are important targets for anti-cancer drugs, since they are situated at crucial positions within the genome. We studied the volumetric properties of the unfolding of three G-quadruplexes in the presence of potassium ion. The unfolding volume changes were determined using high-pressure fluorescence spectroscopy. The c-MYC, KIT, and VEGF sequences unfold with the transition volume of -17, -6 and -18 cm3/mol, respectively. The small magnitude of the unfolding volume of KIT could be explained by its unique structure and the lower amount of void volume. Since the cell interior is highly crowded, the available volume is restricted. Therefore the volumetric changes during the conformational transformations gain biological importance.
Keywords: Biophysics, Bioinformatics, Biophysical chemistry, DNA-Ligand interaction, Spectroscopy, Thermodynamics, G-quadruplex, Fluorescence, High pressure, FRET, Volume
Biophysics; Bioinformatics; Biophysical chemistry; DNA-Ligand interaction; Spectroscopy; Thermodynamics; G-quadruplex; Fluorescence; High pressure; FRET; Volume
1. Introduction
1.1. G-quadruplexes
G-quadruplexes are noncanonical 3D structures that can be formed by short G-rich DNA sequences with a noncanonical 3D structure [1]. They came into the focus of the scientific research when their appearance in the telomere region was first reported. Their telomerase inhibition activity made them a plausible target in cancer research. G-quadruplexes also play an important role in the regulation of several genes, including oncogenes, which underlines their importance in the oncogenesis [2]. G-quadruplex (GQ) forming sequences were also found in several promoter regions, including several important proto-oncogenes like c-MYC, VEGF, HIF-1α, Ret, Bcl-2, c-Kit, and KRAS [3].
Moreover, some GQ-forming aptamers can be used as sensors. The fluorescent labeling of such aptamers allows the detection of different ions or molecules. An example for that is the thrombin-binding aptamer (TBA) [4, 5], which was originally developed for therapeutic applications. Several variants of TBA were later developed to increase its binding affinity.
GQs can be formed if the DNA sequence contains several repeats of guanine bases. The GQ structure is stabilized by the G-tetrads, which are formed by hydrogen bonding interaction of four co-planar guanine residues. Typically, two or three of these G-quartets are on top of each other (Figure 1). As it can be seen in Figure 1, a stabilizing metal ion in the center of the quartet is necessary. Potassium and sodium ions are suitable for stabilizing the 3D structure, the stabilizing effect of K+ is stronger. Li+ is too small for stabilization [6, 7, 8]. The Na+ ion is sitting in the plane of the quartets, while the bigger K+ ion is situated between the guanine plates, and its coordination binding to eight oxygen atoms stabilizes the whole GQ [9].
Figure 1.
The structure of a G-quartet (a) and of a G-quadruplex (b).
1.2. Volumetric effects
The volumetric effects associated to the structural transitions of biological macromolecules are often neglected. The interior of the cell is a highly crowded solution of a series of different macromolecules. The total concentration of such molecules can easily reach 30–40%. This means that the solution is practically divided into small sub-compartments, where the available free volume is small [10]. This underlines the importance of the volume occupied by the macromolecules in a different conformational state. This volume effect has been well studied for proteins [11, 12, 13, 14], but the number of studies on DNA molecules is quite limited [15, 16]. The reason could be the pressure insensitivity of the DNA due to the fact that the double-helical DNA shows little to no volume effect during unwinding.
Volume effects can be ideally studied using high pressure [17, 18, 19, 20, 21, 22, 23]. Although pressurization is technically more complicated than heating, pressure is an equally important thermodynamic parameter as temperature. The mentioned technical difficulty may explain why this technique is available only in a limited number of laboratories. We used a custom-designed high pressure cell, using a diamond window. This allowed us to reach 6 kbar, while maintaining a sample size of a few microliters. If a conformational change (e.g. GQ unfolding) is accompanied by a volume change, pressure will shift the transition. Pressure tries to shift the equilibrium, favoring the state with lower volume.
The definition of volume of molecules is also worthwhile to discuss. There are different volume definitions, but experimentally, we can determine the molar volume νi of a solute molecule. It is defined as the volume change of the solution by the addition of a small amount of the solute over the number of moles of added solute, keeping everything else constant [19].
| (1) |
This definition reflects the fact that we measure experimentally the volume of the whole system (solvent + solution), and the changes in the solvent density induced by the solute are also involved in this volume definition. An example for this is the hydration layer around the solute molecule. The increased density and consequently the reduced volume of this solvent layer appears as a negative contribution to the volume of the solute molecule. In the case of proteins, this hydration layer increases upon unfolding [24, 25, 26], while in the case of GQs, the hydration shell of the released ions may also play a role [27].
1.3. GQs as promising targets for cancer treatment
As GQs turned out as promising targets for cancer treatments, several ligands were investigated in order to stabilize the GQ structure [28, 29, 30]. Porphyrins with several substitutions were tested. Some of them, like TMPyP4, were efficient in stabilizing the structure [31].
In this study, we made a systematic study of several G-quadruplexes present in different positions within the human genome.
C-MYC is an important oncoprotein, its overexpression was observed in several human cancers like breast, cervix, colon, and small-cell lung cancers [3]. The quadruplex-forming sequence occurs in the proximal region of the c-MYC promoter, and has a major role in the control of the transcriptional activity. The sequence in our study is a slightly modified version of this one, which was also used in an earlier NMR study [32].
The c-KIT proto-oncogene codes a receptor tyrosine-kinase. Its overexpression promotes the proliferation of the cells, and can be found in several malignant cancer cells. Two potentially GQ-forming elements were identified in its central promoter region: KIT1 and KIT2. The KIT1 sequence (called thereafter simply as KIT) was studied here.
Vascular endothelial growth factor is very important for the starving tumor cells, since this promotes the growth of new blood vessels [33]. The sequence, which can potentially form GQ structure, can be found in the promoter region in the human VEGF gene. Since this is also a binding site for transcriptional factors, the stabilization of GQ form is a promising target for anticancer drugs.
In order to stabilize the GQs, we must know their structure and behavior in various environments. We have to know what kind of factors can influence their structure. It is important to characterize the effect of different environmental parameters (pH, temperature and pressure) and the presence of various ions on the structure and stability of GQs.
Our technical approach was to use fluorescence spectroscopy. We used Förster Resonance Energy Transfer (FRET) to follow the folding-unfolding of the GQ structure. For these experiments, the DNA oligos were labeled by two fluorophores at the ends. We used FAM and TAMRA as a FRET pair. All the experiments were performed in the presence of K+, which corresponds to the physiological case.
2. Materials and methods
The sequences of the oligos we used are listed in Table 1.
Table 1.
Sequence of the oligos used in this study.
| Oligo name | Sequence (5′-3′) | Number of bases |
|---|---|---|
| c-MYC | TGA GGG TGG GTA GGG TGG GTA A | 22 |
| KIT | AGG GAG GGC GCT GGG AGG AGG G | 22 |
| VEGF | TTG GGG CGG GCC GGG GGC GGG GTT | 24 |
All the oligos were labeled by a FRET pair of FAM and TAMRA. The labeled oligos were purchased from Sigma-Aldrich Kft (Hungary) and from IDT (NY, USA). All other chemicals were purchased from Sigma-Aldrich.
The oligos were obtained from the manufacturers in lyophilized form. First they were dissolved in MilliQ water in a concentration of 100 μM, according to the suggestion of IDT. This stock solution was kept frozen, and diluted with an appropriate buffer during sample preparation. The final concentration of the oligos was 2 μM in K-phosphate buffer (100 mM pH 7.4) for the atmospheric pressure experiments. TRIS buffer (100 mM, pH 7.4) was used for the pressure experiments as it has low pressure dependence, although the GQs are not very pH-sensitive. Both solutions contained 0.1 mM EDTA. The diamond cell used in the pressure experiments had small volume (ca. 0.2 μl), therefore the oligos were used in slightly higher concentration (10 μM).
No annealing procedure was used in our experiments, because the presence of the fluorescence energy transfer confirmed the folded oligo state. Our control experiments show that the energy transfer even decreased after a heat cycle, indicating an incorrect refolding.
Fluorescent spectra were measured with a Fluorolog-FL3 fluorimeter (Horiba Jobin Yvon, France). A custom-made reflection mode diamond cell was adopted into the sample holder. The pressure was measured by recording the ruby fluorescence [19, 34]. A small ruby chip was placed into the pressure cell, and it was excited by a green HeNe laser (Coherent, USA). The emitted light was detected by a CCD camera (Andor, UK) attached to a THR1000 monochromator (Jobin Yvon). The temperature was measured in all the experimental setups by a thermocouple connected directly to the high pressure cell. An HH802U thermometer and the corresponding software from Omega were used to record the temperatures every 30 s (Omega, USA).
The transition temperature was determined by fitting the temperature dependence of spectral parameters by the following sigmoidal function [35]:
| (2) |
Here, y is the physical parameter to be fitted (e.g. fluorescence intensity, or ratio of fluorescence intensities at two different wavelengths), a and b are the parameters describing the linear dependence of y(T) below the transition, T is the thermodynamic temperature, Δa and Δb are the changes of a and b during the transition, ΔH is the enthalpy change, R is the universal gas constant, and Tm is the transition midpoint.
The Clausius-Clapeyron equation was used to calculate the volume changes taking place during the pressure experiments:
| (3) |
where ΔH is the enthalpy change at the transition, Tm is the transition temperature, dTm/dp is the shift of Tm caused by the pressure.
3. Results and discussion
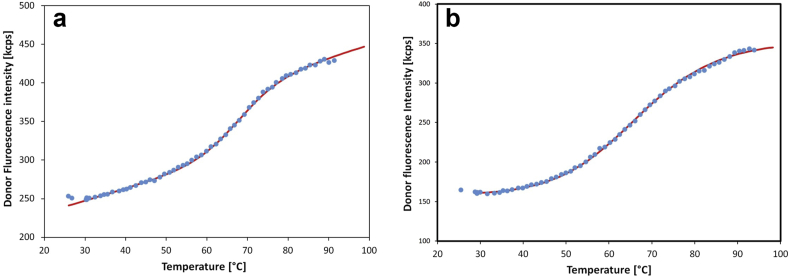
Figure 2a shows the fluorescence spectra of c-MYC measured at atmospheric pressure as function of temperature. At room temperature, the spectrum is dominated by the acceptor peak at around 580 nm, while the intensity of the donor fluorescence at 515 nm increases with the temperature. This means the loss of the energy transfer with increasing temperature. The reason behind the energy transfer decrease is the increasing donor-acceptor distance during the unfolding of the GQ structure. At higher temperatures, the DNA oligo forms a random chain, where the end-to end distance is well above the Förster distance. The Förster distance of the used FAM-TAMRA pair is 5.5 nm, while the contour length of the oligo consisting of 22 bases is above 7 nm. The temperature dependence of the donor fluorescence intensity can be seen in Figure 2b. A sigmoidal fit of Eq. (2) supposing a two-state transition describes the data quite well. This fit gives a transition temperature of 84.1 °C. Similar series of experiments were performed for KIT and VEGF oligos. The transition temperatures are listed in Table 2. As can be seen, the most stable oligo is the c-MYC, which has a parallel structure [32]. This quadruplex is stabilized by stacking interactions with an adenine base on the top and a TA pair at the bottom. Additionally, further stabilization is gained from the two hydrogen bonds between the TA base pairs.
Figure 2.
Fluorescence spectra of c-MYC (a) and fit of donor intensity by Eq. (2) (b).
Table 2.
Stability parameters of the oligos studied.
| Parameter | c-MYC | KIT | VEGF | unit |
|---|---|---|---|---|
| Tm at p = 1bar | 83.4 ± 1.3 | 58.5 ± 0.4 | 78.8 ± 1.1 | °C |
| 129 ± 14 | 119 ± 15 | 146 ± 36 | kJ/mol | |
| dT/dp | -4.66 ± 0.35 | -1.73 ± 0.11 | -4,35 ± 0.27 | °C/kbar |
| -16.9 ± 1.8 | -6.2 ± 0.9 | -18.1 ± 4.6 | cm3/mol |
Figure 3 shows the donor fluorescence intensity of c-MYC as function of the temperature at 2 and 5 kbar. A clear destabilization can be observed under high pressure. This clearly indicates that the volume change during the unfolding process is negative. Since the folded conformation has higher volume, pressure shifts the folded GQ <-> unfolded ssDNA oligo equilibrium into the direction of unfolded state. The volume difference can also be obtained by plotting the pressure dependence of the transition temperature. Figure 4 shows the pressure dependence of the unfolding temperatures of the studied oligos.
Figure 3.
Donor fluorescence intensity of c-MYC vs. temperature at 2 kbar (a) and 5 kbar (b) pressures.
Figure 4.
Pressure dependence of the unfolding temperature for oligos: c-MYC (a), KIT (b) VEGF (c).
Table 2 contains the calculated parameters from the phase diagram curves. The Tm values are calculated from the value of the fitted lines at p = 1bar. The ΔV values were calculated from the slope of the fitted line using the well-known Clausius-Clapeyron equation.
The quadruplexes are stabilized by hydrogen bonds, eight of them in each plane. All the GQs studied here contain three planes, which means that 24 hydrogen bonds stabilize the GQ structure. The obtained ΔH values are in the range of 120–150 kJ/mol, which means 10–12 kJ/(mol guanine base). Naturally, the stability of the GQ form is based not only on the hydrogen bonds, the central stabilizing metal ion and the stacking interaction between the quartets can also considerably contribute to its stability. Additionally, in some cases, the stacking of further bases at the ends can stabilize the structure, or even Watson-Crick base pairs can appear at the ends, like in the case of c-MYC.
Earlier pressure studies were performed on the 2 stage TBA and Htel. To our knowledge, there are no studies on other GQs. Macgregor's lab investigated the Htel oligomer (d[A(GGGTTA)3GGG]). The stabilizing central cation was Na+, which is known to have less stabilizing ability compared to K+. Their results for the ΔV were between -66 and -56 cm3/mol depending slightly on the NaCl concentration [36]. This experimental volume change was the result of the delicate balance of volume changes with different signs. A positive thermal volume change was compensated by negative contributions of change in molecular volume, interaction volume, and the volume change caused by the re-hydration of the sodium ion.
A later study, which included several loop mutants of Htel, resulted in slightly smaller molar volume changes with both sodium and potassium ions upon unfolding of the GQ [37]. Potassium-stabilized GQs had smaller volume, which corresponds well with the higher stability of the K+ containing GQs. -12 cm3/mol out of the -43 cm3/mol volume change was associated to the re-solvation of the potassium ions [37].
Sugimoto's group also investigated Htel with both Na+ and K+ ions, and the effect of PEG as molecular crowder. Their results show a marked pressure dependence of the transition temperature. This effect was lowered upon the effect of the crowding. Their dT/dp was -14.8 °C/kbar and -15.3 °C/kbar for the Na+ and K+ stabilized oligomers respectively [38]. This pressure sensitivity was reduced by more than three-fold in the presence of 40% PEG. The highest unfolding volumes found in these experiments were -83 and -92 cm3/mol. A similar strong destabilizing rate (-8.4 °C/kbar) was found in the case of TBA, which has only two G-quartets [39]. The volume change was calculated as -55 cm3/mol for TBA without crowding agents.
As we can see, the folding parameters found in our experiments were in the same region as the ones for Htel if we consider the temperature stability, but in case of volumetric aspects, we find marked differences. The unfolding volumes in our experiments were ca. -18 cm3/mol, except for KIT, which only had one-third of this volume. -18 cm3/mol is the volume of one water molecule, or it is equal to the hydration volume change associated to three of the K+ ions [40]. Since the ionic radius of a K+ ion does not allow it to fit between the guanine bases [6], the three stage GQs contain only two K+ ions, which are situated between the quartets. This way, the unfolding volume change cannot be explained solely by the re-hydration of the central ions.
Our earlier study on Htel showed a marked concentration dependence of the pressure sensitivity. In the concentration range comparable to the present study, -19 cm3/mol was found [15]. These were also FRET-based experiments. The present volumetric parameters fit well to the ones obtained on Htel by FRET.
The fact that the experimentally determined volumetric parameters of GQs are diverging underlines the delicate balance of opposite volumetric contributions, which govern the stability of these structural forms. One could argue that the presence of the chromophores could influence the obtained results. We consider this, however, quite unlikely. The unfolding volume change could only be influenced by the fluorophores if they interact with the GQ. Such an interaction would influence the unfolding temperature already at atmospheric pressure, by stabilizing the labeled molecule. This was, however, not observed in case of any of GQs studied here.
The reason for the diversity of the ΔV values needs to be revealed by further careful experiments. At the present state, we can only draw attention to the importance of the difference in the experimental conditions. Although GQs are quite pH insensitive, the pH values of some buffers used in the literature (e.g. phosphate buffer) are very pressure-dependent. On the other hand, the effect of the concentration of cations on the volume was not studied extensively either. In our earlier study on TBA, we found the stabilizing effect of increasing KCl concentration to be even well above the Kd of KCl (unpublished results). It has to be mentioned that the used experimental methods were diverging as well. Different experimental methods report about different details of the molecular changes. CD and UV absorption is sensitive to the mutual arrangement of the bases, while FRET is sensitive to the distance between the two ends of the oligo. The unfolding of a GQ structure can happen stepwise, which means that different experimental methods could report on certain steps of this process. This could be the reason behind the differences in the measured experimental values. Since FRET measures the unfolding of the whole GQ, we believe that our values characterize the complete unfolding process.
The exceptionally small magnitude of the unfolding volume of KIT can be explained by its unique structure. Figure 5 shows the 3D structures of the studied oligos schematically. They are based on the following PDB structures: 1XAV [32] (for c-MYC), 2M27 [42] (for VEGF) and 2O3M [41] (for KIT). The KIT structure is clearly a unique one, which contains four loops, while usually three of them are present in most of the GQs [41].
Figure 5.
Schematic representation of the 3D structures of the studied oligos. The schemes were drawn after the following PDB structures: c-MYC 1XAV [32] (a); KIT 2O3M [41] (b) and VEGF 2M27 [42] (c).
It was shown earlier that the high concentration of oligos can also change the volumetric aspects of the unfolding. This could be explained by the concentration-dependent shift of the competing conformations [15, 43], or by the possible intermolecular interactions and a consequent stacking of the GQs [44, 45].
Observing the 3D structures deposited in the PDB database, we can see that both c-MYC and VEGF exhibit a parallel structure with three loops, while KIT has several unique features. Besides the two single-residue chain reversal loops, there is a two-residue group and a five-residue stem-loop in the structure [41]. Interestingly, an isolated guanine is also involved in the formation of one of the G-tetrads, although the sequence contains four GGG tracts. This unique sequence, i.e. the presence of the extra loop might account for the small unfolding volume.
We also calculated the volume of the three GQ structures using the first three structures deposited in the PDB files [46]. It turned out that KIT contains considerably less cavities, which can also explain the lower transition volume. Cavities are present only in the folded GQ, in the unfolded case, the random coil does not enclose any void volume. Consequently, lower void volume can also contribute to the explanation of the lower unfolding volume of KIT GQ.
4. Conclusions
Unfolding volume changes of three GQ forming DNA sequences from different oncogene promoters were measured, using high pressure FRET. The volume of the GQ containing system will be reduced by 17, 6, and 18 cm3/mol in the unfolding of the GQ of c-MYC, KIT, and VEGF sequences. The exceptionally small volume change of the KIT sequence can be explained by its unique structure and smaller cavity volume. Volumetric parameters are important in the crowded environment of the cell, since the available volume is considerably restricted compared to a dilute solution. The determined parameters can be used as reference values in studies characterizing the GQ-drug interactions.
Declarations
Author contribution statement
O. Molnár: Performed the experiments; Analyzed and interpreted the data.
J. Somkuti: Analyzed and interpreted the data; Wrote the paper.
L. Smeller: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Hungarian Scientific Research Fund (K-124697).
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Sugimoto N. Noncanonical structures and their thermodynamics of DNA and RNA under molecular crowding: beyond the Watson-Crick double Helix. Int. Rev. Cell. Mol. Biol. 2014;307:205–273. doi: 10.1016/B978-0-12-800046-5.00008-4. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin Y., Hurley L.H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 5.Deng B., Lin Y., Wang C., Li F., Wang Z., Zhang H., Li X.-F., Le X.C. Aptamer binding assays for proteins: the thrombin example-A review. Anal. Chim. Acta. 2014;837:1–15. doi: 10.1016/j.aca.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya D., Arachchilage G.M., Basu S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016;4 doi: 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venczel E.A., Sen D. Parallel and antiparallel G-DNA structures from a complex telomeric sequence. Biochemistry. 1993;32:6220–6228. doi: 10.1021/bi00075a015. [DOI] [PubMed] [Google Scholar]
- 8.Kankia B.I., Marky L.A. Folding of the thrombin aptamer into a G-quadruplex with Sr2+: stability, heat, and hydration. J. Am. Chem. Soc. 2001;123:10799–10804. doi: 10.1021/ja010008o. [DOI] [PubMed] [Google Scholar]
- 9.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golub M., Martinez N., Michoud G., Ollivier J., Jebbar M., Oger P., Peters J. The effect of crowding on protein stability, rigidity, and high pressure sensitivity in whole cells. Langmuir. 2018;34:10419–10425. doi: 10.1021/acs.langmuir.8b01240. [DOI] [PubMed] [Google Scholar]
- 11.Knop J.M., Winter R. Effects of cosolvents and macromolecular crowding on the phase transitions and temperature-pressure stability of chiral and racemic poly-lysine. Z. Phys. Chemie-Int. J. Res. Phys. Chem. Chem. Phys. 2018;232:1111–1125. [Google Scholar]
- 12.Julius K., Al-Ayoubi S.R., Paulus M., Tolan M., Winter R. The effects of osmolytes and crowding on the pressure-induced dissociation and inactivation of dimeric LADH. Phys. Chem. Chem. Phys. 2018;20:7093–7104. doi: 10.1039/c7cp08242h. [DOI] [PubMed] [Google Scholar]
- 13.Dhar A., Samiotakis A., Ebbinghaus S., Nienhaus L., Homouz D., Gruebele M., Cheung M.S. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikaelsson T., Aden J., Johansson L.B.A., Wittung-Stafshede P. Direct observation of protein unfolded state compaction in the presence of macromolecular crowding. Biophys. J. 2013;104:694–704. doi: 10.1016/j.bpj.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somkuti J., Adanyi M., Smeller L. Self-crowding influences the temperature - pressure stability of the human telomere G-quadruplex. Biophys. Chem. 2019;254 doi: 10.1016/j.bpc.2019.106248. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi S., Sugimoto N. Effect of pressure on the stability of G-quadruplex DNA: thermodynamics under crowding conditions. Angew Chem. Int. Ed. Engl. 2013;52:13774–13778. doi: 10.1002/anie.201307714. [DOI] [PubMed] [Google Scholar]
- 17.Meersman F., McMillan P.F. High hydrostatic pressure: a probing tool and a necessary parameter in biophysical chemistry. Chem. Commun. 2014;50:766–775. doi: 10.1039/c3cc45844j. [DOI] [PubMed] [Google Scholar]
- 18.Meersman F., Heremans K. High pressure induces the formation of aggregation-prone states of proteins under reducing conditions. Biophys. Chem. 2003;104:297–304. doi: 10.1016/s0301-4622(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 19.Heremans K., Smeller L. Protein structure and dynamics at high pressure. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1998;1386:353–370. doi: 10.1016/s0167-4838(98)00102-2. [DOI] [PubMed] [Google Scholar]
- 20.Smeller L. Pressure-temperature phase diagrams of biomolecules Biochim. Biophys. Acta - Protein Struct. Molec. Enzymol. 2002;1595:11–29. doi: 10.1016/s0167-4838(01)00332-6. [DOI] [PubMed] [Google Scholar]
- 21.Cinar H., Cinar S., Chan H.S., Winter R. Pressure-induced dissolution and reentrant formation of condensed, liquid-liquid phase-separated elastomeric alpha-elastin. Chem. Eur J. 2018;24:8286–8291. doi: 10.1002/chem.201801643. [DOI] [PubMed] [Google Scholar]
- 22.Czeslik C., Luong T.Q., Winter R. Enzymatic activity under pressure. MRS Bull. 2017;42:738–742. [Google Scholar]
- 23.Cordeiro Y., Kraineva J., Winter R., Silva J.L. Volume and energy folding landscape of prion protein revealed by pressure. Braz. J. Med. Biol. Res. 2005;38:1195–1201. doi: 10.1590/s0100-879x2005000800006. [DOI] [PubMed] [Google Scholar]
- 24.Chalikian T.V., Macgregor R.B. Origins of pressure-induced protein transitions. J. Mol. Biol. 2009;394:834–842. doi: 10.1016/j.jmb.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Royer C., Winter R. Protein hydration and volumetric properties. Curr. Opin. Colloid Interface Sci. 2011;16:568–571. [Google Scholar]
- 26.Heremans K. Protein dynamics: hydration and cavities. Braz. J. Med. Biol. Res. 2005;38:1157–1165. doi: 10.1590/s0100-879x2005000800002. [DOI] [PubMed] [Google Scholar]
- 27.Li Y.Y., Dubins D.N., Le D., Leung K., Macgregor R.B., Jr. The role of loops and cation on the volume of unfolding of G-quadruplexes related to HTel. Biophys. Chem. 2017;231:55–63. doi: 10.1016/j.bpc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma D.L., Zhang Z.H., Wang M.D., Lu L.H., Zhong H.J., Leung C.H. Recent developments in G-quadruplex probes. Chem. Biol. 2015;22:812–828. doi: 10.1016/j.chembiol.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 30.Chauhan A., Paladhi S., Debnath M., Mandal S., Das R.N., Bhowmik S., Dash J. A small molecule peptidomimetic that binds to c-KIT1 G-quadruplex and exhibits antiproliferative properties in cancer cells. Bioorg. Med. Chem. 2014;22:4422–4429. doi: 10.1016/j.bmc.2014.05.060. [DOI] [PubMed] [Google Scholar]
- 31.Wei C.Y., Wang J.H., Zhang M.Y. Spectroscopic study on the binding of porphyrins to (G(4)T(4)G(4))4 parallel G-quadruplex. Biophys. Chem. 2010;148:51–55. doi: 10.1016/j.bpc.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Ambrus A., Chen D., Dai J.X., Jones R.A., Yang D.Z. Solution structure of the biologically relevant g-quadruplex element in the human c-MYC promoter. implications for g-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 33.Goodsell D.S. The molecular perspective: VEGF and angiogenesis. Stem Cell. 2003;21:118–119. doi: 10.1634/stemcells.21-1-118. [DOI] [PubMed] [Google Scholar]
- 34.Smeller L. 1999. On the Precision of Pressure Determination by Internal Calibrants; pp. 268–271. [Google Scholar]
- 35.Somkuti J., Houska M., Smeller L. Pressure and temperature stability of the main apple allergen Mal d1. Eur. Biophys. J. 2011;40:143–151. doi: 10.1007/s00249-010-0633-8. [DOI] [PubMed] [Google Scholar]
- 36.Fan H.Y., Shek Y.L., Amiri A., Dubins D.N., Heerklotz H., Macgregor R.B., Chalikian T.V. Volumetric characterization of sodium-induced G-quadruplex formation. J. Am. Chem. Soc. 2011;133:4518–4526. doi: 10.1021/ja110495c. [DOI] [PubMed] [Google Scholar]
- 37.Li Y.Y., Dubins D.N., Le D.M.N.T., Leung K., Macgregor R.B. The role of loops and cation on the volume of unfolding of G-quadruplexes related to HTel. Biophys. Chem. 2017;231:55–63. doi: 10.1016/j.bpc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi S., Bhowmik S., Sugimoto N. Volumetric analysis of formation of the complex of G-quadruplex DNA with hemin using high pressure. J. Inorg. Biochem. 2017;166:199–207. doi: 10.1016/j.jinorgbio.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi S., Sugimoto N. Effect of pressure on the stability of G-quadruplex DNA: thermodynamics under crowding conditions. Angew. Chem. Int. Ed. 2013;52:13774–13778. doi: 10.1002/anie.201307714. [DOI] [PubMed] [Google Scholar]
- 40.Marcus Y. The standard partial molar volumes of ions in solution. Part 4. Ionic volumes in water at 0-100 degrees C. J. Phys. Chem. B. 2009;113:10285–10291. doi: 10.1021/jp9027244. [DOI] [PubMed] [Google Scholar]
- 41.Phan A.T., Kuryavyi V., Burge S., Neidle S., Patel D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agrawal P., Hatzakis E., Guo K.X., Carver M., Yang D.Z. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013;41:10584–10592. doi: 10.1093/nar/gkt784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palacky J., Vorlickova M., Kejnovska I., Mojzes P. Polymorphism of human telomeric quadruplex structure controlled by DNA concentration: a Raman study. Nucleic Acids Res. 2013;41:1005–1016. doi: 10.1093/nar/gks1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abu-Ghazalah R.M., Rutledge S., Lau L.W.Y., Dubins D.N., Macgregor R.B., Helmy A.S. Concentration-dependent structural transitions of human telomeric DNA sequences. Biochemistry. 2012;51:7357–7366. doi: 10.1021/bi300689t. [DOI] [PubMed] [Google Scholar]
- 45.Li Y.Y., Abu-Ghazalah R., Zamiri B., Macgregor R.B. Concentration-dependent conformational changes in GQ-forming ODNs. Biophys. Chem. 2016;211:70–75. doi: 10.1016/j.bpc.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Chen C.R., Makhatadze G.I. ProteinVolume: calculating molecular van der Waals and void volumes in proteins. BMC Bioinf. 2015;16 doi: 10.1186/s12859-015-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]