Abstract
MYB (myeloblastosis) transcription factors (TFs) play important roles in controlling various physiological processes in plants, such as responses to biotic and abiotic stress, metabolism, and defense. A previous study identified a gene, Csa6G410090, encoding a plant lipid transfer protein (LTP), as a possible regulator in cucumber (Cucumis sativus L.) of the resistance response to root-knot nematode (RKN) [Meloidogyne incognita Kofoid and White (Chitwood)]. Myb-type DNA-binding TFs were presumed to regulate downstream genes expression, including LTPs, however, the regulation mechanism remained unclear. To elucidate whether and which MYB TFs may be involved in regulation of the resistance response, this study identified 112 genes as candidate members of the CsMYB gene family by combining CDD and SMART databases, using the Hidden Markov Model (HMM) and manual calibration. Within this group, ten phylogenetic subgroups were resolved according to sequence-based classification, consistent with results from comprehensive investigation of gene structure, conserved motifs, chromosome locations, and cis-element analysis. Distribution and collinearity analysis indicated that amplification of the CsMYB gene family in cucumber has occurred mainly through tandem repeat events. Spatial gene expression analysis showed that 8 CsMYB genes were highly expressed at differing levels in ten different tissues or organs. The roots of RKN-resistant and susceptible cucumbers were inoculated with M. incognita, finding that CsMYB (Csa6G538700, Csa1G021940, and Csa5G641610) genes showed up-regulation coincident with upregulation of the “hub” gene LTP (Csa6G410090) previously implicated as a major gene in the resistance response to RKN in cucumber. Results of this study suggest hypotheses regarding the elements and regulation of the resistant response as well as possible RKN resistance-enhancing strategies in cucumber and perhaps more broadly in plants.
Keywords: cucumber, resistant introgression line, MYB, root-knot nematode, resistant mechanism
Introduction
Myeloblastosis proteins, named for their discovery as transcription regulators in myeloblastosis, function in plants as widely distributed and functionally diverse plant genetic transcription factors (TFs). In plants, MYB TFs play crucial roles in regulating plant physiological and biochemical processes (e.g., Daniel et al., 1999; Mengiste et al., 2003,Zhang et al., 2012; Sara et al., 2018). The MYB domain, consisting of a region of about 52 amino acids, bind DNA in a sequence-specific manner (Jin and Martin, 1999). The MYB gene family can be divided into four classes according to the number and type of MYB domains known as 1RMYB (or MYB-related), 2RMYB, 3RMYB, and 4RMYB, respectively. The 2RMYB class is specific to plants and the largest (Dubos et al., 2010), having been annotated genome-wide in many plants. Previous studies showed that there are 190 members of the 2RMYB family in Arabidopsis (Riechmann et al., 2000), 102 members in Oryza sativa (Chen et al., 2006), 117 members in Vitis vinifera (Matus et al., 2008), 192 members in Populus trichocarpa (Wilkins et al., 2009), 244 members in Glycine max (Du et al., 2012), and more than 200 members in Zea mays (Dias et al., 2003). Only 55 2RMYB genes, however, have been identified in the most recent version of cucumber genome sequence (Cucumis sativus L.), the smallest number identified in any plant species to date, owing to the absence of recent gene duplication events (Li et al., 2012).
Functions of these MYB TFs have been explored in many studies on plant defense and response to various biotic and abiotic stresses (Raffaele et al., 2008; Cominelli and Tonelli, 2009; Zhang et al., 2019; Wang H. H. et al., 2020). For example, BOS1 in Arabidopsis encodes a 2RMYB TF that regulates anther development and also responses to biotic stresses, such as biotrophic pathogens (Ogata et al., 1994,Mengiste et al., 2003). PbrMYB5 plays an active role in enhancing tolerance to chilling stresses in tobacco (Nicotiana tabacum) (Xing et al., 2019). AtMYB60 and AtMYB96 in Arabidopsis are required for ABA signaling, stomatal movement, drought stress and disease resistance (Cominelli et al., 2005; Seo et al., 2009, 2011; Seo and Park, 2010). In tomato (Solanum lycopersicum L.), R3-MYB inhibits anthocyanin production (Colanero et al., 2018). In kiwifruit (Actinidia chinensis), MYB7 regulates transcriptional activation of metabolic pathway genes to modulate carotenoid and chlorophyll pigment accumulation (Ampomah-Dwamena et al., 2019). McMYB10 is involved in anthocyanin biosynthesis in crabapple during continuous light treatments (Li et al., 2018). In cotton, GhMYB7, GhMYB9, and GhMYB109 were involved in cotton fiber initial cells, as well as elongating fibers (Suo et al., 2003).
Although new insights regarding the mechanisms by which MYB proteins control various plant processes and several core genes have been identified, the TFs in question are usually encoded by very large multigene families confounding detailed mechanistic studies, consistent with the likelihood that many of the complex regulatory roles played by MYB genes-encoding-proteins remain unknown (Riechmann et al., 2000). Our previous comparative transcriptomic study showed that MYB TFs may participate in regulating the hub gene LTP (lipid transfer protein) of cucumber in resistance to root-knot nematode (RKN) [Meloidogyne incognita Kofoid and White (Chitwood)] (Wang et al., 2018). LTP, a member of plant pathogenesis-related protein (PRs), is involved in lipid transport in giant cells (GCs) membranes and receives signals stimulated by exposure to nematodes (Wang X. et al., 2020). These results from cucumber are consistent with results from Arabidopsis, where LTP3 is positively regulated by the TF MYB96 to mediate freezing and drought stress (Guo et al., 2013). However, the mechanism by which MYB TF(s) regulates LTP in cucumber is still unknown. In the present study, we carried out a systematic analysis of CsMYB genes in cucumber, showing that this family members are highly variable and apparently play a positive role in responding to M. incognita in cucumber. Our results provide more evidence regarding whether and how CsMYB genes regulate LTP, thereby potentially affecting resistance to RKN in cucumber.
Materials and Methods
Identification of Cucumber MYB Gene Family
The cucumber whole genome sequence was downloaded from the cucumber genome database1. The MYB binding domains (PF00249) in the sequence were identified and confirmed using the Hidden Markov Model (HMM) profile from PFam database2 applied as a query to previously annotated cucumber MYB-containing sequences, further identified by HMMSEARCH and manual screened using default parameter (E ≤ 1.2 × 10–8). HMMRESEARCH was applied to establish the cucumber MYB (CsMYB) gene family-specific model (Lozano et al., 2015), which was visualized online by SKylign3. Then all predicted cucumber MYB proteins with an E-value below 0.001 were selected using the cucumber MYB-specific HMM. The candidate genes for the CsMYB family were identified according to cucumber-specific MYB HMM.
All the candidate CsMYB genes initially obtained above were uploaded to BlastP by CDD (Conserved Domain Database) (Marchler-Bauer et al., 2017) and SMART (Simple Modular Architecture Research Tool) (Marchler-Bauer et al., 2015; Letunic and Bork, 2018) using the default parameter E ≤ 0.01. Four classes (1RMYB, 2RMYB, 3RMYB, and 4RMYB) of CsMYB genes were separated according to characteristic conserved domains within the MYB sequence.
All the predicted protein sequences of the candidate CsMYB family members were submitted to ExPASy4 to calculate the number of amino acids (aa), molecular weight (Mw) and isoelectric point (pI). The candidate CsMYB genes were renamed according to the repeat number of conserved domains and the physical position on the chromosome. The subcellular localization analysis of the candidate CsMYB family protein sequences were carried out by CELLO v.2.5 programme (subCELlular LOcalization predictor) at Molecular Bioinformatics Center5 (Yu et al., 2004, 2006).
Gene Structure and Analysis of Conserved Motifs Among Cucumber MYB Gene Family Members
The exon and intron structures of the candidate CsMYB genes were obtained by comparison with the corresponding full-length genome sequences. Conserved motifs in the gene family were determined using the following parameters: arbitrary number of repetitions, up to 10; length, 6–200 amino acid residues via MEME software and predicted protein sequence (Bailey et al., 2009; Letunic et al., 2015). The gene structures and conserved motifs of CsMYB genes were displayed by TBTools (Chen et al., 2020).
Sequence Alignment and Phylogenetic Analysis of CsMYB
The sequences of candidate CsMYB proteins obtained were used to create Clustal V multiple sequence alignment using MEGA7 with AtMYBs in Arabidopsis thaliana. MEGA7 was used to analyze the phylogeny and evolution of all the predicted protein sequences by using the adjacency method (neighbor-joining, NJ) criterion. According to the results of the alignment, all the nodes were analyzed by bootstrap under the parameters, No. of Differences, Uniform rates, Complete deletion, and then repeateds 1000 times. The maximum likelihood (maximum likelihood, ML) method was used to verify the phylogenetic tree. Besides, phylogenetic tree between cucumber and other species was constructed based on MYB gene sequences from Gcorn database6.
Orthologous Analysis and Chromosome Localization
The candidate CsMYB genes were located on cucumber chromosomes by Mapchart (Voorrips, 2002). The duplication events within the gene family were analyzed by Multiple Collinearity Scan toolkit (MCScanX) (Wang et al., 2012) and Circos (Krzywinski et al., 2009). In order to reveal the homology of MYB gene between cucumber and Arabidopsis thaliana, the homology analysis of MYB gene between Arabidopsis thaliana and cucumber was constructed by using the Dual Synteny Plotter software.
Cis-Elements Analysis and Prediction of CsMYB
The upstream sequence (1.5 kb) of CsMYB coding sequences were extracted from the cucumber genome sequence and submitted to PlantCARE7 (Magali et al., 2002) for identification of cis-elements. The Gene Structure Display Server (GSDS)8 (Li et al., 2012; Hu et al., 2014) is used to visualize all the regulatory elements involved in defense and stress response or defense factors, and to further analyze the regulatory elements related to resistance defense.
Response to Meloidogyne incognita
In order to study changes in gene expression correlated with expression of resistance to Meloidogyne incognita in cucumber, gene expression in a RKN-resistant line, IL10-1, and RKN-susceptible line, “Beijingjietou” (CC3), that display marked differences in against M. incognita (Ye et al., 2011), were obtained directly from NCBI by SRP125669 (Wang et al., 2018) at 0, 1, 2 and 3 days after inoculation (dpi) with approximately 50 fresh and motile second-stage juveniles (J2) M. incognita. The readings per kilobyte million enzyme (RPKM) for each MYB gene were downloaded and calculated from NCBI9. The RPKM values were converted through log2 transformation value (Li et al., 2011), and the cucumber genome database HEATMAP was used to draw the heatmap online.
RNA Isolation and RT-qPCR Analyses
Seeds of IL10-1 and CC3, provided by the State Key Lab of Cucurbit Genetics and Germplasm Enhancement of Nanjing Agricultural College, were surface sterilized (70% ethanol for 15 s and 1% sodium hypochlorite solution for 10 min, then rinsing with distilled water three times). Sterilized seeds were germinated on wet filter paper in a growth chamber at 28°C. After 3–4 days, seedlings were transplanted onto pluronic F-127 (Sigma-Aldrich, United States) gel to assess RKN infectivity (Wang et al., 2009), under controlled condition (28°C/16 h light, 24°C/8 h dark).
Inoculated root samples were harvested at 0, 1, 2, and 3 dpi as follows. Root tips from five different seedlings were mixed into a single biological sample, then 3 replicates were made from each sample. Total RNA from cucumber roots was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States). Extracted RNA was quantified by Thermo NanoDrop 2000 (Thermo Fisher Scientific, MA, United States). cDNA was synthesized from the total RNA used for RNA-seq using the PrimeScript RT reagent Kit (Takara, Japan), and then qPCR, repeated three times, was performed on Bio-Rad CFX96 thermocycler using the 2 × SYBR green PCR master mix (Applied Biosystems). Seven DEGs were selected to verify the accuracy of the transcriptome results by qPCR. A housekeeping gene Csa6M484600 encoding ACTIN was used as the internal control (Wilson et al., 2015). The primers for qPCR listed in Supplementary Table S3 were designed using Primer 6 software. The relative expression of each gene was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). Each expression profile was independently verified in three replicated experiments performed under identical conditions. Primer sequences used are listed in Supplementary Table S3.
Tissue-Specific Expression
In order to study the gene-specific expression of CsMYB genes in different tissues of cucumber, the serial number PRJNA80169 was used from NCBI to obtain cucumber RNA samples from different tissues and organs (leaves, stems, female flowers, male flowers, enlarged ovary (unfertilized), dilated ovary (fertilized), ovary, root, RNA-Seq data for tendrils and the base of tendrils (Harris et al., 2008).
Results
Identification of the Cucumber CsMYB Gene Family
A cucumber-specific model (Supplementary Figure S1) was built for typical MYB or MYB-like domains using the HMM profile (PF00249) of the MYB-conserved domain. A total of 197 CsMYB genes were selected from the cucumber genome database10. Combining the CDD and SMART databases using manual calibration, 215 genes were identified for the CsMYB gene family, further divided into the four subclasses according to the number of repeats of the conserved protein domain. Predicted CsMYB proteins (listed in Table 1) showed considerable redundancy, e.g., 33, 67, and 3 redundant proteins in the group MYB-related, R2R3-MYB, and R1R2R3-MYB, respectively. The number of unique proteins is 112, corresponding to the actual number of 112 genes.
TABLE 1.
The characteristics of CsMYB -encoded proteins in cucumber.
| Rename | gene ID | Chr | length | MV (Da) | pI | Predicted location | Classification |
| Cs1RMYB-1 | Csa1G004120 | 1 | 89 | 10203.8 | 9.08 | Extracellular | MYB_related |
| Cs1RMYB-2 | Csa1G024160 | 1 | 248 | 27448.6 | 4.89 | Nuclear | MYB_related |
| Cs1RMYB-3 | Csa1G033200 | 1 | 280 | 31195.2 | 5.57 | Nuclear | MYB_related |
| Cs1RMYB-4 | Csa1G042350 | 1 | 252 | 27974.9 | 5.29 | Nuclear | MYB_related |
| Cs1RMYB-5 | Csa1G071840 | 1 | 313 | 34927.7 | 5.17 | Nuclear | MYB_related |
| Cs1RMYB-6 | Csa1G420880 | 1 | 299 | 33745.2 | 5.5 | Nuclear | MYB_related |
| Cs1RMYB-7 | Csa1G488750 | 1 | 103 | 11966.7 | 10.01 | Chloroplast | MYB_related |
| Cs1RMYB-8 | Csa2G007440 | 2 | 164 | 18840.4 | 4.34 | Nuclear | MYB_related |
| Cs1RMYB-9 | Csa2G035350 | 2 | 305 | 34400.3 | 7.44 | Nuclear | MYB_related |
| Cs1RMYB-10 | Csa2G049890 | 2 | 117 | 13463.8 | 9.42 | Plasma Membrane | MYB_related |
| Cs1RMYB-11 | Csa2G169770 | 2 | 160 | 18460.9 | 6.6 | Cytoplasmic | MYB_related |
| Cs1RMYB-12 | Csa2G229940 | 2 | 240 | 28644.2 | 10.61 | Nuclear | MYB_related |
| Cs1RMYB-13 | Csa2G229950 | 2 | 62 | 6881.8 | 8.55 | Nuclear | MYB_related |
| Cs1RMYB-14 | Csa2G352940 | 2 | 90 | 10047.6 | 9.89 | Nuclear | MYB_related |
| Cs1RMYB-15 | Csa2G370470 | 2 | 660 | 70029.6 | 8.11 | Nuclear | MYB_related |
| Cs1RMYB-16 | Csa3G045200 | 3 | 59 | 6782.7 | 9.69 | Nuclear | MYB_related |
| Cs1RMYB-17 | Csa3G122510 | 3 | 95 | 10392.6 | 5.2 | Nuclear | MYB_related |
| Cs1RMYB-18 | Csa3G134010 | 3 | 109 | 12886.9 | 10.84 | Mitochondrial | MYB_related |
| Cs1RMYB-19 | Csa3G135050 | 3 | 100 | 11579.5 | 10.09 | Nuclear | MYB_related |
| Cs1RMYB-20 | Csa3G165690 | 3 | 93 | 10737.2 | 9.34 | Nuclear | MYB_related |
| Cs1RMYB-21 | Csa3G740800 | 3 | 116 | 13176.1 | 9.07 | Nuclear | MYB_related |
| Cs1RMYB-22 | Csa3G742860 | 3 | 205 | 24306.1 | 6.94 | Nuclear | MYB_related |
| Cs1RMYB-23 | Csa3G828990 | 3 | 109 | 12185.8 | 9.32 | Nuclear | MYB_related |
| Cs1RMYB-24 | Csa3G848130 | 3 | 280 | 30712.4 | 9.85 | Nuclear | MYB_related |
| Cs1RMYB-25 | Csa3G891670 | 3 | 386 | 42250.2 | 8.67 | Nuclear | MYB_related |
| Cs1RMYB-26 | Csa4G000700 | 4 | 329 | 36786.4 | 8.62 | Cytoplasmic | MYB_related |
| Cs1RMYB-27 | Csa4G290190 | 4 | 674 | 76072.4 | 10.06 | Nuclear | MYB_related |
| Cs1RMYB-28 | Csa4G645320 | 4 | 51 | 5651.51 | 8.54 | Nuclear | MYB_related |
| Cs1RMYB-29 | Csa5G171690 | 5 | 109 | 12436.6 | 9.67 | Mitochondrial | MYB_related |
| Cs1RMYB-30 | Csa5G524690 | 5 | 158 | 17462.2 | 4.87 | Nuclear | MYB_related |
| Cs1RMYB-31 | Csa5G524720 | 5 | 83 | 9198.4 | 5.54 | Plasma Membrane | MYB_related |
| Cs1RMYB-32 | Csa6G105150 | 6 | 68 | 8080.3 | 11.56 | Nuclear | MYB_related |
| Cs1RMYB-33 | Csa6G136580 | 6 | 275 | 31613.5 | 4.44 | Nuclear | MYB_related |
| Cs1RMYB-34 | Csa6G303240 | 6 | 191 | 22007.6 | 4.85 | Nuclear | MYB_related |
| Cs1RMYB-35 | Csa6G311520 | 6 | 107 | 12124.7 | 9.75 | Nuclear | MYB_related |
| Cs1RMYB-36 | Csa6G495710 | 6 | 307 | 33757.5 | 10.25 | Cytoplasmic | MYB_related |
| Cs1RMYB-37 | Csa6G496960 | 6 | 56 | 6084 | 8.75 | Cytoplasmic | MYB_related |
| Cs1RMYB-38 | Csa6G499890 | 6 | 315 | 35014.2 | 6.03 | Nuclear | MYB_related |
| Cs1RMYB-39 | Csa7G179630 | 7 | 559 | 63088.6 | 6.91 | Nuclear | MYB_related |
| Cs2RMYB-1 | Csa1G004900 | 1 | 472 | 54883.7 | 6.3 | Nuclear | R2R3-MYB |
| Cs2RMYB-2 | Csa1G008430 | 1 | 368 | 41942.7 | 6.19 | Nuclear | R2R3-MYB |
| Cs2RMYB-3 | Csa1G009700 | 1 | 332 | 37603.3 | 9.14 | Nuclear | R2R3-MYB |
| Cs2RMYB-4 | Csa1G021940 | 1 | 308 | 33313 | 7.41 | Nuclear | R2R3-MYB |
| Cs2RMYB-5 | Csa1G029570 | 1 | 233 | 26589.2 | 8.84 | Nuclear | R2R3-MYB |
| Cs2RMYB-6 | Csa1G046820 | 1 | 378 | 42093.5 | 6.92 | Nuclear | R2R3-MYB |
| Cs2RMYB-7 | Csa1G109320 | 1 | 319 | 36229 | 8.2 | Nuclear | R2R3-MYB |
| Cs2RMYB-8 | Csa1G529120 | 1 | 432 | 48469.4 | 5.67 | Nuclear | R2R3-MYB |
| Cs2RMYB-9 | Csa1G561370 | 1 | 342 | 37757.4 | 5.94 | Nuclear | R2R3-MYB |
| Cs2RMYB-10 | Csa1G575180 | 1 | 249 | 27776.7 | 6.73 | Nuclear | R2R3-MYB |
| Cs2RMYB-11 | Csa1G682630 | 1 | 517 | 59414.2 | 6.97 | Nuclear | R2R3-MYB |
| Cs2RMYB-12 | Csa2G100550 | 2 | 265 | 29982.6 | 8.85 | Nuclear | R2R3-MYB |
| Cs2RMYB-13 | Csa2G174670 | 2 | 108 | 12650.5 | 10.2 | Nuclear | R2R3-MYB |
| Cs2RMYB-14 | Csa2G270220 | 2 | 232 | 26160.6 | 7.61 | Nuclear | R2R3-MYB |
| Cs2RMYB-15 | Csa2G302300 | 2 | 211 | 23544.3 | 10.95 | Nuclear | R2R3-MYB |
| Cs2RMYB-16 | Csa2G352410 | 2 | 205 | 23037 | 8.29 | Nuclear | R2R3-MYB |
| Cs2RMYB-17 | Csa2G355030 | 2 | 291 | 31804.4 | 9.57 | Nuclear | R2R3-MYB |
| Cs2RMYB-18 | Csa2G360620 | 2 | 243 | 27199.1 | 9.27 | Nuclear | R2R3-MYB |
| Cs2RMYB-19 | Csa2G427310 | 2 | 302 | 32296.8 | 8.13 | Nuclear | R2R3-MYB |
| Cs2RMYB-20 | Csa2G439230 | 2 | 300 | 33891.8 | 6.37 | Nuclear | R2R3-MYB |
| Cs2RMYB-21 | Csa3G076520 | 3 | 520 | 57033.1 | 7.78 | Nuclear | R2R3-MYB |
| Cs2RMYB-22 | Csa3G168940 | 3 | 306 | 33693.1 | 5.69 | Nuclear | R2R3-MYB |
| Cs2RMYB-23 | Csa3G182040 | 3 | 300 | 34029.5 | 10.12 | Nuclear | R2R3-MYB |
| Cs2RMYB-24 | Csa3G199590 | 3 | 220 | 25251.4 | 6.78 | Nuclear | R2R3-MYB |
| Cs2RMYB-25 | Csa3G264750 | 3 | 355 | 40501.9 | 9.6 | Nuclear | R2R3-MYB |
| Cs2RMYB-26 | Csa3G303630 | 3 | 268 | 30096.4 | 5.81 | Nuclear | R2R3-MYB |
| Cs2RMYB-27 | Csa3G386830 | 3 | 170 | 19348.4 | 4.48 | Nuclear | R2R3-MYB |
| Cs2RMYB-28 | Csa3G535090 | 3 | 225 | 25603.6 | 7.86 | Nuclear | R2R3-MYB |
| Cs2RMYB-29 | Csa3G592130 | 3 | 220 | 24887.1 | 6.61 | Nuclear | R2R3-MYB |
| Cs2RMYB-30 | Csa3G732480 | 3 | 269 | 30010.8 | 9.76 | Extracellular | R2R3-MYB |
| Cs2RMYB-31 | Csa3G812750 | 3 | 349 | 39131.2 | 9.07 | Nuclear | R2R3-MYB |
| Cs2RMYB-32 | Csa3G816030 | 3 | 295 | 32662.3 | 6.53 | Nuclear | R2R3-MYB |
| Cs2RMYB-33 | Csa3G824850 | 3 | 234 | 26130.2 | 9.77 | Nuclear | R2R3-MYB |
| Cs2RMYB-34 | Csa3G826690 | 3 | 314 | 35125.9 | 6.35 | Nuclear | R2R3-MYB |
| Cs2RMYB-35 | Csa3G901090 | 3 | 105 | 12050.7 | 8.76 | Nuclear | R2R3-MYB |
| Cs2RMYB-36 | Csa3G914600 | 3 | 254 | 28517.6 | 9.38 | Nuclear | R2R3-MYB |
| Cs2RMYB-37 | Csa4G022940 | 4 | 315 | 36057.3 | 6.51 | Nuclear | R2R3-MYB |
| Cs2RMYB-38 | Csa4G305350 | 4 | 378 | 40500 | 5.31 | Nuclear | R2R3-MYB |
| Cs2RMYB-39 | Csa4G308500 | 4 | 300 | 34358.2 | 6.81 | Nuclear | R2R3-MYB |
| Cs2RMYB-40 | Csa4G638510 | 4 | 230 | 25192.6 | 9.56 | Nuclear | R2R3-MYB |
| Cs2RMYB-41 | Csa4G641690 | 4 | 264 | 29140.4 | 8.35 | Nuclear | R2R3-MYB |
| Cs2RMYB-42 | Csa5G148680 | 5 | 261 | 28837.7 | 4.74 | Nuclear | R2R3-MYB |
| Cs2RMYB-43 | Csa5G152790 | 5 | 324 | 36414.6 | 6.44 | Nuclear | R2R3-MYB |
| Cs2RMYB-44 | Csa5G160120 | 5 | 259 | 29457.7 | 6.17 | Nuclear | R2R3-MYB |
| Cs2RMYB-45 | Csa5G198240 | 5 | 296 | 33799.6 | 8.84 | Mitochondrial | R2R3-MYB |
| Cs2RMYB-46 | Csa5G209490 | 5 | 285 | 31234.3 | 6.88 | Nuclear | R2R3-MYB |
| Cs2RMYB-47 | Csa5G353650 | 5 | 296 | 31215.5 | 6.92 | Nuclear | R2R3-MYB |
| Cs2RMYB-48 | Csa5G425400 | 5 | 301 | 33770.6 | 5.11 | Nuclear | R2R3-MYB |
| Cs2RMYB-49 | Csa5G579040 | 5 | 287 | 30876.7 | 6.17 | Nuclear | R2R3-MYB |
| Cs2RMYB-50 | Csa5G604410 | 5 | 465 | 52285.8 | 7.27 | Nuclear | R2R3-MYB |
| Cs2RMYB-51 | Csa5G605730 | 5 | 370 | 41397.2 | 7.08 | Nuclear | R2R3-MYB |
| Cs2RMYB-52 | Csa5G637660 | 5 | 399 | 44517.7 | 6.99 | Nuclear | R2R3-MYB |
| Cs2RMYB-53 | Csa5G641610 | 5 | 231 | 27272.2 | 9.77 | Nuclear | R2R3-MYB |
| Cs2RMYB-54 | Csa5G651640 | 5 | 295 | 33129 | 8.51 | Nuclear | R2R3-MYB |
| Cs2RMYB-55 | Csa6G040640 | 6 | 287 | 32771 | 6.87 | Nuclear | R2R3-MYB |
| Cs2RMYB-56 | Csa6G046240 | 6 | 234 | 26439.4 | 8.87 | Nuclear | R2R3-MYB |
| Cs2RMYB-57 | Csa6G046460 | 6 | 149 | 16821.1 | 7.68 | Cytoplasmic | R2R3-MYB |
| Cs2RMYB-58 | Csa6G121970 | 6 | 270 | 31333.9 | 6.92 | Nuclear | R2R3-MYB |
| Cs2RMYB-59 | Csa6G187960 | 6 | 316 | 35118.2 | 9.91 | Nuclear | R2R3-MYB |
| Cs2RMYB-60 | Csa6G344210 | 6 | 310 | 35826.2 | 9.56 | Nuclear | R2R3-MYB |
| Cs2RMYB-61 | Csa6G491690 | 6 | 302 | 32449.4 | 9.71 | Nuclear | R2R3-MYB |
| Cs2RMYB-62 | Csa6G495660 | 6 | 240 | 26841.1 | 10.28 | Nuclear | R2R3-MYB |
| Cs2RMYB-63 | Csa6G538700 | 6 | 290 | 33192 | 6.95 | Nuclear | R2R3-MYB |
| Cs2RMYB-64 | Csa7G043580 | 7 | 501 | 54622.8 | 6.31 | Nuclear | R2R3-MYB |
| Cs2RMYB-65 | Csa7G045590 | 7 | 255 | 29446.4 | 5.21 | Nuclear | R2R3-MYB |
| Cs2RMYB-66 | Csa7G046120 | 7 | 338 | 37544.7 | 6.39 | Nuclear | R2R3-MYB |
| Cs2RMYB-67 | Csa7G375780 | 7 | 255 | 29393.9 | 4.78 | Nuclear | R2R3-MYB |
| Cs2RMYB-68 | Csa7G413890 | 7 | 277 | 31884.7 | 4.97 | Nuclear | R2R3-MYB |
| Cs2RMYB-69 | Csa7G431360 | 7 | 1503 | 169045 | 4.87 | Nuclear | R2R3-MYB |
| Cs3RMYB-1 | Csa2G375240 | 2 | 1000 | 111544 | 5.54 | Nuclear | R1R2R3-MYB |
| Cs3RMYB-2 | Csa3G734230 | 3 | 551 | 61163.7 | 8.53 | Nuclear | R1R2R3-MYB |
| Cs3RMYB-3 | Csa3G889150 | 3 | 961 | 105806 | 5.3 | Nuclear | R1R2R3-MYB |
| Cs4RMYB | Csa3G113280 | 3 | 1005 | 113684 | 7.06 | Nuclear | 4RMYB |
Predicted physicochemical properties of the CsMYB gene family proteins, including molecular weight (MW), isoelectric point (pI) and length (aa), were analyzed through ExPASy online. CsMYB genes encoded proteins ranging from 56 to 1503 aa in length, pI values ranging from 4.34 to 11.56 and molecular weights from 6084 to 169044.6 Da. Protein subcellular location was predicted through CELLO program (v.2.5). It was found that the vast majority genes (89.72%) encoded proteins that were predicted to be intranuclear, while others were predicted to be located in mitochondria, chloroplasts, cytoplasm, and the plasma membrane (Table 1). Members of the CsMYB gene family were then renamed according to their classification (subclasses 1–4) and their chromosome location (Cs1RMYB-1 to Cs1RMYB-39, Cs2RMYB-1 to Cs2RMYB-69, Cs3RMYB-1 to Cs3RMYB-3, and Cs4RMYB).
Evolutionary Relationships, Gene Structure and Conserved Motif Analyses of the CsMYB Gene Family in Cucumber
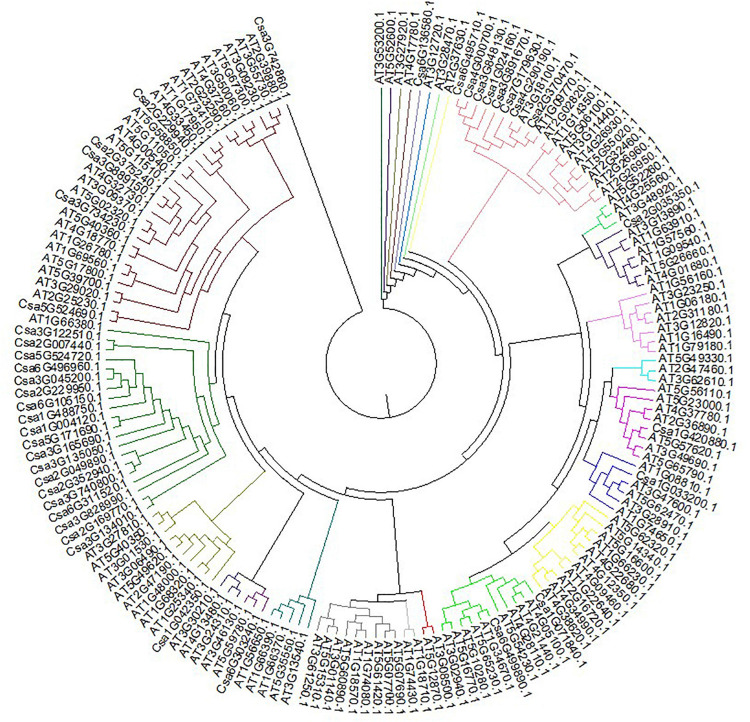
Clustal V was applied to generate multiple sequence alignment of the conserved structural domains of all cucumber CsMYB genes (Figure 1). Results were used to construct a phylogenetic tree (Figure 2), illustrating distribution and extent of conserved structural elements (Figure 3, Supplementary Table S1, and Supplementary Figure S2). As shown in Figure 3, the number of exons in CsMYB gene family members varied from 1 to 19 (18 with one exon, 34 with two exons, 41 with three exons, 6 with four exons, 1 with five exons, 3 with six exons, 3 with seven exons, 3 with eleven exons, 1 with twelve exons, 1 with thirteen exons and 1 with nineteen exons). Intron-exon structure of CsMYB genes was also analyzed after exton identification (Supplementary Figure S4). The results showed that exon 1 (133 bp) and exon 2 (130 bp) appeared to be more consistent in length, while exon 3 was more variable (31–850 bp). Phylogenetic analysis of the predicted CsMYB protein sequences revealed differences between cucumber and Arabidopsis, with eight sub-groups in common. Two cucumber subgroups were missing from Arabidopsis; ten sub-groups from Arabidopsis were absent in the cucumber genome, indicating CsMYB genes may evolve or be lost in a given taxon, following divergence. Some cucumber CsMYB proteins were clustered into Arabidopsis functional clades, which provided an excellent reference to explore the putative functions of the cucumber CsMYB genes. The comparative evolutionary analysis between Arabidopsis and cucumber suggests both species are derived from a common ancestor, no longer extant, and represent independent paths defined by duplication, divergence and loss of gene family members (Figure 4).
FIGURE 1.
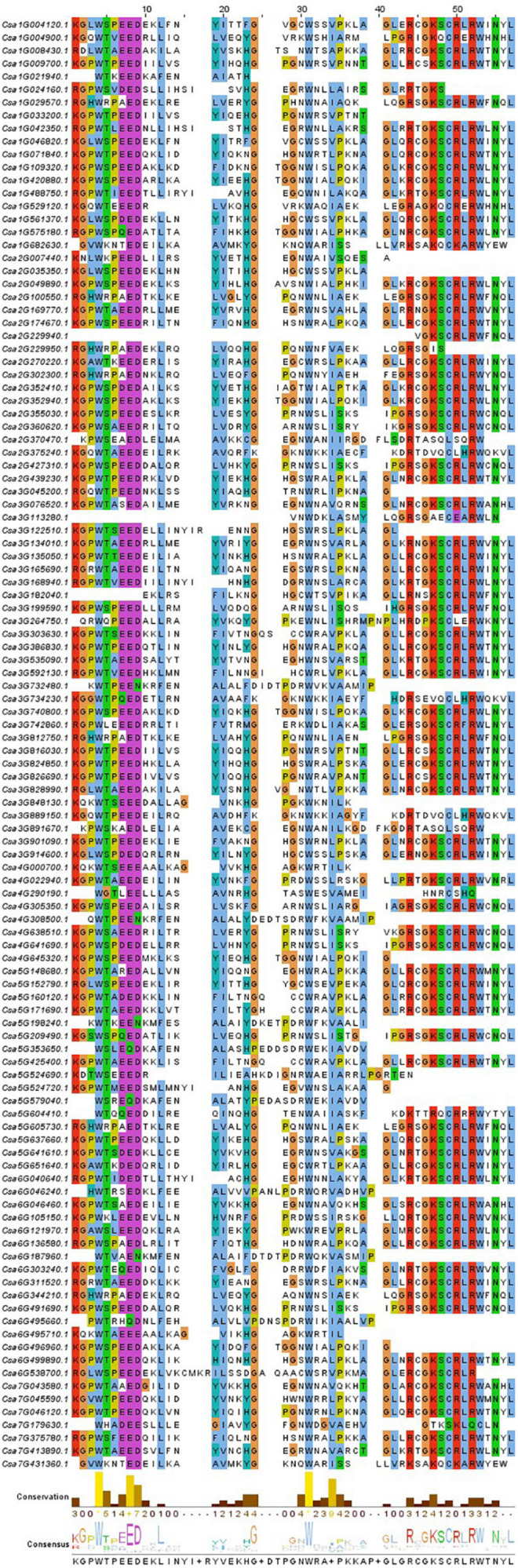
Alignment, degree of conservation and consensus of the conserved domain of CsMYB genes. The sequence logos of the cucumber special model was built based on full-length alignments of the MYB-conserved domain. The overall height of each stack indicates the conservation of the sequence at that position, whereas the height of letters within each stack represents the relative frequency of the corresponding amino acid.
FIGURE 2.
Phylogenetic relationships between CsMYB genes in cucumber and AtMYB genes in Arabidopsis. CsMYB proteins in were divided into ten groups based on sequence similarity grouped by color.
FIGURE 3.
Conserved protein motifs and gene structure of CsMYB genes in cucumber. The left column shows conserved motifs in all the proteins encoded by CsMYB gene. There are 10 different color boxes with numbers 1–10 indicted 10 subclasses defined by conserved motifs. The right column shows exons, introns, and UTRs (untranslated regions) structures with yellow boxes, single lines and green boxes, respectively. Introns phases 0, 1 and 2 are indicated by numbers 0, 1 and 2, respectively. The length of each CsMYB gene can be estimated using the scale at the bottom.
FIGURE 4.
Phylogenetic tree for cucumber and other species based on MYB gene sequences from Gcorn database.
The obvious redundancy of CsMYB gene at the level of amino acid sequence is partly due to the similar molecular functions of CsMYB gene, but different biological phenotypes can still be displayed by these loci when mutated, due to different temporal or spatial expression characteristics. Studies have shown that homologous members of this gene family are not redundant in terms of plant development (Jin and Martin, 1999).
Distribution and Collinearity Analysis of CsMYB Genes in Cucumber
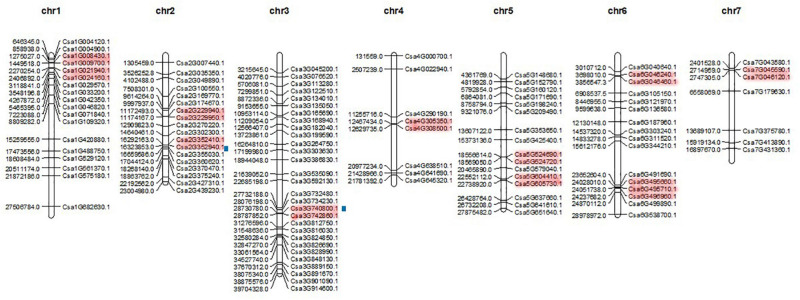
Members of the CsMYB gene family were mapped to all seven chromosomes with Mapchart (Figure 5). Results showed that chromosome 3 had the largest number of gene family members, 29 genes, while chromosome 7 had the fewest number of genes, containing only seven. The other five chromosomes 1, 2, 4, 5 and 6 each contained 18, 18, 8, 16, and 16 gene family members, respectively. The chromosome distribution of CsMYB gene family members is non-regular and does not correlate with overall chromosome length.
FIGURE 5.
The chromosomal distribution of CsMYB genes positions. Chromosomal positions of all the CsMYB genes are indicated by the full name of the genes (all the genes assigned in Table 1). The red color regions in chromosomes indicate tandem duplication; Blue squares along chromosome indicate segmental duplication.
To further analyze divergence of CsMYB gene family members in cucumber, the tandem repeat events and fragment repeat events of the gene family were analyzed by using KAKS_Calculator2.0. In this analysis, if two or more genes are present within 200 kb, the elements are considered a tandem repeat event. In total, 23 CsMYB genes were involved in 11 tandem repeats as follows: (Csa1G008430.1 & Csa1G009700.1, Csa1G021940.1 & Csa1G024160.1, Csa2G229940.1 & Csa2G229950.1, Csa2G352410.1 & Csa2G352940.1, Csa3G740800.1 & Csa3G742860.1, Csa4G305350.1 & Csa4G308500.1, Csa5G524690.1 & Csa5G524720.1, Csa5G604410.1 & Csa5G605730.1, Csa6G046240.1 & Csa6G046460.1, Csa6G495660.1 & Csa6G495710.1 & Csa6G496960.1, Csa7G045590.1 & Csa7G046120.1). Gene family fragment duplication events were determined by BLASTP, which showed that only one pair of genes (Csa3G740800.1, Csa2G352940.1) had experienced fragment duplication. Based on the results above, it is inferred that tandem repeat events play an important role in the amplification of gene families rather than duplication of gene fragments.
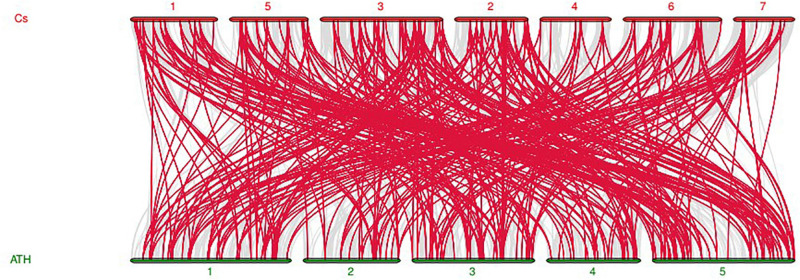
In order to uncover the linkage relationships of CsMYB genes throughout the cucumber genome, collinearity analysis was carried out between cucumber and Arabidopsis thaliana MYB genes (Figure 6) showing that high homology of the MYB gene family existed between the two species. Twenty-five pairs of genes showed collinearity and conserved linkage relationships (Supplementary Figure S3).
FIGURE 6.
Synteny analysis of MYB genes between cucumber and Arabidopsis thaliana. The red lines indicate MYB collinear genes, and the gray indicated the collinear genes of both species. Cs represents cucumber, while ATH represents Arabidopsis thaliana.
Analysis of cis-Elements of CsMYB Genes in Cucumber
In order to study associated cis-elements of CsMYB promoters, the sequences (1.5 kb) upstream of CsMYB coding sequence were selected from cucumber genome, and then were submitted to PlantCARE for further identification of regulatory elements. Using the online site Genetic Structure Display Server (GSDS), visualization all regulatory elements was conducted involving TF binding sites in defense and stress responses or defense responses (Figure 7). Three genes, Csa6G538700, Csa1G021940, and Csa5G641610 which had shown higher expression level in responses of cucumber to M. incognita, showed specific differences in the upstream cis-elements associated with the homolog in question. All three genes show associated upstream sequence that contains the cis-element G-box (TACGTG) and cis-element ABRE (ACGTG). The former sequence is associated with light-regulated gene expression; the latter is involved in regulation of gene expression by abscisic acid.
FIGURE 7.

Analysis of cis-elements in the upstream regions of CsMYB genes. Predicted cis-elements in CsMYB promoters. Promoter sequences (–1500 bp) of 112 CsMYB genes were analysis by PlantCARE for further identification of regulatory elements. The upstream length to the translation start site can be inferred according to the scale at the bottom.
Expression Analysis of MYB Gene Detected From Transcriptome
In order to analyze tissue-specific expression of the CsMYB gene family in cucumber, expression levels of gene family members were assayed in 10 tissues and organs obtained by PRJNA80169 (Figure 8). Results showed that most of the genes were expressed in all the different tissues sampled, with similar level of expression between tissues. Some genes, such as Csa6G495710, Csa3G848130, Csa2G355030, Csa4G308500, Csa5G198240, Csa5G579040, Csa6G491690, and Csa5G353650, were highly expressed in all 10 tissues and organs. Expression of Csa3G076520, Csa2G229940, Csa2G229950, Csa1G529120, and Csa1G008430 were low in almost all tissues. Csa6G303240, Csa5G524720, Csa5G160120, Csa6G136580, Csa3G168940, Csa6G496960, Csa2G007440, Csa3G824850, Csa1G029570, Csa1G488750, Csa2G169770, Csa5G152790, Csa5G605730, Csa3G740800, Csa6G344210, Csa6G495660, Csa2G049890, Csa6G499890, Csa6G121970, Csa3G134010, Csa1G420880, Csa6G538700, Csa4G645320, and Csa2G3522410 (21.4%) were only highly expressed in the root, showing lower levels of expression in other tissues and organs.
FIGURE 8.
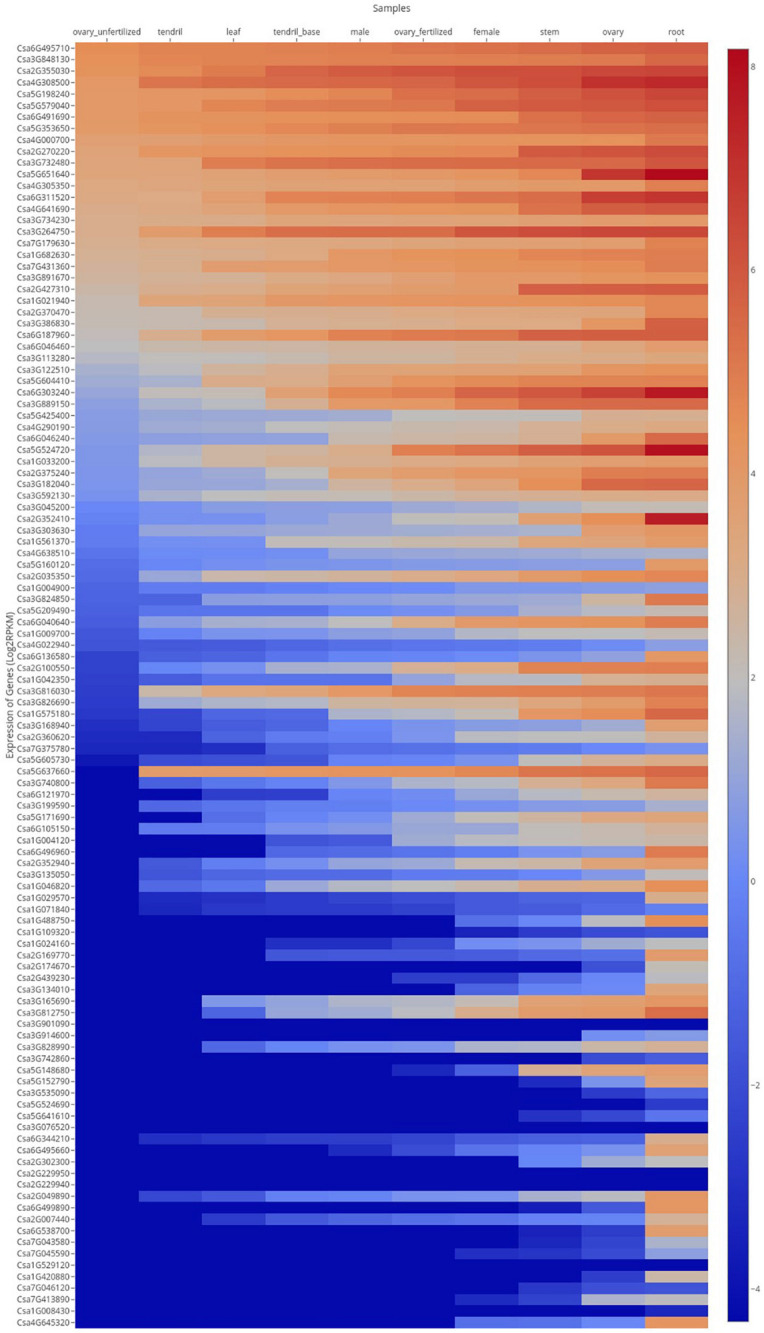
Expression profiles of CsMYB genes in different tissues or organs. FPKM values of CsMYB genes were transformed by log2 and a heatmap was constructed using the cucumber genome database HEATMAP to draw the heatmap online. Red color indicates a high level. Blue color indicates a low level of gene expression.
The post-RKN-inoculation transcriptome data extracted from resistant IL10-1 and susceptible CC3 lines were acquired using SRP125669 to ascertain whether members of the CsMYB gene family were subject to altered regulation during susceptible or resistant response to RKN (Figure 9). Six genes were randomly picked to validate the RNA-seq data. Additionally, to gain more insight into the role of CsMYB genes in the response to RKN in cucumber, relative real-time qPCR was performed to evaluate the transcript abundance of these CsMYB genes previously identified as differentially regulated during the resistant response to M. incognita (Supplementary Figure S5), which showed the similar expression trends with their RNA-seq data, respectively.
FIGURE 9.
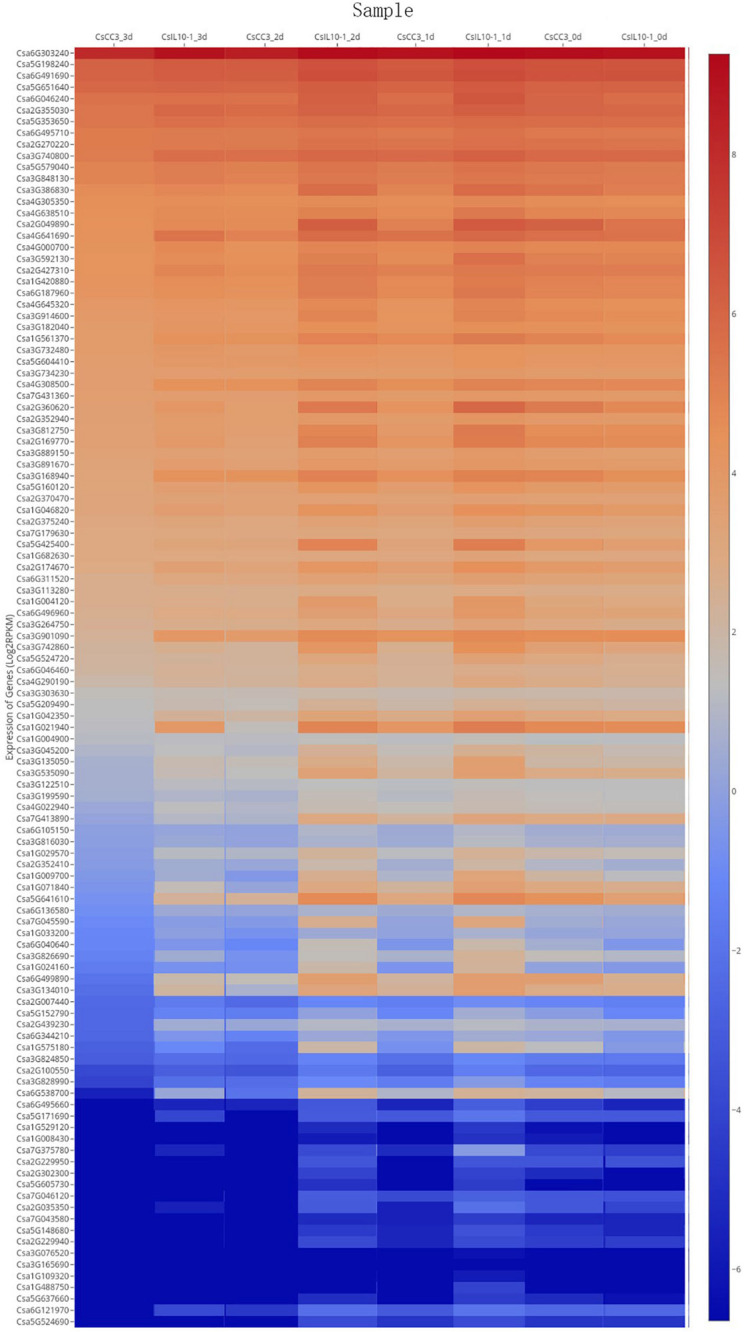
Expression profile of CsMYB genes through time after infection with M. incognita in resistant and susceptible cucumber lines, IL10-1 and CC3. Expression profile of CsMYB genes in response to the inoculation of M. incognita on published data RNA-seq SRP125669 (Wang et al., 2018). The color scale represents relative gene expression levels.
The homolog Csa6G303240 was highly expressed in both resistant and susceptible cucumber genotypes, but at different time points, suggesting that expression of this gene was not specifically correlated with resistance to RKN in cucumber. Further comparative analysis of the transcriptomes from RKN-resistant and susceptible genotypes revealed that the MYB gene family member, Csa5G641610, shown to affect expression of the core gene in resistance to RKN, LTP, gradually decreased in CC3, while in IL10-1, transcript levels initially increased, then decreased. The expression level of Csa5G641610 was analyzed, similar to the expression patterns of Csa1G021940, and Csa6G538700. Thus, it is speculated that Csa6G538700, Csa1G021940, and Csa5G641610 may regulate gene LTP to some extent, further affecting resistance to RKN in cucumber.
Discussion
Organization and Characterization of the Cucumber CsMYB Gene Family
The MYB family is the largest TF gene family in plants and has been shown to play an important role in secondary metabolite formation, sexual differentiation, cell differentiation, plant morphogenesis, and biotic and abiotic stress (Jung et al., 2003; Hsu et al., 2005; Guo et al., 2013; Li et al., 2018). The MYB TFs are ubiquitous to all plant species and have been surveyed in several species with sequenced genomes, such as Arabidopsis (Riechmann et al., 2000), grape (Matus et al., 2008), poplar (Wilkins et al., 2009), soybean (Du et al., 2012), maize (Dias et al., 2003). Additionally, 55 members of the cucumber R2R3MYB gene family were identified and characterized from annotated genes in the 26682 cucumber genome (Li et al., 2012). In the current study, 112 CsMYB genes were identified in the last and previous versions of the genome assembly of the cucumber “Chinese Long” (v3). Compared to the draft genome sequence of Cucumis sativus var. sativus L. used by Li et al. (2012), the reference genome sequence has been improved, and therefore, our 112 CsMYB genes family, included those 55 R2R3MYB genes in which 36 genes show one to one correspondence (Supplementary Table S2).
The 112 CsMYB genes family was consist of 39 1RMYB, 69 R2R3-MYB, 3 R1R2R3-MYB and 1 4RMYB according to the number and type of MYB domains. All the details of the gene structure, conserved motifs, and cis-element were analyzed and illustrated. Structural analysis found that the lengths of exons 1 and 2 are very conserved, similar to previous results reported for cucumber R2R3MYB work (Li et al., 2012). However, in our study, exon 1 and 2 were not as highly conserved, perhaps because we investigated all the MYB genes instead of just the subclass, R2R3MYB, and the other subclasses of MYB genes increased the variation in structural conservation. All 112 CsMYB genes have been located on seven chromosomes with uneven distribution. The largest number of homologs are found chromosome 3 and the smallest number on chromosome 7 (Figure 5). The locations are generally consistent with the more limited results in a previous study (Li et al., 2012). It is interesting to note that after the correspondence was identified between the published MYB gene (R2R3-MYB_v1) (Li et al., 2012) and CsMYB_v2 (Supplementary Table S2), two inversions were found in chromosome 5 and 7, (Csa 5G148680.1, Csa5G152790.1 and Csa5G160120.1 on chr.5, Csa7G046120.1 and Csa7G045590.1 on chr.7), compared with the chromosomal locations of Li et al. (2012). These chromosomal inversions might due to the differences between the improved cucumber genome and the old version released in 2009, which been reported (Lou et al., 2013).
Although whole genome duplication events occur commonly in many angiosperms, especially the recent gene duplications which are crucial for the rapid expansion and evolution of gene families (Cannon et al., 2004; Taylor and Raes, 2004), whole-genome duplication events and tandem duplications are rare in cucumber genome (Huang et al., 2009). In this study of the CsMYB gene family, tandem repeat events were detected more frequently than duplication of gene fragments in further analysis of the divergence of CsMYB gene family members, thus duplication may be the main driving force in the amplification of gene families.
Expression Analysis of CsMYB Genes Expressed in Response to M. incognita
Studies show that selection of favorable interactions between stress tolerant genes and their TFs can be an effective method to improve the comprehensive stress tolerance (Sun et al., 2010). The MYBS1 gene of Arabidopsis thaliana is involved in salt stress of plants in response to various plant hormones (such as auxin, gibberellin, jasmonic acid, salicylic acid). In cucumber, the TF, Csa1G021940, is the homolog of MYBS1, and also responds to various phytohormones, suggesting that, similar to its homolog in Arabidopsis, this gene could condition a response to biological stress in cucumber, such as RKN infection (Gutierrez et al., 2009; Molinari et al., 2014; Zhao et al., 2018). In Arabidopsis, the MYB67TF (cucumber homolog, Csa6G538700) is involved in cell differentiation (Gheysen and Mitchum, 2011), thus in cucumber, perhaps the homolog may also affect the differentiation of giant cells which would hinders the development of M. incognita, thereby causing resistance to M. incognita in IL10-1. Finally, in Arabidopsis, AT3G30210.1 (cucumber homolog, Csa5G641610) is a TF that responds to abscisic acid (ABA) which in cucumber, could inhibit cell elongation, affecting giant cell elongation, thereby enhancing the resistance to RKN. The results from our previous study of physiological observations of abnormal development of giant cells (GCs) between IL10-1 (resistant line) and CC3 (susceptible line) indicated that a series of reactions may occurred after the infection of RKN in the resistant line. Also, multi-omics study showed high levels of expression of the LTP gene which has been identified as the “hub gene” involved in lipid transport in GCs membranes and receives signals stimulated by nematodes (Wang et al., 2019).
In this study, the expression data for all 112 CsMYB genes in the two lines, IL10-1 and CC3, contrasting for RKN resistance were obtained directly from NCBI by SRP125669 (Wang et al., 2018) at 0, 1, 2 and 3 days after inoculation (dpi) with M. incognita. Further comparative analysis of the transcriptomes from IL10-1 (RKN-resistant) and CC3 (RKN-susceptible) revealed that three genes (Csa5G641610, Csa1G021940, and Csa6G538700) in CsMYB gene family, showed the same expression pattern where they initially increased, then decreased in IL10-1, but only decreased through time in the susceptible line, CC3. It further affected the expression of LTP, which assumed to be the core gene in response to RKN (Wang et al., 2018). In our previous comparative study on transcriptional events, combining with the physiological responses that occurs in resistant line IL10-1 and susceptible line CC3 during M. incognita infection, potential effector-targeted host genes were identified, while their regulation network was illuminated in the IL10-1 against M. incognita. Through the construction of a co-expression network, 2 out of 8 genes with the highest edges, considered the core genes of the network and assumed to have consequential roles in the resistant response against M. incognita perhaps as a result of abnormal development of GCs, were related to plant lipid transfer proteins (LTPs). One of the two most significantly induced LTP genes, which had the most edges, was considered as the hub gene in the resistance to RKN (Wang et al., 2018). More and more studies have indicated that LTPs are associated with plant resistance and defense (Thoma et al., 1994; Jung et al., 2003), for instance, LTPs can inhibit α-amylase, thereby contributing to resistance to biotic stress in wheat (Oda et al., 1997). However, reports on the relationships between LTPs and plant nematodes are still limited. As a result, further studies are necessary to elucidate the exact role of LTPs in plant resistance to nematodes. Studies in Arabidopsis show that the Myb-type DNA-binding transcriptional factor, MYB96, positively regulated the downstream gene LTP3 to mediate freezing and drought stress (Guo et al., 2013). Whether there is a correlation between MYB TFs and LTP genes during plant-pathogen interaction process remains unknown.
Conclusion
In summary, this paper carried out the basic bioinformatics analysis including the characterization, the phylogenetic analysis and evolutionary analysis of the 112 CsMYB genes newly identified in cucumber. Additionally, comparative analysis of the transcriptomes discovered three CsMYB genes (Csa5G641610, Csa1G021940, and Csa6G538700) might positively regulate predicted hub gene LTP (Csa6G410090) in resistance to RKN in cucumber. The exact relationship between these three MYB TFs and the LTP gene during the plant-pathogen interactions, as well as the mechanisms how CsMYB genes regulate LTP to work will require further study.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at NCBI under the accession PRJNA80169, https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA80169.
Author Contributions
JC and MJ designed and managed the project. QL, JL, and XW handled the samples and performed the experiments. QL and CC performed the data analyses and wrote the manuscript. All authors reviewed and approved this submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Wan Hongjian (Associate researcher of Institute of Vegetable, Zhejiang Academy of Agricultural Sciences) for his insightful comments and pertinent suggestions to improve earlier version of the manuscript. We also thank Professor Wei Lihui (Researcher of Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences) for providing the origin source of southern root-knot nematode.
Funding. This work was supported by the Key Project of Natural Science Foundation of China (Nos. 31902007 and 31672168), National Key Research and Development Program of China (2018YFD1000804), the Fundamental Research Funds for the Central Universities of China (KJQN202030), and Jiangsu Agricultural Innovation of New Cultivars Innovation and application of cucumber cultivars with high quality and multi disease resistances (No. PZCZ201719).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.550677/full#supplementary-material
Special Hidden Markov Model (HMM) of CsMYB gene in cucumber. The special HMM of CsMYB gene in cucumber was built according to the MYB HMM (PF00249) from PFam with the parameter (E ≤ 1.2 × 10–8). Then the logo of special model was visualizated online by SKylign.
Sequence logo of conserved Motif 1 to Motif 10.
Circos diagram of CsMYB genes in cucumber. Twenty-five pairs of CsMYB genes showed collinearity and linkage relationships in cucumber.
Exon length distribution analysis of the cucumber MYB genes. (A) Exon length values were analyzed using Boxplot. Each box represents the exon size range in which 50% of the values for a particular exon are grouped. The mean value is shown as a dotted line and the median as a continuous line. The number of exons in CsMYB gene family members varied from 1 to 19. (B) First, second and third exon lengths distribution of CsMYB genes using 3D Scatter Plot.
Relative RT-qPCR assay of six CsMYB family genes randomly picked from RNA-seq data in CC3 and IL10-1 at 0, 1, 2, 3 days post inoculation with M. incognita. (A–F) represented Cs1RMYB-15, Cs2RMYB-7, Cs2RMYB-4, Cs2RMYB-65, Cs1RMYB-18, and Cs2RMYB-63, respectively. The default expression value for each gene was Actin gene. The default expression value for each gene was from 0 dpi plants. Error bars indicate the standard deviation from the mean (three replicates).
The sequence and characterize of motif.
Correspondence between the published MYB gene (R2R3-MYB_v1) in cucumber (Li et al., 2012) and identified genes (CsMYB_v2) in cucumber.
Primers Sequencs of the three random picked genes.
References
- Ampomah-Dwamena C., Thrimawithana A. H., Dejnoprat S., Lewis D., Espley R. V. (2019). A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 221 309–325. 10.1111/nph.15362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucl. Acids Res. 37 202–208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D., May G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. 10.1186/1471-2229-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y. H., et al. (2020). TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang X. Y., He K., Liu M. H., Li J. G., Gao Z. F., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60 107–124. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- Colanero S., Perata P., Gonzali S. (2018). The atroviolacea gene encodes an R3-MYB protein repressig anthocyanin syntheis in tomato plants. Front. Plant Sci. 9:830. 10.3389/fpls.2018.00830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E., Tonelli C. (2009). A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res. 19, 1231–1232. 10.1038/cr.2009.123 [DOI] [PubMed] [Google Scholar]
- Cominelli E., Galbiati M., Vavasseur A., Conti L., Sala T., Vuylsteke M., et al. (2005). A Guard-Cell-Specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15 1196–1200. 10.1016/j.cub.2005.05.048 [DOI] [PubMed] [Google Scholar]
- Daniel X., Lacomme C., Morel J. B., Roby D. (1999). A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 20 57–66. 10.1046/j.1365-313x.1999.00578.x [DOI] [PubMed] [Google Scholar]
- Dias A. P., Braun E. L., McMullen M. D., Grotewold E. (2003). Recently Duplicated Maize R2R3 Myb Genes Provide Evidence for Distinct Mechanisms of Evolutionary Divergence after Duplication. Plant Physiol. 131 610–620. 10.1104/pp.012047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Yang S. S., Feng B. R., Liu L., Huang Y. B., Tang Y. X. (2012). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12:106. 10.1186/1471-2229-12-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Gheysen G., Mitchum M. G. (2011). How nematodes manipulate plant development pathways for infection. Curr. Opin. Plant Biol. 14 415–421. 10.1016/j.pbi.2011.03.012 [DOI] [PubMed] [Google Scholar]
- Guo L., Yang H., Zhang X., Yang S. H. (2013). Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J Exp. Bot. 64 1755–1767. 10.1093/jxb/ert040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez O. A., Wubben M. J., Howard M., Roberts B., Hanlon E., Wilkinson J. R. (2009). The role of phytohormones ethylene and auxin in plant-nematode interactions. Russ. J Plant Physiol. 56 1–5. 10.1134/s1021443709010014 [DOI] [Google Scholar]
- Harris T. D., Buzby P. R., Babcock H., Beer E., Bowers J., Braslavsky I., et al. (2008). Single-molecule DNA sequencing of a viral genome. Science 320, 106–109. [DOI] [PubMed] [Google Scholar]
- Hsu C. Y., Jenkins J. N., Saha S., Ma D. P. (2005). Transcriptional regulation of the lipid transfer protein gene LTP3 in cotton fibers by a novel MYB protein. Plant Sci. 168 167–181. 10.1016/j.plantsci.2004.07.033 [DOI] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Gao G. (2014). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. W., Li R. Q., Zhang Z. H., Li L., Gu X. F., Fan W., et al. (2009). The genome of the cucumber. Cucumis sativus L. Nat Genet. 475 1–7. [DOI] [PubMed] [Google Scholar]
- Jin H., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41 577–585. 10.1023/A:1006319732410 [DOI] [PubMed] [Google Scholar]
- Jung H. W., Kim W., Hwang B. K. (2003). Three pathogen-inducible genes encoding lipid transfer protein from pepper are differentially activated by pathogens, abiotic, and environmental stresses. Plant Cell Environ. 26:915. 10.1046/j.1365-3040.2003.01024.x [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. T., Zhang J., Kang Y. H., Chen C. M., Song T. T., Geng H., et al. (2018). McMYB10 modulates the expression of a Ubiquitin Ligase. McCOP1 during leaf coloration in crabapple. Front. Plant Sci. 9 704–717. 10.3389/fpls.2018.00704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhang C., Li J., Wang L. N., Ren Z. H. (2012). Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS One. 7:e47576. 10.1371/journal.pone.0047576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang Z. H., Yan P. C., Huang S. W., Lin K. (2011). RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genomics. 12 540–550. 10.1186/1471-2164-12-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lou Q. F., He Y. H., Cheng C. Y., Zhang Z. H., Li J., Huang S. W., et al. (2013). Integration of high-resolution physical and genetic map reveals differential recombination frequency between chromosomes and the genome assembling quality in cucumber. PLoS One. 8:e62676. 10.1371/journal.pone.0062676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R., Hamblin M. T., Prochnik S., Jannink J. (2015). Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics 16:360 10.1186/s12864-015-1554-1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magali L., Patrice D., Gert T., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 30 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L. Y., He J., Lanczycki C. J., Lu S. N., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucl. Acids Res. 45 200–203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S. N., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucl. Acids Res. 43 222–226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus J. T., Aquea F., Arce-Johnson P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 8:83. 10.1186/1471-2229-8-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 Gene Encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 15 2551–2565. 10.1105/tpc.014167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari S., Fanelli E., Leonetti P. (2014). Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 15 255–264. 10.1111/mpp.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Matsunaga T., Fukuyama K., Miyazaki T., Morimoto T. (1997). Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution. Biochemistry 36:13503. 10.1021/bi971307m [DOI] [PubMed] [Google Scholar]
- Ogata K., Morikawa S., Nakamura H., Sekikawa A., Inoue T., Kanai H., et al. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79 639–648. 10.1016/0092-8674(94)90549-5 [DOI] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767. 10.1105/tpc.107.054858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- Sara C., Pierdomenico P., Silvia G. (2018). The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 9 830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Lee S. B., Suh M. C., Park M. J., Go Y. S., Park C. M. (2011). The MYB96 Transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 23 1138–1152. 10.1105/tpc.111.083485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Park C. M. (2010). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186 471–483. 10.1111/j.1469-8137.2010.03183.x [DOI] [PubMed] [Google Scholar]
- Seo P. J., Xiang F., Qiao M., Park J. Y., Park C. M. (2009). The MYB96 Transcription Factor Mediates Abscisic Acid Signaling during Drought Stress Response in Arabidopsis. Plant Physiol. 151 275–289. 10.1104/pp.109.144220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. L., Shao W. C., Cai H., Zhu Y. M. (2010). Regulation of AtbZIP1 gene expression by t-dna insertion in Arabidopsis thaliana. J. North. Agricul. Unive. 17 19–23. [Google Scholar]
- Suo J., Liang X., Pu L., Zhang Y. S., Xue Y. B. (2003). Identification of GhMYB109 encoding a R2R3MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim. Biophys. Acta 1630 25–34. 10.1016/j.bbaexp.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Taylor J. S., Raes J. (2004). Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 38 615–643. 10.1146/annurev.genet.38.072902.092831 [DOI] [PubMed] [Google Scholar]
- Thoma S., Hecht U., Kippers A., Botella J., Vries S. D., Somerville C. (1994). Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol. 105:35. 10.1104/pp.105.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93 77–78. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- Wang C. L., Lower S., Williamson V. M. (2009). Application of Pluronic gel to the study of root-knot nematode behaviour. Nematology 11 453–464. 10.1163/156854109x447024 [DOI] [Google Scholar]
- Wang H. H., Wang X. Q., Yu C. Y., Wang C. T., Zhang H. X. (2020). MYB transcription factor PdMYB118 directly interacts with bHLH transcription factor PdTT8 to regulate wound-induced anthocyanin biosynthesis in poplar. BMC Plant Biol 20:173. 10.1186/s12870-020-02389-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cheng C. Y., Li Q. R., Zhang K. J., Chen J. F. (2019). Multi-omics analysis revealed that MAPK signaling and flavonoid metabolic pathway contributed to resistance against Meloidogyne incognita in the introgression line cucumber. J Proteomics. 220:103675. 10.1016/j.jprot.2020.103675 [DOI] [PubMed] [Google Scholar]
- Wang X., Cheng C. Y., Zhang K. J., Tian Z., Yang S. Q., Lou Q. F., et al. (2018). Comparative transcriptomics reveals suppressed expression of genes related to auxin and the cell cycle contributes to the resistance of cucumber against Meloidogyne incognita. BMC Genomics. 19 583–596. 10.1186/s12864-018-4979-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li Q. R., Cheng C. Y., Zhang K. J., Lou Q. F., Li J., et al. (2020). Genome-wide analysis of putative lipid transfer protein LTP_2 gene family reveals CsLTP_2 genes involved in response of cucumber against root-knot nematode (Meloidogyne incognita). Genome 63 225–238. 10.1139/gen-2019-0157 [DOI] [PubMed] [Google Scholar]
- Wang Y. P., Tang H. B., Debarry J. D., Tan X., Li J. P., Wang X. Y., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids Res. 40:e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N. J., Campbell M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149 981–993. 10.1104/pp.108.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H. T., Xu K., Taylor A. G. (2015). Transcriptome analysis of gelatin seed treatment as a biostimulant of cucumber plant growth. Sci. World J. 2015:391234. 10.1155/2015/391234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C. H., Liu Y., Zhao L. Y., Zhang S. L., Huang X. S. (2019). A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 42, 832–845. [DOI] [PubMed] [Google Scholar]
- Ye D. Y., Qian C. T., Chen J. F. (2011). Screening and identification of cucumber-sour cucumber introgression lines resistant to the root-knot nematode Meloidogyne incognita. Acta Horticult. Sin. 12:16. [Google Scholar]
- Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. (2006). Prediction of protein subcellular localization. Prot. Struct. Funct. Bioinform. 64 643–651. 10.1110/ps.03479604 [DOI] [PubMed] [Google Scholar]
- Yu C. S., Lin C. J., Hwang J. K. (2004). Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Prot. Sci. 13 1402–1406. 10.1110/ps.03479604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. C., Zhao G. Y., Jia J. Z., Liu X., Kong X. Y. (2012). Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 63 203–214. 10.1093/jxb/err264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. L., Zhang C. L., Wang G. L., Wang Y. X., Hao Y. J. (2019). The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 19:362. 10.1186/s12870-019-1918-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhou X., Lei H., Fan J., Yang R., Li Z., et al. (2018). Transcriptional evidence for crosstalk between JA and ET or SA during root-knot nematode invasion in tomato. Physiol. Genomics. 50 197–207. 10.1152/physiolgenomics.00079.2017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Special Hidden Markov Model (HMM) of CsMYB gene in cucumber. The special HMM of CsMYB gene in cucumber was built according to the MYB HMM (PF00249) from PFam with the parameter (E ≤ 1.2 × 10–8). Then the logo of special model was visualizated online by SKylign.
Sequence logo of conserved Motif 1 to Motif 10.
Circos diagram of CsMYB genes in cucumber. Twenty-five pairs of CsMYB genes showed collinearity and linkage relationships in cucumber.
Exon length distribution analysis of the cucumber MYB genes. (A) Exon length values were analyzed using Boxplot. Each box represents the exon size range in which 50% of the values for a particular exon are grouped. The mean value is shown as a dotted line and the median as a continuous line. The number of exons in CsMYB gene family members varied from 1 to 19. (B) First, second and third exon lengths distribution of CsMYB genes using 3D Scatter Plot.
Relative RT-qPCR assay of six CsMYB family genes randomly picked from RNA-seq data in CC3 and IL10-1 at 0, 1, 2, 3 days post inoculation with M. incognita. (A–F) represented Cs1RMYB-15, Cs2RMYB-7, Cs2RMYB-4, Cs2RMYB-65, Cs1RMYB-18, and Cs2RMYB-63, respectively. The default expression value for each gene was Actin gene. The default expression value for each gene was from 0 dpi plants. Error bars indicate the standard deviation from the mean (three replicates).
The sequence and characterize of motif.
Correspondence between the published MYB gene (R2R3-MYB_v1) in cucumber (Li et al., 2012) and identified genes (CsMYB_v2) in cucumber.
Primers Sequencs of the three random picked genes.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at NCBI under the accession PRJNA80169, https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA80169.