Abstract
Interferon‐induced transmembrane proteins (IFITMs) restrict infections by many viruses, but a subset of IFITMs enhance infections by specific coronaviruses through currently unknown mechanisms. We show that SARS‐CoV‐2 Spike‐pseudotyped virus and genuine SARS‐CoV‐2 infections are generally restricted by human and mouse IFITM1, IFITM2, and IFITM3, using gain‐ and loss‐of‐function approaches. Mechanistically, SARS‐CoV‐2 restriction occurred independently of IFITM3 S‐palmitoylation, indicating a restrictive capacity distinct from reported inhibition of other viruses. In contrast, the IFITM3 amphipathic helix and its amphipathic properties were required for virus restriction. Mutation of residues within the IFITM3 endocytosis‐promoting YxxФ motif converted human IFITM3 into an enhancer of SARS‐CoV‐2 infection, and cell‐to‐cell fusion assays confirmed the ability of endocytic mutants to enhance Spike‐mediated fusion with the plasma membrane. Overexpression of TMPRSS2, which increases plasma membrane fusion versus endosome fusion of SARS‐CoV‐2, attenuated IFITM3 restriction and converted amphipathic helix mutants into infection enhancers. In sum, we uncover new pro‐ and anti‐viral mechanisms of IFITM3, with clear distinctions drawn between enhancement of viral infection at the plasma membrane and amphipathicity‐based mechanisms used for endosomal SARS‐CoV‐2 restriction.
Keywords: COVID‐19, IFITM, IFITM3, interferon, SARS‐CoV‐2
Subject Categories: Microbiology, Virology & Host Pathogen Interaction
While IFITM‐family host‐cell antiviral factors generally restrict SARS‐CoV‐2 infection, human IFITM3 surprisingly exhibits both anti‐ and pro‐viral effects, inhibiting endocytic entry of the virus while also enhancing virus fusion at the plasma membrane.
Introduction
Severe acute respiratory syndrome (SARS) coronavirus (CoV)‐2 is the causative agent of the respiratory and multi‐organ‐associated disease known as COVID‐19 (Wu et al, 2020; Zhu et al, 2020). Understanding virus–host interactions and immune responses will be critical for explaining the high variation in severity of COVID‐19 observed in humans (Zhang et al, 2020a). Type I interferon (IFN) is a critical component of the innate immune system that induces expression of hundreds of genes, many of which encode proteins with anti‐viral activities that block specific steps in viral replication cycles (Schoggins et al, 2011; Chemudupati et al, 2019). Among these are genes that encode the IFN‐induced transmembrane proteins (IFITMs), including IFITM1, IFITM2, and IFITM3. IFITMs have been shown to block membrane fusion of diverse enveloped viruses by using an amphipathic helix domain to mechanically alter membrane lipid order and curvature in a manner that disfavors virus fusion (Li et al, 2013; Lin et al, 2013; Chesarino et al, 2017; preprint: Guo et al, 2020; Rahman et al, 2020). While IFITMs have been shown in most instances to inhibit CoV infections (Huang et al, 2011; Bertram et al, 2013; Wrensch et al, 2014; Zheng et al, 2020), in some cases they enhance infections (Zhao et al, 2014; Zhao et al, 2018). For example, IFITM2 and IFITM3 enhance cellular entry of human CoV‐OC43 (Zhao et al, 2014). Similarly, infections by virus‐like particles containing SARS‐CoV or MERS‐CoV glycoprotein (pseudovirus) are increased upon expression of an IFITM3 variant with enhanced plasma membrane localization versus endosomal localization (Zhao et al, 2018). Given that polymorphisms in the genes encoding IFITM2 and IFITM3 have been proposed to increase their plasma membrane localization (Everitt et al, 2012; Jia et al, 2014; Chesarino et al, 2014a; Wu et al, 2017), and that an additional polymorphism in the IFITM3 gene promoter results in significant loss of IFITM3 expression (Allen et al, 2017), determining whether IFITMs affect SARS‐CoV‐2 cellular infections should be informative for understanding susceptibility and resistance of human cells to infection.
Published experimental results are contradictory with regard to the effect of IFITMs on SARS‐CoV‐2 cell entry. SARS‐CoV‐2 Spike‐pseudotyped vesicular stomatitis virus infection was inhibited by human IFITM2 and IFITM3, but not IFITM1, in an overexpression screen of IFN effector proteins (Zang et al, 2020). In contrast, IFITM3 overexpression did not inhibit infection of Spike‐containing pseudovirus in a separate study (Zhao et al, 2020). IFITMs have also been reported to inhibit cell‐to‐cell fusion induced by SARS‐CoV‐2 Spike protein (Buchrieser et al, 2020), though a conflicting report did not observe inhibition of Spike‐mediated cell fusion when IFITM2 or IFITM3 was expressed in target cells (Zang et al, 2020). Overall, the impact of IFITMs on SARS‐CoV‐2 infections remains unclear and has yet to be examined with bona fide SARS‐CoV‐2. Herein, we measure the effects of mouse and human IFITMs on SARS‐CoV‐2 infections using both pseudovirus and genuine virus systems. We further utilize a series of mutants affecting critical functional amino acid motifs within IFITM3 and identify pro‐ and anti‐viral effects that we delineate in terms of distinct cellular locations and mechanisms of action.
Results
IFITMs inhibit SARS‐CoV‐2 Spike‐pseudotyped lentivirus infection
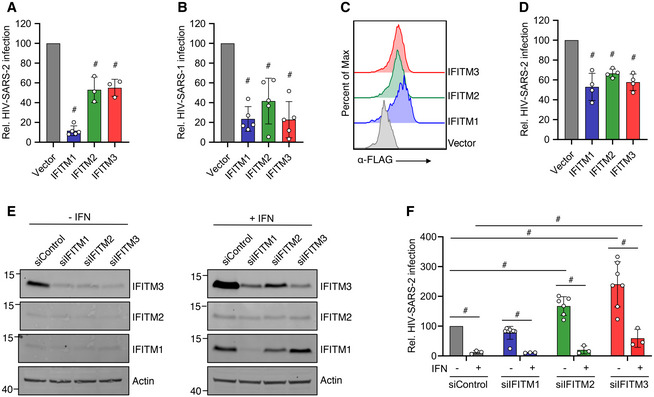
To explore the effects of human IFITM1, IFITM2, and IFITM3 on SARS‐CoV‐2 infection, we transfected the SARS‐CoV‐2 receptor, ACE2, into HEK293T cells stably expressing FLAG‐tagged IFITM constructs. We then infected the transfected cells with SARS‐CoV‐2 Spike‐pseudotyped HIV‐1‐expressing luciferase (HIV‐SARS‐2) and measured luciferase production at 48 hpi. Compared to vector control cells, each of the IFITM‐expressing lines showed significantly less luciferase production, indicating inhibition of Spike‐mediated infection, with IFITM1 expression exhibiting particularly strong inhibition (Fig 1A). We also observed that all three of the IFITMs inhibited SARS‐CoV Spike‐pseudotyped lentivirus infection (HIV‐SARS‐1) (Fig 1B), with IFITM3 appearing to exhibit relatively more anti‐viral activity against HIV‐SARS‐1 than HIV‐SARS‐2 (Fig 1A and B). We confirmed uniform expression of each of the FLAG‐tagged IFITM constructs by flow cytometry (Fig 1C). We further confirmed the ability of these IFITM constructs to limit HIV‐SARS‐2 infection in transiently transfected Caco‐2 cells, which naturally express ACE2 (Fig 1D). Furthermore, Caco‐2 cells express basal IFITM3 protein at relatively high levels, while IFITM1 and IFITM2 are nearly absent. Knockdown of IFITM3 by siRNA transfection increased HIV‐SARS‐2 infection (Fig 1E). IFNβ treatment of Caco‐2 cells strongly inhibited HIV‐SARS‐2 infection, which could be partially reversed with siRNA‐mediated knockdown of IFITM3 (Fig 1E and F). These results indicate that basal, IFN‐induced, and ectopically expressed IFITM3 inhibits HIV‐SARS‐2 infection of Caco‐2 cells, and that, while multiple IFN stimulated genes are likely involved in repression of infection, IFITM3 is among those critical factors. In sum, our experiments indicate that IFITMs, both when expressed endogenously or ectopically, are restrictive of SARS‐CoV‐2 Spike‐mediated virus infection.
Figure 1. Human IFITMs inhibit SARS‐CoV‐2 Spike‐pseudotyped lentivirus infections.
-
A, BHEK293T cells transduced with the indicated FLAG‐tagged IFITM expression constructs or vector control plus transient transfection of ACE2 were infected with HIV‐1 expressing a luciferase reporter gene pseudotyped with (A) SARS‐CoV‐2 Spike (HIV‐SARS‐2) or (B) SARS‐CoV Spike (HIV‐SARS‐1). Measurement of luciferase activity 72 h post‐infection served as a readout for infection. Luciferase readings were normalized to vector control, which was set to a value of 100. Bars in (A) and (B) represent averages with individual data points from 3 to 5 independent experiments shown as circles. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test. Not indicated in (A), IFITM1 was also significantly different from IFITM2 and IFITM3.
-
CRepresentative example flow cytometry plots from cells transfected as in (A) and (B) demonstrate high transfection efficiency and expression of each of the FLAG‐tagged IFITMs.
-
DCaco‐2 cells were transfected as in (A) and (B) and infected with HIV‐SARS‐2. Luciferase readings at 72 h post‐infection were normalized to vector control, which was set to a value of 100. Bars represent averages with individual data points from 4 independent experiments shown as circles. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test.
-
ECaco‐2 cells were transfected with the indicated siRNAs for 48 h followed by treatment with IFNβ for 24 h. Western blotting of cell lysates is shown.
-
FCells as in (E) were infected with HIV‐SARS‐2. Luciferase readings at 48 h post‐infection were normalized to vector control, which was set to a value of 100. Bars represent averages with individual data points from 3 to 7 independent experiments shown as circles. Error bars represent SD. # P < 0.05 by ANOVA followed by Tukey’s multiple comparisons test for the comparisons indicated by lines.
Source data are available online for this figure.
IFITMs inhibit SARS‐CoV‐2 infections
We next sought to validate results from pseudotyped virus infections using genuine SARS‐CoV‐2 at Biosafety Level 3. We utilized HEK293T cell lines stably expressing IFITMs (Fig 2A) that were transiently transfected with ACE2‐GFP and measured percent infection via staining for viral N protein and flow cytometry. IFITM proteins are unique from many classical interferon effectors in that they affect virus entry processes rather than intracellular virus replication (Feeley et al, 2011; Bailey et al, 2014). Analysis of virus protein production early in infection as done by flow cytometry thus provides a measure of whether virus was able to fuse with cellular membranes and continue the replicative process. Consistent with past reports suggesting low endogenous ACE2 expression in HEK293T cells, we found that infection was limited to cells that were ACE2‐GFP‐positive (Fig 2B). We thus employed a gating strategy that focused on ACE2‐GFP‐positive cells for measuring percent infection. Similar to results with Spike‐pseudotyped lentivirus, we found that human IFITM1, IFITM2, and IFITM3 were each capable of inhibiting SARS‐CoV‐2 infections (Fig 2C and D). Additionally, we examined a cell line expressing a palmitoylation‐deficient IFITM3 triple cysteine mutant (C71A, C72A, C105A; termed Palm∆) that has been shown to lose activity against influenza virus and other viruses (Yount et al, 2010; Yount et al, 2012; Hach et al, 2013; John et al, 2013; Percher et al, 2016; McMichael et al, 2017; Benfield et al, 2020). Remarkably, IFITM3‐Palm∆ maintained restriction of SARS‐CoV‐2 (Fig 2D). Given the highly unusual result with the IFITM3‐Palm∆ cell line, we sought to authenticate the cell line by performing an influenza virus infection alongside one of our SARS‐CoV‐2 infections. Indeed, we confirmed the well‐established results that IFITM3 strongly restricts influenza virus infection in a palmitoylation‐dependent manner (Fig 2E; Yount et al, 2010; Yount et al, 2012; Hach et al, 2013; Chesarino et al, 2014b; Melvin et al, 2015; Percher et al, 2016; Chesarino et al, 2017; McMichael et al, 2017). These results suggest that IFITMs provide palmitoylation‐independent restriction of SARS‐CoV‐2 that is distinct from other viruses.
Figure 2. Human IFITMs inhibit infection by SARS‐CoV‐2.
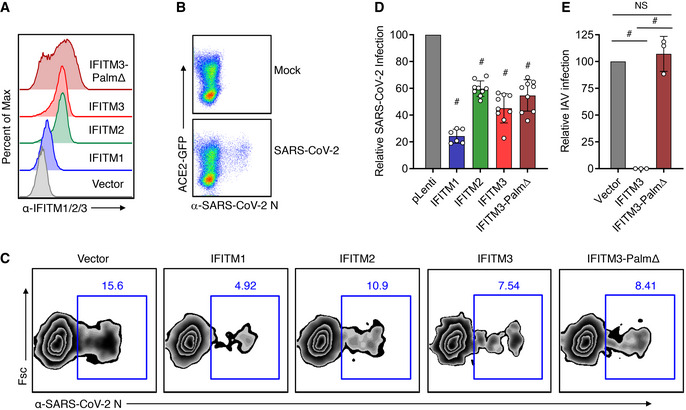
- Flow cytometry histograms showing stable expression of the indicated IFITMs in HEK293T cells.
- Flow cytometry plots of HEK293T cells transfected with ACE2‐GFP and infected with SARS‐CoV‐2 (MOI 0.5) for 24 h showing SARS‐CoV‐2 infection occurs in ACE2‐GFP‐positive cells.
- Stable HEK293T lines as in (A) were infected with SARS‐CoV‐2 (MOI 1) for 24 h and analyzed for percent infection by flow cytometry. Representative example plots are shown for each cell line.
- Graph depicts normalized infection percentage measurements from 2 to 3 independent experiments, each performed in triplicate, from cells infected as in (C). Bars represent averages with individual data points shown as circles. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test.
- The indicated cell lines were infected with influenza A virus (IAV) for 24 h, and percent infection was determined by flow cytometry. Graph depicts percent infection of triplicate samples. Bars represent averages with individual data points shown as circles. Error bars represent SD. # P < 0.05. NS, not significant, by ANOVA followed by Tukey’s multiple comparisons test.
Source data are available online for this figure.
Human IFITM3 with mutations in its endocytic YxxФ motif enhances SARS‐CoV‐2 infection
We additionally sought to confirm the effects of IFITMs on SARS‐CoV‐2 by transfecting IFITM constructs into stable HEK293T‐ACE2‐GFP cells. To measure SARS‐CoV‐2 infection, we employed a flow cytometry gating strategy in which cells that were expressing the highest levels of ACE2‐GFP as well as IFITMs were analyzed for percent infection (Fig 3A and B). We again found that human IFITMs inhibit SARS‐CoV‐2 infection, though effects of transiently transfected IFITM2 did not reach statistical significance (Fig 3C and D). We also confirmed via transient transfection that IFITM3‐Palm∆ is active in restricting the virus (Fig 3C and D). We further found that, consistent with past results examining endogenous ACE2 (Huang et al, 2011), IFITM expression did not affect levels of exogenously expressed ACE2‐GFP (Fig 3E and F). We additionally examined mouse IFITM1‐3 through transient transfection of HEK293T‐ACE2‐GFP cells and found that restriction of SARS‐CoV‐2 infection is a conserved activity of mouse and human IFITMs, and that restriction by mouse IFITM3 occurred independently of palmitoylation like its human counterpart (Fig 4A–C). As an additional investigation of restriction of SARS‐CoV‐2 by endogenous IFITMs, WT, IFITM3 KO, and IFITM‐locus‐deficient (IFITMdel) mouse embryonic fibroblasts (MEFs) were transduced with adenovirus expressing both ACE2 and GFP, followed by infection with SARS‐CoV‐2 (Fig 5A–D). IFITMdel MEFs were significantly more susceptible to SARS‐CoV‐2 compared to WT or IFITM3 KO cells (Fig 5D). Additionally, both IFITM3 KO and IFITM‐locus deletion prevented type I IFN from inhibiting SARS‐CoV‐2 infections to the level observed in WT cells (Fig 5C and D).
Figure 3. Mutation of the human IFITM3 endocytosis motif converts IFITM3 into an enhancer of SARS‐CoV‐2 infections.
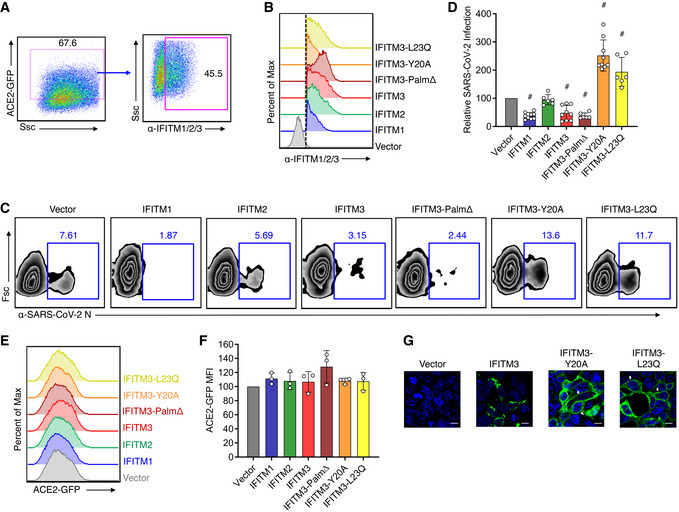
- Flow cytometry gating strategy for analyzing cells with high ACE2‐GFP expression and IFITM expression.
- Flow cytometry histograms showing expression of each of the IFITM constructs, and the gating as indicated in (A) and by the dotted line, which was used for examining percent infections in IFITM‐expressing cells.
- Transfected HEK293T‐ACE2‐GFP cells were infected with SARS‐CoV‐2 (MOI 1) for 24 h and analyzed for percent infection by flow cytometry. Representative example plots are shown for each transfection condition.
- Graph depicts normalized infection percentage measurements from 2 to 3 independent experiments, each performed in triplicate, from cells infected as in (C). Bars represent averages with individual data points shown as circles. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test.
- Representative example flow cytometry histograms of ACE2‐GFP from non‐infected samples.
- Normalized measurements of ACE2‐GFP mean fluorescence intensity (MFI) of non‐infected samples from three separate experiments. Bars represent averages with individual data points shown as circles. Error bars represent SD. Statistical significance (P < 0.5 by ANOVA followed by Tukey’s multiple comparisons test) was not observed for comparisons between any samples.
- Confocal microscopy of HEK293T cells transfected with empty vector, hIFITM3, hIFITM3‐Y20A, or hIFITM3‐L23Q. Green, anti‐IFITM3; blue, DAPI. White arrows highlight plasma membrane localization of IFITM3‐Y20A and IFITM3‐L23Q. Scale bars represent 10 µm.
Source data are available online for this figure.
Figure 4. Mouse IFITMs inhibit SARS‐CoV‐2 infection.
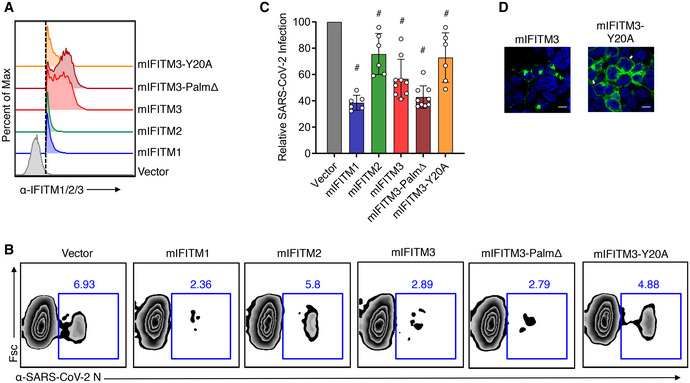
- Flow cytometry histograms showing expression of each of the IFITM constructs with the dotted line indicating gating that was used for examining percent infections in IFITM‐expressing cells.
- Transfected HEK293T‐ACE2‐GFP cells were infected with SARS‐CoV‐2 (MOI 1) for 24 h and analyzed for percent infection by flow cytometry. Representative example plots are shown for each transfection condition.
- Graph depicts normalized infection percentage measurements from 2 to 3 independent experiments, each performed in triplicate, from cells infected as in (B). Bars represent averages with individual data points shown as circles. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test.
- Confocal microscopy of HEK293T cells transfected with WT mIFITM3 or mIFITM3‐Y20A. Green, anti‐IFITM3; blue, DAPI. White arrows highlight plasma membrane localization of IFITM3‐Y20A. Scale bars represent 10 µm.
Source data are available online for this figure.
Figure 5. Deficiency of endogenous mouse IFITMs enhances SARS‐CoV‐2 infection.
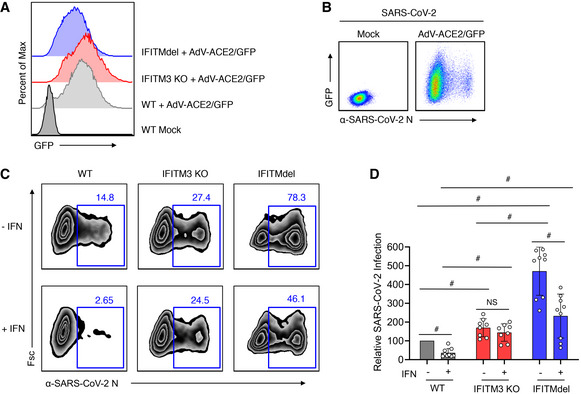
- WT, IFITM3 KO, or IFITMdel MEFs were transduced with adenovirus expressing human ACE2 and GFP (AdV‐ACE2/GFP) at an MOI of 100 for 48 h. Flow cytometry histograms show expression of GFP indicating near‐complete transduction in each MEF line.
- Flow cytometry plots of WT MEF cells with or without transduction with AdV‐ACE2/GFP for 48 h followed by infection with SARS‐CoV‐2 (MOI 1) for 24 h demonstrating that SARS‐CoV‐2 infection of MEFs can occur when transduced with AdV‐ACE2‐GFP.
- MEF cells transduced with AdV‐ACE2/GFP as in (B) were mock treated (−IFN) or treated with mouse IFNβ (+IFN) for 24 h followed by infection with SARS‐CoV‐2 (MOI 1) for 24 h. Representative example flow cytometry plots measuring percent infection are shown for each cell line and treatment condition.
- Graph depicts normalized infection percentage measurements from two experiments with cells infected as in (C) and one experiment with MOI 0.1. Bars represent averages with individual data points shown as circles. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test.
Source data are available online for this figure.
We additionally examined mouse and human IFITM3 mutants that impact protein localization. Y20 is a critical amino acid within a YxxФ endocytosis motif (20‐YEML‐23 in human IFITM3) and has been shown in several studies to be required for active endocytosis of human and mouse IFITM3 (Jia et al, 2012; Jia et al, 2014; Chesarino et al, 2014a; McMichael et al, 2018). Mutation of Y20 or modifying this residue via phosphorylation by Src kinases results in accumulation of IFITM3 at the plasma membrane (Figs 3G and 4D) (Jia et al, 2012; Jia et al, 2014; Chesarino et al, 2014a; Compton et al, 2016; McMichael et al, 2018). Human IFITM3‐Y20A caused an increase in SARS‐CoV‐2 infection as compared to vector control cells, suggesting that IFITM3 at the plasma membrane promotes, rather than inhibits, infection (Fig 3C and D). In contrast, mouse IFITM3‐Y20A did not increase infection compared to vector control cells (Fig 4B and C), revealing a species‐specific distinction between mouse and human IFITM3 in their ability to promote SARS‐CoV‐2 infection. To confirm that enhancement of infection was due to disruption of the 20‐YEML‐23 endocytosis motif, we tested an additional construct, human IFITM3‐L23Q, which similarly accumulates at the plasma membrane due to disruption of the critical Ф residue of the endocytosis signal (Fig 3G; Chesarino et al, 2014a). IFITM3‐L23Q expression also increased SARS‐CoV‐2 infection, confirming the unusual ability of IFITM3 to enhance infection of this virus (Fig 3C and D). Overall, these results demonstrate that while mouse and human IFITMs generally restrict infection, human IFITM3 can promote infection when its localization is shifted toward the plasma membrane.
Human IFITM3 with mutations in its endocytic YxxФ motif enhances SARS‐CoV‐2 Spike‐mediated cell‐to‐cell fusion
We next examined whether IFITM3 affects SARS‐CoV‐2 Spike‐mediated membrane fusion in a cell‐to‐cell fusion assay. U2OS cells co‐transfected with Spike and mCherry expression plasmids were mixed with Calu‐3 target cells that were co‐transfected with plasmids encoding EGFP and IFITMs. After 24‐h co‐culture, mCherry and EGFP double‐positive multi‐nucleated cell syncytia could be observed dependent upon the presence of Spike protein, demonstrating Spike‐mediated fusion between the two cell types (Fig 6A). Upon transfection of IFITM3 into Calu‐3 target cells, double‐positive syncytia remained readily observable upon visual inspection, though quantification of nuclei number within these double‐positive syncytia suggested that IFITM3 may have a modest ability to limit S‐mediated cell‐to‐cell fusion (Fig 6A and B). In contrast, transfection of IFITM3‐Y20A or IFITM3‐L23Q resulted in an increase in the size of double‐positive syncytia as compared to vector control or WT IFITM3 transfections, which was confirmed by quantification of nuclei per syncytia (Fig 6A and B). These results provide confirmation of infection experiments indicating that IFITM3 enriched at the plasma membrane due to endocytosis‐impairing mutations enhances SARS‐CoV‐2 Spike‐mediated membrane fusion.
Figure 6. Mutation of the human IFITM3 endocytosis motif converts IFITM3 into an enhancer of SARS‐CoV‐2 Spike‐mediated cell‐to‐cell membrane fusion.
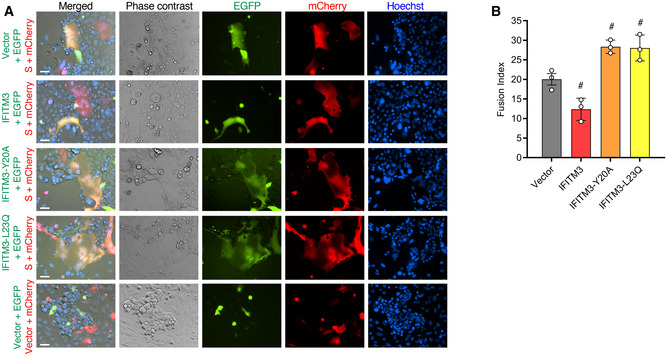
- Representative example fluorescent imaging of the indicated co‐cultures showing the presence of EGFP/mCherry double‐positive cell syncytia dependent upon S expression as well as varying effects of the IFITM3 constructs. Scale bar indicates 50 µm.
- Quantification of nuclei per EGFP/mCherry double‐positive cell syncytia in co‐cultures involving the indicated IFITM3 constructs as compared to vector control. Bars depict averages of three independent experiments with circles representing the individual data points. For each condition, nuclei within 50–75 individual syncytia were counted. Error bars represent SD. # P < 0.05 compared to vector control by ANOVA followed by Tukey’s multiple comparisons test.
Source data are available online for this figure.
Anti‐SARS‐CoV‐2 activity of IFITM3 requires its amphipathic helix domain and is counteracted by TMPRSS2 overexpression
Our results suggest that IFITM3 is able to inhibit SARS‐CoV‐2 infection overall, but that shifting human IFITM3 localization away from the endosomal system and toward the plasma membrane results in enhancement of infection (Figs 2 and 3). Given that SARS‐CoV‐2 has dual cellular entry pathways involving Spike proteolytic activation either at the plasma membrane or within endosomes (Hoffmann et al, 2020; Shang et al, 2020), these findings may indicate that IFITM3 has distinct effects on the virus dependent upon the location at which virus fuses with cell membranes. Plasma membrane‐associated processing of Spike involves TMPRSS2 protease, and it has been reported that TMPRSS2 overexpression increases the proportion of virus able to fuse at the cell surface (Hoffmann et al, 2020). Further, it is reported that high TMPRSS2 levels allow human coronaviruses to avoid restriction by IFITMs (Bertram et al, 2013; Zheng et al, 2020). In line with these data, we found that TMPRSS2 overexpression decreased the ability of IFITM3 to inhibit SARS‐CoV‐2 infection (Fig 7A–C). Interestingly, IFITM3 slightly enhanced infection in the presence of TMPRSS2 overexpression in 2 out of 3 experiments (Fig 7B). Inhibition of infection by IFITM3‐Palm∆ and IFITM1 was also counteracted by TMPRSS2, though enhancement of infection was not observed for these constructs (Fig 7A–C).
Figure 7. IFITM3 restriction of SARS‐CoV‐2 infection requires the IFITM3 amphipathic helix while enhancement of infection does not.
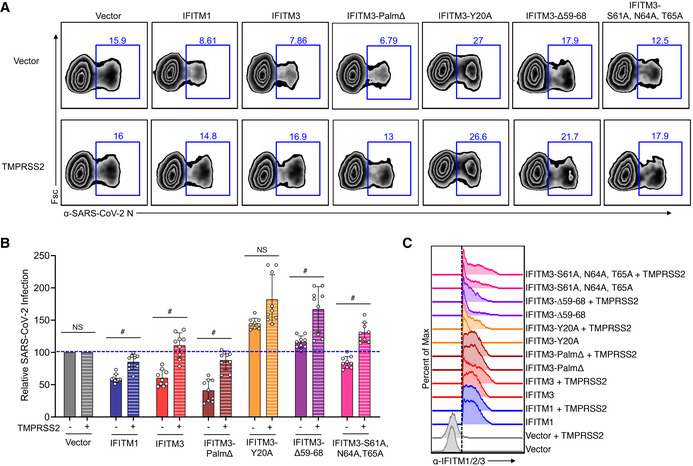
- Transfected HEK293T‐ACE2‐GFP cells were infected with SARS‐CoV‐2 (MOI 1) for 24 h and analyzed for percent infection by flow cytometry. Representative example plots are shown for each transfection condition.
- Graph depicts normalized infection percentage measurements from 3 independent experiments, each performed in triplicate, from cells infected as in (B). Bars represent averages with individual data points shown as circles. Error bars represent SD. The blue dotted line is added to the graph to assist with comparisons to vector control levels of infection. # P < 0.05 by ANOVA followed by Tukey’s multiple comparisons test. NS, not significant. Not indicated, additional comparisons of interest with P < 0.05 include IFITM3 (−) versus IFITM3‐Y20 (−), IFITM3∆59‐68 (−), and IFITM3‐S61A,N64A,T65A (−).
- Flow cytometry histograms showing expression of each of the IFITM constructs with the dotted line indicating gating that was used for examining percent infections in IFITM‐expressing cells.
Source data are available online for this figure.
The IFITM3 amphipathic helix domain located at amino acid positions 59–68 is required for inhibition of influenza virus infection (Chesarino et al, 2017). This helix was recently shown to be able to alter mechanical properties of lipid membranes, such as curvature and lipid order, thus inhibiting viral membrane fusion (preprint: Guo et al, 2020; Rahman et al, 2020). In the context of SARS‐CoV‐2, we found that expression of an amphipathic helix‐deleted mutant of IFITM3 (∆59–68) resulted in a modest enhancement rather than inhibition of infection (Fig 7A–C). Likewise, decreasing amphipathicity of the helix by mutation of its three hydrophilic residues (S61A, N64A, and T65A) decreased restriction of infection (Fig 7A–C), further demonstrating that the amphipathicity of IFITM3, and likely its associated membrane‐altering properties, underlies restriction of SARS‐CoV‐2. Notably, both of the amphipathic helix mutants were increased in their ability to enhance SARS‐CoV‐2 infection when co‐expressed with TMPRSS2 (Fig 7A–C), providing the first clear evidence that IFITM3 enhancement of CoV infection occurs via a mechanism distinct from its amphipathicity‐based membrane alterations. These results support a model wherein IFITM3 exerts opposing activities on SARS‐CoV‐2, including amphipathicity‐dependent restriction of virus at endosomes and amphipathicity‐independent enhancement of infection at the plasma membrane.
Discussion
In the present study, we have identified IFITM1, IFITM2, and IFITM3 as restrictors of SARS‐CoV‐2 infection of cells. Our result showing that IFITM3 inhibits SARS‐CoV‐2 infection independently of S‐palmitoylation is surprising since intact palmitoylation sites are necessary for inhibition of a number of viruses, including influenza virus, dengue virus, CoV‐NL63, and CoV‐229E (Yount et al, 2010; Yount et al, 2012; Hach et al, 2013; John et al, 2013; Percher et al, 2016; McMichael et al, 2017; Zhao et al, 2018; Benfield et al, 2020). This conclusion is supported by experiments with three distinct expression systems (stable transduction of human IFITM3‐Palm∆ in the pLenti vector and transient transfections of human and mouse IFITM3‐Palm∆ in pCMV) (Figs 2, 3, 4). Previous work suggests that S‐palmitoylation increases the rate at which IFITM3 traffics to influenza virus‐containing endosomes during infection (Spence et al, 2019). Our results suggest possible differences between influenza virus and SARS‐CoV‐2 either in their endocytic entry pathways or fusion kinetics, and IFITM constructs may prove useful in dissecting such differences.
Mechanistically, we found that the IFITM3 amphipathic helix is necessary for its SARS‐CoV‐2 restrictive capacity (Fig 7). We previously showed that this helix was also required for restriction of influenza virus and proposed that the helix affected virus fusion by wedging into the inner leaflet of the membrane bilayer, thus generating local membrane curvature that is disfavorable to virus‐to‐cell fusion (Chesarino et al, 2017). More recent research has shown that the amphipathic helix is indeed able to mechanically induce membrane curvature and increase lipid order (preprint: Guo et al, 2020; Rahman et al, 2020). Overall, IFITMs likely restrict SARS‐CoV‐2 infection by mechanically disfavoring viral membrane fusion reactions occurring at endosomes.
Remarkably, we observed that IFITM3‐Y20A and IFITM‐L23Q constructs enhance SARS‐CoV‐2 infection (Fig 3). Since these constructs are well characterized to accumulate at the plasma membrane of cells due to loss of interaction with the AP‐2 endocytic adaptor protein complex (Jia et al, 2014; Chesarino et al, 2014a; McMichael et al, 2018), these data suggest that IFITM3 localization to the plasma membrane enhances infection. Given that WT IFITM3 traffics to the plasma membrane prior to its endocytosis, and since phosphorylation of IFITM3 Y20 by Src kinases also blocks endocytosis (Jia et al, 2012; Chesarino et al, 2014a), there are likely opposing restrictive and enhancing activities occurring in cells expressing WT IFITM3. It will be interesting to determine whether these balances are shifted in different cell types with distinct endocytic or phosphorylation capacities. Dueling enhancing and inhibiting activities of IFITM3 in SARS‐CoV‐2 infection are also consistent with the partial restriction of SARS‐CoV‐2 infection observed in cells stably expressing IFITM3 as compared to the near‐complete inhibition of influenza virus in the same cells (Fig 2D and E).
Enhancement of SARS‐CoV‐2 infection at the plasma membrane does not require the IFITM3 amphipathic helix, as demonstrated by our observation that TMPRSS2 overexpression converts amphipathic helix mutants into strong enhancers of infection (Fig 7). We did not observe an enhancing effect of human IFITM1, which also has a conserved and functional amphipathic helix domain (Chesarino et al, 2017), further supporting that enhancement of infection at the plasma membrane is not occurring via amphipathic helix‐mediated mechanical membrane alterations (Fig 6). These data are in alignment with past reports that IFITM3, but not IFITM1, enhances CoV‐OC43 infections (Zhao et al, 2014). Removal of the IFITM1 extended C‐terminal tail converted IFITM1 into an enhancer of specific CoV infections (Zhao et al, 2014; Zhao et al, 2018). Likewise, and similar to our results with SARS‐CoV‐2, mutation of human IFITM3 Y20 was also reported to enhance SARS‐CoV and MERS‐CoV pseudotype virus infections, but not CoV‐229E or CoV‐NL63 pseudotype infections (Zhao et al, 2018). Since enhancement of infection does not correlate with specific receptor usage of the different coronaviruses, it is unlikely that IFITMs directly affect the virus receptors. In that regard, we did not observe an effect of IFITM proteins on levels of exogenously expressed ACE2‐GFP (Fig 3E and F). It may be possible that the short IFITM3 C‐terminal tail, predicted to be exposed at the cell surface, directly interacts with specific features of CoV surface glycoproteins or with entry factors. It is also interesting to note that mouse IFITM3 has an extended C‐terminal tail compared to human IFITM3, consistent with results that mouse IFITM3‐Y20A did not enhance infection (Fig 4B and C). Overall, our results indicate that IFITMs are not likely exerting a broad mechanism of amphipathicity‐based membrane alteration to promote infection, but rather are more specifically coopted by specific CoVs, such as SARS‐CoV‐2, under certain conditions. An early study on a limited cohort suggests that the human IFITM3 rs12552‐C SNP, which has been proposed to result in IFITM3 plasma membrane localization (Everitt et al, 2012; Jia et al, 2012; Chesarino et al, 2014a), is a risk factor for severe COVID‐19 (Zhang et al, 2020b). Larger studies are warranted to examine whether this and other polymorphisms in IFITM3 (Zani & Yount, 2018) positively or negatively influence COVID‐19 severity.
Materials and Methods
Biosafety
All work with live SARS‐CoV‐2 was performed at Biosafety Level 3 (BSL3) according to standard operating procedures approved by the Ohio State University BSL3 Operations Group and Institutional Biosafety Committee. Samples were removed from the BSL3 facility for flow cytometry analysis only after decontamination with 4% paraformaldehyde for a minimum of 1 h according to an in‐house validated and approved method of sample decontamination.
Cell lines and cell culture
All cells were cultured at 37°C with 5% CO2 in a humidified incubator and were grown in Dulbecco’s modified Eagle’s medium (DMEM) or minimal medium supplemented with 10–20% FBS (Atlas Biologicals). HEK293T, Calu‐3, and Caco‐2 were purchased from ATCC. The origin of our U2OS cells is unknown, but the identity and purity of the cells were verified by STR profiling. Mouse embryonic fibroblast (MEF) cells were a kind gift from Dr. Michael S. Diamond (Washington University in St. Louis). Vero E6 cells were a kind gift from Dr. Mark Peeples (Nationwide Children’s Hospital) and were treated with Mycoplasma Removal Agent (MP Biomedical) for 1 month to ensure lack of contamination before virus propagation. All other cell lines were routinely treated with Mycoplasma Removal Agent for 1 week upon thawing of new cultures. HEK293T cells stably expressing human IFITM1, IFITM2, IFITM3, IFITM3‐PalmΔ, or empty pLenti‐puro vector that were used in genuine virus infections were generated and validated as described previously (McMichael et al, 2018), with the exception that the IFITM3‐PalmΔ cell line required three sequential transductions to achieve expression comparable to WT IFITM3 in the majority of the cells. HEK293T cells stably expressing GFP‐tagged human ACE2 (OriGene) were generated using the same methodology. HEK293T cell lines stably transduced with FLAG‐tagged human IFITM1, IFITM2, IFITM3, or pQCXIP vector control for HIV‐SARS‐1 and HIV‐SARS‐2 infection experiments were described previously (Shi et al, 2018). Type I interferon (human recombinant interferon beta, NR‐3085, BEI Resources) was added to Caco‐2 cells at 48 h post‐siRNA transfection at a concentration of 250 IU/ml. Mouse IFNα2 (eBioscience) was added to MEFs at a concentration of 0.1 µg/ml. Cells were incubated with interferon for a 24 h period prior to being removed and replaced with virus inoculum.
SARS‐CoV‐2 propagation and titering
SARS‐CoV‐2 USA‐WA1/2020 stock from BEI Resources was diluted 1:10,000 in DMEM and added to confluent Vero cells for 1 h at 37°C. Following the 1‐h infection period, virus‐containing media was replaced with DMEM containing 4% FBS and incubated at 37°C for 72 h at which point significant cytopathic effects were observed. Cell supernatant containing infectious virus was centrifuged at 1,000 g for 10 min to remove cell debris, and was aliquoted, flash‐frozen in liquid nitrogen, and stored at −80°C. Virus titer was determined by TCID50 assay in Vero cells using the presence of cytopathic effect coupled with viral N protein staining for identification of infected wells. The viral stock titer was further confirmed by standard plaque assay with 0.3% agarose (Sigma) overlay and visualization with 0.25% Crystal Violet (Sigma) in Vero cells. Full‐genome sequencing of the virus stock by the Ohio State University Medical Center Clinical Diagnostics Laboratory confirmed a matching nucleotide consensus sequence with the reported sequence of the USA‐WA1/2020 strain.
Plasmids, transfections, transductions, and siRNA knockdowns
Plasmid encoding untagged ACE2 was a gift from Hyeryun Choe (Addgene plasmid #1786). Transfection of HEK293T cell lines stably expressing human IFITM1, IFITM2, or IFITM3 (35) with ACE2 was performed with TransIT‐293 reagent (Mirus). Transfection of Caco‐2 cells with empty pQCXIP or pQCXIP‐IFITM1, pQCXIP‐IFITM2, pQCXIP‐IFITM3 (gifts from Chen Liang) was performed using Lipofectamine 2000 (Thermo Fisher). For SARS‐CoV‐2 infection experiments, pCMV expression plasmid for N‐terminally HA‐tagged human and mouse IFITM1, IFITM2, IFITM3, IFITM3‐PalmΔ, and IFITM3‐Y20A as well as human IFITM3‐L23Q, IFITM3‐Δ59‐68, and IFITM3‐S61A,N64A,T65A was previously described (Yount et al, 2010; Hach et al, 2013; Chesarino et al, 2014a; Percher et al, 2016; Chesarino et al, 2017). TMPRSS2 in pCAGGS was a kind gift from Dr. Stefan Pöhlmann (German Primate Center). ACE2‐GFP in pLenti‐puro was purchased from OriGene. Adenovirus expressing ACE2 and GFP was acquired from the Iowa University Viral Vector Core Facility. All transient transfections for SARS‐CoV‐2 infection experiments were performed using LipoJet transfection reagent (Signagen Laboratories). siRNAs targeting IFITM1 (s16192), IFITM2 (s20771), IFITM3 (s195035), and a non‐targeting siRNA control pool (4390844) were purchased from Ambion. siRNA‐mediated knockdown was performed by transfecting Caco‐2 cells with 20 nM amounts of siRNA and Lipofectamine RNAiMAX (Thermo Fisher). Knockdown efficiency was assessed by Western blot analysis at 72 h post‐transfection.
Pseudotyped virus infections and luciferase assays
Virus‐like particles pseudotyped with SARS‐CoV‐2 or SARS‐CoV‐1 Spike protein were produced by transfecting HEK293T cells with pNL4‐3.Luc.R‐E‐ (obtained from NIH AIDS Reagent Resource; 3418) and pcDNA‐SARS‐CoV2‐C9 or pcDNA‐SARS‐CoV1‐C9 (gifts from Thomas Gallagher) using TransIT‐293 reagent. Cells were incubated with virus‐containing medium for 72 h, after which cells were lysed with Passive Lysis Buffer (Promega). Luciferase assays were performed using Luciferase Assay System (Promega). Luciferase values were measured with a Tecan Infinite M1000 plate reader.
SARS‐CoV‐2 infections and flow cytometry
Cells were infected with SARS‐CoV‐2 for 24 h, after which cells were fixed with 4% paraformaldehyde (Thermo Scientific) for 1 h at room temperature, permeabilized with PBS containing 0.1% Triton X‐100, and blocked with PBS/2% FBS. Cells were then stained with mouse‐produced anti‐SARS‐CoV‐2 N (Sino Biological, 40143‐MM08) and a cocktail of rabbit‐produced anti‐IFITM2/3 (ProteinTech, 11714‐1‐AP) and rabbit‐produced anti‐IFITM1 (Cell Signaling Technologies, 13126S). Primary antibody labeling was followed by labeling with goat anti‐mouse Alexa Fluor 555 (Life Technologies, A‐21424) and donkey anti‐rabbit Alexa Fluor 647 (Life Technologies, A‐31573) secondary antibodies. Flow cytometry data were collected using a FACSCanto II flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software. Infected cell gates were set using non‐infected control samples.
Cell fusion assay
Using Lipofectamine 3000 (Life Technologies), U2OS cells were co‐transfected with pmCherry and either empty pCAGGS vector or SARS‐CoV‐2 Spike Glycoprotein (BEI Resources, deposited by Dr. Florian Krammer, Icahn School of Medicine at Mount Sinai); Calu‐3 cells were co‐transfected with pEGFP and either empty pCMV vector, IFITM3, IFITM3‐Y20A, or IFITM3‐L23Q. Transfected Calu‐3 and U2OS cells were mixed and co‐incubated for 24 h. Following incubation, the co‐cultures were contra‐stained with Hoechst 33342 and imaged using a Nikon Eclipse Ti‐E inverted microscope (Nikon Instruments). Nikon NIS Elements AR software was used to quantify fusion indices calculated as number of nuclei per double‐positive (expressing both EGFP and mCherry) cell fusion cluster.
Confocal microscopy
To examine IFITM3 localization, HEK293T cells grown on coverslips were transiently transfected with empty vector, IFITM3, IFITM3‐Y20A, IFITM3‐L23Q, mIFITM3, or mIFITM3‐Y20A. Twenty‐four h post‐transfection, cells were fixed with 4% paraformaldehyde in PBS, were permeabilized with 0.1% Triton X‐100 in PBS, and were incubated with anti‐IFITM3 (ProteinTech), followed by washing and incubation with donkey anti‐rabbit Alexa 488 secondary antibody (Life Technologies, A‐11029). Coverslips were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher). Fluorescence images were captured using an Olympus FV 1000 Spectral Confocal system.
Author contributions
GS, ADK, EK, AZ, and LZ performed experiments, analyzed data, and generated figures. KKL provided assistance and expertise in generating pseudotyped lentiviruses. LH‐S and RTR provided critical assistance and expertise in generating and titering SARS‐CoV‐2 stocks in BSL3 containment, and in developing methodology for analysis of infected cells. Experiments were supervised by DSK, AAC, and JSY. GS, ADK, AAC, and JSY wrote an initial manuscript draft that was critically edited and approved by all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We would like to thank Alan Rein for facilitating the production of lentiviral pseudotypes containing SARS‐CoV‐1 and SARS‐CoV‐2 Spike proteins. We thank Dr. Eugene Oltz for critical reading of the manuscript. Work in the laboratory of AAC is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research and by an Intramural Targeted Anti‐COVID‐19 Award from the National Institute of Allergy and Infectious Diseases. Work in the laboratory of JSY is supported by NIH Grants AI130110, AI151230, AI142256, AI146690, and HL154001. This work was also partially supported by COVID‐19 Seed Grants from the Ohio State University to JSY and DSK. Work in the laboratory of DSK is supported by NIH grant GM114666. ADK was supported by an NIH T32 fellowship funded by NIH grant GM068412 and the Ohio State University Presidential Fellowship. Ashley Zani was supported by an NIH T32 fellowship funded by NIH grant AI112542 and the National Science Foundation Graduate Research Fellowship Program. The synopsis figure was created using Biorender software with an institutional license provided by the NIH.
The EMBO Journal (2021) 40: e106501.
Contributor Information
Alex A Compton, Email: alex.compton@nih.gov.
Jacob S Yount, Email: Jacob.Yount@osumc.edu.
Data availability
This study includes no data deposited in external repositories.
References
- Allen EK, Randolph AG, Bhangale T, Dogra P, Ohlson M, Oshansky CM, Zamora AE, Shannon JP, Finkelstein D, Dressen A et al (2017) SNP‐mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med 23: 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, Zhong G, Huang IC, Farzan M (2014) IFITM‐Family proteins: the cell's first line of antiviral defense. Annu Rev Virol 1: 261–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfield CT, MacKenzie F, Ritzefeld M, Mazzon M, Weston S, Tate EW, Teo BH, Smith SE, Kellam P, Holmes EC et al (2020) Bat IFITM3 restriction depends on S‐palmitoylation and a polymorphic site within the CD225 domain. Life Sci Alliance 3: e201900542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S, Dijkman R, Habjan M, Heurich A, Gierer S, Glowacka I, Welsch K, Winkler M, Schneider H, Hofmann‐Winkler H et al (2013) TMPRSS2 activates the human coronavirus 229E for cathepsin‐independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol 87: 6150–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel‐Benhassine F, Van der Werf S et al (2020) Syncytia formation by SARS‐CoV‐2‐infected cells. EMBO J 39: e106267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemudupati M, Kenney AD, Bonifati S, Zani A, McMichael TM, Wu L, Yount JS (2019) From APOBEC to ZAP: diverse mechanisms used by cellular restriction factors to inhibit virus infections. Biochim Biophys Acta Mol Cell Res 1866: 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, McMichael TM, Hach JC, Yount JS (2014a) Phosphorylation of the antiviral protein interferon‐inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J Biol Chem 289: 11986–11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, McMichael TM, Yount JS (2014b) Regulation of the trafficking and antiviral activity of IFITM3 by post‐translational modifications. Future Microbiol 9: 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino NM, Compton AA, McMichael TM, Kenney AD, Zhang L, Soewarna V, Davis M, Schwartz O, Yount JS (2017) IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep 18: 1740–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Roy N, Porrot F, Billet A, Casartelli N, Yount JS, Liang C, Schwartz O (2016) Natural mutations in IFITM3 modulate post‐translational regulation and toggle antiviral specificity. EMBO Rep 17: 1657–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ et al (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ et al (2011) IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7: e1002337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Steinkühler J, Marin M, Li X, Lu W, Dimova R, Melikyan GB (2020) Interferon‐induced transmembrane protein 3 blocks fusion of diverse enveloped viruses by locally altering mechanical properties of cell membranes. bioRxiv 10.1101/2020.06.25.171280 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Hach JC, McMichael T, Chesarino NM, Yount JS (2013) Palmitoylation on conserved and nonconserved cysteines of murine IFITM1 regulates its stability and anti‐influenza A virus activity. J Virol 87: 9923–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine‐Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L et al (2011) Distinct patterns of IFITM‐mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 7: e1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Pan Q, Ding S, Rong L, Liu SL, Geng Y, Qiao W, Liang C (2012) The N‐terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol 86: 13697–13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng YM, Liu SL, Guo F, Geng Y, Qiao W et al (2014) Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol 16: 1080–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John SP, Chin CR, Perreira JM, Feeley EM, Aker AM, Savidis G, Smith SE, Elia AE, Everitt AR, Vora M et al (2013) The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J Virol 87: 7837–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC et al (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9: e1003124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Chin CR, Everitt AR, Clare S, Perreira JM, Savidis G, Aker AM, John SP, Sarlah D, Carreira EM et al (2013) Amphotericin B increases influenza A virus infection by preventing IFITM3‐mediated restriction. Cell Rep 5: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael TM, Zhang L, Chemudupati M, Hach JC, Kenney AD, Hang HC, Yount JS (2017) The palmitoyltransferase ZDHHC20 enhances interferon‐induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity. J Biol Chem 292: 21517–21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael TM, Zhang Y, Kenney AD, Zhang L, Zani A, Lu M, Chemudupati M, Li J, Yount JS (2018) IFITM3 restricts human metapneumovirus infection. J Infect Dis. 218: 1582–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin WJ, McMichael TM, Chesarino NM, Hach JC, Yount JS (2015) IFITMs from mycobacteria confer resistance to influenza virus when expressed in human cells. Viruses 7: 3035–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A, Ramakrishnan S, Thinon E, Yuan X, Yount JS, Hang HC (2016) Mass‐tag labeling reveals site‐specific and endogenous levels of protein S‐fatty acylation. Proc Natl Acad Sci USA 113: 4302–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman K, Coomer CA, Majdoul S, Ding S, Padilla‐Parra S, Compton AA (2020) Homology‐guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state. Elife 9: e58537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472: 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F (2020) Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci USA 117: 11727–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Ozog S, Torbett BE, Compton AA (2018) mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc Natl Acad Sci USA 115: E10069–E10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JS, He R, Hoffmann HH, Das T, Thinon E, Rice CM, Peng T, Chandran K, Hang HC (2019) IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol 15: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch F, Winkler M, Pohlmann S (2014) IFITM proteins inhibit entry driven by the MERS‐coronavirus spike protein: evidence for cholesterol‐independent mechanisms. Viruses 6: 3683–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WL, Grotefend CR, Tsai MT, Wang YL, Radic V, Eoh H, Huang IC (2017) Delta20 IFITM2 differentially restricts X4 and R5 HIV‐1. Proc Natl Acad Sci USA 114: 7112–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY et al (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Karssemeijer RA, Hang HC (2012) S‐palmitoylation and ubiquitination differentially regulate interferon‐induced transmembrane protein 3 (IFITM3)‐mediated resistance to influenza virus. J Biol Chem 287: 19631–19641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC (2010) Palmitoylome profiling reveals S‐palmitoylation‐dependent antiviral activity of IFITM3. Nat Chem Biol 6: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R, Case JB, Gomez Castro MF, Liu Z, Zeng Q, Zhao H, Son J, Rothlauf PW, Hou G, Bose S et al (2020) Cholesterol 25‐hydroxylase suppresses SARS‐CoV‐2 replication by blocking membrane fusion. Proc Natl Acad Sci USA 10.1073/pnas.2012197117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani A, Yount JS (2018) Antiviral protection by IFITM3 in vivo . Curr Clin Microbiol Rep 5: 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, Jia X, Wu M, Shi B, Xu S et al (2020a) Viral and host factors related to the clinical outcome of COVID‐19. Nature 583: 437–440 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Qin L, Zhao Y, Zhang P, Xu B, Li K, Liang L, Zhang C, Dai Y, Feng Y et al (2020b) Interferon‐induced transmembrane protein 3 genetic variant rs12252‐C associated with disease severity in coronavirus disease 2019. J Infect Dis 222: 34–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo JT (2014) Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci USA 111: 6756–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sehgal M, Hou Z, Cheng J, Shu S, Wu S, Guo F, Le Marchand SJ, Lin H, Chang J et al (2018) Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J Virol 92: e01535‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zheng S, Chen D, Zheng M, Li X, Li G, Lin H, Chang J, Zeng H, Guo JT (2020) LY6E restricts the entry of human coronaviruses, including the currently pandemic SARS‐CoV‐2. J Virol 94: e00562‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Zhao X, Zheng S, Chen D, Du P, Li X, Jiang D, Guo JT, Zeng H, Lin H (2020) Bat SARS‐Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion. Emerg Microbes Infect 9: 1567–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R et al (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
This study includes no data deposited in external repositories.


