Abstract
Proteomics offers a wide collection of methodologies to study biological systems at the finest granularity. Faced with COVID‐19, the most worrying pandemic in a century, proteomics researchers have made significant progress in understanding how the causative virus hijacks the host's cellular machinery and multiplies exponentially, how the disease can be diagnosed, and how it develops, as well as its severity predicted. Numerous cellular targets of potential interest for the development of new antiviral drugs have been documented. Here, the most striking results obtained in the proteomics field over this first semester of the pandemic are presented. The molecular machinery of SARS‐CoV‐2 is much more complex than initially believed, as many post‐translational modifications can occur, leading to a myriad of proteoforms and a broad heterogeneity of viral particles. The interplay of protein–protein interactions, protein abundances, and post‐translational modifications has yet to be fully documented to provide a full picture of this intriguing but lethal biological threat. Proteomics has the potential to provide rapid detection of the SARS‐CoV‐2 virus by mass spectrometry proteotyping, and to further increase the knowledge of severe respiratory syndrome COVID‐19 and its long‐term health consequences.
1. The Unanticipated COVID‐19 Tsunami
The world is dealing with one of the most pernicious respiratory diseases ever seen, COVID‐19. Over 51 million diagnosed cases were logged as of November 9th, 2020, alongside a dramatic and disheartening more than 1.2 million deaths. The first description of COVID‐19 was linked to its emergence in December 2019 in Wuhan, China.[ 1 ] Its subsequent fulminant spread around the world within just a few weeks led the WHO (www.who.int) to qualify it as a pandemic on March 11, 2020. This scenario was facilitated by i) unusual infectious features such as a diversity of symptoms, a high ratio of asymptomatic but nevertheless infectious cases, and infectiousness before the onset of symptoms, ii) a lack of knowledge about the disease and of tools to detect the causative pathogen, but also iii) a vast global playground with overdensified habitats and extensive travel facilities which contributed to the multiplication of human‐to‐human contacts. Due to its relatively high fatality rate and effective infectious properties, COVID‐19 quickly raised huge concerns, triggered unprecedented measures including long‐term lockdowns around the world, and had an enormous impact on human society, with dramatic economic and social consequences. The pathogen causing COVID‐19, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), was isolated and its genome sequence was published on February 3rd, 2020.[ 1 , 2 ] SARS‐CoV‐2 is a Sarbecovirus belonging to the coronavirus family lineage, which was known from previous outbreaks of SARS‐CoV and MERS‐CoV to often cause severe human disease. Once its genome sequence had been established, diagnostic RT‐qPCR assays were promptly designed to detect the virus and facilitate diagnosis, helping to curtail chains of contamination. Unprecedented efforts were rapidly implemented by the scientific community to better characterize the infectious mechanisms of SARS‐CoV‐2, to understand the disease and its consequences, propose new antiviral solutions, and more importantly to develop vaccines.[ 3 ] A collective mass spectrometry effort was launched to study COVID‐19 and propose alternative solutions for the detection of SARS‐CoV‐2. In their letter of intent,[ 4 ] this coalition of nearly 600 primed scientists from sixty countries anticipated that proteomics could play a key role in understanding COVID‐19. Table 1 shows the first 26 proteomics datasets produced by this community, which are currently publicly available through the PRIDE repository (https://www.ebi.ac.uk/pride/), thus offering new opportunities for data scientists. These results illustrate the different aspects to which proteomics can contribute. Some of these works were commented in a recent review focussed on olfactory proteomics[ 5 ] and the most common methodologies used for these works were presented.[ 6 ] Now, six months on from the start of the pandemic, we propose an analysis of the main findings gathered by proteomists over this first semester, and discuss how they could represent game‐changers in the COVID‐19 battlefield.
Table 1.
List of COVID‐19‐related PRIDE datasets available on 2020, September 30th
| PXD accession number | Submission date | Subject | Instrument | Reference |
|---|---|---|---|---|
| PXD017710 | 2020‐02‐25 | Infection mechanisms‐shotgun | Q Exactive HF | [ 25 ] |
| PXD018117 | 2020‐03‐20 | Infection mechanisms‐shotgun | Q Exactive Plus | [ 31 ] |
| PXD018241 | 2020‐03‐29 | Infection mechanisms‐shotgun | Fusion Lumos | ‐ |
| PXD018357 | 2020‐04‐04 | Infection mechanisms‐shotgun | Fusion Lumos | [ 25 ] |
| PXD018581 | 2020‐04‐15 | Infection mechanisms‐shotgun & PTMs | Q Exactive HF X | ‐ |
| PXD018594 | 2020‐04‐16 | Infection mechanisms | Q Exactive HF | [ 9 ] |
| PXD018682 | 2020‐04‐20 | Virus detection | Fusion | [ 15 ] |
| PXD018760 | 2020‐04‐23 | Virus detection | Eclipse & Fusion Lumos | ‐ |
| PXD018804 | 2020‐04‐26 | Virus detection | Q Exactive HF | [ 13 ] |
| PXD018983 | 2020‐06‐26 | Infection mechanisms‐interactants | Fusion Lumos | [ 52 ] |
| PXD019113 | 2020‐05‐12 | Infection mechanisms‐shotgun & PTMs | Exploris 480 | [ 26 ] |
| PXD019163 | 2020‐05‐13 | Analytical control | Q Exactive Plus | [ 53 ] |
| PXD019423 | 2020‐06‐25 | Virus detection | Fusion | [ 15 ] |
| PXD019645 | 2020‐07‐15 | Infection mechanisms‐shotgun | Fusion Lumos | [ 10 ] |
| PXD019648 | 2020‐06‐08 | Virus detection | timsTOF Pro | [ 49 ] |
| PXD019686 | 2020‐06‐10 | Virus detection | Q Exactive HF | [ 14 ] |
| PXD019937‐40 | 2020‐06‐22 | Infection mechanisms‐shotgun | Orbitrap Fusion Lumos | [ 54 ] |
| PXD020019 | 2020‐06‐26 | Infection mechanisms‐PTMs | Q Exactive HF | ‐ |
| PXD020222 | 2020‐07‐06 | Infection mechanisms‐interactants | Q Exactive HF | ‐ |
| PXD020394 | 2020‐07‐16 | Virus detection | Q Exactive Plus | ‐ |
| PXD020601 | 2020‐07‐28 | COVID‐19 pathology | timsTOF Pro | [ 55 ] |
| PXD020644 | 2020‐07‐30 | Virus detection | Orbitrap Eclipse | ‐ |
| PXD021388 | 2020‐09‐10 | Virus detection | Microflex/Autoflex | [ 45 ] |
2. The SARS‐CoV‐2 Machinery Landscape
Based on the genomic sequences of the first cultivated isolates and viruses present in bronchoalveolar lavage fluids from patients, similarities were found between SARS‐CoV‐2 and other coronaviruses.[ 7 ] Since then, a total of 113 252 genomes have been sequenced and shared (www.gisaid.org), providing a unique resource for tracking and tracing the ongoing outbreak. The SARS‐CoV‐2 single‐strand RNA genome has a median GC percentage of 38% and a median total length of 29 882 nucleotides, which ranks it among the largest known RNA genomes. Its 14 open reading frames (ORFs) are quite similar to those of the closest known sarbecoviruses in terms of sequence and length.[ 8 ] The four structural proteins encoded at the 3′ end of the genome—the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins—make up the protective shell surrounding the RNA molecule. The remaining 25 proteins help to hijack the molecular machinery of the host to ultimately assemble myriad viral particles. ORF1a encodes the replicase polyprotein 1a. From the huge polyprotein 1ab, protease cleavage produces 16 smaller non‐structural proteins (Nsp1‐16). In addition, nine accessory proteins have been delineated, but their roles remain to be established (Figure 1 ). While structural proteins N, S, and M can be robustly detected by mass spectrometry, the number of peptides for other viral proteins and their detection levels depend on their abundance, size, and the sample preparation method used.[ 9 , 10 ] However, the architecture of the viral transcriptome and translatome can be much more complex, as indicated by non‐canonical transcripts, RNA modifications, and polypeptides generated by unannotated viral ORFs revealed by deep sequencing technologies.[ 11 ] Moreover, a recent N‐terminomics analysis identified a variety of SARS‐CoV‐2 proteolytic proteoforms in the context of viral infection,[ 12 ] thus further increasing the molecular complexity of this biological entity.
Figure 1.
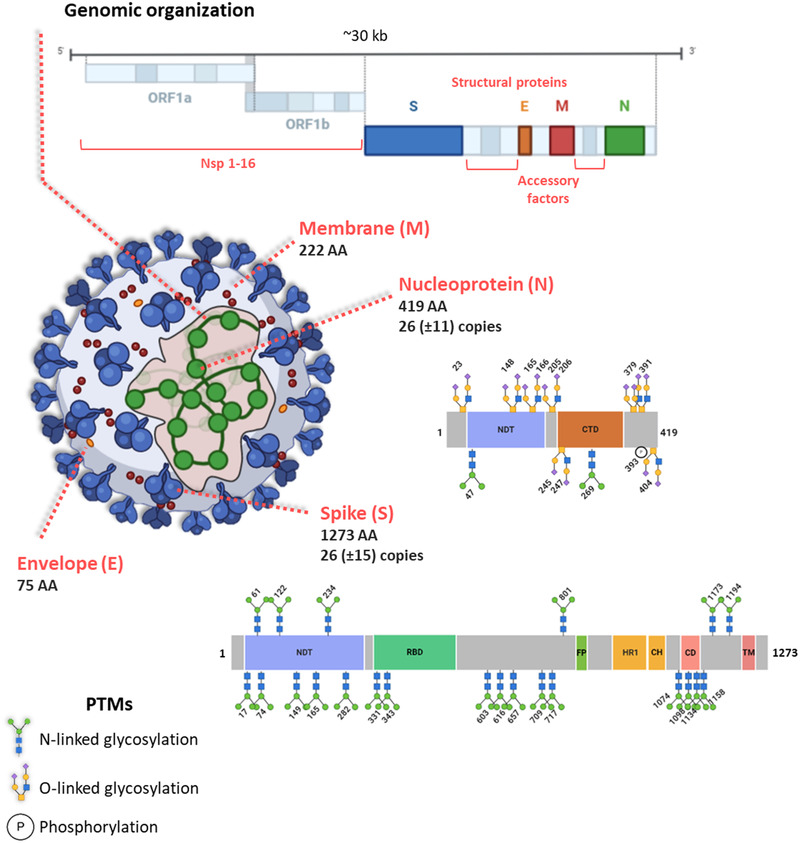
SARS‐CoV‐2 machinery. Genomic organization and main characteristics of the SARS‐CoV‐2 structural proteins S, M, E, and N. A schematic overview of the protein domains and identified PTMs on N and S as described in Supekar et al., 2020 and Watanabe et al., 2020, respectively, is illustrated. N‐terminal domain, C‐terminal domain (CTD), RBD, fusion peptide (FP), heptad repeat 1 (HR1), central helix (CH), connector domain (CD), and transmembrane domain (TM).
In line with variations in the number of SARS‐CoV‐2 RNA molecules measurable by RT‐qPCR and the number of infectious virus particles, determined by plaque assay titration, virus profiling by mass spectrometry can be used to monitor the kinetics of SARS‐CoV‐2 infection, thus contributing to the optimization of the production of whole viral particle antigens for vaccines.[ 9 ] In combination with this, efficient detection of almost all 29 annotated possible proteolytically mature SARS‐CoV‐2 proteins by mass spectrometry provided a library of high‐quality virus peptide spectra for targeted proteomics approaches.[ 13 , 14 , 15 ] Tandem mass spectrometry, by allowing the identification of peptides unique to variants of SARS‐CoV‐2 proteins has notably started to reveal significant findings with regard to protein cleavage, cell tropism, and infectivity.[ 16 ] Post‐translational modifications of structural proteins have also been studied. The trimeric S protein, which binds to the host's cell surface receptor, is heavily glycosylated. The exact structures of at least 22 N‐linked glycans were successfully established by mass spectrometry[ 17 ] and independently confirmed.[ 18 ] In addition, the nucleocapsid protein is decorated with O‐glycans and N‐glycans, at 7 and 2 confirmed sites, respectively.[ 19 ] The glycans of the spike protein act as a shield to thwart the host immune response, but a recent study has shown that N‐glycans at two sites (N165 & N234) modulate the conformational dynamics of the receptor‐binding domain (RBD).[ 20 ] The structure of the viral particle, established in exceptional detail using cryo‐electron tomography, revealed its extensive heterogeneity.[ 21 ] For example, the S protein is randomly distributed on each virion in 26 (±15) copies. Similarly, 26 (±11) copies of the ribonucleoprotein N are present, but how it contributes to the packing of the ≈30 kb RNA within the ≈80 nm diameter viral lumen, and whether it is involved in virus assembly remain to be clarified (Figure 1).
Due to their rather small number, to fulfill a variety of critical functions during the viral life cycle, viral proteins form various combinations of protein complexes. Li et al.[ 22 ] characterized 58 distinct intraviral protein–protein interactions (PPIs) among the 28 SARS‐CoV‐2 proteins potentially involved in virus replication. Among them, 20 overlap those of the SARS‐CoV PPI network, suggesting critical roles for these specific interactions in the Sarbecovirus family.
3. How Does SARS‐CoV‐2 Hijack the Host Cell Machinery so Efficiently?
Efforts to diagnose, treat, and produce a vaccine to prevent SARS‐CoV‐2 infection all benefit from an improved understanding of the basic biology of this pathogen. In a matter of months, the molecular examination of infected cells by unbiased proteomics approaches (Figure 2 ), including analysis of the infectome, phosphoproteome, ubiquitome, and interactome of SARS‐CoV‐2, unraveled the mechanisms employed by SARS‐CoV‐2 to bind, enter, hijack, and exit host cells. Similar to SARS‐CoV, the spike protein of SARS‐CoV‐2 interacts directly with the human ACE2 receptor to facilitate virus entry into host cells.[ 22 , 23 ] Interestingly, ACE2 and other potential entry factors such as TMPRSS2, TMPRSS4, CTSB, CTSL, BSG, and FURIN are expressed at different levels across cell lines. ACE2 and TMPRSS2 were reported not to be co‐expressed, even though these two proteins have been reported to act in concert to facilitate viral entry.[ 10 ] Very recently, neuropilin‐1 (NRP1) was also shown as a facilitator of SARS‐CoV‐2 cell entry and infectivity,[ 24 ] and maybe other alternative receptors/facilitators could be uncovered.
Figure 2.
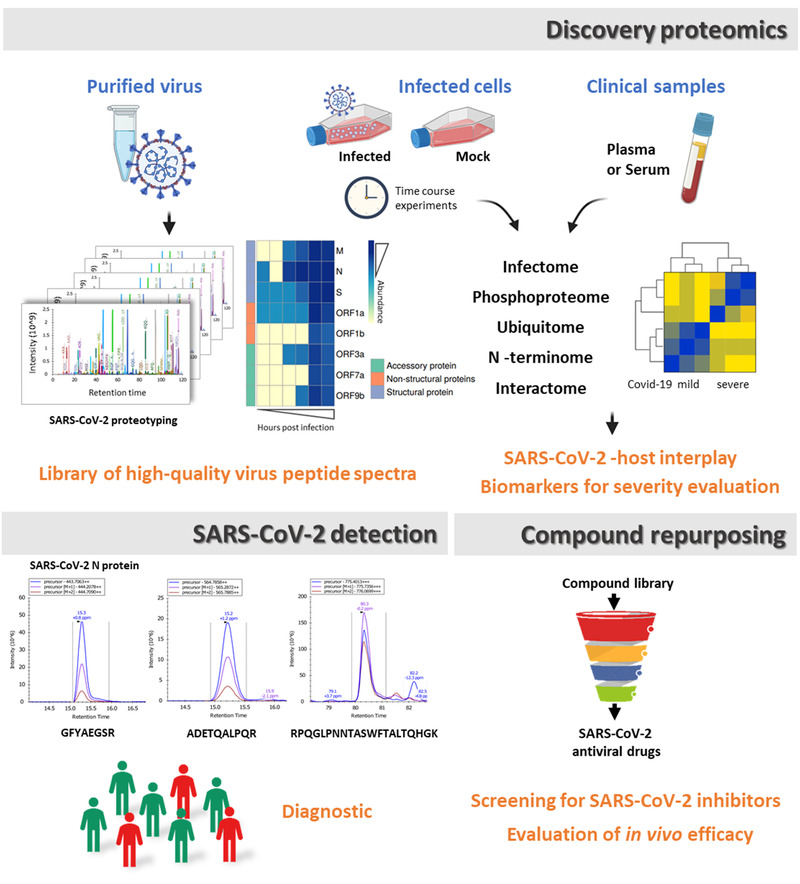
Proteomics in the COVID‐19 battlefield. Examination of infected cell models and clinical samples by system‐wide unbiased discovery proteomics approaches is used to unravel the mechanisms employed by SARS‐CoV‐2 to bind, enter, hijack, and exit the host. Together with data from the SARS‐CoV‐2 profiling by mass spectrometry, these analyses provided a library of high‐quality viral peptide spectra that can be used for targeted proteomics detection. In addition, insights from the various multi‐level proteomics datasets represent a valuable resource for the identification of promising targets for therapeutic intervention. Besides, quantitative proteomics represents a promising tool to support the screening of drug repurposing libraries and for the understanding of cellular changes in metabolism occurring following initiation of anti‐SARS‐CoV‐2 treatment.
Quantitative mass spectrometry with or without the use of stable isotopes has started to uncover the intricate mechanisms underlying SARS‐CoV‐2 infection, revealing details of several key processes used by the virus to adapt their host hijacking approach. Widespread changes observed in multiple metabolic pathways and biological processes, such as those related to innate immunity, RNA metabolism, and the cell cycle, led to the identification of signatures defining and driving COVID‐19, including a reduced antiviral response.[ 9 , 10 , 25 , 26 , 27 ] The extent to which SARS‐CoV‐2 suppresses the interferon (IFN) response is a key characteristic of COVID‐19.[ 28 ] Compared to other coronaviruses, SARS‐CoV‐2 fails to counteract phosphorylation of signal transducers and activators of transcription (STAT1 and STAT2) and expression of IFN‐stimulated genes proteins. The consequences of a dramatic rewiring of phosphorylation on host proteins were highlighted also by Bouhaddou et al.[ 26 ] Their quantitative mass spectrometry‐based phosphoproteomics survey of SARS‐CoV‐2 infected cells revealed how viral infection promotes casein kinase II (CK2) and p38 MAPK activation, production of diverse cytokines, shutdown of mitotic kinases, resulting in cell cycle arrest as well as a marked induction of CK2‐containing filopodial protrusions possessing budding viral particles. As an additional strategy to inhibit the innate immune response, once inside host cells, not only does the virus hijack the cell translational machinery, but the virulence factor non‐structural protein 1 (Nsp1) also shuts down translation of host messenger RNA.[ 29 ] Nonetheless, the extent to which SARS‐CoV‐2 uses these or other strategies and how they may be executed at a molecular level remains unclear.
Further insights into virus–host interplay and protein function during viral infections were obtained by the ambitious implementation of the complementary affinity purification (AP‐MS) and proximity‐dependent biotinylation (BioID) ‐based proteomics approaches. A successful intracellular viral life cycle relies indeed on molecular interactions with host proteins that are repurposed to support viral replication. The identification of high confidence PPIs, and their comparison with the SARS‐CoV interactome, brought to light potential virus‐specific interactions and the multitude of biological processes involved, for example, DNA replication, vesicle trafficking, signaling, and mitochondria‐related pathways.[ 27 , 30 , 31 , 32 ] Besides, the identification of common SARS‐CoV‐2 protein and human protein interactions following different approaches in several cell models suggest that these cellular processes are vital molecular targets of SARS‐CoV‐2 infection (Figure 3 ). Among these, associations between viral proteins and host ubiquitin pathway components like the E3 ligases TRIM59 and MYCBP2 suggest the potential modulation of the host ubiquitin system by SARS‐CoV‐2. Interestingly, the existence of an interplay between various functional control modes (e.g., PPI, protein abundance, and post‐translational modifications) is emerging as a strategy used by SARS‐CoV‐2 for concerted fine‐tuning of its regulation of these pathways. Additional links between viral polypeptides and host factors involved in multiple COVID19‐associated mechanisms, such as avoiding the innate immune response and manipulation of lipid trafficking, were revealed by BioID analyses which complement the potential lack of detection of poorly soluble protein partners or low‐affinity interactors of AP‐MS. Similar analyses in the context of human viral infection, and at various times during infection, could help to circumvent the limitations associated with artificial overproduction of individual viral proteins, paving the way for the discovery of time‐specific interactions to further refine our understanding of the SARS‐CoV‐2 infection profile.
Figure 3.
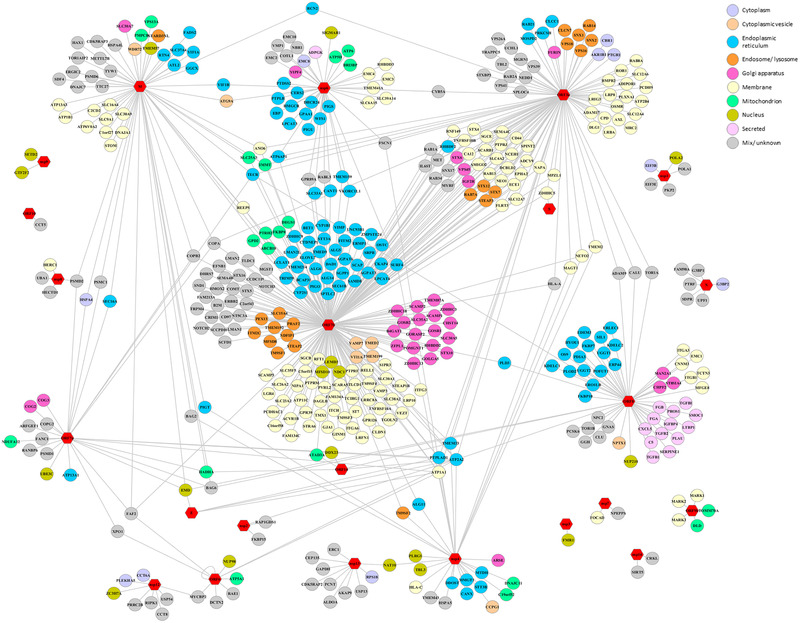
SARS‐CoV‐2‐host interactome. SARS‐CoV‐2‐Human PPIs commonly identified by both affinity purification (AP‐MS) and proximity‐dependent biotinylation (BioID) ‐based proteomics approaches. The network depicts PPIs described in[ 22 , 27 , 30 , 31 ] and,[ 32 , 56 ] respectively. The data are derived from BioGRID COVID‐19 Coronavirus Project.[ 57 ] The subcellular localization of human proteins is labeled with the indicated colors. Viral proteins are represented by red hexagons. PPIs described only following one of the proteomics approaches[ 27 , 31 , 32 ] are omitted.
Overall, through the insights they provide into how SARS‐CoV‐2 proteins operate to hijack host cells, these multi‐level proteomics datasets represent a valuable resource which could be mined to identify attractive targets for therapeutic intervention. As an example, clinically actionable drugs targeting human interactors of SARS‐CoV‐2 proteins and inhibiting mRNA translation or regulating the activity of Sigma1/2 receptors were identified following a chemo‐proteomic analysis.[ 31 ] 87 additional drugs and compounds, representing potential COVID‐19 therapies, were shortlisted by mapping global phosphorylation profiles to dysregulated kinases like p38, CK2, CDK, AXL, and PIKFYVE and their pathways.[ 26 ]
One of the most practicable strategies for the rapid identification and deployment of treatments for COVID‐19 is to reposition clinically evaluated drugs (Figure 2). To this end, an extraordinary number of investigational programs and clinical trials have been initiated since January 2020. While approved antiviral therapies, including inhibitors of HIV‐1, and hepatitis C virus proteases or the viral RNA polymerase inhibitor remdesivir, have been the focus of clinical investigations,[ 33 ] the elucidation of additional candidate therapies has been proposed to enable the development of combinatorial regimens. Toward this end, the high‐throughput analysis of nearly 12 000 known drugs either FDA approved or at different stages of clinical development foregrounded 21 molecules, including remdesivir, that inhibit SARS‐CoV‐2 in mammalian cells and with a dose–response relationship with their antiviral activity.[ 34 ] As part of these programs, quantitative proteomics analyses represent a new and promising tool with the potential to contribute to our understanding of the cellular changes in metabolism that occur following initiation of anti‐SARS‐CoV‐2 treatment.[ 35 ]
4. COVID‐19 Translational Proteomics
Obtaining an efficient vaccine to prevent infection was quickly a priority for public and private specialized laboratories. At the earliest stage of this development, in silico predictions based on immunoinformatics and structural analysis were proposed to identify the most probable immunogenic peptide targets.[ 36 ] The serological response of patient sera to SARS‐CoV‐2 infection can be probed with microarrays,[ 37 ] but mass spectrometry could be an interesting complementary tool in this quest.
Most COVID‐19 studies have focused on its epidemiological and clinical characteristics. Transcriptomics and proteomics of human lung tissue of fatal cases confirmed that neutrophil activation and pulmonary fibrosis are major upregulated pathways.[ 38 ] About 80% of patients infected with SARS‐CoV‐2 display mild symptoms and have good prognosis. Up to 10% suffer from respiratory distress which rapidly progresses to clinically severe disease. According to Zhang et al.,[ 39 ] disease severity appears to stem mostly from host factors, and viral genetic variation does not significantly affect outcomes. Even though our understanding of the pathophysiology of COVID‐19 is continuously improving, the clinical management of patients infected with SARS‐CoV‐2 remains challenging. To apply preventive measures and reduce the severity of outcomes, we crucially need to be able to stratify patients at diagnosis to better manage those who will develop critical disease. In this context, although COVID‐19 can be diagnosed by nucleic acid‐based methods at an early stage, the need to identify patients who will develop a severe disease form before their symptoms become manifest was rapidly raised by numerous researchers. With the help of machine learning, proteomic and metabolomic profiling of molecular changes induced by SARS‐CoV‐2 shed some light on the indicators associated with severe cases, based on expression levels for serum and urinary proteins and metabolites.[ 40 ] The potential of the various biomarkers shortlisted was further confirmed in a richer molecular compendium resulting from a large‐scale multi‐omics analysis.[ 41 ] The medical relevance of this predictive approach remains to be confirmed because these studies include restricted sample size, lack of longitudinal severity correlation, show older median age, and comorbidities of severe patients compared to patients with less severe disease. The challenges for establishing valuable protein biomarkers to assess COVID‐19 disease progression were recently discussed.[ 42 ]
Because disease severity varies considerably between patients, it is also essential to be able to identify infected individuals that develop no obvious symptoms. Serology may be a promising way to assess SARS‐CoV‐2 infection, but it remains unclear if all asymptomatic patients produce measurable antibody titers upon infection,[ 43 ] further stressing the need for biomarkers reflecting the healthy or diseased status.
5. The Potential of Mass Spectrometry for a Quick Detection of SARS‐CoV‐2
RT‐qPCR, the current reference method to identify SARS‐CoV‐2, is reliable, robust, and widely used in clinical settings. However, it requires specific oligonucleotide primers and is relatively expensive and time‐consuming as it requires RNA isolation and sample processing. The perfect diagnostic tool should give an accurate response within minutes of sampling, allowing immediate isolation of the affected patient and implementation of therapeutic actions. Protein mass profiling by MALDI‐TOF mass spectrometry became the gold standard in bacteriology a decade ago, but this approach is limited to pure isolate material.[ 44 ] An adaptation of the method was developed, acquiring MALDI‐TOF mass fingerprints for many nasal swab samples without prior sample purification.[ 45 ] In these data, the intensities of 88 peaks detected in the 3000–15500 m/z range were shown to discriminate between positive and negative SARS‐CoV‐2 samples but the methodology is not based on detection of SARS‐CoV‐2 proteins but rather host response, and has not been challenged against other infections. Interestingly, this approach had the same sensitivity as RT‐qPCR in the most informative range (Cycle threshold values below 37). Two other reports documented the potential of MALDI‐TOF for COVID‐19 diagnostics.[ 46 ] As recently commented,[ 47 ] further ongoing studies including a large number of samples should help establish this methodology and prove its performance capacity. Alternatively, viral peptide biomarkers could be monitored by tandem mass spectrometry, and this approach is likely to be relevant due to its high precision and sensitivity (Figure 2). A pioneering study short‐listed SARS‐CoV‐2 peptides for targeted proteomics detection, based on their abundance, ionizability, and detectability, and conservation across SARS‐CoV‐2 genome variants.[ 13 ] Targeted proteomics assays can be rapid, as discriminating peptides could be resolved and detected within a 3‐min chromatographic window.[ 14 ] This performance has recently been further improved, and the potential throughput of targeted proteomics has been convincingly documented.[ 48 ] Tandem mass spectrometry detection of SARS‐CoV‐2 has been applied successfully to nasal swabs and gargle samples[ 9 , 15 , 49 ] but on a limited number of clinical samples. Significantly, tandem mass spectrometry‐based proteotyping can easily be adapted to new variants of SARS‐CoV‐2 that might appear as the pandemic progresses,[ 50 ] and protocols could be rapidly adjusted to detect other respiratory viruses that may raise concerns in the future. The major current drawbacks of the methodology—sample preparation time, mass spectrometry expertise, throughput, and cost of high‐resolution instruments—were recently discussed,[ 51 ] but in our opinion could be mitigated through investments, as the current outlook for the diagnostics market is excellent. Following targeted investment, proteomics‐based diagnosis could become much more common in the near future.
6. Perspectives
The COVID‐19 pandemic has been the most serious infectious threat in a century. After a difficult first semester, with unprecedented lockdowns and massive changes in social behavior around the world, the current situation shows that viral resurgence can be swift and devastating. Better and earlier detection of the SARS‐CoV‐2 virus in infected patients is necessary to break chains of transmission. New antiviral drugs will also be needed to improve the status of patients whose bodies cannot cope with the infection. We believe that proteomics has a lot to offer in this fight. In this review, we have highlighted the most recent works in this area as summarized in Figure 1. In the near future, other approaches may provide complementary information related to the SARS‐CoV‐2 virus, the disease itself COVID‐19, and its long‐term consequences on human health. Comparative proteomics could help to establish which types of cells are susceptible to viral infection, depending on the mode of infection. The many facets of proteomics could provide new information to understand and more efficiently treat COVID‐19. Thus, top‐down proteomics approaches would be useful to better assess post‐translational modifications of viral components while also shedding light on their heterogeneity and roles. Thermal proteome profiling could be used to further describe the various complexes in which viral proteins are involved, and to determine how viral proteins hijack the host cell's metabolism. Metaproteomics could help decipher the long‐term consequences on the lung microbiota in severe or mild cases. More than ever, international collaborative pluridisciplinary efforts must be considered if we are to win this crucial battle.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank the Commissariat à l'Energie Atomique et aux Energies Alternatives (France) and the ANR program “Phylopeptidomics” (ANR‐17‐CE18‐0023‐01) for financial support.
Biographies
Lucia Grenga is a researcher in the laboratory of Innovative Technologies for Detection and Diagnostics at the French Alternative Energies and Atomic Energy Commission (CEA). She is interested in the development of tandem mass spectrometry‐based methodologies and their integration with other omics approaches for the characterization of clinically relevant microbiomes. She received her Ph.D. in Cellular and Molecular Biology in 2010 at the University of Rome “Tor Vergata”. From 2016, she is specialist in Clinical Pathology.

Jean Armengaud is Chief Deputy of the laboratory of Innovative technologies for Detection and Diagnostics located near Avignon in France. He is also Director of the ProGénoMIX platform, specialized in proteogenomics and metaproteomics. He wishes to contribute to a better understanding of the functioning of complex biological systems and exploit this knowledge for medical and environmental purposes. He received his Ph.D. in Biochemistry in 1994 at the University of Grenoble.

Grenga L., Armengaud J., Proteomics in the COVID‐19 Battlefield: First Semester Check‐Up. Proteomics 2020, 21, 2000198. 10.1002/pmic.202000198
References
- 1. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., Zhang Y. Z., Nature 2020, 579, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armengaud J., Expert Rev. Proteomics 2020, 17, 507. [DOI] [PubMed] [Google Scholar]
- 4. Struwe W., Emmott E., Bailey M., Sharon M., Sinz A., Corrales F. J., Thalassinos K., Braybrook J., Mills C., Barran P., Lancet 2020, 395, 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lachén‐Montes M., Corrales F. J., Fernández‐Irigoyen J., Santamaría E., Frontiers in Microbiology 2020, 11, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Coombs K. M., Expert Rev. Proteomics 2020, 17, 513. [DOI] [PubMed] [Google Scholar]; b) Sperk M., van Domselaar R., Rodriguez J. E., Mikaeloff F., Sa Vinhas B., Saccon E., Sonnerborg A., Singh K., Gupta S., Vegvari A., Neogi U., J. Proteome Res. 2020, 19, 4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W., Tan W., Lancet 2020, 395, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R., Cells 2020, 9, 1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grenga L., Gallais F., Pible O., Gaillard J. C., Gouveia D., Batina H., Bazaline N., Ruat S., Culotta K., Miotello G., Debroas S., Roncato M. A., Steinmetz G., Foissard C., Desplan A., Alpha‐Bazin B., Almunia C., Gas F., Bellanger L., Armengaud J., Emerging Microbes Infect. 2020, 9, 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zecha J., Lee C. Y., Bayer F. P., Meng C., Grass V., Zerweck J., Schnatbaum K., Michler T., Pichlmair A., Ludwig C., Kuster B., Mol. Cell. Proteomics 2020, 19, 1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Finkel Y., Mizrahi O., Nachshon A., Weingarten‐Gabbay S., Morgenstern D., Yahalom‐Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth‐Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern‐Ginossar N., Nature 2020, In press. [DOI] [PubMed] [Google Scholar]; b) Kim D., Lee J. Y., Yang J. S., Kim J. W., Kim V. N., Chang H., Cell 2020, 181, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meyer B., Chiaravalli J., Brownridge P., Bryne D. P., Daly L. A., Agou F., Eyers C. E., Eyers P. A., Vignuzzi M., Emmott E., bioRxiv 2020, 10.1101/2020.09.16.297945. [DOI] [Google Scholar]
- 13. Gouveia D., Grenga L., Gaillard J. C., Gallais F., Bellanger L., Pible O., Armengaud J., Proteomics 2020, 20, 2000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gouveia D., Miotello G., Gallais F., Gaillard J. C., Debroas S., Bellanger L., Lavigne J. P., Sotto A., Grenga L., Pible O., Armengaud J., J. Proteome Res. 2020, 19, 4407. [DOI] [PubMed] [Google Scholar]
- 15. Ihling C., Tanzler D., Hagemann S., Kehlen A., Huttelmaier S., Arlt C., Sinz A., J. Proteome Res. 2020, 19, 4389. [DOI] [PubMed] [Google Scholar]
- 16. Davidson A. D., Williamson M. K., Lewis S., Shoemark D., Carroll M. W., Heesom K. J., Zambon M., Ellis J., Lewis P. A., Hiscox J. A., Matthews D. A., Genome Med. 2020, 12, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe Y., Allen J. D., Wrapp D., McLellan J. S., Crispin M., Science 2020, 369, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y., Zhao W., Mao Y., Chen Y., Wang S., Zhong Y., Su T., Gong M., Du D., Lu X., Cheng J., Yang H., Mol. Cell. Proteomics 2020, 19, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Supekar N. T., Shajahan A., Gleinich A. S., Rouhani D., Heiss C., Azadi P., bioRxiv 2020, 10.1101/2020.08.26.269043. [DOI] [Google Scholar]
- 20. Casalino L., Gaieb Z., Goldsmith J. A., Hjorth C. K., Dommer A. C., Harbison A. M., Fogarty C. A., Barros E. P., Taylor B. C., McLellan J. S., Fadda E., Amaro R. E., ACS Cent. Sci. 2020, 6, 1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., Cheng L., Shi D., Lu X., Lei J., Crispin M., Shi Y., Li L., Li S., Cell 2020, 183, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li G., He X., Zhang L., Ran Q., Wang J., Xiong A., Wu D., Chen F., Sun J., Chang C., J. Autoimmun. 2020, 112, 102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffmann M., Kleine‐Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Muller M. A., Drosten C., Pohlmann S., Cell 2020, 181, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cantuti‐Castelvetri L., Ojha R., Pedro L. D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio‐Kokko H., Osterlund P., Joensuu M., Meunier F. A., Butcher S. J., Winkler M. S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Science 2020, 370, 856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Munch C., Nature 2020, 583, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bouhaddou M., Memon D., Meyer B., White K. M., Rezelj V. V., Correa Marrero M., Polacco B. J., Melnyk J. E., Ulferts S., Kaake R. M., Batra J., Richards A. L., Stevenson E., Gordon D. E., Rojc A., Obernier K., Fabius J. M., Soucheray M., Miorin L., Moreno E., Koh C., Tran Q. D., Hardy A., Robinot R., Vallet T., Nilsson‐Payant B. E., Hernandez‐Armenta C., Dunham A., Weigang S., Knerr J., Modak M., et al., Cell 2020, 182, 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stukalov A., Girault V., Grass V., Bergant V., Karayel O., Urban C., Haas D. A., Huang Y., Oubraham L., Wang A., Hamad S. M., Piras A., Tanzer M., Hansen F. M., Enghleitner T., Reinecke M., Lavacca T. M., Ehmann R., Wölfel R., Jores J., Kuster B., Protzer U., Rad R., Ziebuhr J., Thiel V., Scaturro P., Mann M., Pichlmair A., bioRxiv 2020, 10.1101/2020.06.17.156455. [DOI] [Google Scholar]
- 28. Lokugamage K. G., Hage A., de Vries M., Valero‐Jimenez A. M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V. D., J. Virol. 2020, 94, e01410‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.a) Schubert K., Karousis E. D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L. A., Leibundgut M., Thiel V., Muhlemann O., Ban N., Nat. Struct. Mol. Biol. 2020, 27, 959; [DOI] [PubMed] [Google Scholar]; b) Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens‐Kiani T., Cheng J., Straub J. H., Sturzel C. M., Frohlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K. M. J., Beckmann R., Science 2020, 369, 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davies J. P., Almasy K. M., McDonald E. F., Plate L., bioRxiv 2020. [Google Scholar]
- 31. Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., O'Meara M. J., Rezelj V. V., Guo J. Z., Swaney D. L., Tummino T. A., Huttenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B. J., Braberg H., Fabius J. M., Eckhardt M., Soucheray M., Bennett M. J., Cakir M., McGregor M. J., Li Q., et al., Nature 2020, 583, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.a) Laurent E. M. N., Sofianatos Y., Komarova A., Gimeno J.‐P., Tehrani P. S., Kim D.‐K., Abdouni H., Duhamel M., Cassonnet P., Knapp J. J., Kuang D., Chawla A., Sheykhkarimli D., Rayhan A., Li R., Pogoutse O., Hill D. E., Calderwood M. A., Falter‐Braun P., Aloy P., Stelzl U., Vidal M., Gingras A.‐C., Pavlopoulos G. A., Van Der Werf S., Fournier I., Roth F. P., Salzet M., Demeret C., Jacob Y., Coyaud E., bioRxiv 2020, 10.1101/2020.08.28.272955; [DOI] [Google Scholar]; b) St‐Germain J. R., Astori A., Samavarchi‐Tehrani P., Abdouni H., Macwan V., Kim D.‐K., Knapp J. J., Roth F. P., Gingras A.‐C., Raught B., bioRxiv 2020, 10.1101/2020.08.28.269175. [DOI] [Google Scholar]
- 33. Li G., De Clercq E., Nat. Rev. Drug Discovery 2020, 19, 149. [DOI] [PubMed] [Google Scholar]
- 34. Riva L., Yuan S., Yin X., Martin‐Sancho L., Matsunaga N., Pache L., Burgstaller‐Muehlbacher S., De Jesus P. D., Teriete P., Hull M. V., Chang M. W., Chan J. F., Cao J., Poon V. K., Herbert K. M., Cheng K., Nguyen T. H., Rubanov A., Pu Y., Nguyen C., Choi A., Rathnasinghe R., Schotsaert M., Miorin L., Dejosez M., Zwaka T. P., Sit K. Y., Martinez‐Sobrido L., Liu W. C., White K. M., Chapman M. E., Lendy E. K., Glynne R. J., Albrecht R., Ruppin E., Mesecar A. D., Johnson J. R., Benner C., Sun R., Schultz P. G., Su A. I., Garcia‐Sastre A., Chatterjee A. K., Yuen K. Y., Chanda S. K., Nature 2020, 586, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamamoto K. A., Blackburn K., Migowski E., Goshe M. B., Brown D. T., Ferreira D. F., Soares M. R., Sci. Rep. 2020, 10, 14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tilocca B., Britti D., Urbani A., Roncada P., J. Proteome Res. 2020, 19, 4233. [DOI] [PubMed] [Google Scholar]
- 37. Zamecnik C. R., Rajan J. V., Yamauchi K. A., Mann S. A., Loudermilk R. P., Sowa G. M., Zorn K. C., Alvarenga B. D., Gaebler C., Caskey M., Stone M., Norris P. J., Gu W., Chiu C. Y., Ng D., Byrnes J. R., Zhou X. X., Wells J. A., Robbiani D. F., Nussenzweig M. C., DeRisi J. L., Wilson M. R., Cell Rep Med 2020, 1, 100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu M., Chen Y., Xia H., Wang C., Tan C. Y., Cai X., Liu Y., Ji F., Xiong P., Liu R., Guan Y., Duan Y., Kuang D., Xu S., Cai H., Xia Q., Yang D., Wang M. W., Chiu I. M., Cheng C., Ahern P. P., Liu L., Wang G., Surana N. K., Xia T., Kasper D. L., Proc Natl Acad Sci U. S. A. 2020, 117, 28336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang X., Tan Y., Ling Y., Lu G., Liu F., Yi Z., Jia X., Wu M., Shi B., Xu S., Chen J., Wang W., Chen B., Jiang L., Yu S., Lu J., Wang J., Xu M., Yuan Z., Zhang Q., Zhao G., Wang S., Chen S., Lu H., Nature 2020, 583, 437. [DOI] [PubMed] [Google Scholar]
- 40.a) Di B., Jia H., Luo O. J., Lin F., Li K., Zhang Y., Wang H., Liang H., Fan J., Yang Z., Signal Transduction Targeted Ther. 2020, 5, 217; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Messner C. B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., Textoris‐Taube K., Vernardis S. I., Egger A. S., Kreidl M., Ludwig D., Kilian C., Agostini F., Zelezniak A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., von Kalle C., Campbell A., Hayward C., Porteous D. J., Marioni R. E., Langenberg C., Lilley K. S., Kuebler W. M., Mulleder M., Drosten C., Suttorp N., Witzenrath M., Kurth F., Sander L. E., Ralser M., Cell Syst. 2020, 11, 11; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Guo T., Cell 2020, 182, 59; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wendt R., Kalbitz S., Lubbert C., Kellner N., Macholz M., Schroth S., Ermisch J., Latosisnka A., Arnold B., Mischak H., Beige J., Metzger J., Proteomics 2020, 2000202. [DOI] [PubMed] [Google Scholar]
- 41. Overmyer K. A., Shishkova E., Miller I. J., Balnis J., Bernstein M. N., Peters‐Clarke T. M., Meyer J. G., Quan Q., Muehlbauer L. K., Trujillo E. A., He Y., Chopra A., Chieng H. C., Tiwari A., Judson M. A., Paulson B., Brademan D. R., Zhu Y., Serrano L. R., Linke V., Drake L. A., Adam A. P., Schwartz B. S., Singer H. A., Swanson S., Mosher D. F., Stewart R., Coon J. J., Jaitovich A., Cell Syst. 2020, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Whetton A. D., Preston G. W., Abubeker S., Geifman N., J. Proteome Res. 2020, 19, 4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tuaillon E., Bollore K., Pisoni A., Debiesse S., Renault C., Marie S., Groc S., Niels C., Pansu N., Dupuy A. M., Morquin D., Foulongne V., Bourdin A., Le Moing V., Van de Perre P., J. Infect. 2020, 81, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grenga L., Pible O., Armengaud J., Clinical Mass Spectrometry 2019, 14, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nachtigall F. M., Pereira A., Trofymchuk O. S., Santos L. S., Nat. Biotechnol. 2020, 38, 1168. [DOI] [PubMed] [Google Scholar]
- 46.a) Iles R. K., Zmuidinaite R., Iles J. K., Carnell G., Sampson A., Heeney J. L., Diagnostics 2020, 10, 746; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rocca M. F., Zintgraff J. C., Dattero M. E., Santos L. S., Ledesma M., Vay C., Prieto M., Benedetti E., Avaro M., Russo M., Nachtigall F. M., Baumeister E., J. Virol. Methods 2020, 286, 113991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. SoRelle J. A., Patel K., Filkins L., Park J. Y., Clin. Chem. 2020, 66, 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh P., Chakraborty R., Marwal R., Radhakrishan V. S., Bhaskar A. K., Vashisht H., Dhar M. S., Pradhan S., Ranjan G., Imran M., Raj A., Sharma U., Singh P., Lall H., Dutta M., Garg P., Ray A., Dash D., Sivasubbu S., Gogia H., Madan P., Kabra S., Singh S. K., Agrawal A., Rakshit P., Kumar P., Sengupta S., J. Proteins Proteomics 2020, 11, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nikolaev E. N., Indeykina M. I., Brzhozovskiy A. G., Bugrova A. E., Kononikhin A. S., Starodubtseva N. L., Petrotchenko E. V., Kovalev G. I., Borchers C. H., Sukhikh G. T., J. Proteome Res. 2020, 19, 4393. [DOI] [PubMed] [Google Scholar]
- 50. Armengaud J., Delaunay‐Moisan A., Thuret J. Y., van Anken E., Acosta‐Alvear D., Aragon T., Arias C., Blondel M., Braakman I., Collet J. F., Courcol R., Danchin A., Deleuze J. F., Lavigne J. P., Lucas S., Michiels T., Moore E. R. B., Nixon‐Abell J., Rossello‐Mora R., Shi Z. L., Siccardi A. G., Sitia R., Tillett D., Timmis K. N., Toledano M. B., van der Sluijs P., Vicenzi E., Environ. Microbiol. 2020, 22, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.a) Grossegesse M., Hartkopf F., Nitsche A., Schaade L., Doellinger J., Muth T., J. Proteome Res. 2020, 19, 4380; [DOI] [PubMed] [Google Scholar]; b) Mahmud I., Garrett T. J., J. Am. Soc. Mass Spectrom. 2020, 31, 2013. [DOI] [PubMed] [Google Scholar]
- 52. Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A. R., Tascher G., Geurink P. P., Wilhelm A., van der Heden van Noort G. J., Ovaa H., Muller S., Knobeloch K. P., Rajalingam K., Schulman B. A., Cinatl J., Hummer G., Ciesek S., Dikic I., Nature 2020, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Juno J. A., Tan H. X., Lee W. S., Reynaldi A., Kelly H. G., Wragg K., Esterbauer R., Kent H. E., Batten C. J., Mordant F. L., Gherardin N. A., Pymm P., Dietrich M. H., Scott N. E., Tham W. H., Godfrey D. I., Subbarao K., Davenport M. P., Kent S. J., Wheatley A. K., Nat. Med. 2020, 26, 1428. [DOI] [PubMed] [Google Scholar]
- 54. Zhao P., Praissman J. L., Grant O. C., Cai Y., Xiao T., Rosenbalm K. E., Aoki K., Kellman B. P., Bridger R., Barouch D. H., Brindley M. A., Lewis N. E., Tiemeyer M., Chen B., Woods R. J., Wells L., Cell Host Microbe 2020, 28, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. D'Alessandro A., Thomas T., Dzieciatkowska M., Hill R. C., Francis R. O., Hudson K. E., Zimring J. C., Hod E. A., Spitalnik S. L., Hansen K. C., J. Proteome Res. 2020, 19, 4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Samavarchi‐Tehrani P., Abdouni H., Knight J. D. R., Astori A., Samson R., Lin Z.‐Y., Kim D.‐K., Knapp J. J., St‐Germain J., Go C. D., Larsen B., Wong C. J., Cassonnet P., Demeret C., Jacob Y., Roth F. P., Raught B., Gingras A.‐C., bioRxiv preprint 2020, 10.1101/2020.09.03.282103. [DOI] [Google Scholar]
- 57. Oughtred R., Rust J., Chang C., Breitkreutz B. J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Zhang F., Dolma S., Coulombe‐Huntington J., Chatr‐Aryamontri A., Dolinski K., Tyers M., Protein Sci. 2020, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]


