Abstract
SARS‐CoV‐2 is a novel human coronavirus responsible for the Coronavirus disease 2019 (COVID‐19) pandemic. Pneumonia and acute respiratory distress syndrome are the major complications of COVID‐19. SARS‐CoV‐2 infection can activate innate and adaptive immune responses and result in massive inflammatory responses later in the disease. These uncontrolled inflammatory responses may lead to local and systemic tissue damage. In patients with severe COVID‐19, eosinopenia and lymphopenia with a severe reduction in the frequency of CD4+ and CD8+ T cells, B cells and natural killer (NK) cells are a common feature. COVID‐19 severity hinges on the development of cytokine storm characterized by elevated serum levels of pro‐inflammatory cytokines. Moreover, IgG‐, IgM‐ and IgA‐specific antibodies against SARS‐CoV‐2 can be detected in most patients, along with the viral RNA, forming the basis for assays that aid in patient diagnosis. Elucidating the immunopathological outcomes due to COVID‐19 could provide potential targets for immunotherapy and are important for choosing the best clinical management by consultants. Currently, along with standard supportive care, therapeutic approaches to COVID‐19 treatment involve the use of antiviral agents that interfere with the SARS‐CoV‐2 lifecycle to prevent further viral replication and utilizing immunomodulators to dampen the immune system in order to prevent cytokine storm and tissue damage. While current therapeutic options vary in efficacy, there are several molecules that were either shown to be effective against other viruses such as HIV or show promise in vitro that could be added to the growing arsenal of agents used to control COVID‐19 severity and spread.
Keywords: acute respiratory distress syndrome, coronavirus disease 2019, COVID‐19, cytokine storm, hyperinflammation, immunopathology
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has been declared as a new pandemic by the World Health Organization (WHO) 1 on 11 March 2020. COVID‐19 causes both pneumonia and acute respiratory distress syndrome (ARDS). Other COVID‐19 complications may include acute liver, cardiac and kidney injury, as well as secondary infection and inflammatory response. It was revealed that there is no protective immunity against the virus and this later is capable of escaping innate immune responses. 2 The mechanism of innate immune sensing serves as the first line of antiviral defence, which constitutes an essential aspect of immunity to viruses. 3 The pathway is initiated through the engagement of pattern recognition receptors (PRRs), which upon activation triggers cytokine secretion (most importantly being type I/III IFNs). However, SARS‐CoVs have developed several mechanisms to inhibit IFN‐I induction and signalling. SARS‐CoV‐2 has also been suggested to lack robust IFN‐I/III signatures from infected cell lines, primary bronchial cells and a ferret model. 4 Therefore, primary infected tissues are characterized by virus proliferation resulting in cell death and virus release followed by the recruitment of immune cells, the immune complex generation and associated organ damage. 2
Both innate and adaptive immunity can be activated by SARS‐CoV‐2 infection. It was revealed that infection of mononuclear cells in addition to the immune cells recruitment can result in massive inflammatory responses in the later phase of the disease. 2 These uncontrolled inflammatory immune responses may lead to local and systemic tissue damage. In patients with severe COVID‐19, eosinopenia and lymphopenia with a severe reduction in the frequency of CD4+ and CD8+ T cells, B cells and natural killer (NK) cells are a common feature. 5 , 6 Several mechanisms may likely contribute to the lymphopenia observed in COVID‐19 patients, including effects from the cytokine milieu. Diao et al and Wan et al showed a correlation between lymphopenia and IL‐6, IL‐10 and TNF‐α, while restored bulk T cells frequencies paired with overall lower pro‐inflammatory cytokine levels were observed in convalescent patients. 7 , 8 T cell recirculation in the blood may be inhibited by cytokines such as IFN‐I and TNF‐α through the promotion of retention in lymphoid organs and attachment to the endothelium. 9 , 10 Additionally, a robust humoral immune response (B cell response) is also triggered as evidenced by the near‐universal detection of virus‐specific IgM, IgG and IgA, and neutralizing IgG‐antibody (nAbs) in the days following infection. Several studies 11 , 12 , 13 have demonstrated seroconversion occurring in COVID‐19 patients between 7 and 14 days after the onset of symptoms with persistent antibody titres in the weeks following viral clearance. However, long‐term memory response has not yet been established as a result of the timing of the outbreak, but a study by Thevaranjan et al 14 has demonstrated the induction of CD38Hi and CD27Hi antibody‐secreting cells (ASCs) concomitant with an increase with circulating follicular T helper cells (Tfh). Another study by Guo et al 15 using the scRNA‐seq study of peripheral blood mononuclear cell (PBMCs) from critically ill and recently recovered patients demonstrated an increase in the plasma cell population. In addition, RBD‐specific memory cells (IgG) have been identified in COVID‐19 patients. 16 Neutrophilia and an increase in the neutrophil/lymphocyte ratio usually are accompanied by advanced severity and poor clinical outcome. 17 Most severe patients show ‘cytokine storm’ (CS) characterized by higher pro‐inflammatory cytokines in the serum. 5 , 18 Moreover, IgG‐, IgM‐ and IgA‐specific antibodies against SARS‐CoV‐2 can be detected in most patients. Elucidating these immunopathological changes could provide possible targets for immunotherapy and are imperative for choosing the best clinical management by consultants. 19
In this review, we will provide an overview of the interaction between SARS‐CoV‐2 and the immune system and the consequences as a result of dysfunctional immune responses to organ damages. Finally, we will briefly state the implications of these approaches for COVID‐19 serological diagnosis and potential immunotherapy that target viral replication as well as boosting or suppression of immune response wherever necessary.
2. COVID‐19 IMMUNOPATHOLOGY
The effective response of the human innate and adaptive immunity against viruses includes the secretion of several pro‐inflammatory cytokines and the activation of several subsets of T cells which are vital for controlling the viral replication, restraining the spread of the virus, restricting inflammation and ‘cleaning’ the infected cells. 20 , 21 , 22 T cell response in the healthy state is a finely balanced set of events composed of the principal populations of reactive T cells. 23
Persistent viral antigen stimulation results in CD8+ T cell exhaustion, which reflects a decline in effector functions and also the proliferative capacity; these exhausted cells are denoted—Tex. 24 Tex manifest over‐expression of inhibitory receptors, including CD279 (PD‐1), a lymphoid cell surface protein of the Ig superfamily, and a member of the extended CD28/CTLA‐4 family of T cell regulators, which acts as a mature T cell checkpoint for the modulation of apoptosis. PD‐1 interaction with either of its ligands (PD‐1L1 or PD‐1L2—both members of the B7 family of T cell co‐receptors that includes CD28) constitutes significant negative immune checkpoints in the pathway responsible for blunting cell‐mediated immune responses, specifically CD8+ responses and for upregulating resulting pathologies. 25 , 26 In addition, exhaustion markers, such as NKG2A, are upregulated in NK cells and cytotoxic T lymphocytes in patients with COVID‐19. 27
2.1. Hyperinflammation in COVID‐19
The tissue injury caused by SARS‐CoV‐2 leads to the excessive secretion of pro‐inflammatory cytokines and the recruitment of other pro‐inflammatory cells such as granulocytes and macrophages (Figure 1). 28 This results in a snowballing of cytokine secretion and leucocytes recruitment causing a systemic inflammatory response termed as a macrophage activation syndrome (MAS) or secondary hemophagocytic lymphohistiocytosis (sHLH) popularly called cytokine storm (CS). 28 Published data obtained from SARS‐CoV‐2 infected patients have indicated that severe cases are usually depicted by a CS usually progressing to ARDS. 5 , 18 Various aspects of COVID‐19, such as the immune profile, serological markers and clinical features, have been reported to be similar to those of other viral infections. 29 Interestingly, it was shown that the severity of COVID‐19 is related to the level of the pro‐inflammatory cytokines and cellular immune profile (Figure 2). 30
Figure 1.
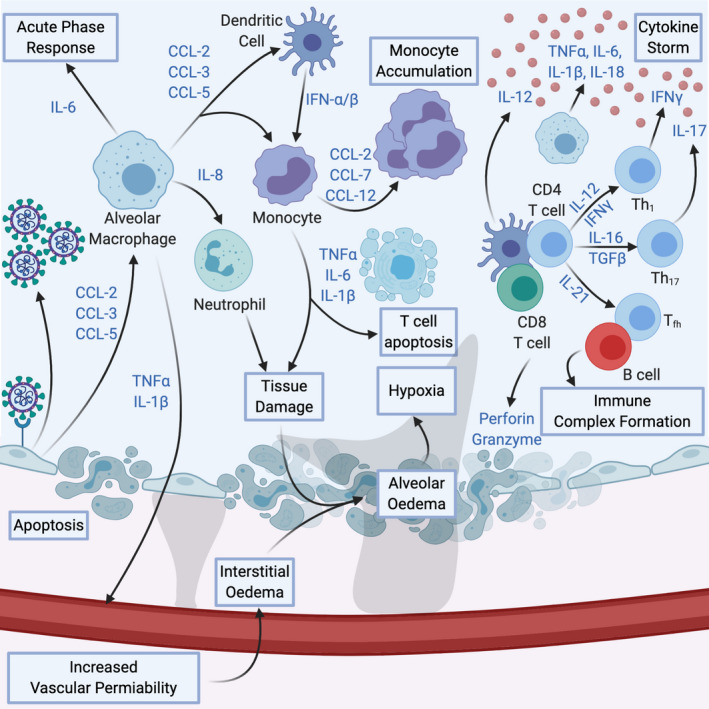
COVID‐19 Immunopathology. Cytokines produced by infected cells recruit alveolar macrophages, which in turn increase vascular permeability, recruit other components of the immune system and mount the acute phase response. Tissue damage caused by death of infected cells and killed cells through immune cells causes’ alveolar oedema, leading to hypoxia. Hyperactivation of both innate and adaptive immune responses induces cytokine storm. These factors converge into the progressive development of ARDS
Figure 2.
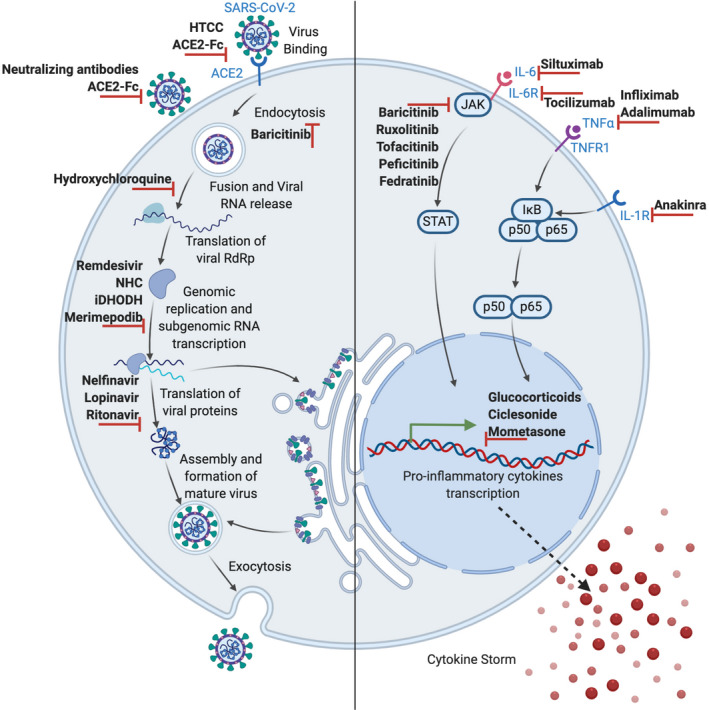
Therapeutic approaches in COVID‐19. Treatments involve either interference with the SARS‐CoV‐2 life cycle (left) or suppression of the hyperactive inflammatory immune response during the course of SARS‐CoV‐2 infection using immunomodulatory and anti‐inflammatory agents (right)
In COVID‐19 patients, several cytokines and chemokines have been shown to have different levels from the mild to severe stage of the disease. 29 A study analysed several cytokines and found the following to be increased in the plasma of patients infected with SARS‐COV‐2: IL‐1β, IL‐1RA, IL‐7, IL‐8, IL‐10, IFN‐ɣ, MCP‐1, MIP‐1α, G‐CSF and TNF‐α. 18 Moreover, these elevated levels correlate with disease severity in another study, higher levels of IL‐2 and IL‐6 in plasma, in addition to the aforementioned was observed in severe infection. 5 Both studies have reported the correlation between these increased cytokine levels and worsening lung injury. An elevated level of IL‐6, a significant contributor to CS, has been observed both in mild and severe SARS‐CoV‐2 infected patients, with a substantial increase of this cytokine level in patients with severe disease progression than those with mild or non‐severe SARS‐CoV‐2. 5 , 31
2.2. Mechanism of cytokine storm and ARDS in COVID‐19
The role of CS in ARDS development highlights the similarity of COVID‐19 to other human coronavirus infections including severe acute respiratory syndrome coronavirus (SARS‐CoV) and middle east respiratory syndrome coronavirus (MERS‐CoV). 32 Experiments using respiratory epithelial cells demonstrate that the release of immune modulators such as cytokines and chemokines is delayed which hinders DCs and macrophage recruitment during the initial stages of SARS‐CoV infection. 33 Subsequently, it was observed that the cells discharge low levels of the antiviral IFNs and increased levels of pro‐inflammatory cytokines primarily IL‐1β, IL‐6 and TNF, and chemokines CCL‐2, CCL‐3 and CCL‐5. 33 Like SARS‐CoV and MERS‐CoV, the SARS‐CoV‐2 virus infects human airway epithelial cells and possibly other ACE‐2 expressing tissues. THP‐1 cells, macrophages and DCs sustain delayed but elevated levels of pro‐inflammatory cytokines and chemokines. 34 Studies in MERS‐CoV infection showed that the major producers of interferons are pDCs and not mononuclear macrophages or other dendritic cells. 35
In MERS patients, the elevated number of neutrophils and monocytes in the alveolar tissues and peripheral blood of patients correlated with the elevated cytokine and chemokine levels implying that these cells may contribute to lung pathology. 36 Similar observations were seen in patients with SARS‐CoV infection. 37 The production of the IFN‐I or IFN‐α/β is the crucial natural immunity against most viral infections, and IFN‐I is vital in the early stages of viral infection. 38 In SARS‐CoV and MERS‐CoV infection, delayed release of IFNs hampers the body's antiviral response in the initial phases of infection. 39 The rapid and sustained rise in cytokines and chemokines invites many other inflammatory cells, such as neutrophils and monocytes, leading to a disproportionate infiltration of the inflammatory cells into the alveolar tissue and thus causes lung injury. It seems from these reports that either a dysregulated or exaggerated cytokine and chemokine responses or both by virus‐infected cells could play an important role in the progression and pathological features of SARS or MERS. Thus, the same can be inferred to SARS‐CoV‐2. 32 , 40
The amassed mononuclear macrophages are activated via the IFN‐α/β receptors present on their surface and secrete more monocyte chemo‐attractants (such as CCL2, CCL7 and CCL12), resulting in the further attraction and aggregation of mononuclear macrophages. These cells yield significantly high levels of TNF, IL‐6, IL1‐β and reactive free radicals such as iNOS, thus, increasing the disease severity. Also, the IFN‐α/β and other pro‐inflammatory cytokines secreted by mononuclear macrophages encourage T cells apoptosis, which further impedes viral clearance. 41 Another end result of rapid viral replication and a robust pro‐inflammatory cytokine/chemokine response is the initiation of apoptosis in respiratory epithelial and endothelial cells. The mechanism of this apoptosis is Fas–Fas ligand (FasL) or TRAIL‐death receptor 5 as a result of IFN‐αβ and IFN‐γ ‐induced inflammatory cell infiltration. 42 , 43 Apoptosis of respiratory endothelial cells leads to vascular leakage and alveolar oedema. On the other hand, the wearing‐out of epithelial cells causes injuries to the pulmonary microvascular and alveolar epithelial cell barriers and eventually leads to hypoxia. 32 , 40
2.3. Leucocytes in COVID‐19
Upon exposure to a virus or viral antigen, both innate and adaptive immune cells participate synergistically in the antiviral response. 2 Early studies have shown the implication of various immune cell populations in COVID‐19. A study involving 41 individuals linked severe disease culminating in the intensive care unit (ICU) admission and mortality, with neutrophilia and lymphopenia. 44 Another study reported significant frequencies of lymphopenia (77.6%) and thrombopenia (41.2%) in a cohort of 85 patients who died from the disease. 45 A feature of severe SARS‐CoV‐2 infection is lymphopenia with severely reduced CD4+ T cells, CD8+ T cells, B cells and NK cells counts. It was also reported that reduced percentage of monocytes, eosinophils and basophils were observed. 45 Another feature seen in severe cases is increased neutrophil count and the neutrophil‐to‐lymphocyte ratio. Patients with these features correlated with higher severity of disease and a poorer clinical outcome. 19
The significant increase in the number of neutrophils and of the neutrophil‐lymphocyte‐ratio (NLR) unsurprisingly was not seen in mild cases. The prominent lymphopenia, indicates an impairment of the immune system, is observed to develop in most severe cases of COVID‐19. 5 , 46 Therefore, it can be said that in COVID‐19, neutrophils and leucocytes as well as lymphocytes contribute and strengthen the CS. Several studies reported eosinopenia among hospitalized COVID‐19 patients 47 , 48 with about 82% of fatal cases showing marked eosinopenia. However, early‐stage disease and mild cases did not show eosinopenia. 49 In addition, a study by Lucas et al 50 demonstrated an increase in monocytes, low‐density neutrophils and eosinophils correlating with disease severity. It also demonstrated a similar report among COVID‐19 patients establishing a relation between increased basophils and eosinophils to the severity of COVID‐19, and both cells were among the most dynamic cell population during severe disease, suggestive of an important contribution to antiviral defence and immunopathology. 51
2.4. Mononuclear phagocytes in COVID‐19
scRNA‐seq analysis of broncho‐alveolar lavage fluid (BALF) collected from COVID‐19 patients showed that mononuclear phagocytes (MNPs) account for about 80% of the total number of BALF cells in patients with severe SARS‐COV‐2, 60% in mild disease and 40% in healthy controls. The composition was further characterized by the increased availability of inflammatory‐derived macrophages and relatively fewer tissue‐resident alveolar macrophages in patients with increased severity of the disease. 52 In addition, macrophages seen in patients with SARS‐CoV‐2 infection possess genes that are upregulated and as a result, linking it with tissue repair and with genes that promote fibrosis generation such as in hepatic cirrhosis. 53 This might provide further explanation of the fibrotic complications seen in patients placed under mechanical ventilation and also that the damaging effect of infiltrating macrophages could spread beyond the progression of acute inflammation. 54
Post‐mortem examination on patients who had died from SARS‐CoV‐2 complications revealed that the SARS‐CoV‐2 entry receptor—ACE2 is expressed by a subset of macrophages—CD169+ lymph node subcapsular and splenic marginal zone macrophages, as well as the SARS‐CoV‐2 nucleoprotein. 52 , 55 These macrophages express IL‐6, and their presence was connected to severe depletion of lymphocytes from secondary lymphoid organs such as the spleen and lymph nodes. 56 All these data go to show that type I interferon continuous activation of infiltrating monocytes and monocyte‐derived macrophages, 41 oxidative stress, 57 anti‐spike protein IgG immune complexes 58 and NLRP3 inflammasome activation 58 , 59 could explain the pathophysiology of COVID‐19 as it does for SARS and MERS.
2.5. Other complications seen in COVID‐19
Thrombocytopenia, increased levels of D‐dimers 60 and defective coagulation functions are more and more linked to poor prognosis and might contribute to several organ failures and death in COVID‐19 patients. 61 Thrombi and microthrombi of the lungs, extremities—lower limbs and hands, the brain, 62 heart, liver and kidneys 62 have been observed in patients with COVID‐19. Coagulation abnormalities and disseminated intravascular coagulation are hallmarks of organ injury in conditions like sepsis, where it is mainly facilitated by pro‐inflammatory cytokines. 63 In experimental acute lung injury, oxidized phospholipids elicit macrophage activation through the TLR4–TRIF–TRAF6–NF‐κB pathway which might be a similar mechanism in this disease. 64 The SARS‐CoV‐2 entry receptor (ACE2) is constitutively expressed on arterial and venous endothelial cells 65 ; however, it is important to note that it plays an anti‐inflammatory/ protective effect. 66
From the available data, children are particularly less susceptible to COVID‐19. 67 However, an emerging concern is a novel severe Kawasaki‐like disease seen in children with about a 30‐fold increase in incidence. 68 , 69 It is an acute and usually self‐limiting vasculitis of the medium calibre vessels, which almost exclusively affects children, with coronary artery aneurysms as its main complication. 70 It is characterized by a persistent fever, exanthema, lymphadenopathy, conjunctival injection and changes to the mucosae and extremities. 71 Some patients progressed to hemodynamic instability; a disorder termed as Kawasaki disease shock syndrome. 72 These patients might fulfil the criteria of MAS, resembling sHLH 73 although the relationship with COVID‐19 is yet to be completely unravelled.
3. THERAPEUTIC APPROACH IN COVID‐19
Despite the challenges associated with COVID‐19 therapy, there are still several approaches currently being undertaken which show significant outcomes. Some medications are recommended for exhibiting some clinically positive impacts on COVID‐19 patients, though there are also several drugs in clinical trials, some of which are already demonstrating a significant promise in addressing COVID‐19. Table 1 shows some examples of these drugs, and their mechanism of actions, which makes them potential candidates for the treatment of COVID‐19.
Table 1.
Potential candidates drugs for the treatment of COVID‐19
| S/N | Class of therapy | Example(s) | Mechanism of action | Reference(s) |
|---|---|---|---|---|
| 1 | Broad‐spectrum antivirals |
β‐D‐N4‐hydroxycytidine (NHC), Dihydroorotate dehydrogenase (DHODH) Merimepodib N‐(2‐hydroxypropyl)‐3‐trimethylammonium chitosan chloride (HTCC) |
Known activity against a number of human RNA viruses; reduces viral titre by introducing mutations in the viral RNA genome; Non‐competitive inhibitor of the enzyme‐ Inosine‐5´‐monophosphate dehydrogenase (IMPDH), which is involved in the biosynthesis of host guanosine and is capable of reducing the replication of SARS‐CoV‐2 in vitro; Show efficacy on less pathogenic human coronavirus HCoV‐NL63, pseudotyped SARS‐CoV‐2 and MERS‐CoV, in the airway of human epithelial cells |
|
| 2 | Protease inhibitors |
Peptidomimetic inhibitors (11a and 11b); Nelfinavir |
Target the SARS‐CoV‐2 main protease (Mpro) | 93, 94 |
| 3 | RNA‐dependent RNA polymerase (RdRp) inhibitors | Remdesivir | Adenosine triphosphate analog that prevents RdRp as a result of binding to RNA strands and inhibiting nucleotides addition, bringing about the termination of viral RNA transcription | 95 |
| 4 | Glucocorticoids | Ciclesonide, mometasone and lopinavir | Reducing the function of certain aspects of the immune system such as inflammation and therefore, used in the treatment of diseases caused by an overactive immune system | 96 |
| 5 | JAK inhibitors (JAKinibs) |
Baricitinib Ruxolitinib, memolitinib and oclacitinib |
Interrupts the passage as well as the intracellular assembly of SARS‐CoV‐2 into target cells through disruption of AAK1 signalling and also reduces inflammation in patients with ARDS; target both JAK1 and JAK2 which further affects signalling pathways downstream of the receptors, involved in the development of COVID‐19; its possibility of hindering a range of inflammatory cytokines including IFN‐α, which plays a key role in reducing virus activity |
|
| 6 | Recombinant monoclonal antibody | Tocilizumab (TCZ) | Binds both soluble and membrane‐bound IL‐6 receptors (IL‐6R) of immunoglobulin IgG1 subtype where such binding inhibits sIL‐6R and mIL‐6R‐mediated signal transduction | 99, 100 |
| 7 | Chimeric monoclonal antibody | Siltuximab | Binding to IL‐6 and blocking its effect | 101 |
4. OTHER POSSIBLE COVID‐19 REMEDIES
4.1. ACE2 immunoadhesin
Rather than shielding cells from being infected, a better approach could be to generate an antibody‐like molecule that would have the ability to bind to the coronavirus itself. This approach is suggested to use a soluble version of the ACE2 receptor that binds to the S‐protein of SARS‐CoV‐2 and eventually neutralizes the virus. A study conducted by Li et al 74 demonstrated that the soluble ACE2 receptor can block the SARS virus from infecting cells in culture. The affinity of the soluble ACE2 receptor for the SARS S‐protein was reported to be 1.70 nM, which is equivalent to the affinities of monoclonal antibodies, 75 suggesting that SARS‐CoV‐2 may have a similar affinity for ACE2. It would be desirable to transform soluble ACE2 into an immunoadhesin form fused to an immunoglobulin Fc domain (ACE2‐Fc) when using it as a therapy to treat patients. By doing so, it will extend the lifespan of the circulating molecule, while employing the effector functions of the immune system against the virus. Furthermore, Imai et al 76 demonstrated that the administration of recombinant ACE2 can recover acute lung injury by reducing angiotensin II levels. Thus, SARS‐CoV‐2 could be trapped with an evolutionary strategy when confronted with a potential ACE2‐Fc therapy, leading towards a more benign clinical course.
4.2. Cell‐based therapy
A number of clinical researches on cell‐based therapies have begun for SARS‐CoV‐2 diseases and complications that target a wide range of patient groups using different approaches. 77 Several cell types are included, most of which are directed at the use of mesenchymal stromal cells (MSCs), MSC‐derived conditioned media (CM) or extracellular vesicles (EVs). A pilot study by Leng et al 78 in Beijing administered MSCs to 7 patients with COVID‐19 pneumonia with different grades of severity, including one patient who is critically ill and requiring ICU care. A single dose of MSCs (certified by the National Institute for Food and Drug Control of China) was administered intravenously (1 × 106 cells/kg body weight in a total volume of 100 mL saline) at different time points after the early symptomatic presentation. 78 All the patients including severely critical demonstrated clinical improvements within 2‐4 days, including resolution of clinical symptoms (cough, fever, elevated respiratory rates) and improvements in oxygen saturation. CRP was also observed to have increased on day 1 after the initial administration of MSC and later decline over the 14‐day observational period.
4.3. Plasma therapy (Convalescent plasma)
While scientists are in search of a vaccine that can educate the human immune system to make their own neutralizing antibodies (nAb) against SARS‐CoV‐2, the adoptive transfer of nAb is an approach that is gaining much interest. Convalescent plasma (CP) has been suggested and approved for the treatment of COVID‐19 based on the experience gathered treating influenza, Ebola and SARS (PMID: 32 113 510; PMID: 20 154 602). WHO interim guidelines developed for the 2014 Ebola outbreak outlined the advantages of CP over other proposed treatments: it can be produced independently of pharmaceutical companies (requires low technology); it is low cost and its production is easily scalable as long as there are sufficient donors. 79 This approach used the fact that SARS‐CoV and SARS‐CoV‐2 are essentially identical and share the same cleavage junction, 96% sequence similarity in their main protease, 76% similarity in the amino acid sequence of their S‐protein, a similar S2´ cleavage site, a similarity in the spectrum of cells they can access, and the similarity of the most residues essential for binding ACE2. 80 A recent pilot study by Duan et al 81 investigated the safety and effectiveness of CP with high antibody titres (>1:640) in combination with regular antivirals and standard supportive care on clinical outcome of patients with severe SARS‐CoV‐2 infection. Clinical improvements were recorded in all patients (increased lymphocyte count and a decreased CRP). Furthermore, following transfusion with CP, all the patients who tested positive for SARS‐CoV‐2 before transfusion turned SARS‐CoV‐2 negative. In addition to all these and the aforementioned strategies, recent reports have shown some positive findings supporting the use of ReciGen (IFN‐β‐1a) in combination with hydroxychloroquine and lopinavir/nitonavir in the management of COVID‐19; further indicating the breadth of strategies that can be used in the mangement of this disease. 82
5. COVID‐19 SEROLOGICAL DIAGNOSIS
The diagnosis of COVID‐19 is an important step in tracing the virus and understanding its epidemiology. The key consideration in the diagnosis of COVID‐19 is the early detection of symptoms in clinical situations. Currently, molecular techniques are more suitable than syndromic testing and computed tomography (CT) scans for accurate diagnoses because they can target and identify specific pathogens. However, molecular techniques are not applicable in asymptomatic infections and epidemiological studies, while serological tests can be used for diagnosis, population surveillance and vaccine responses. 83 Serological testing has been reported to be significant and applicable for the diagnosis of suspected cases with negative RT‐PCR and for the detection of asymptomatic infections. 84 Another serologic test application is serological surveillance at the local, regional, state and national levels, and identification of those who have already had the virus and thus may be immune. The assays are also important for contact tracing and to check humoral protective immunity in recovered patients and recipients of vaccine candidates. 85 , 86 Assuming there is protective immunity, serologic information may be used to guide return‐to‐work decisions, including healthcare workers who work in environments where they can potentially be re‐exposed to viruses. Screening of individuals who may be a source for prophylactic or therapeutic neutralizing antibodies is another application of serologic testing. In addition, antibody testing can be used in research studies to evaluate the sensitivity of molecular assays for detecting viruses and be employed retrospectively to determine the true scope of the pandemic and epidemiological assessment. Moreover, serologic testing can be used diagnostically to test RNA‐negative suspected patients who presented late in their illness. 87 Serologic tests have some limitations. A serologic test may not be able to show if an individual has a current infection, because seroconversion can take 1‐3 weeks after viral infection to make antibodies. On the other hand, the development of an antibody response can be host‐dependent and take time. In the case of SARS‐CoV‐2, the majority of patients seroconvert between seven (7) and eleven (11) days post‐exposure, although some patients may develop antibodies sooner. 87 Numerous serological immunoassays have been developed for the detection of SARS‐CoV‐2 viral proteins and/or antibodies in human serum or plasma. The IgM and IgG antibodies are the most widely used biomarkers for the detection of SARS‐CoV‐2 infection in commercial immunoassays. Table 2 shows the currently available serologic tests along with their FDA status for the diagnosis of COVID‐19.
Table 2.
The currently available serologic tests for diagnosis of COVID‐19
| SN | Technique | Comment | FDA approved | References |
|---|---|---|---|---|
| Enzyme‐linked immunosorbent assay (ELISA) | The SARS‐CoV‐2 N and S proteins have been reported to be important antigenic sites for the development of ELISA assays. | Yes | 102 | |
|
A recent meta‐analysis has reported 14 ELISA‐based COVID‐19 tests, which detected anti‐N or anti‐S IgG, IgM antibodies or both. The S‐based ELISAs performed better than N‐based. The different approaches had specificities ranging from 96.1% to 99.5%. |
103 | |||
|
Virus Neuralization Test (VNT) |
Virus Neutralization Test is the current gold standard serological test, which detects neutralizing antibodies in a patient's blood. The assay requires a live virus to handle and thus a biosafety level 3 containment facility. |
Yes | ||
|
The use of the live virus is avoided in another protocol called the surrogate virus neutralization test, which also detects neutralizing antibodies. |
105 | |||
| Rapid Lateral Flow Immunoassay (LFIA) Tests |
These methods are more convenient in resource‐limited settings and can be handled by personnel with low technical skills. LFIA detects the IgM and IgG produced by individuals in response to SARS‐CoV‐2 infection. |
No | ||
|
The detection sensitivity is higher in the IgG‐IgM combined antibody test than in individual IgG or IgM antibody test. The use of the lanthanide‐doped polystyrene nanoparticles‐based LFIA was reported to be better than the colloidal gold‐based LFIAs, to detect anti‐SARS‐CoV‐2 IgG in human serum, in terms of optimal performance. |
6. CONCLUSION
The COVID‐19 continues to expand in the form of pandemic and will be responsible for significant morbidity and mortality. Respiratory infections in COVID‐19 can cause pathology as a result of a poor immune response leading to virus‐induced pathology, or a hyperactive immune response that leads to immunopathology in pulmonary tissues. Cytokine storms associated with the uncontrolled inflammation resulted in the release of pro‐inflammatory cytokines and chemokines which severely damages pulmonary tissues and even lead to death in severe COVID‐19 patients. The progress in COVID‐19 immunopathology may lead to the development of vaccines and blocking antibodies for the management of patients.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Anka AU, Tahir MI, Abubakar SD, et al. Coronavirus disease 2019 (COVID‐19): An overview of the immunopathology, serological diagnosis and management. Scand J Immunol.2021;93:e12998. 10.1111/sji.12998
Funding informationThis work was supported by CinnaGen Medical Biotechnology Company.
Contributor Information
Araz Sabzevari, Email: azizi@abzums.ac.ir, Email: sabzvari.a@orchidpharmed.com.
Gholamreza Azizi, Email: azizi@abzums.ac.ir.
REFERENCES
- 1. WHO . World Health Organization. WHO Director‐General’s opening remarks at the media briefing on COVID‐19 ‐ 11 March 2020. 2020. Available from: https://www.who.int/dg/speeches/detail/who‐director‐general‐s‐opening‐remarks‐at‐the‐media‐briefing‐on‐covid‐19–‐11‐march‐2020
- 2. Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID‐19: Immunology and treatment options. Clin Immunol. 2020;215:108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vabret N, Britton GJ, Gruber C, et al. Immunology of COVID‐19: Current State of the Science. Immunity. 2020;52(6):910‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced Host Response to SARS‐CoV‐2 Drives Development of COVID‐19. Cell. 2020;181(5):1036‐45 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tan M, Liu Y, Zhou R, et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160(3):261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diao B, Wang C, Tan Y, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID‐19). Front Immunol. 2020;11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wan S, Xiang Y, Fang W, et al. Clinical features and treatment of COVID‐19 patients in northeast Chongqing. J Med Virol. 2020;92(7):797‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kamphuis E, Junt T, Waibler Z, Forster R, Kalinke U. Type I interferons directly regulate lymphocyte recirculation and cause transient blood lymphopenia. Blood. 2006;108(10):3253‐3261. [DOI] [PubMed] [Google Scholar]
- 10. Shiow LR, Rosen DB, Brdickova N, et al. CD69 acts downstream of interferon‐alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440(7083):540‐544. [DOI] [PubMed] [Google Scholar]
- 11. Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS‐CoV‐2 infection: the first case study in Finland, January to February 2020. Eurosurveillance. 2020;25(11). pii=2000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lou B, Li TD, Zheng SF, et al. Serology characteristics of SARS‐CoV‐2 infection after exposure and post‐symptom onset. Eur Respir J. 2020;56(2):2000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu YC, Chen CS, Chan YJ. The outbreak of COVID‐19: An overview. J Chinese Med Assoc. 2020;83(3):217‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thevarajan I, Nguyen THO, Koutsakos M, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med. 2020;26(4):453‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo C, Li B, Ma H, et al. Single‐cell analysis of two severe COVID‐19 patients reveals a monocyte‐associated and tocilizumab‐responding cytokine storm. Nat Commun. 2020;11(1):3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature. 2020;584(7819):115‐119. [DOI] [PubMed] [Google Scholar]
- 17. Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cao X. COVID‐19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020;20(5):269‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92(4):424‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. WHO . World Health Organization Clinical Management of Severe Acute Respiratory Infection when Novel Coronavirus (nCoV) Infection is Suspected. 2020.
- 23. Chiappelli F, Balenton N, Khakshooy A. Future Innovations in Viral Immune Surveillance: A Novel Place for Bioinformation and Artificial Intelligence in the Administration of Health Care. Bioinformation. 2018;14(5):201‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winkler F, Bengsch B. Use of Mass Cytometry to Profile Human T Cell Exhaustion. Front Immunol. 2019;10:3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. David P, Megger DA, Kaiser T, et al. The PD‐1/PD‐L1 Pathway Affects the Expansion and Function of Cytotoxic CD8(+) T Cells During an Acute Retroviral Infection. Front Immunol. 2019;10:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chiappelli F, Khakshooy A, Greenberg G. CoViD‐19 Immunopathology and Immunotherapy. Bioinformation. 2020;16(3):219‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yaqinuddin A, Kashir J. Innate immunity in COVID‐19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med Hypotheses. 2020;140:109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McGonagle D, Sharif K, O'Regan A, Bridgewood C. The Role of Cytokines including Interleukin‐6 in COVID‐19 induced Pneumonia and Macrophage Activation Syndrome‐Like Disease. Autoimmun Rev. 2020;19(6):102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tufan A, Avanoglu Guler A, Matucci‐Cerinic M. COVID‐19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turkish J Med Sci. 2020;50(SI‐1):620‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang F, Nie J, Wang H, et al. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID‐19 Pneumonia. J Infect Dis. 2020;221(11):1762‐1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crayne CB, Albeituni S, Nichols KE, Cron RQ. The immunology of macrophage activation syndrome. Front Immunol. 2019;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘Cytokine Storm' in COVID‐19. J Infect. 2020;80(6):607‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Law HK, Cheung CY, Ng HY, et al. Chemokine up‐regulation in SARS‐coronavirus‐infected, monocyte‐derived human dendritic cells. Blood. 2005;106(7):2366‐2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tynell J, Westenius V, Ronkko E, et al. Middle East respiratory syndrome coronavirus shows poor replication but significant induction of antiviral responses in human monocyte‐derived macrophages and dendritic cells. J Gen Virol. 2016;97(2):344‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scheuplein VA, Seifried J, Malczyk AH, et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol. 2015;89(7):3859‐3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim ES, Choe PG, Park WB, et al. Clinical progression and cytokine profiles of middle east respiratory syndrome coronavirus infection. J Korean Med Sci. 2016;31(11):1717‐1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang CH, Liu CY, Wan YL, et al. Persistence of lung inflammation and lung cytokines with high‐resolution CT abnormalities during recovery from SARS. Respir Res. 2005;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garcia‐Sastre A, Biron CA. Type 1 interferons and the virus‐host relationship: a lesson in detente. Science. 2006;312(5775):879‐882. [DOI] [PubMed] [Google Scholar]
- 39. Channappanavar R, Fehr AR, Zheng J, et al. IFN‐I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest. 2019;130:3625‐3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID‐19: An overview of the involvement of the chemokine/chemokine‐receptor system. Cytokine Growth Fac Rev. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated Type I Interferon and Inflammatory Monocyte‐Macrophage Responses Cause Lethal Pneumonia in SARS‐CoV‐Infected Mice. Cell Host Microbe. 2016;19(2):181‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hogner K, Wolff T, Pleschka S, et al. Macrophage‐expressed IFN‐beta contributes to apoptotic alveolar epithelial cell injury in severe influenza virus pneumonia. PLoS Pathog. 2013;9(2):e1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodrigue‐Gervais IG, Labbe K, Dagenais M, et al. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 2014;15(1):23‐35. [DOI] [PubMed] [Google Scholar]
- 44. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du Y, Tu L, Zhu P, et al. Clinical Features of 85 Fatal Cases of COVID‐19 from Wuhan. A Retrospective Observational Study. Am J Respir Crit Care Med. 2020;201(11):1372‐1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus‐Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li YX, Wu W, Yang T, et al. Characteristics of peripheral blood leukocyte differential counts in patients with COVID‐19. Zhonghua Nei Ke Za Zhi. 2020;59:E003. [PubMed] [Google Scholar]
- 49. Lindsley AW, Schwartz JT, Rothenberg ME. Eosinophil responses during COVID‐19 infections and coronavirus vaccination. J Allergy Clin Immunol. 2020;146(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lucas C, Wong P, Klein J, et al. Longitudinal analyses reveal immunological misfiring in severe COVID‐19. Nature. 2020;584(7821):463‐469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tabachnikova A, Chen ST. Roles for eosinophils and basophils in COVID‐19? Nat Rev Immunol. 2020;20(8):461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Merad M, Martin JC. Pathological inflammation in patients with COVID‐19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramachandran P, Dobie R, Wilson‐Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single‐cell level. Nature. 2019;575(7783):512‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cabrera‐Benitez NE, Laffey JG, Parotto M, et al. Mechanical ventilation‐associated lung fibrosis in acute respiratory distress syndrome: a significant contributor to poor outcome. Anesthesiology. 2014;121(1):189‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feng Z, Diao B, Wang R, et al. The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) Directly Decimates Human Spleens and Lymph Nodes. medRxiv. 2020. Preprint. 10.1101/2020.03.27.20045427 [DOI] [Google Scholar]
- 56. Jérôme H, Nader Y, Laura B, et al. Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science. 2020;369(6504):718–724. 10.1126/science.abc6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vijay R, Hua X, Meyerholz DK, et al. Critical role of phospholipase A2 group IID in age‐related susceptibility to severe acute respiratory syndrome‐CoV infection. J Exp Med. 2015;212(11):1851‐1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu L, Wei Q, Lin Q, et al. Anti‐spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS‐CoV infection. JCI Insight. 2019;4(4).e123158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi CS, Nabar NR, Huang NN, Kehrl JH. SARS‐Coronavirus Open Reading Frame‐8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thachil J, Tang N, Gando S, et al. ISTH interim guidance on recognition and management of coagulopathy in COVID‐19. J Thromb Haemost. 2020;18(5):1023‐1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xiang‐Hua Y, Le‐Min W, Ai‐Bin L, et al. Severe acute respiratory syndrome and venous thromboembolism in multiple organs. Am J Respir Crit Care Med. 2010;182(3):436‐437. [DOI] [PubMed] [Google Scholar]
- 62. Zhang Y, Xiao M, Zhang S, et al. Coagulopathy and antiphospholipid antibodies in patients with Covid‐19. N Engl J Med. 2020;382(17):e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Levi M, Nieuwdorp M, van der Poll T, Stroes E. Metabolic modulation of inflammation‐induced activation of coagulation. Semin Thromb Hemost. 2008;34(1):26‐32. [DOI] [PubMed] [Google Scholar]
- 64. Imai Y, Kuba K, Neely GG, et al. Identification of oxidative stress and Toll‐like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133(2):235‐249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS‐CoV‐2. Cardiovasc Res. 2020;116(6):1097‐1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Team CC‐R . Coronavirus Disease 2019 in Children ‐ United States, February 12‐April 2, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(14):422‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Verdoni L, Mazza A, Gervasoni A, et al. An outbreak of severe Kawasaki‐like disease at the Italian epicentre of the SARS‐CoV‐2 epidemic: an observational cohort study. Lancet. 2020;392(10239):1771–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Viner RM, Whittaker E. Kawasaki‐like disease: emerging complication during the COVID‐19 pandemic. Lancet. 2020;395(10239):1741–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kato H, Sugimura T, Akagi T, et al. Long‐term consequences of Kawasaki disease. A 10‐ to 21‐year follow‐up study of 594 patients. Circulation. 1996;94(6):1379‐1385. [DOI] [PubMed] [Google Scholar]
- 71. Dietz SM, van Stijn D, Burgner D, et al. Dissecting Kawasaki disease: a state‐of‐the‐art review. Eur J Pediatr. 2017;176(8):995‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kanegaye JT, Wilder MS, Molkara D, et al. Recognition of a Kawasaki disease shock syndrome. Pediatrics. 2009;123(5):e783‐e789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang W, Gong F, Zhu W, Fu S, Zhang Q. Macrophage activation syndrome in Kawasaki disease: more common than we thought? Semin Arthritis Rheum. 2015;44(4):405‐410. [DOI] [PubMed] [Google Scholar]
- 74. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sui J, Li W, Murakami A, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A. 2004;101(8):2536‐2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Imai Y, Kuba K, Rao S, et al. Angiotensin‐converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Khoury M, Rocco PRM, Phinney DG, et al. Cell‐Based Therapies for COVID‐19: Proper Clinical Investigations are Essential. Cytotherapy. 2020;22(11):602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Leng Z, Zhu R, Hou W, et al. Transplantation of ACE2(‐) Mesenchymal Stem Cells Improves the Outcome of Patients with COVID‐19 Pneumonia. Aging Dis. 2020;11(2):216‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pérez‐Cameo C, Marín‐Lahoz J. Serosurveys and convalescent plasma in COVID‐19. EClinicalMedicine. 2020;100370. [DOI] [PMC free article] [PubMed]
- 80. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271‐80 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kai D, Bende L, Cesheng L, et al.. Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proceedings of the National Academy of Sciences. 2020;117(17):9490–9496. 10.1073/pnas.2004168117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dastan F, Nadji SA, Saffaei A, et al. Subcutaneous administration of interferon beta‐1a for COVID‐19: A non‐controlled prospective trial. International Immunopharmacology. 2020;85106688. 10.1016/j.intimp.2020.106688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID‐19: The Disease and Tools for Detection. ACS Nano. 2020;14(4):3822‐3835. [DOI] [PubMed] [Google Scholar]
- 84. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020. [DOI] [PubMed] [Google Scholar]
- 85. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wah TC, Ni CW, Xijian Q, et al. A SARS‐CoV‐2 surrogate virus neutralization test based on antibody‐mediated blockage of ACE2–spike protein–protein interaction. Nature Biotechnology. 2020;38 (9):1073–1078. 10.1038/s41587-020-0631-z [DOI] [PubMed] [Google Scholar]
- 87. Patel R, Babady E, Theel ES, et al. Report from the American Society for Microbiology COVID‐19 International Summit, 23 March 2020: Value of Diagnostic Testing for SARS‐CoV‐2/COVID‐19. MBio. 2020;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sheahan TP, Sims AC, Graham RL, et al. Broad‐spectrum antiviral GS‐5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396).eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30(3):269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xiong R, Zhang L, Li S, et al. Novel and potent inhibitors targeting DHODH are broad‐spectrum antivirals against RNA viruses including newly‐emerged coronavirus SARS‐CoV‐2. Protein Cell. 2020;11(10):723‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Natalya B, Sattler RA., Mantlo EK., et al. The IMPDH inhibitor merimepodib provided in combination with the adenosine analogue remdesivir reduces SARS‐CoV‐2 replication to undetectable levels in vitro. F1000Research. 2020;9:361. 10.12688/f1000research.23639.1 [DOI] [Google Scholar]
- 92. Aleksandra M, Ying C, Artur S, et al. HTCC as a Polymeric Inhibitor of SARS‐CoV‐2 and MERS‐CoV. Journal of Virology. 2020; 10.1128/jvi.01622-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An Overview of severe acute respiratory syndrome‐coronavirus (SARS‐CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59(14):6595‐6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dai W, Zhang B, Jiang XM, et al. Structure‐based design of antiviral drug candidates targeting the SARS‐CoV‐2 main protease. Science. 2020;368(6497):1331‐1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Agostini ML, Andres EL, Sims AC, et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS‐5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018;9(2).e00221‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Saghazadeh A, Rezaei N. Towards treatment planning of COVID‐19: Rationale and hypothesis for the use of multiple immunosuppressive agents: Anti‐antibodies, immunoglobulins, and corticosteroids. Int Immunopharmacol. 2020;84:106560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019‐nCoV acute respiratory disease. Lancet. 2020;395(10223):e30‐e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Seif F, Aazami H, Khoshmirsafa M, et al. JAK Inhibition as a New Treatment Strategy for Patients with COVID‐19. Int Arch Allergy Immunol. 2020;181(6):467‐475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Castilletti C, Bordi L, Lalle E, et al. Coordinate induction of IFN‐alpha and ‐gamma by SARS‐CoV also in the absence of virus replication. Virology. 2005;341(1):163‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zheng Y, Wang QY. [Bioinformatics analysis on molecular mechanism of ribavirin and interferon‐alpha in treating MERS‐CoV]. Zhonghua Liu Xing Bing Xue Za Zhi. 2016;37(2):291‐293. [DOI] [PubMed] [Google Scholar]
- 101. Gritti GRF, Ripamonti D, Riva I, Landi F, Alborghetti L, Frigeni M, Damiani M, Micò C, Fagiuoli S, Cosentini R, Luca Lorini F, Fabretti F, Morgan J, Owens BM, Kanhai K, Cowburn J, Tonkovivic Reljanovic G, Rizzi M, Di Marco F.Use of siltuximab in patients with COVID‐19 pneumonia requiring ventilatory support. 2020.
- 102. Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory Diagnosis of COVID‐19: Current Issues and Challenges. J Clin Microbiol. 2020;58(6).e00512‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. Antibody Tests in Detecting SARS‐CoV‐2 Infection: A Meta‐Analysis. Diagnostics. 2020;10(5):319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tan CW, Chia WN, Qin X, et al. A SARS‐CoV‐2 surrogate virus neutralization test based on antibody‐mediated blockage of ACE2‐spike protein‐protein interaction. Nat Biotechnol. 2020;38(9):1073‐1078. [DOI] [PubMed] [Google Scholar]
- 105. Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS‐CoV‐2. Emerg Microbes Infect. 2020;9(1):680‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Vashist SK. In vitro diagnostic assays for COVID‐19: recent advances and emerging trends. Diagnostics. 2020;10(4):202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]


