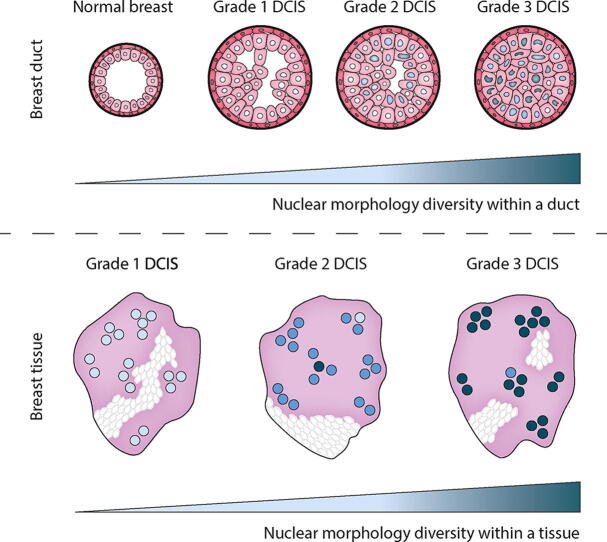
Graphical abstract
Keywords: Nuclear morphology, Heterogeneity, Image analysis, Breast cancer, Pathology
Highlights
-
•
Abnormalities in cell nuclear morphology is a hallmark of cancer.
-
•
Changes in nuclear morphology are utilized by pathologists in tumor grading.
-
•
Heterogeneity in DCIS grade may influence invasive progression.
-
•
Current pathological assessment of DCIS does not incorporate heterogeneity.
-
•
I mage analysis is a supplemental tool for pathologists to quantitatively grade DCIS.
Abstract
Abnormalities in cell nuclear morphology are a hallmark of cancer. Histological assessment of cell nuclear morphology is frequently used by pathologists to grade ductal carcinoma in situ (DCIS). Objective methods that allow standardization and reproducibility of cell nuclear morphology assessment have potential to improve the criteria needed to predict DCIS progression and recurrence. Aggressive cancers are highly heterogeneous. We asked whether cell nuclear morphology heterogeneity could be incorporated into a metric to classify DCIS. We developed a nuclear heterogeneity image index to objectively, and quantitatively grade DCIS. A whole-tissue cell nuclear morphological analysis, that classified tumors by the worst ten percent in a duct-by-duct manner, identified nuclear size ranges associated with each DCIS grade. Digital image analysis further revealed increasing heterogeneity within ducts or between ducts in tissues of worsening DCIS grade. The findings illustrate how digital image analysis comprises a supplemental tool for pathologists to objectively classify DCIS and in the future, may provide a method to predict patient outcome through analysis of nuclear heterogeneity.
1. Introduction
Changes in cell nuclear morphology occur across a range of diseases, including cancer. Pathologists assess these observable changes in nuclear morphology to make a cancer diagnosis and inform tumor grading [1]. Preinvasive breast cancer, ductal carcinoma in situ (DCIS), is characterized by a proliferation of neoplastic epithelial cells confined to the ductal-lobular structures that has not invaded beyond the myoepithelial-basement membrane barrier into the surrounding stroma. Pathological input into grading DCIS has led to a number of classification systems based on nuclear morphology, among other features, to broadly group DCIS into low (1), intermediate (2) and high (3) grade [2]. Low grade DCIS is characterized by evenly spaced small, regular cells with centrally placed, round monotonous nuclei, and high grade DCIS is made up of pleomorphic, irregularly spaced cells with large, irregular nuclei. Intermediate grade DCIS show features between both grades [2]. Though all DCIS is treated as potentially invasive, it is established that not all DCIS lesions progress into invasive disease within the patient’s lifetime [3], [4]. Some studies suggest that all grades have equal potential to progress however, high grade DCIS is likely to progress more rapidly and lead to metastatic disease and death [3], [4], [5].
Histological grading of DCIS is subjective, due to the limitations of a categorical grading system to convey the heterogeneity that exists within a tissue, and the inter-observer variability between pathologists in assigning scores to features of the grading system [6]. These issues are clinically important, since to be of value, such classification systems need to be reproducible, and must account for heterogeneity [7], [8], which is likely to influence tumor progression and resistance to therapy [9]. To circumvent this subjectivity, digital image analysis of nuclear morphology aims to provide quantitative measurements of nuclear morphometric information such as nuclear size and shape for classifying histopathology images. In this work, we demonstrate the value of image analysis in classifying in situ breast cancer grade through quantification of nuclear parameters, and show quantitative analysis of the heterogeneity of nuclear grade that exists within ducts and between ducts of individual cases of DCIS.
2. Materials and methods
2.1. Human breast samples
Breast tissue samples were obtained from women with DCIS undergoing breast surgery at Duke University Medical Center, USA or Barts Health NHS Trust London, UK between 1998 and 2016. Tissue samples and associated clinical data were analyzed with deidentified labels to protect patient data in accordance with the UCSF IRB protocol (#10-03832), the Duke University IRB protocol (Pro00054515) and Barts Cancer Institute Breast Tissue Bank protocol (REC no: 15/EE/0192). 90 cases of DCIS (without evidence of invasion) were included in the study (average, 1.2 blocks per case). Formalin fixed and paraffin embedded (FFPE) tissue was cut into 5 µm sections and stained with hematoxylin and eosin (H&E) in a CLIA-certified laboratory as part of standard clinical care, and not restained for the purposes of this study.
2.2. Conventional histological grading
DCIS tissues were reviewed for nuclear grade by two highly experienced breast pathologists (JLJ and AH), and only cases with a consensus grade to the clinical pathology report were included for analysis. Briefly, the features and general scoring criteria included (a) the degree of nuclear atypia or pleomorphism and (b) the presence or absence of necrosis [10]. Tissues were assigned to the highest grade present. DCIS tissues were then grouped for analysis by grade as low (1), n = 12; intermediate (2), n = 34 and high (3), n = 44 grade. Clinicopathologic details are provided in Supplementary Table 1.
2.3. Tissue imaging and analysis
H&E-stained whole-slide images (WSI) were obtained using an Aperio ScanScope slide scanner at 20x magnification with an image resolution of 0.5 µm/pixel. Image analysis was performed in a semi-automated manner using QuPath 0.2.3 open-source software [11]. Tissues were analyzed on a duct-by-duct basis. Within each sample, all ducts were numbered and identified as normal or DCIS by a pathologist. A small number of ducts were excluded from the analysis, these included benign or hyperplastic ducts, as well as ducts presenting technical artefacts, such as; damage secondary to surgical diathermy, extensively cross cut ducts and disrupted ducts showing loss of epithelium. In each identified duct, the epithelium was manually selected by an expert, and nuclear segmentation was performed by automated-detection based on hematoxylin optical density. A size filter of 50–500 pixels was applied to exclude objects of extremely small or large size to improve the specificity of nuclear detection. Nuclear morphology features extracted from segmented nuclei included; cross-sectional area, perimeter, major and minor axis. Features were compared between tissues of different pathological groups, and within a duct and between ducts of the same tissue.
2.4. Validation of DCIS analysis
Validation of DCIS annotation was assessed by both inter- and intra-observer agreement, and evaluation of QuPath segmentation, and were performed on 10% of tissues within the series, randomly selected with at least 1 case of each pathological grade. For intra-observer agreement, the number of ducts identified as DCIS in two separate observations on the same slide at different times (unknown to the pathologist) were compared. For inter-observer agreement, the number of ducts identified as DCIS on the same slide were compared between three independent pathologists. The accuracy of QuPath segmentation was also compared in three ways. (1) Ground truth annotations for nuclei were hand-marked as a collective by an expert and two pathologists, and all DCIS ducts in tissues were annotated in full. QuPath analysis was then performed, and the number of nuclei detected were compared. (2) QuPath analysis was also performed at two different times (on different days) on the same slides with the epithelium identically selected, and the number of nuclei detected were compared. (3) QuPath analysis was also performed on the same slides with the epithelium selected separately by three independent experts, and the number of nuclei detected were compared.
2.5. Public dataset for analysis
A public dataset of 100 breast biopsy H&E images of DCIS was selected due to availability of DCIS grade and annotation of images [12]. DCIS was graded as low (1), n = 24; intermediate (2), n = 43 and high (3), n = 33, based on the degree of nuclear atypia. If a case was reported to be between two grades, we classified the case as the higher grade. H&E-stained WSI were obtained using a Philips Ultra Fast 1.6 slide scanner at 40x magnification with an image resolution of 0.25 µm/pixel. Diagnostic ROIs (1–4 per case) were available from the WSI. For each ROI, the epithelium was manually selected by an expert, and nuclear segmentation was performed as described previously.
2.6. Statistical analysis
For comparing whole-tissue averages between pathological groups, statistical significance was determined by one-way ANOVA. For comparing the equality of diversity between ducts of the same tissue, the Browns-Forsythe test was used. For comparing the diversity within a single duct, the Simpsons diversity index was determined using: where n is the total number of cells belonging to a grade category and N is the total number of cells within the duct. This statistical analysis was performed using the statistical software GraphPad Prism 9. For intra- and inter-observer agreement, rater values for duct and cell identification were converted to nominal outcomes based on quartile categories of rater 1 (gold-standard used in analysis) and assessed using percent agreement and weighted kappa (ĸw) statistic with Cicchetti-Allison weights and exact tests. Level of agreement was defined as 0–0.20 (none), 0.21–0.39 (minimal), 0.40–0.59 (weak), 0.60–0.79 (moderate), 0.80–0.90 (strong) and above 0.90 (almost perfect). For assessing over-scoring and under-scoring duct identification between raters, the difference in raw values between rater 3 and rater 1 was compared to the difference between rater 2 and rater 1 using a one-sided Wilcoxon rank-sum exact test. This statistical analysis was performed using the statistical software SAS version 9.4. Statistical significance was indicated: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
3. Results
3.1. Image analysis to quantify nuclear morphological features between DCIS grades
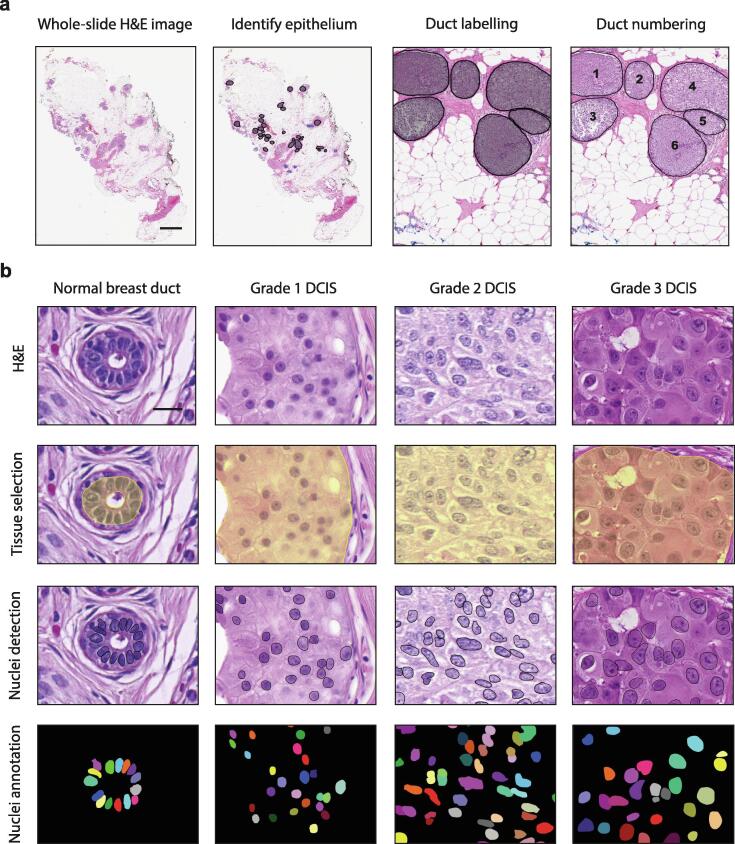
To assess nuclear morphological characteristics associated with DCIS grade, we analyzed epithelial cell nuclei in normal and DCIS ducts present in a series of DCIS tissues. These tissues were selected to represent each pathological grade used to classify DCIS (low (1), n = 12; intermediate (2), n = 34; high (3), n = 44) (Supplemental Table 1), and pathological examination confirmed the absence of invasive disease in all samples. To analyze nuclear morphology, we used QuPath software, which allows for a semi-automated image analysis of hematoxylin and eosin (H&E)-stained whole-slide images (WSI) [11]. This involved the identification and numbering of all normal and DCIS ducts within a tissue (Fig. 1a). For each individual duct, we manually selected the epithelium within the duct, segmented nuclei within the selected epithelium by automated-detection and extracted features of the detected nuclei using QuPath (Fig. 1b). We analyzed 405 normal and 1455 DCIS ducts (average of 5 normal and 16 DCIS ducts per case), containing 6009 and 797,296 epithelial nuclei, respectively (average 67 normal and 8858 DCIS epithelial nuclei per case). 41 features were extracted per cell using an automated pipeline, and of these, 4 nuclear size features (cross-sectional area, perimeter, major and minor axis) were analyzed further to quantify nuclear morphology. We excluded features that quantified cell morphology due to the absence of defined cell boundaries in H&E images, as well as features quantifying nuclear shape descriptors (circularity, eccentricity, solidity) which are an extension of nuclear size features that are less accurate due to the use of 2D images obtained from thin tissue sections in routine histopathology. Additionally, although densitometric and texture features have been shown to have value in classifying H&E images of DCIS [13], [14], we did not pursue these further, retaining our focus on carefully quantifying nuclear morphology.
Fig. 1.
Image analysis workflow for nuclear morphology in DCIS tissues. a, Hematoxylin and eosin (H&E)-stained whole slide image (WSI) of a representative DCIS tissue. Within each tissue, all ducts were identified and numbered as either; normal (blue) or DCIS (black). Scale bar, 2 mm. b, For each individual duct, epithelium was manually selected, nuclei were segmented by automated-detection and nuclear features were extracted using QuPath. Segmented images of nuclei were compared to manually annotated images. Scale bar, 20 µm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To validate the accuracy of our annotation and nuclear segmentation method, we assessed; 1) intra- and inter-observer agreement of DCIS identification by pathologists’; 2) accuracy of nuclei detection by comparing to ground truth annotations, 3) reproducibility of nuclei detection in the same epithelium identically selected and 4) variability of nuclei detection in the same epithelium differently selected. The intra-observer agreement of DCIS identification was perfect (ĸw 1.00; 100% agreement). The inter-observer agreement of DCIS identification between rater 1 and 2 was moderate (ĸw 0.75; 95% CI 0.42–1.00; p < 0.01; 77.8% agreement), while the agreement between rater 1 and rater 3 was perfect (ĸw 1.00; 100% agreement). This difference is due to the under-scoring of ducts by rater 2 (p < 0.05), and these unidentified ducts were small. The accuracy of QuPath in comparison to ground-truth annotations was strong (ĸw 0.83; 95% CI 0.59–1.00; p < 0.001; 77.8% agreement). This included errors of under and over segmentation, which was commonly due to high cell density and clustering, image contrast or out-of-focal-plane noise. The reproducibility and variability in nuclei detection by QuPath was perfect (ĸw 1.00; 100% agreement).
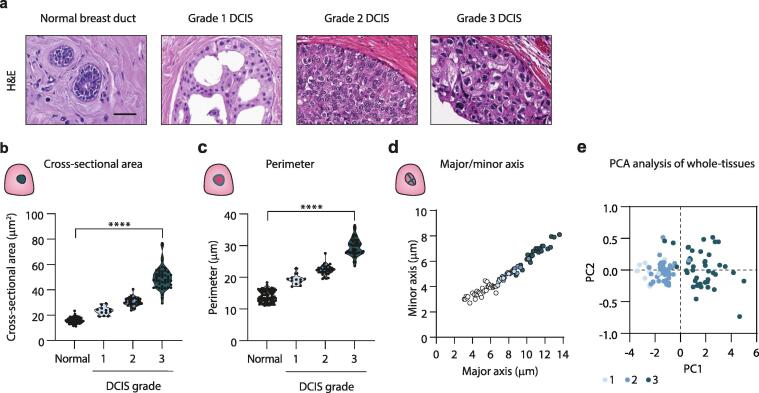
To quantify nuclear morphological features associated with DCIS grade, we used whole-tissue averages for each nuclear feature extracted from normal or DCIS nuclei for each case. For normal epithelial nuclei analysis, all tissues were grouped together, while for DCIS, tissues were grouped by their conventional histopathological grade (Fig. 2a). Nuclear quantification identified a significant increase in nuclear cross-sectional area from normal (15.9 µm ± 2.1) with worsening DCIS grade (1, 23.3 µm ± 3.4; 2, 30.6 µm ± 3.3 and 3, 49.6 µm ± 9.2) (Fig. 2b). Quantification of nuclear perimeter further supported an increase in nuclear size from normal (14.0 µm ± 1.8) with increasing DCIS grade (1, 19.3 µm ± 1.6; 2, 22.3 µm ± 1.5; 3, 29.6 µm ± 2.8) (Fig. 2c). Similarly, quantification of the major/minor axis demonstrated a significant increase in nuclear size from normal (major/minor, 5.0/3.7 µm) with higher DCIS grade (1, 7.3/4.6 µm; 2, 8.4/5.2 µm; 3, 11.0/6.7 µm) (Fig. 2d). Principal component analysis (PCA) shows a shift with increasing DCIS grade (Fig. 2e), highlighting differences in nuclear morphology associated with grade.
Fig. 2.
Nuclear morphological features of normal and DCIS epithelial cells. a, H&E staining of a representative normal duct, and DCIS ducts from a DCIS tissue pathologically defined as grade 1–3. Scale bar, 20 µm. b-c, Violin plots of whole-tissue averages of cross-sectional area (b) and perimeter (c) of normal and DCIS cell nuclei. Each dot represents a tissue. d, Dot plot of whole-tissue averages of major and minor axis of normal and DCIS cell nuclei. Each dot represents a tissue. e, PCA analysis comparing nuclear features of DCIS with grade. Grade 1 (light blue), 2 (medium blue), and 3 (dark blue). For plots quantifying nuclear features, normal nuclear averages from each tissue are grouped (n = 90), while DCIS nuclear averages from each tissue are grouped by pathological grade. All plots represent grade 1, n = 12; 2, n = 34; 3, n = 44. All violin plots have a p-value of < 0.0001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Quantitative grading of DCIS tissues based on nuclear morphological features
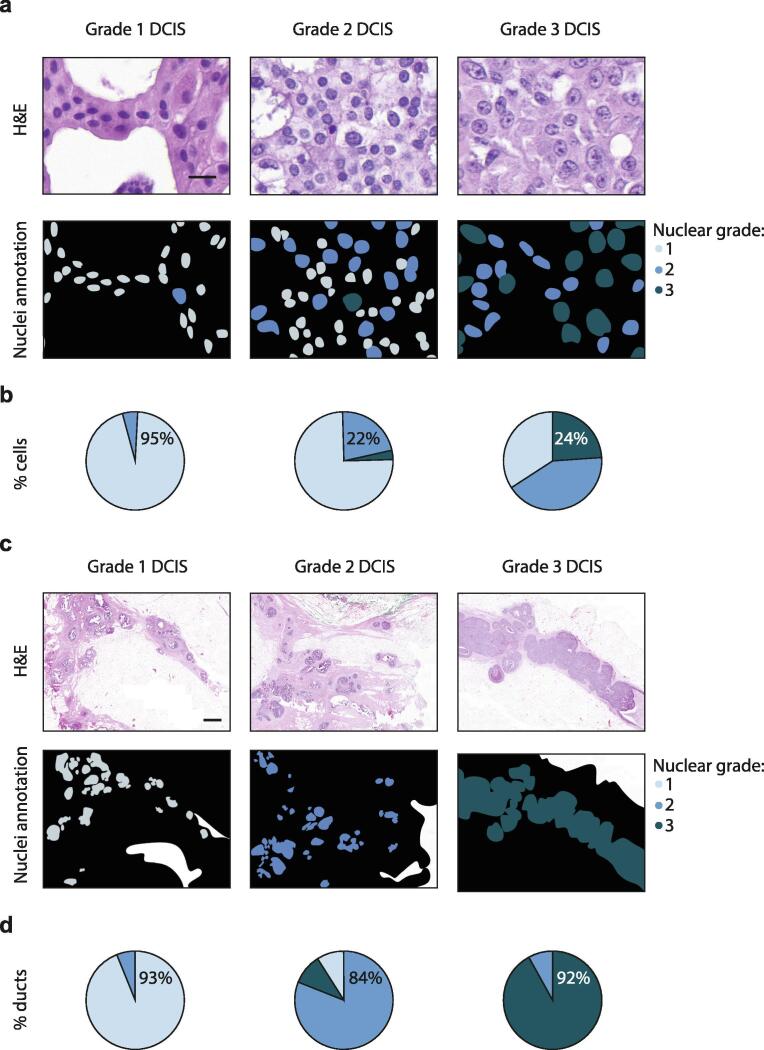
To quantitatively grade DCIS tissues, we used whole-tissue analysis of nuclear cross-sectional area to identify ranges of nuclear size to categorize grade 1 (20–30 µm), 2 (30–40 µm) and 3 (40 µm and over) cells within these tissues. These nuclear size categories were based on the ranges of whole-tissue averages of nuclear cross-sectional area from each tissue within a pathological group. For all tissues, we then categorized each DCIS cell present to one of these groups. We first scored tissues for nuclear size according to the highest-grade comprising at least 1%, 5%, 10% or 20% of cells present within the whole-tissue, in order to mimic the pathological defined grade. For example, for a 10% threshold, tumors with 10% to 100% of cells showing high nuclear size would score as high grade. In this manner, a 1% or 5% threshold incorrectly assigned 34% or 38% of cases, respectively, to a higher grade than that pathologically reported. In contrast, a 20% threshold incorrectly assigned 33% of cases to a lower grade than that pathologically reported. However, a 10% threshold correctly assigned all tissues to their conventional histopathological grade, with no tissue scoring over 10% of cells with a worse tumor grade than that clinically reported (Fig. 3a) (Supplemental Table 2). We also assessed the ability of this method to grade an independent dataset of publicly available DCIS H&E images, with known DCIS grade. A highly experienced breast pathologist reviewed these DCIS images for grading, according to our conventional histological grading method. For cases that were reported in the dataset to be between two grades, we reported the higher of the grades, and we excluded cases in this analysis with a different grade to that clinically reported in dataset. With this, our quantitative grading method utilizing a 10% threshold, accurately classified nuclear grade of 97% of DCIS cases in this dataset (Supplemental Table 3).
Fig. 3.
Quantitative grading of DCIS using nuclear morphological features. a, Annotations categorizing each DCIS cell within a tissue to a nuclear grade based on their cross-sectional area from a H&E image. Scale bar, 20 µm. b, Average number of cells belonging to grade 1–3 within a DCIS tissue grouped by pathological grade. c, Annotations categorizing each DCIS duct within a tissue to a nuclear grade based on the worst 10% of cells according to their cross-sectional area from a H&E image. Scale bar, 1 mm. d, Average number of ducts belonging to grade 1–3 within a DCIS tissue grouped by pathological grade. For all annotations, grade 1 (light blue), 2 (medium blue), and 3 (dark blue). All plots represent grade 1, n = 12; 2, n = 34; 3, n = 44. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We then scored our DCIS cases according to the worst 10% of cells present within each duct across a tissue (Fig. 3b) (Supplemental Table 4). In this manner, we identified in grade 1 DCIS tissues, 33% of cases had ducts scoring as grade 2, while the remaining cases scored all ducts as grade 1. In grade 2 DCIS tissues, 29% of cases (which had no ducts scoring as grade 3) had ducts scoring as grade 1, while 32% of cases (which had no ducts scoring as grade 1) had ducts scoring as grade 3 and the remaining 39% of cases scored all ducts as grade 2. In grade 3 DCIS tissues, 34% of cases had ducts scoring as grade 2, while the remaining cases scored all ducts as grade 3.
3.3. Quantitative assessment of heterogeneity within and between ducts of a tissue
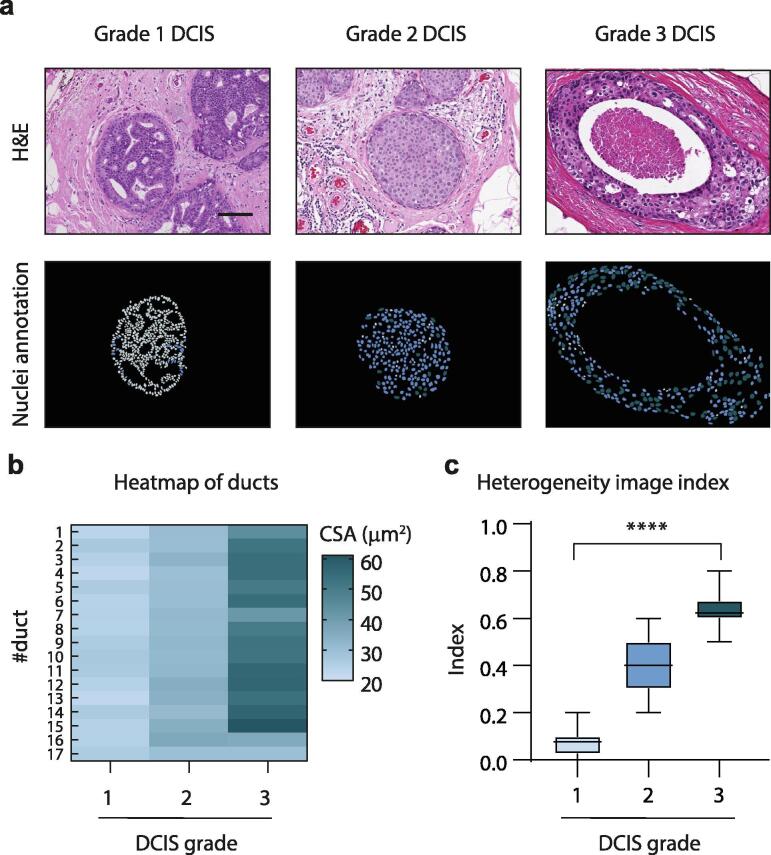
To quantitatively study heterogeneity in DCIS tissues, we assessed diversity in nuclear cross-sectional area within a single duct and between ducts of the same tissue (Fig. 4a, b). Using the proportion of cells belonging to each grade on a duct-by-duct basis, as determined previously by our classification of grade by nuclear cross-sectional area, the heterogeneity within each duct was assessed using Simpsons diversity index. In this index, 1 represents infinite diversity and 0 represents no diversity. Infinite diversity is reached when all abundances are equal to the number of species observed in the sample, and therefore it is not possible to achieve this score. We identified increasing diversity with worsening DCIS grade; with grade 1 ducts scoring a low diversity index of 0.1 (range, 0.0–0.2), grade 2 ducts scoring a moderate diversity index of 0.4 (0.2–0.6) and grade 3 ducts scoring a high diversity index of 0.6 (0.5–0.8) (Fig. 4c). We then compared the heterogeneity between ducts of the same tissue using a test for equality of variance between groups, where significance demonstrates unequal variance and hence, diversity. We similarly identified increasing diversity with worsening DCIS grade; with all grade 1 DCIS cases demonstrating equal variance between ducts in the same tissue (no diversity), while 38% of grade 2 cases demonstrated equal variance and no grade 3 cases demonstrated equal variance between ducts (high diversity).
Fig. 4.
Quantifying heterogeneity within and between DCIS ducts of single tissues. a, Annotations categorizing each DCIS cell within a duct based on their cross-sectional area from a H&E image. Scale bar, 100 µm. b, Heat map analysis of DCIS duct averages of cross-sectional area (CSA) of nuclei. A single tissue with a matched-number of DCIS ducts from each pathological grade 1–3 was used for this analysis. c, Box plot analysis of the heterogeneity image index. Each dot represents a tissue, and tissues are grouped by pathological grade (grade 1, n = 12; 2, n = 34; 3, n = 44). Box plot has a p-value of < 0.0001. For all annotations, grade 1 (light blue), 2 (medium blue), and 3 (dark blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
High DCIS nuclear grade has been associated with an increased risk of disease recurrence [5], [15], [16] and progression to invasion [4], [16], [17], and thus, is a required component of the pathological classification of DCIS [18]. Pathological assignment of nuclear grade depends on the visual inspection and subjective judgement of several criteria, including the degree of pleomorphism, nuclear–cytoplasmic ratio, nucleolar prominence and mitotic frequency. Pathologists assign a score to nuclear pleomorphism based on the highest grade that comprises at least 10% of tumor cells (1; mild, 2; moderate, 3; severe) [2]. Here, we utilized image analysis of cell nuclear morphological features to objectively and quantitatively grade DCIS. The assessment of nuclear size and shape alone is not sufficient for DCIS classification, for example many nuclei may not be large enough to classify as high grade but show other features such as lack of orientation, coarse chromatin and prominent nucleoli. However, our study illustrates that digital image analysis of cell nuclear morphology in DCIS could provide a supportive tool to conventional pathological analysis for the objective and reproducible assessment of nuclear grade [19]. Importantly, this study also quantifies heterogeneity of nuclear morphology within DCIS, which is currently not formally acknowledged in the pathological reporting of DCIS. The degree of nuclear heterogeneity may have clinical implications, and this study provides a robust method to address this. The ability to use standardized digital image analysis algorithms, such as QuPath, is an important consideration for clinical utility. These issues are particularly important when considering the currently subjective nature of assessing DCIS heterogeneity in grade.
We identified a nuclear size range that classified nuclei belonging to each DCIS grade. Pathological guidelines for size comparison with red blood cells or normal epithelial cell nuclei supports our data, with low grade DCIS demonstrating nuclear sizes 1.5-fold larger and high grade DCIS demonstrating nuclear sizes 2.5-fold larger, with intermediate grade DCIS demonstrating nuclear sizes between this range [2]. Indeed, the high accuracy of our image analysis method also supports that these nuclear features are representative of real differences between DCIS grade. Using these ranges of nuclear size, our whole-tissue analysis aimed to classify tissues according to the highest-scoring 10% of cells present. This threshold was selected to represent a significant proportion of the tumor. In the clinical setting, a threshold of 10% is commonly used to determine hormone receptor positivity (in Europe) and to define a mixed subtype in invasive breast cancers [20], [21]. This analysis correctly graded all tissues according to their conventionally defined grade. Additionally, we were able to demonstrate the ability of this method to accurately grade 97% of DCIS images to their clinically-reported grade in an independent, public dataset. Importantly, our study performed whole-tissue analysis in a duct-by-duct manner to quantitatively assess all DCIS nuclei present, while many other studies using image analysis to establish nuclear features associated with grade frequently analyzed only a small proportion of cells or ducts from a representative region [22], [23], [24], often utilizing clinical biopsies or tissue microarrays that do not reflect the entire lesion [12], [25], [26].
By studying tissues in a duct-by-duct manner, we demonstrated that scoring ducts by the worst 10% of cells present, rather than across the whole tissue, may be used to identify subgroups within the current DCIS classification system. We showed that a proportion of low grade DCIS tissues had ducts scoring as intermediate grade (33%). An almost equal proportion of intermediate grade DCIS tissues had ducts scoring as low and intermediate grade (29%), intermediate grade alone (39%), or intermediate and high grade (32%). In contrast, a small proportion of high grade DCIS tissues had ducts scoring as intermediate grade (34%). We showed that for 44% of cases evaluated (40 out of 90), DCIS tissues do not exhibit a single grade, and instead demonstrate a range of nuclear grades with varying proportions. This proportion of cases is consistent with a conventional pathological analysis, assessing the presence of multiple histological nuclear grades in DCIS (46% of 120 DCIS cases evaluated) [27]. We suggest that while whole-tissue analysis may assign an average overall grade, the quantitative data obtained by scoring duct-by-duct may identify different clinical risk groups. For example, in cases of intermediate grade DCIS where some ducts score as low grade, these cases may have an improved prognosis and lower risk of progression, compared with intermediate grade DCIS where some ducts score as high grade. In the era of personalized medicine, an objective and quantitative assessment of the degree of clinical risk posed by a patient’s DCIS could prove a valuable tool to aid clinical decision making. For example, whether or not to irradiate the breast following surgery. In addition, given there are concerns regarding the potential overtreatment of women with breast tumors deemed to be of low risk, having an objective score to support a more conservative monitoring-based approach for these individuals would be useful. Moreover, having an objective standardized score of DCIS nuclear grade could help refine the entry criteria for existing surveillance DCIS trials, such as COMET (USA), LORIS (UK), LORD (Europe) and LARRIKIN (Australia and New Zealand), which aim to focus exclusively on low-risk disease [28].
The intratumoral diversity we found suggests that the current classification of DCIS into 3 distinct grades may be an oversimplification, and highlights the need to have reasonable inter-observer agreement in clinical diagnosis. We propose a nuclear heterogeneity image index as a means to integrate heterogeneity into routine pathological reporting of DCIS, in addition to composite grading. This index would score tissues in a duct-by-duct manner to determine the extent of heterogeneity. Future studies are required to assess the potential clinical utility of this method of analysis in adding prognostic value to histopathology reports. Most importantly, this study highlights image analysis as an adjunctive tool for pathologists to standardize grading.
Other studies suggest that DCIS exhibits considerable histological and biological heterogeneity within a single sample [27], [29], [30]. Our detailed analysis verified and expanded upon this conclusion by objectively and reproducibly quantifying the level of heterogeneity in nuclear morphology that exists within a duct and between ducts of a single tissue sample. We demonstrated that with worsening DCIS grade, more heterogeneity was seen within a duct and between ducts of that tissue. These data support the notion that DCIS exhibits broad diversity, and indicate that this diversity is greater with increasing grade. Together, these data suggest that the proposed models of DCIS evolution, which indicate that low grade and high grade DCIS have distinct pathways of progression, may not be sufficiently comprehensive to reflect the true complexity of tumor evolution [31], [32]. In fact, differentiation of low grade to high grade seems more likely for the majority of cases, although the rate of sequential change may differ, and some cases may evolve in a more direct manner. An accurate understanding of breast cancer evolution is essential to inform breast cancer prevention strategies. In the future, we hope that image analysis can identify small numbers of defective cells in ‘healthy’ tissue biopsies from women at high risk of breast cancer. Thus, image analysis methods to identify morphological alterations may prove to be an effective tool in early cancer diagnosis and prevention.
CRediT authorship contribution statement
Mary-Kate Hayward: Conceptualization, Formal analysis, Visualization, Writing - original draft. J. Louise Jones: Validation, Resources. Allison Hall: Validation, Resources. Lorraine King: Resources. Alastair J. Ironside: Validation. Andrew C. Nelson: Validation. E. Shelley Hwang: Resources. Valerie M. Weaver: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to acknowledge the role of the Breast Cancer Now Tissue Bank in collecting and making available the samples used in the generation of this publication, and all the patients who donated. We thank both Jason J. Northey and Connor Stashko for their editorial input. We also thank the Biostatistics core, which is funded by the UCSF Department of Surgery, for their expert statistical support. This work was supported by US National Institutes of Health/National Cancer Institute grants R01CA192914 (V.M.W), 1R01CA222508-01 (V.M.W., E.S.H.), 1R35CA242447-01A1 (V.M.W., E.S.H.), R01CA185138-01 (E.S.H.), US Department of Defense (DOD) grant BC132057 (E.S.H.), Cancer Research UK ECMC grant C28106/A25175 (A.J.I) and American Cancer Society grant 132574-CSDG-18-139-01-CSM: Clinical Scientist Development Award (A.C.N.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.040.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Uhler C., Shivashankar G.V. Nuclear mechanopathology and cancer diagnosis. Trends Cancer. 2018;4(4):320–331. doi: 10.1016/j.trecan.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Lester S.C. Protocol for the examination of specimens from patients with ductal carcinoma in situ of the breast. Arch Pathol Lab Med. 2009;133(1):15–25. doi: 10.5858/133.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Sanders M.E., Schuyler P.A., Dupont W.D., Page D.L. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer. 2005;103(12):2481–2484. doi: 10.1002/cncr.21069. [DOI] [PubMed] [Google Scholar]
- 4.Collins L.C., Tamimi R.M., Baer H.J., Connolly J.L., Colditz G.A., Schnitt S.J. Outcome of patients with ductal carcinoma in situ untreated after diagnostic biopsy: results from the Nurses' Health Study. Cancer. 2005;103(9):1778–1784. doi: 10.1002/cncr.20979. [DOI] [PubMed] [Google Scholar]
- 5.Badve S., A'hern R.P., Ward A.M., Millis R.R., Pinder S.E., Ellis I.O., Gusterson B.A., Sloane J.P. Prediction of local recurrence of ductal carcinoma in situ of the breast using five histological classifications: a comparative study with long follow-up. Hum Pathol. 1998;29(9):915–923. doi: 10.1016/S0046-8177(98)90196-4. [DOI] [PubMed] [Google Scholar]
- 6.Onega T., Weaver D.L., Frederick P.D., Allison K.H., Tosteson A.N.A., Carney P.A., Geller B.M., Longton G.M., Nelson H.D., Oster N.V., Pepe M.S., Elmore J.G. The diagnostic challenge of low-grade ductal carcinoma in situ. Eur J Cancer. 2017;80:39–47. doi: 10.1016/j.ejca.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokbel K., Cutuli B. Heterogeneity of ductal carcinoma in situ and its effects on management. Lancet Oncol. 2006;7(9):756–765. doi: 10.1016/S1470-2045(06)70861-0. [DOI] [PubMed] [Google Scholar]
- 8.Allegra C.J., Aberle D.R., Ganschow P., Hahn S.M., Lee C.N., Millon-Underwood S., Pike M.C., Reed S.D., Saftlas A.F., Scarvalone S.A., Schwartz A.M., Slomski C., Yothers G., Zon R. National institutes of health state-of-the-science conference statement: diagnosis and management of ductal carcinoma in situ September 22-24, 2009. JNCI J National Cancer Inst. 2010;102(3):161–169. doi: 10.1093/jnci/djp485. [DOI] [PubMed] [Google Scholar]
- 9.Badve S., Gökmen-Polar Y. Tumor Heterogeneity in Breast Cancer. Adv Anat Pathol. 2015;22(5):294–302. doi: 10.1097/PAP.0000000000000074. [DOI] [PubMed] [Google Scholar]
- 10.Consensus Conference on the classification of ductal carcinoma in situ. The Consensus Conference Committee. Cancer, 1997;80(9):1798–802. [DOI] [PubMed]
- 11.Bankhead P. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong F. Computational pathology to discriminate benign from malignant intraductal proliferations of the breast. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klimov S. A whole slide image-based machine learning approach to predict ductal carcinoma in situ (DCIS) recurrence risk. Breast Cancer Res. 2019;21(1):83. doi: 10.1186/s13058-019-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axelrod D.E. Effect of quantitative nuclear image features on recurrence of Ductal Carcinoma In Situ (DCIS) of the breast. Cancer Inform. 2008;6:99–109. doi: 10.4137/cin.s401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burstein H.J. Ductal carcinoma in situ of the breast. N Engl J Med. 2004;350(14):1430–1441. doi: 10.1056/NEJMra031301. [DOI] [PubMed] [Google Scholar]
- 16.Lagios M.D. Heterogeneity of duct carcinoma in situ (DCIS): relationship of grade and subtype analysis to local recurrence and risk of invasive transformation. Cancer Lett. 1995;90(1):97–102. doi: 10.1016/0304-3835(94)03683-a. [DOI] [PubMed] [Google Scholar]
- 17.Kerlikowske K. Characteristics associated with recurrence among women with ductal carcinoma in situ treated by lumpectomy. J Natl Cancer Inst. 2003;95(22):1692–1702. doi: 10.1093/jnci/djg097. [DOI] [PubMed] [Google Scholar]
- 18.Lester S.C., Connolly J.L., Amin M.B. College of American Pathologists protocol for the reporting of ductal carcinoma in situ. Arch Pathol Lab Med. 2009;133(1):13–14. doi: 10.5858/133.1.13. [DOI] [PubMed] [Google Scholar]
- 19.Hipp J. Computer aided diagnostic tools aim to empower rather than replace pathologists: Lessons learned from computational chess. J Pathol Inform. 2011;2:25. doi: 10.4103/2153-3539.82050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond M.E. American society of clinical oncology/college of American pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract. 2010;6(4):195–197. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinn HP, Kreipe H, A Brief Overview of the WHO Classification of Breast Tumors, 4th Edition, Focusing on Issues and Updates from the 3rd Edition. Breast Care (Basel), 2013;8(2):149–54. [DOI] [PMC free article] [PubMed]
- 22.Chapman J.A. Ductal carcinoma in situ of the breast (DCIS) with heterogeneity of nuclear grade: prognostic effects of quantitative nuclear assessment. BMC Cancer. 2007;7:174. doi: 10.1186/1471-2407-7-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan P.H. Correlation of nuclear morphometry with pathologic parameters in ductal carcinoma in situ of the breast. Mod Pathol. 2001;14(10):937–941. doi: 10.1038/modpathol.3880415. [DOI] [PubMed] [Google Scholar]
- 24.Miller N.A. Heterogeneity Between Ducts of the Same Nuclear Grade Involved by Duct Carcinoma In Situ (DCIS) of the Breast. Cancer Inform. 2010;9:209–216. doi: 10.4137/cin.s5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veta M. Prognostic value of automatically extracted nuclear morphometric features in whole slide images of male breast cancer. Mod Pathol. 2012;25(12):1559–1565. doi: 10.1038/modpathol.2012.126. [DOI] [PubMed] [Google Scholar]
- 26.Axelrod D.E., Miller N., Chapman J.A. Avoiding Pitfalls in the Statistical Analysis of Heterogeneous Tumors. Biomed Inform Insights. 2009;2:11–18. doi: 10.4137/bii.s2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allred D.C. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14(2):370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 28.Groen E.J. Finding the balance between over- and under-treatment of ductal carcinoma in situ (DCIS) Breast. 2017;31:274–283. doi: 10.1016/j.breast.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Lennington W.J. Ductal carcinoma in situ of the breast. Heterogeneity of individual lesions. Cancer. 1994;73(1):118–124. doi: 10.1002/1097-0142(19940101)73:1<118::aid-cncr2820730121>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 30.Gerdes M.J. Single-cell heterogeneity in ductal carcinoma in situ of breast. Mod Pathol. 2018;31(3):406–417. doi: 10.1038/modpathol.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson P.T. Molecular evolution of breast cancer. J Pathol. 2005;205(2):248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 32.Sgroi D.C. Preinvasive breast cancer. Annu Rev Pathol. 2010;5:193–221. doi: 10.1146/annurev.pathol.4.110807.092306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.