Summary
Whole-genome sequencing data were produced from a single flathead grey mullet female and assembled into a draft genome sequence, whereas publicly available sequence data were used to obtain a male draft sequence. Two pools, each consisting of 60 unrelated individuals, respectively, of male and female fish were analyzed using Pool-Sequencing. Mapping and analysis of Pool-Seq data against the draft genome(s) revealed >30 loci potentially associated with sex, the most promising locus of which, encoding the follicle-stimulating hormone receptor (fshr) and harboring two missense variants, was genotyped on 245 fish from four Mediterranean populations. Genotype data showed that fshr represents a previously unknown sex-determining locus, although the incomplete association pattern between fshr genotype and sex-phenotype, the variability of such pattern across different populations, and the presence of other candidate loci reveal that a greater complexity underlies sex determination in the flathead grey mullet.
Subject areas: Genetics, Evolutionary Biology, Genomics
Graphical abstract
Highlights
-
•
Three SNPs with sex-specific allele frequencies found in the grey mullet fshr gene
-
•
Two missense variants at the fshr locus are significantly associated with male sex
-
•
fshr male genotypes show divergent distribution across geographical populations
-
•
Incomplete penetrance of fshr variants suggests that additional factors exist
Genetics; Evolutionary Biology; Genomics
Introduction
Fish roe is a highly sought seafood consisting of raw or dried female gonads. The flathead grey mullet (Mugil cephalus, Linnaeus, 1758; Mugilidae) salted and dried roe is a highly priced food delicacy, traditionally produced in many regions of the world. In some Mediterranean areas (e.g., Tyrrhenian [west] coast of Italy, Northern and Western Greece), it represents an important value product for the local economy as it is sold at “extravagantly high price” (Crosetti 2016): top-quality mullet egg roes, “bottarga,” from Orbetello or Cabras lagoons in Italy are sold at 280€/kg retail price. The Greek roe from Messolongi lagoon complex, “Avgotaraxo Messolongiou,” is a protected designation of origin product (Koutrakis, 2016). In the Mediterranean region, mullet roe is obtained from the gonads of at least 4- to 5-year-old females, which are normally captured at coastal lagoon fish barriers during their spawning migration back to sea in late summer/early autumn. A renewed interest for grey mullets is coming out among fish farmers, aiming at developing grey mullet-intensive rearing for egg roe production, as already performed in Taiwan (Crosetti, 2016).
A quick non-lethal method for early sorting of males and females, even before sexual maturation, would be greatly desirable to culture monosex female stocks for their egg roe. Such a method would require reliable sex-associated genetic markers, which is not always possible in fish. In teleosts, sex determination mechanisms seem to be extremely diverse, ranging from the presence of evident heterochromatic sex chromosomes to simultaneous hermaphroditism. When genetic sex determination (GSD) is observed, the underlying mechanisms might be diverse even between closely related species or within the same species, and environmental factors (environmental sex determination [ESD]) could affect sex determination (Heule et al., 2014; Geffroy and Douhard, 2019; Geffroy and Wedekind, 2020). The flathead grey mullet is gonochoristic with genetic determination of sex (McDonough et al., 2005). However, several cytogenetic studies showed that there is no evidence for heterochromatic sex chromosomes in all the different tested populations (e.g., Rossi et al., 1996; Gornung et al., 2001), although a major sex-linked locus was shown to segregate in a single family (Dor et al., 2016), therefore sex-associated genetic markers could be identified in this species.
The identification of sex-determining genomic regions has been recently boosted by the advent of next-generation sequencing technologies, which provide several options toward this goal (Palmer et al., 2019). Genome-wide association mapping is one of such options. Unrelated individuals of both sexes are analyzed using either a large set of genetic markers or whole-genome sequencing.
In the present study, a cost-effective genome-wide sequencing approach for association mapping, Pool Sequencing (Pool-Seq), (Schlötterer et al., 2014) was implemented with the goal of finding sex-associated genetic markers, which were then validated on four wild populations.
Results
Mugil cephalus genome draft
Whole-genome sequencing yielded 394.4 million reads corresponding to 119 Gb and an estimated coverage higher than 110×. The assembly consisted of 175,167 scaffolds, with an average length of 3,750 nt (nucleotides) and N50 equal to 11,134 nt. The quality of the genome assessed with BUSCO using Actinopterygii dataset (4,584 well-conserved genes) showed 81.4% BUSCO completeness. The assembly of a male genome using publicly available data had a lower coverage (50×), was more fragmented (234,577 scaffolds, with an average length of 2,580 nt and N50 equal to 4,584 nt), and had lower completeness estimated with BUSCO (71.9%). Therefore, the female draft genome was used for most analyses.
Sequencing sex-specific pools
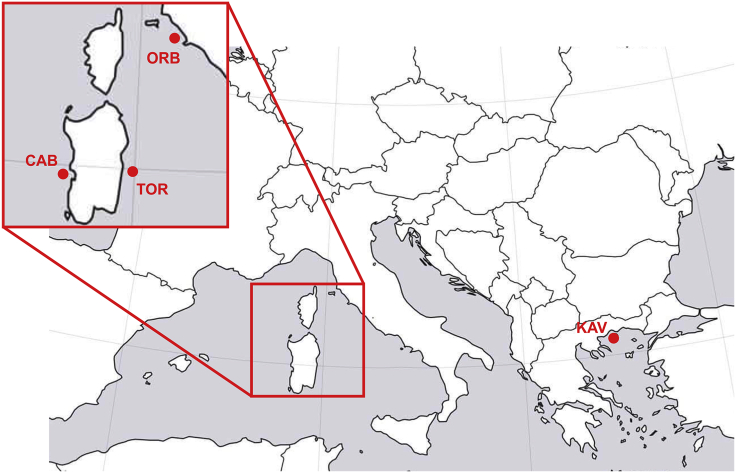
A pool of 60 females and another pool of 60 males were analyzed. Each pool included 30 individuals for each of the two distinct Sardinian populations (CAB and TOR, Figure 1).
Figure 1.
Sampling sites
(ORB: Orbetello Lagoon, CAB: Cabras Lagoon, TOR: Tortolì Lagoon, KAV: Kavala).
Three replicates for each pool were prepared and sequenced independently to minimize technical variation. A total of 185 million reads were obtained for the female pool and 181.5 for the male one, which correspond to an estimated 63× and 62× coverage, respectively, which fulfills the suggested sequencing depth for Pool-Seq analysis (Schlötterer et al., 2014). A dedicated bioinformatics pipeline, Popoolation2 (Kofler et al., 2011), yielded 613,379 SNPs that showed significantly different allele frequencies between the male and the female pools with nominal p value≤0.05. After false discovery rate correction, significantly divergent SNPs were 181, located on 135 contigs. To further refine the analysis, we looked for loci showing a single (fixed) allele in one sex and two alleles at frequency ranging between 0.4 and 0.6 in the other sex, which is the expected allele frequency pattern for sex-specific variants.
This analysis led to the identification of 114 genomic regions harboring SNPs with significant allele frequency association with either sex (62 with one allele fixed or nearly fixed for the female pool and 52 SNPs for the male pool). Of these SNPs, 49 showed significant (1e-10) match with the Nile tilapia genome and 32 were located within an annotated protein/coding gene.
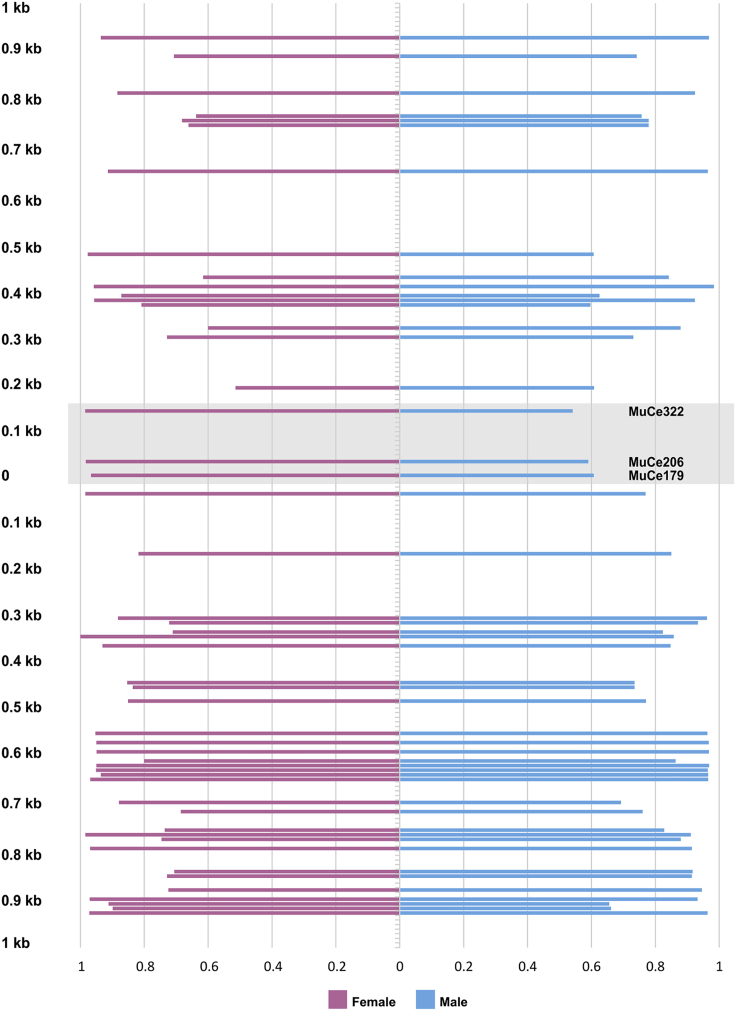
Among candidate sex-determining variants, three SNPs at position 179 (MuCe179), 206 (MuCe206), and 322 (MuCe322) of the assembled contig_111122 were found (see Data S1). The first two variants were always found in cis based on single sequence read analysis, defining a single allele; they had approximately 0.5 frequency in males but were almost absent in females (m1 allele frequency = 0.02). They are the only three SNPs with sex-specific allele frequencies in the candidate genomic region, as all other neighboring SNPs did not show such evidence (Figure 2).
Figure 2.
Female- and male-specific allele frequencies at variable nucleotide positions surrounding the three significant variants
The grey square indicates the three variants associated with sex. Distance from MuCe179 variant is showed in y axis; x axis reports variants' allele frequency. On the X-axis, observed frequency of the major allele is shown for each SNP. On the Y-axis, the relative distance in kilobases (kb), respectively upstream and downstream of SNP MuCe179.
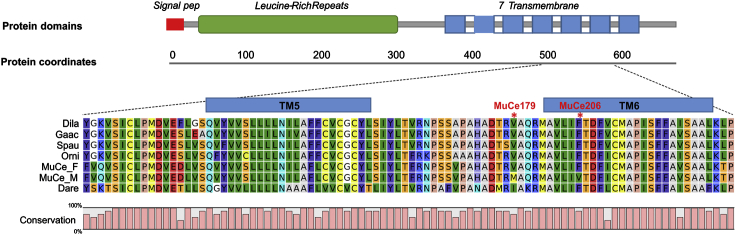
Comparative genomic analysis against the Nile tilapia genome (Oreochromis_niloticus_UMD_NMBU) showed that the three variants are located on the putative exon 14 of the fshr gene (Figure 3). The presence of a conserved gene (fbx011) upstream of fshr in both Nile tilapia and M. cephalus suggests that the identified fshr gene in the flathead grey mullet is the ortholog of Nile tilapia fshr. Extensive similarity searches against the two assembled genomes did not find any evidence for a duplicated fshr copy. Although the female and male M. cephalus genomes presented here are not complete, the probability of missing a potential second fshr copy is less than 0.05, based on the completeness BUSCO scores. Furthermore, the identified fshr gene was included in the sex-linked region reported by Dor et al. (2016). Mutations MuCe179 and MuCe206 correspond to positions on tilapia LG8:g.10686582 G > A (ENSONIP00000043437:p.Val557Met) and LG8:g.10686555T > G (ENSONIP00000043437:p.Phe566Val), and both represent missense variants. Mutation MuCe322 is a synonymous variant. Protein structural mapping revealed that both missense SNPs are located in the fshr transmembrane region, MuCe206 in the transmembrane helix 6 (TM6) and MuCe179 in the intracellular loop between TM5 and TM6 (Figure 3), a highly conserved region of great functional relevance (Ulloa-Aguirre et al., 2018).
Figure 3.
Position of the two non-synonymous variants on the fshr sequence and multiple fish species sequence alignment
(Dila: Dicentrarchus labrax, Gaac: Gasterosteus aculeatus, Spau: Sparus aurata, Orni: Oreochromis niloticus, MuCe_F: Mugil cephalus wt allele, MuCe_M: m1 allele, Dare: Danio rerio). Nucleotide sequence of the fshr contig and exact positions of MuCe179, MuCe206 and MuCe322 variants are reported in Data S1.
The effect of MuCe179 and MuCe206 mutations on fshr function was predicted using the Ensembl variant effect predictor VEP (McLaren et al., 2016), which reported both variants to be functionally deleterious with SIFT code 0.03 and 0, respectively.
fshr is known to have a key role in folliculogenesis in fish and mammal female gonads (Li and Cheng, 2018). Although fshr was never reported before as a sex-determining locus, all the above-mentioned evidence suggested that it might be one of the genes involved in GSD in flathead grey mullet.
Validation of fshr variants in four flathead grey mullet populations
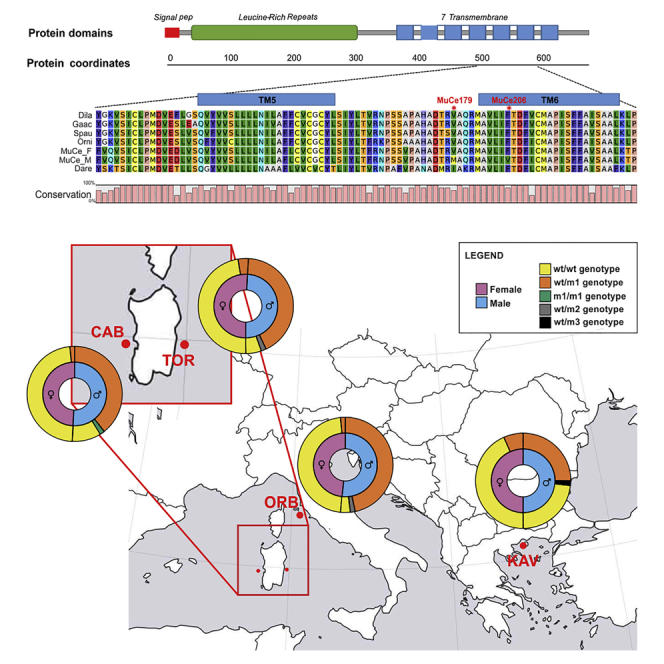
To validate Pool-Seq results, a region of 308 nt encompassing MuCe179, MuCe206, and MuCe322 variants was sequenced in 245 individuals collected from the wild at four different sites (Figure 1) representing the Western Mediterranean (Cabras Lagoon), the Tyrrhenian (Tortolì and Orbetello Lagoons), and the North Aegean Sea (Kavala). Individuals included in TOR and CAB populations were used both for Pool-Seq analysis and validation. Results of individual genotyping are summarized in Table 1.
Table 1.
Genotype distribution in males and females of the four wild populations
| Males |
Females |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| wt/wt | wt/m1a | m1/m1 | wt/m2b | wt/m3c | wt/m4d | wt/wt | wt/m1 | wt/m2 | wt/m3 | wt/m4 | |
| ORB | 6% (2/31) | 91% (28/31) | 3% (1/31) | 93% (28/30) | 3.5% (1/30) | 3.5% (1/30) | |||||
| TOR | 11% (3/28) | 86% (24/28) | 3% (1/28) | 84.5% (27/32) | 6% (2/32) | 9.5% (3/32) | |||||
| CAB | 18% (6/33) | 73% (24/33) | 3% (1/33) | 6% (2/33) | 97% (29/30) | 3% (1/30) | |||||
| KAV | 45% (14/31) | 48% (15/31) | 3.5% (1/31) | 3.5% (1/31) | 84% (26/31) | 13% (4/31) | 3% (1/31) | ||||
For a detailed description of individual sampling site, sex, phenotyping method, and genotype see Table S1.
allele m1: MuCe179, MuCe206, MuCe322
m2: MuCe179, MuCe206
m3: MuCe179
m4: MuCe322.
The most frequent fshr allele, which also showed conserved amino acid residues compared with other fish species at positions MuCe179 and MuCe206 (Figure 3), was considered wild-type (wt), m1 was the name attributed to the allele harboring all three variants (MuCe179, MuCe206, MuCe322), m2 to a second mutant allele only containing the two missense mutations, m3 to a third one with the missense mutation MuCe179 alone, and m4 the allele with the synonymous mutation MuCe322 alone.
Overall, individual fshr genotype data confirmed that alleles m1 and m2 are significantly associated with the male sex (Fisher exact test p < 0.00001). However, the association was not complete, as sex phenotype and fshr genotype did not match the expected pattern in 3%-13% of females, depending on the population. Individuals with mismatched sex-phenotype/genotype cannot be explained as the consequence of erroneous sex identification, as all sampled individuals were sexed during the reproductive season, when sex, especially for males, is macroscopically evident (an example of the highly different gonad morphology is shown in Figure 4); in case of dubious sex, gonad histological examination was carried out (see Figure S1). Finally, the presence of a fshr m1/m1 male represents genetic evidence that wt/m1 females do exist. The observed frequency of m1 homozygotes also conforms with the one expected from the observed frequency of wt/m1 females and wt/m1 males and Mendelian proportions in F1 homozygotes. The highest expected frequency for m1/m1 is 0.02 [highest wt/m1 males frequency (0.93) × highest wt/m1 female frequency (0.095) × 0.25], which is comparable with present observations (0.03).
Figure 4.
Macroscopic gonad morphology of male and female M. cephalus individuals
(A–D) Male individual (A), macroscopic appearance of male (B) and female (C) gonads, and female individual (D).
The difference between populations in the frequency of wt/m1 females was not significant (p > 0.5). Wild-type homozygous (wt/wt) males were present at variable frequencies in the four populations (6%–45%). Differences across populations were highly significant (chi-square test p < 0.01), with the lowest frequency (6%) in ORB and the highest in KAV (45%). Analysis of the observed “recombinant” genotypes, which either contain at least one missense mutation (m2, m3) or the synonymous variant (m4) alone, suggests that MuCe322 might not be relevant for sex determination as it has a significant bias toward females (Fisher exact test p < 0.05) compared with non-synonymous variants.
Discussion
In the present study, conclusive evidence that two missense single-nucleotide variants at the fshr locus are significantly associated with male sex was provided. The observed pattern of wt/m1 and wt/m2 heterozygous males suggests an “XX-XY” GSD type. In other fish species, the sex-determining gene is represented by a duplicated copy (e.g., amhy in the Patagonian pejerrey, Hattori et al., 2012; dmrt1b in medaka, Matsuda et al., 2002), which has evolved sex-specific sequence variants, whereas the “original” copy (amh, dmrt1b) conserved its ancestral function. There was no evidence for two copies of the fshr locus in the flathead grey mullet. How variant alleles of the single copy fshr might act as a switch toward male differentiation still remains to be clarified. In several fish species fshr is reported to be expressed in the ovary theca and granulosa cells, and the testis Leydig and Sertoli cells (Li and Cheng, 2018). Deleterious mutations at the fshr locus in mice and humans cause severe female infertility and milder effects on males. Likewise, in zebrafish females fshr knockout (KO) induces female sterility with impaired folliculogenesis in the ovary, whereas fshr KO males only show delayed puberty; sex reversal (female to male) might occur in part of putative fshr KO females (Chu et al., 2015). A similar outcome was observed in fshr KO in medaka (Oryzias latipes), where fshr KO females have small ovaries and are infertile, whereas males have normal testes and are fertile (Murozumi et al., 2014). As in zebrafish, masculinization (sex reversal) of genetic (XX) female medakas was observed in 10% of KO fish. The major fshr variant in the flathead grey mullet might act as negative dominant, causing the loss of function of the wild-type copy and inducing male development as in zebrafish and medaka fshr KO. Negative dominant fshr mutations were also reported in humans (Zariñán et al., 2010). The alternative explanation of wt/m1 fshr showing haploinsufficiency is unlikely as heterozygous wt/KO fshr medaka females are fertile (Murozumi et al., 2014). The observed incomplete penetrance of the mutations observed in the flathead grey mullet (3%–10% of wt/m1 females) agrees with the evidence found for dominant negative fshr mutations in humans, where the mutant form expression level correlated with the wild-type functional suppression level (Zariñán et al., 2010). Variation in the m1/m2 allele expression across females could be responsible for the incomplete penetrance. This remains a working hypothesis that awaits functional evidence, for instance, overexpressing a transgenic m1/m2 allele copy in a wt/wt background. Unfortunately, this experimental approach is currently impossible in a non-model species as the flathead grey mullet, although zebrafish or medaka might be used as proxy.
A third hypothesis is that the observed variants at the fshr locus act indirectly on sex determination, by increasing the sensitivity of the sex-determining signaling pathway to environmental factors, for instance, to water temperature. Sex-associated genetic variants that are variably influenced by temperature have been already reported in the Nile tilapia (Wessels et al., 2014) and zebrafish (Ribas et al., 2017).
Irrespective of the functional mechanisms that underlie genotype-phenotype interaction in wt/m1 (wt/m2) individuals, it appears that fshr is a new entry in the ever-growing list of loci putatively involved in GSD in teleost fish. However, this is not the most interesting point conveyed by the present study. The classic paradigm of a single master gene involved in GSD is becoming increasingly challenged both theoretically and on the basis of experimental evidence. This is particularly true in teleost fish, which harbor an overwhelming diversity of sex-determining mechanisms. A new paradigm was proposed where sex determination in fish does not rely on a single genetic cascade, but the process is regulated along a continuum of environmental and genetic factors (Heule et al., 2014). Furthermore, the traditional genetic architecture with a single sex-determining locus might not be the only model for GSD. As predicted by Moore and Roberts (2013), polygenic genetic sex determination (PSD) was proposed for an increasing number of fish species, in parallel with the wider application of genome-wide association mapping (e.g., Wilson et al., 2014; Li et al., 2020; Junker et al., 2020). Even more intriguingly, in the European sea bass (Dicentrarchus labrax), a species where both ESD and GSD are present, several loci with minor effects were recently reported to contribute to sex determination (i.e., PSD), but in different geographic populations these loci are partially different (Faggion et al., 2019). Such evidence provides strong support to the hypothesis of population-specific evolution of PSD systems in relation to different environments. In this perspective, flathead grey mullet might represent an excellent model, as it has a global distribution in tropical, subtropical, and temperate seas worldwide, with several evolutionary lineages and eventual cryptic species (Durand and Borsa 2015) and likely a large number of distinct populations within these lineages. For instance, M. cephalus shows at least three genetic clusters in the Atlantic-Mediterranean-Black Sea area, and an even finer population structure might be present (Durand et al., 2013). In particular, populations in the North Aegean and Black seas were clearly distinct from those collected in the Western Mediterranean. In light of such evidence, the divergent distribution of fshr genotypes in males between KAV (North Aegean) and TOR-CAB-ORB (Western Mediterranean) appears quite suggestive. It is possible that in the KAV population fshr role is less relevant and other genetic loci, possibly linked to local conditions, substantially contribute to sex determination. Additionally, as already mentioned, environmental factors such as temperature might interplay with genetic variation, at the fshr locus and possibly at other loci. The broad distribution of the flathead grey mullet in marine habitats characterized by large differences in environmental factors (e.g., temperature, salinity) might provide a unique opportunity to explore the interaction between PSD and ESD. The incomplete penetrance of the fshr variants also suggests that additional genetic factors might exist. This is confirmed by the evidence of over 100 putative sex-associated SNPs reported here, more than 30 located on protein-coding genes. Preliminary sequence analysis on additional candidate loci confirmed that for two variants at the nuclear receptor NR2F5 almost all females were homozygous, whereas half of the males were heterozygous, indicating a partial sex association (see Transparent Methods). Extensive validation of all putative sex-associated variants would, however, require a higher-quality genome sequence.
In conclusion, the original goal of identifying a simple genetic test to sex fish to culture monosex grey mullet stocks and egg roe has only been partially achieved as the sex determination system appears more complex than the one with a single master gene with complete association with sex. Nevertheless, the high-frequency association of m1 and m2 alleles with male sex in Sardinian population might allow to develop population-specific genetic tools suitable for early sex sorting in grey mullets. In addition, the presence of fshr wt/wt males might enable to obtain predominantly female F1 by crossing them with wt/wt females, as the two genotypes could be analyzed without sacrificing the broodstock. More generally, its distribution and population structure, the presence of a major sex-determining locus (fshr) already identified and showing population-specific variation, and the existence of other candidate sex-associated variants make the flathead grey mullet an ideal playground for evolutionary biologists to study the variable complex architecture of sex determination in teleost fish.
Limitations of the study
The study provides genetic evidence that fshr variants are significantly associated with male sex development, but how one single copy of the variant fshr alleles might act as a switch toward male differentiation still remains to be clarified by means of controlled functional experiments. Variation at the fshr locus was assessed across several populations, although additional samples representing other geographic populations (e.g., Atlantic and Black Sea ones) would provide a broader view of such variation. Evidence for additional sex-associated loci should be further characterized.
Resource availability
Lead contact
Information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Luca Bargelloni (luca.bargelloni@unipd.it).
Material availability
This study did not generate unique reagents. Primers for PCR amplification of the fshr fragment are available upon request to the Lead Contact. Primer sequences are reported in the Transparent Methods section. Biological materials will be available, although for several samples the amount is limited.
Data and code availability
The Bioproject accession number for the sequence data reported in this paper is SRA:PRJNA657721, BioSample SAMN15835396 to SAMN15835401. The sequence of the fshr contig is included in the Data S1.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
Authors thank fishermen who helped collecting samples, in particular the “Cooperativa Pescatori di Orbetello” for ORB sampling, “Vistonis Lagoon Fishermen Cooperative” for KAV sampling, and “Cooperativa Pescatori di Tortolì” and “Cooperativa Pescatori di Cabras,” respectively, for TOR and CAB sampling. Authors acknowledge the funding of the Italian Ministry of Education, University and Research (MIUR) for the project “Centro di Eccellenza per la Salute degli Animali Acquatici ECCE AQUA.” In-house funding from the University of Cagliari and University of Padova are also acknowledged.
Author contributions
R.C., A.C., M.C.F., R.M., and L.C. collected and analyzed CAB and TOR samples. M.K. and A.S. collected and analyzed KAV samples. D.C. collected and analyzed ORB samples. S.F. performed all genomic and genetic experiments. M.B. and S.F. carried out all bioinformatics analyses. L.B., T.P., and A.C. designed the experiments. S.F. and L.B. drafted the paper. D.C., M.K., T.P., and R.C. commented and edited the manuscript. All authors revised and approved the paper.
Declaration of interests
The authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101886.
Supplemental Information
Sampling ID (ORB Orbetello, TOR Tortolì, KAV Kavala, CAB Cabras), sex, phenotyping method, and individual genotype (for allele nomenclature see Table 1 in the Results section).
References
- Chu L., Li J., Liu Y., Cheng C.H. Gonadotropin signaling in zebrafish ovary and testis development: insights from gene knockout study. Mol. Endocrinol. 2015;29:1743–1758. doi: 10.1210/me.2015-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetti D. Current state of grey mullet fisheries and culture. In: Crosetti D., Blaber S.J.M., editors. Biology, Ecology and Culture of Grey Mullet (Mugilidae) CRC Press; 2016. pp. 398–450. [Google Scholar]
- Dor L., Shirak A., Rosenfeld H., Ashkenazi I.M., Band M.R., Korol A., Ronin Y., Seroussi E., Weller J.I., Ron M. Identification of the sex-determining region in flathead grey mullet (Mugil cephalus) Anim. Genet. 2016;47:698–707. doi: 10.1111/age.12486. [DOI] [PubMed] [Google Scholar]
- Durand J.D., Borsa P. Mitochondrial phylogeny of grey mullets (Acanthopterygii: Mugilidae) suggests high proportion of cryptic species. C R. Biol. 2015;2338:266–277. doi: 10.1016/j.crvi.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Durand J.D., Blel H., Shen K.N., Koutrakis E.T., Guinand B. Population genetic structure of Mugil cephalus in the Mediterranean and Black Seas: a single mitochondrial clade and many nuclear barriers. Mar. Ecol. Prog. Ser. 2013;474:243–261. [Google Scholar]
- Faggion S., Vandeputte M., Chatain B., Gagnaire P.A., Allal F. Population-specific variations of the genetic architecture of sex determination in wild European sea bass Dicentrarchus labrax L. Heredity. 2019;122:612–621. doi: 10.1038/s41437-018-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffroy B., Douhard M. The adaptive sex in stressful environments. Trends Ecol. Evol. 2019;34:628–640. doi: 10.1016/j.tree.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Geffroy B., Wedekind C. Effects of global warming on sex ratios in fishes. J. Fish Biol. 2020 doi: 10.1111/jfb.14429. [DOI] [PubMed] [Google Scholar]
- Gornung E., Cordisco C.A., Rossi A.R., De Innocentiis S., Crosetti D., Sola L. Chromosomal evolution in mugilids: karyotype characterization of Liza saliens and comparative localization of major and minor ribosomal genes in the six Mediterranean grey mullets. Mar. Biol. 2001;139:55–60. [Google Scholar]
- Hattori R.S., Murai Y., Oura M., Masuda S., Majhi S.K., Sakamoto T., Fernandino J.I., Somoza G.M., Yokota M., Strüssmann C.A. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U S A. 2012;109:2955–2959. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heule C., Salzburger W., Böhne A. Genetics of sexual development: an evolutionary playground for fish. Genetics. 2014;196:579–591. doi: 10.1534/genetics.114.161158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker J., Rick J.A., McIntyre P.B., Kimirei I., Sweke E.A., Mosille J.B., Wehrli B., Dinkel C., Mwaiko S., Seehausen O., Wagner C.E. Structural genomic variation leads to genetic differentiation in Lake Tanganyika's sardines. Mol. Ecol. 2020 doi: 10.1111/mec.1555. [DOI] [PubMed] [Google Scholar]
- Kofler R., Pandey R.V., Schlötterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutrakis E. Biology and ecology of fry and juveniles. In: Crosetti D., Blaber S., editors. Biology, Ecology and Culture of Grey Mullets (Mugilidae) Taylor & Francis Group; 2016. pp. 264–291. [Google Scholar]
- Li J., Cheng C.H.K. Evolution of gonadotropin signaling on gonad development: insights from gene knockout studies in zebrafish. Biol. Reprod. 2018;99:686–694. doi: 10.1093/biolre/ioy101. [DOI] [PubMed] [Google Scholar]
- Li Y.L., Xing T.F., Liu J.X. Genome-wide association analyses based on whole-genome sequencing of Protosalanx hyalocranius provide insights into sex determination of Salangid fishes. Mol. Ecol. Resour. 2020;20:1038–1049. doi: 10.1111/1755-0998.13172. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., Morrey C.E., Shibata N., Asakawa S., Shimizu N. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- McDonough C.J., Roumillat W.A., Wenner C.A. Sexual differentiation and gonad development in striped mullet (Mugil cephalus L.) from South Carolina estuaries. Fish Bull. 2005;103:601–619. [Google Scholar]
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie C.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E.C., Roberts R.B. Polygenic sex determination. Curr. Biol. 2013;23:R510–R512. doi: 10.1016/j.cub.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Murozumi N., Nakashima R., Hirai T., Kamei Y., Ishikawa-Fujiwara T., Todo T., Kitano T. Loss of follicle-stimulating hormone receptor function causes masculinization and suppression of ovarian development in genetically female medaka. Endocrinology. 2014;155:3136–3145. doi: 10.1210/en.2013-2060. [DOI] [PubMed] [Google Scholar]
- Palmer D.H., Rogers T.F., Dean R., Wright A.E. How to identify sex chromosomes and their turnover. Mol. Ecol. 2019;28:4709–4724. doi: 10.1111/mec.15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas L., Liew W.C., Díaz N., Sreenivasan R., Orbán L., Piferrer F. Heat-induced masculinization in domesticated zebrafish is family-specific and yields a set of different gonadal transcriptomes. Proc. Natl. Acad. Sci. U S A. 2017;114:E941–E950. doi: 10.1073/pnas.1609411114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Crosetti D., Gornung E., Sola L. Cytogenetic analysis of global populations of Mugil cephalus (striped mullet) by different staining techniques and fluorescent in situ hybridization. Heredity. 1996;76:77–82. doi: 10.1038/hdy.1997.125. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Tobler R., Kofler R., Nolte V. Sequencing pools of individuals - mining genome-wide polymorphism data without big funding. Nat. Rev. Genet. 2014;15:749–763. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- Ulloa-Aguirre A., Zariñán T., Jardón-Valadez E., Gutiérrez-Sagal R., Dias J.A. Structure-function relationships of the follicle-stimulating hormone receptor. Front. Endocrinol. (Lausanne) 2018;9:707. doi: 10.3389/fendo.2018.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels S., Sharifi R.A., Luehmann L.M., Rueangsri S., Krause I., Hoerstgen-Schwark G., Knorr C. Allelic variant in the anti-Müllerian hormone gene leads to autosomal and temperature-dependent sex reversal in a selected Nile tilapia line. PLoS One. 2014;9:e104795. doi: 10.1371/journal.pone.0104795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.A., High S.K., McCluskey B.M., Amores A., Yan Y.-L., Titus A., Anderson J.L., Batzel P., Carvan M.J., Schartl M., Postlethwait J.H. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics. 2014;198:1291–1308. doi: 10.1534/genetics.114.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariñán T., Perez-Solís M.A., Maya-Núñez G., Casas-González P., Conn P.M., Dias J.A., Ulloa-Aguirre A. Dominant negative effects of human follicle-stimulating hormone receptor expression-deficient mutants on wild-type receptor cell surface expression. Rescue of oligomerization-dependent defective receptor expression by using cognate decoys. Mol. Cell. Endocrinol. 2010;321:112–122. doi: 10.1016/j.mce.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling ID (ORB Orbetello, TOR Tortolì, KAV Kavala, CAB Cabras), sex, phenotyping method, and individual genotype (for allele nomenclature see Table 1 in the Results section).
Data Availability Statement
The Bioproject accession number for the sequence data reported in this paper is SRA:PRJNA657721, BioSample SAMN15835396 to SAMN15835401. The sequence of the fshr contig is included in the Data S1.