Abstract
Background
The paucity of hepatitis B virus (HBV) DNA measurement in low-/middle-income countries hinders the identification of HBV-infected pregnant women at risk of perinatal transmission. This study evaluates the validity of an algorithm selecting HBeAg-positive women and HBeAg-negative women with alanine aminotransferase (ALT) ≥40 IU/L as a predictor of high HBV DNA level.
Methods
All women with reactive samples for hepatitis B surface antigen (HBsAg) were assessed with an SD BIOLINE HBeAg rapid test and HBV DNA quantification was performed. Validities of HBeAg and of the algorithm to identify HBV DNA >2 thresholds (5.3 and 7.3 log10 IU/mL) were evaluated.
Results
For the 515 HBsAg-positive women, median age was 29 years, 92 (17.9%) were HBeAg positive, 47 (9.1%) were HBeAg negative with ALT ≥40 IU/L, and 144 (28.0%) had an HBV DNA >5.3 log10 IU/mL. Sensitivity and specificity of HBeAg were 61.8% and 99.2% for HBV DNA >5.3 log10 IU/mL and 81.3% and 96.7% for HBV DNA >7.3 log10 IU/mL. For the algorithm, sensitivity and specificity were 79.2% and 93.3% for HBV DNA level >5.3 log10 IU/mL and 92.7% and 88.1% for HBV DNA >7.3 log10 IU/mL. The AUCs for the algorithm (0.92 and 0.94 for HBV DNA >5.3 and 7.3, respectively) were significantly greater (P < .001) than the AUCs for HBeAg (0.81 and 0.89 for HBV DNA >5.3 and 7.3, respectively).
Conclusions
An algorithm using HBeAg and ALT level could be an effective strategy to identify HBV-infected pregnant women at risk of perinatal transmission in countries where HBV DNA quantification is not routinely available.
Keywords: hepatitis B, mother-to-child transmission, HBeAg rapid test, public health, Western-Pacific area
An algorithm selecting Hepatitis B e Antigen (HBeAG)-positive and HBeAg-negative women with ALT ≥40 IU/L could be an effective strategy to identify pregnant women with hepatitis B virus (HBV) eligible for antiviral preventive treatment in countries where HBV DNA quantification is not routinely available.
Among the estimated 257 million people living with hepatitis B virus (HBV) infection, 45% are in the Western Pacific Region [1, 2]. The World Health Organization (WHO) aims to eliminate viral hepatitis as a public health threat by 2030 by reducing the incidence and mortality of chronic HBV infection by 90% and 65%, respectively [3]. Most of the HBV epidemic is driven by perinatal transmission—infection transmitted from mother to child during the third trimester of pregnancy in utero or at delivery [4]. Universal immunization remains the first-line prevention against perinatal infection for all infants. The WHO recommends at least 3 doses of HBV vaccination for all children worldwide, with the first dose to be administered at birth. However, mother-to-child transmission (MTCT) still occurs in infants born from women with a high HBV DNA viral load and/or with a positive hepatitis B e antigen (HBeAg) test [5–7]. International guidelines (American Association for the Study of Liver Diseases, Asian Pacific Association for the Study of the Liver, European Association for the Study of the Liver) recommend the use of antiviral drugs during the third trimester of pregnancy if maternal HBV DNA viral load is greater than 5.3 log10 IU/mL [8–10], in addition to HBV vaccine with or without anti-hepatitis B immunoglobulin (HBIg). However, the paucity of HBV DNA measurement in many low- and middle-income countries (LMICs) presents a major obstacle to identifying women at risk of MTCT who require these additional preventive strategies. A positive HBeAg test is associated with high HBV DNA levels [6] and, therefore, can act as a surrogate marker of HBV replication, including in women of childbearing age [11].
In Cambodia, HBV prevalence ranges from 3% to 5% in the general population [12, 13], but the vast majority of pregnant women are not screened for HBV infection during antenatal care (ANC). For those who are hepatitis B surface antigen (HBsAg) positive, HBV DNA quantification is costly and accessible in only a few laboratories located in Phnom Penh, the capital city of Cambodia. As recently described in the Agence Nationale de Recherche sur le Sida (ANRS) 12328 study in Cambodia, the performance of the SD Bioline HBeAg rapid diagnosis test (RDT) has a 76.5% sensitivity and a 96% specificity to predict a threshold of 5.3 log10 IU/mL of HBV DNA [14]. Sensitivity issues for this RDT seem relate to mutations in the basal core promoter (BCP) and precore (PC) genes [14] responsible for down-regulation of HBeAg production despite HBV DNA and alanine aminotransferase (ALT) levels remaining elevated. Therefore, we questioned whether we could improve the sensitivity of HBeAg RDT to predict HBV DNA viral load thresholds if ALT level was added to the algorithm for women with a negative HBeAg RDT.
The objective of the study was to evaluate the performance of an algorithm selecting HBeAg-positive women and HBeAg-negative women with ALT of 40 IU/L or higher as a predictor of high HBV DNA viral load to identify pregnant women most at risk of HBV MTCT in Cambodia.
METHODS
Data Collection
The present analysis is part of the ANRS 12345-TA PROHM study (ClinicalTrials.gov Identifier: NCT02937779), which has been implemented in Cambodia since October 2017. It is a phase IV, multicenter, observational and interventional prospective study conducted in 5 maternity wards (2 in Phnom Penh and 3 in province) aimed at assessing the effectiveness of a strategy to prevent HBV MTCT based on the following: (1) use of an HBsAg and HBeAg RDT serial algorithm to identify women at risk of MTCT, (2) tenofovir disoproxil fumarate (TDF) treatment from 24 weeks of pregnancy in women positive for both HBsAg and HBeAg RDT, and (3) early vaccination at birth (<2 hours of life) for all newborns. Women with a positive HBeAg RDT are treated with TDF, with a daily administration of one 300-mg pill, from 24 weeks of amenorrhea until 8 weeks postpartum. For women with their first ANC visit after 24 weeks of amenorrhea, treatment begins on the day of inclusion. For women needing HBV treatment for themselves, TDF is funded by the study until 24 weeks postpartum, followed by referral to the hepatology unit of 1 national hospital involved in the study to continue care and treatment. Women with a negative HBeAg RDT women are not treated with TDF but remain in the study for follow-up. In all cases, vaccination of the newborn is performed according to the Cambodian national protocol—1 injection at birth and 6, 10, and 14 weeks of age, with the first dose of vaccine provided within the first 2 hours of life.
The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected by a priori approval by the Cambodian National Ethics Committee for Health Research (NECHR). Each participant signs an informed-consent form.
Algorithms
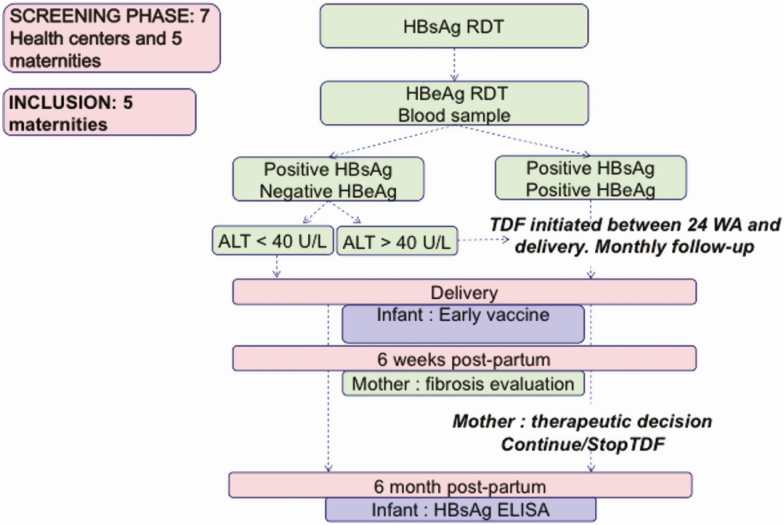
In the current ANRS 12345-TA PROHM study, a positive HBeAg RDT identifies pregnant women eligible for the TDF preventive strategy. Here, we proposed a new algorithm using 2 biological markers (HBeAg RDT and ALT) to identify women eligible for TDF (Figure 1). The threshold of 40 IU/L for ALT level was chosen based on the publication of the TREAT-B score in Africa [15].
Figure 1.
Description of the evaluated algorithm. Abbreviations: ALT, alanine aminotransferase; ELISA, enzyme-linked immunoassay; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; RDT, rapid diagnosis test; TDF, tenofovir disoproxil fumarate; WA, weeks of amenorrhea.
Biological Analysis
Two HBV RDTs were performed following a sequential testing algorithm. First, all pregnant women were tested with the SD BIOLINE HBsAg RDT (Standard Diagnostics [SD], Inc, Kyonggi-do, Korea). All samples found positive for HBsAg RDT were further assayed with the SD BIOLINE HBeAg RDT. HBV DNA quantification was performed in the Virology Unit of the Institut Pasteur du Cambodge using a quantitative polymerase chain reaction (PCR) assay targeting the S gene of HBV (PUMA HBV kit; Omunis, Clapiers, France). ALT level was measured on the ABX PENTRA C400 by using the IFCC method (UV without pyridoxal phosphate).
Statistical Analysis
Summary statistics are presented as percentages, medians, and interquartile ranges (IQRs). Wilcoxon test and Fisher tests were applied to test differences. Spearman correlation was used to assess the association between 2 quantitative variables.
The validity of HBeAg RDT to identify HBV DNA viral load higher than 2 thresholds (5.3 and 7.3 log10 IU/mL) was evaluated using sensitivity, specificity, and positive and negative predictive values obtained with asymptotic 95% confidence intervals (95% CIs). Interpretation of positive and negative predictive values is related to the prevalence of high HBV DNA among HBsAg-positive women calculated in this population. The threshold of 7.3 log10 IU/mL was chosen to identify highly viremic women, those most at risk of MTCT [6].
The validity of the new algorithm for selecting not only an HBeAg-positive RDT but also an HBeAg-negative RDT with ALT of 40 IU/L or greater as a predictor of high HBV DNA viral load level was evaluated with the same thresholds (5.3 and 7.3 log10 IU/mL).
Receiver operating characteristic (ROC) curves were obtained to assess the reference cutoff value of 40 IU/L for ALT. Cutoff values were characterized by the usual criteria based on the Youden index (ie, specificity + specificity − 1) and the minimal-distance-to-0,1 (ie √[1 − sensitivity]2 + [1 – specificity]2). A nonparametric approach was used to compare the areas under the ROC curve (AUC) of the current and the new algorithms with DeLong’s method [16] in the LOGISTIC procedure in SAS/STAT. All analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC).
RESULTS
Participants’ Characteristics at Inclusion
From 4 October 2017 to 19 December 2018, 519 HBsAg-positive women were enrolled in the ANRS 12345-TA PROHM study and 515 were retained in the analysis. Median age was 29 years (26–33 years), 92 (17.9%) were HBeAg positive, and 47 (9.1%) were HBeAg negative with an ALT of 40 IU/L or greater. HBV DNA viral load greater than 5.3 log10 IU/mL was found in 144 (28.0%), and 96 (18.6%) had an HBV DNA level greater than 7.3 log10 IU/mL. Characteristics of the participants are reported in Table 1.
Table 1.
Characteristics of Participants at Inclusion
| Total (N = 515) | HBV DNA >5.3 log10 IU/mL (n = 144; 28.0%) | HBV DNA ≤5.3 log10 IU/mL (n = 371; 72.0%) | P | |
|---|---|---|---|---|
| Age, median (IQR), years | 29 (26–33) | 27 (24–32) | 30 (26–34) | <.001 |
| Weeks of amenorrhea at inclusion, median (IQR) | 26 (19–34) | 25 (18–31.5) | 26 (20–34) | .049 |
| Known HBV status, n (%) | 237 (46.0) | 57 (39.6) | 180 (48.5) | .076 |
| Prior HBV treatment, n (%) | 22 (4.3) | 10 (6.9) | 12 (3.2) | .086 |
| Positive HBeAg, n (%) | 100 (18.5) | 97 (63) | 3 (0.8) | <.001 |
| ALT ≥40 IU/L at inclusion, n (%) | 62 (12.0) | 40 (27.8) | 22 (5.9) | <.001 |
| APRI score ≥1.5, n (%) | 11 (2.1) | 6 (4.2) | 5 (1.4) | .082 |
N = 515.
Abbreviations: ALT, alanine aminotransferase; APRI, AST to Platelet Ratio Index; AST, aspartate aminotransferase; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; IQR, interquartile range.
Overall, HBsAg-positive women with HBV DNA greater than 5.3 log10 IU/mL were significantly younger, had a higher frequency of HBeAg RDT positivity, and a higher frequency of ALT level of 40 IU/L or greater.
Correlation Between HBV DNA Levels and ALT Values in HBeAg- Positive and -Negative Women
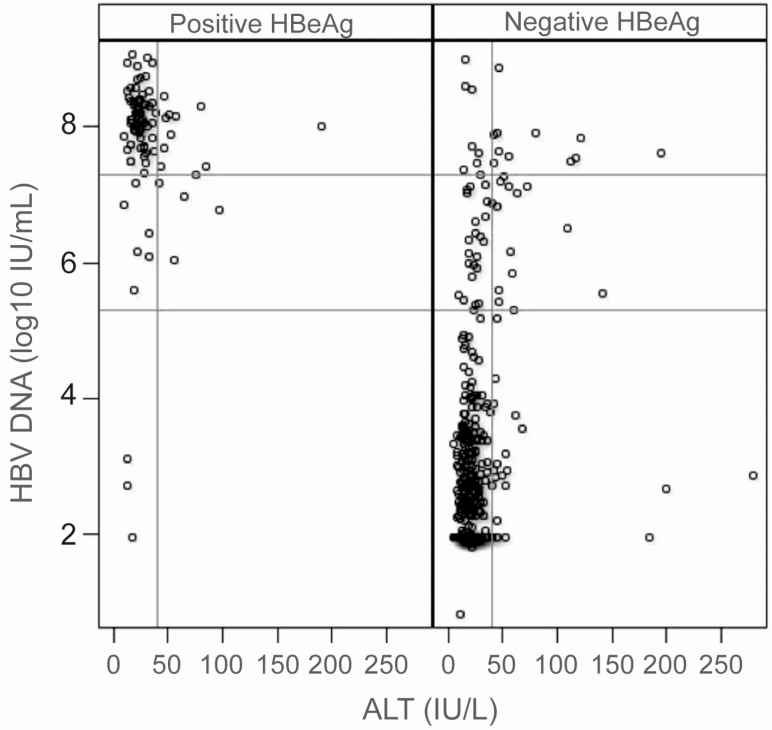
A moderate correlation was observed between HBV DNA levels and ALT in HBeAg-negative women (r = 0.28, P < .0001); however, this association was not observed in HBeAg-positive participants (r = −0.08074, P = .44) (Figure 2).
Figure 2.
HBV DNA serum levels and ALT values in HBeAg-positive and -negative women (N = 515). Reference lines at HBV DNA = 5.3 and 7.3 log10 IU/mL and ALT = 40 IU/L. Abbreviations: ALT, alanine aminotransferase; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus.
Performance of the HBeAg RDT and Novel Algorithm for Identifying HBV DNA Viral Load at Variable Thresholds (5.3 and 7.3 Log10 IU/mL)
The sensitivity and specificity of the HBeAg RDT for identifying viremic samples were 61.8% (95% CI, 53.9–69.7%) and 99.2% (95% CI, 98.3–100%) for HBV DNA levels greater than 5.3 log10 IU/mL and 81.3% (95% CI, 73.4–89.1%) and 96.7% (95% CI, 94.9–98.4%) for HBV DNA levels greater than 7.3 log10 IU/mL, respectively. On an individual level, with a 28% and 18% prevalence of HBV DNA level greater than 5.3 or 7.3 log10 IU/mL among HBsAg-positive women, positive predictive values were 96.7% (95% CI, 93.1–100%) and 84.8% (95% CI, 77.4–92.1%) and negative predictive values were 87% (95% CI, 83.8–90.2%) and 95.7% (95% CI, 93.8–97.7%), respectively. Results are presented in Table 2.
Table 2.
Performance of HBeAg Rapid Diagnosis Test Alone to Predict Hepatitis B Virus DNA >5.3 and 7.3 Log10 IU/mL
| HBV DNA Thresholds (log10 IU/mL) | HBeAg+ | HBeAg− | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|---|
| >5.3 | 89 | 55 | 61.8 (53.9–69.7) | 99.2 (98.3–100) | 96.7 (93.1–100) | 87.0 (83.8–90.2) |
| ≤5.3 | 3 | 368 | … | … | … | … |
| >7.3 | 78 | 18 | 81.3 (73.4–89.1) | 96.7 (94.9–98.4) | 84.8 (77.4–92.1) | 95.7 (93.8–97.7) |
| ≤7.3 | 14 | 405 | … | … | … | … |
Abbreviations: CI, confidence interval; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; NPV, negative predictive values; PPV, positive predictive value; +, positive; –, negative.
The sensitivity and specificity of the novel algorithm were 79.2% (95% CI, 72.5–85.8%) and 93.3% (95% CI, 90.7–95.8%) for HBV DNA levels greater than 5.3 log10 IU/mL and 92.7% (95% CI, 87.5–97.9%) and 88.1% (95% CI, 85.0–91.2%) for HBV DNA levels greater than 7.3 log10 IU/mL, respectively. On an individual level, with a 28% and 18% prevalence of HBV DNA levels greater than 5.3 and 7.3 log10 IU/mL among HBsAg-positive women, positive predictive values of the novel algorithm were 82% (95% CI, 75.6–88.4%) and 64% (95% CI, 56.1–72.0%) and negative predictive values were 92% (95% CI, 89.3–94.8%) and 98.1% (95% CI, 96.8–99.5%), respectively. Results are presented in Table 3.
Table 3.
Performance of Novel Algorithm to predict Hepatitis B Virus DNA >5.3 and 7.3 Log10 IU/mL
| HBV DNA Thresholds (log10 IU/mL) | Eligible for TDF With New Algorithm | Noneligible for TDF With New Algorithm | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|---|---|
| >5.3 | 114 | 30 | 79.2 (72.5–85.8) | 93.3 (90.7–95.8) | 82.0 (75.6–88.4) | 92.0 (89.3–94.8) |
| ≤5.3 | 25 | 346 | … | … | … | … |
| >7.3 | 89 | 7 | 92.7 (87.5–97.9) | 88.1 (85.0–91.2) | 64.0 (56.1–72.0) | 98.1 (96.8–99.5) |
| ≤7.3 | 50 | 369 | … | … | … | … |
Abbreviations: CI, confidence interval; HBV, hepatitis B virus; NPV, negative predictive values; PPV, positive predictive value; TDF, tenofovir disoproxil fumarate.
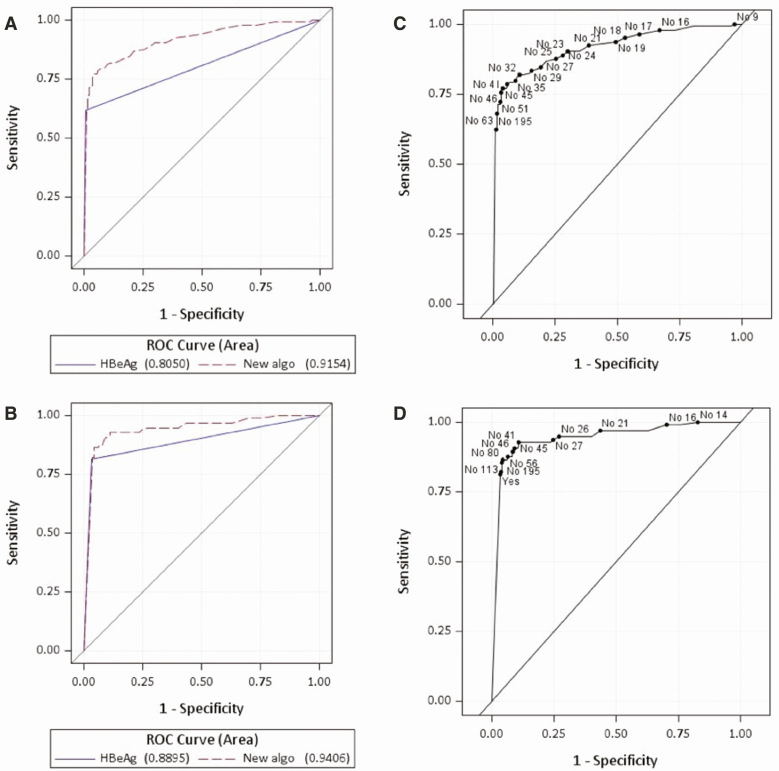
The ROC curves showed high AUCs to assess viremic samples (Figure 3). The AUCs for the novel algorithm (0.92 and 0.94 for HBV DNA viral load >5.3 and 7.3, respectively) were significantly greater (P < .001) than the AUC for the current algorithm (0.81 and 0.89 for HBV DNA viral load >5.3 and 7.3, respectively).
Figure 3.
ROC curves for (A) the current algorithm (HBeAg only) and the novel algorithm (HBeAg and ALT) for HBV DNA >5.3 log10 IU/mL, (B) for HBV DNA >7.3 log10 IU/mL, (C) for the novel algorithm (HBeAg and ALT) for HBV DNA >5.3 log10 IU/mL, and (D) for HBV DNA >7.3 log10. Points are labeled with HBeAg positivity and ALT. Abbreviations: algo, algorithm; ALT, alanine aminotransferase; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; ROC, receiver operating characteristic.
Similar sensitivity and specificity were observed for the reference level of ALT (40 IU/mL) and for the optimal cutoffs, as well as for the Youden index and the minimum “distance to 0,1” (Table 4).
Table 4.
ROC Curve Analysis—Reference and Optimal Cutoffs
| Criterion | ALT Cutoff | Sensitivity | Specificity | Youden Value | Distance to 0,1 Value |
|---|---|---|---|---|---|
| HBV DNA >5.3 | |||||
| Reference | 40 | 0.792 | 0.933 | 0.725 | 0.219 |
| Distance to 0,1 | 32 | 0.819 | 0.889 | 0.708 | 0.212 |
| Youden | 45 | 0.771 | 0.960 | 0.730 | 0.232 |
| HBV DNA >7.3 | |||||
| Reference | 40 | 0.927 | 0.881 | 0.808 | 0.140 |
| Distance to 0,1 | 41 | 0.927 | 0.89 | 0.817 | 0.132 |
| Youden | 80 | 0.865 | 0.955 | 0.81924 | 0.142 |
N = 515.
Abbreviations: ALT, alanine aminotransferase; HBV, hepatitis B virus; ROC, receiver operating characteristic.
Characteristics of Participants at Inclusion According to the Outcomes of the Novel Algorithm
Of the 515 pregnant women evaluated, 30 had an HBV DNA level greater than 5.3 log10 IU/mL but were not identified by the novel algorithm (false negative) and 25 were identified by the novel algorithm but did not have an HBV DNA level greater than 5.3 log10 IU/mL (false positive). Characteristics of participants at inclusion according to the outcomes of the novel algorithm are reported in Table 5. For the 30 false-negative women, median HBV DNA was 6.5 log10 IU/mL compared with 7.95 log10 IU/mL in the true-positive women and 23% had HBV DNA greater than 7.3 log10 IU/mL compared with 78% in true-positive women.
Table 5.
Characteristics of Participants at Inclusion According to the Outcomes of the Novel Algorithm
| False Negative (n = 30) | False Positive (n = 25) | True Negative (n = 346) | True Positive (n = 114) | |
|---|---|---|---|---|
| Age, median (IQR), years | 32 (26–34) | 32 (30–34) | 29.5 (26–33) | 27 (23–30) |
| Weeks of amenorrhea at inclusion, median (IQR) | 25.5 (19–30) | 28 (18–35) | 26 (20–34) | 25 (18 – 32) |
| Known HBV status, n (%) | 10 (33.3) | 12 (48) | 168 (48.5) | 47 (41.2) |
| Prior HBV treatment, n (%) | 0 (0) | 1 (4) | 11 (3.2) | 10 (8.8) |
| Positive HBeAg, n (%) | 0 (0) | 3 (12) | 0 (0) | 89 (78.1) |
| ALT at inclusion, U/L, median (IQR) | 23 (18–27) | 44 (42–52) | 17 (14–23) | 28 (21–46) |
| ALT ≥40 IU/L at inclusion, n (%) | 0 (0) | 22 (88) | 0 (0) | 40 (35) |
| HBV DNA at inclusion, median (IQR) | 6.5 (6–7.3) | 2.8 (1.9–3.1) | 2.3 (1.95–3) | 7.95 (7.5–8.3) |
| HBV DNA >7.3 log10 IU/mL, n (%) | 7 (23.3) | 0 (0) | 0 (0) | 89 (78.1) |
Abbreviations: ALT, alanine aminotransferase; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; IQR, interquartile range.
DISCUSSION
A simple algorithm selecting HBeAg RDT–positive pregnant women and HBeAg RDT–negative pregnant women with ALT of 40 IU/L or greater as a predictor of high HBV DNA viral load level has a 79% sensitivity and 93% specificity to predict an HBV DNA level greater than 5.3 log10 IU/mL, which is the threshold used to recommend antiviral preventive strategies. Algorithm sensitivity improved to 92.7% for women with an HBV DNA level greater than 7.3 log10 IU/mL. These women are considered as highly viremic and most at risk for perinatal transmission [6].
When compared with using the HBeAg RDT alone, sensitivity improves by 17% at 5.3 log10 IU/mL and by 11% at 7.3 log10 IU/mL thresholds, respectively. These results are specifically relevant to public health response. Indeed, increasing the ability to detect pregnant women at high risk of HBV MTCT is critical to implement preventive strategies like HBIg and/or antiviral preventive treatment. In 2016, a randomized clinical trial conducted in China in 200 mothers with HBV DNA viral load greater than 5.3 log10 IU/mL demonstrated that short-course TDF administration in addition to immunoprophylaxis of newborns at birth led to a significant decrease in the HBV MTCT compared with placebo [16]. Conversely, a double-blind clinical trial conducted in Thailand reported that maternal use of TDF did not significantly lower the rate of transmission in conjunction with HBIg and HBV vaccine administration very early after birth [17]. However, the 2% rate of perinatal transmission with placebo was much lower than the initially expected 12%, leaving the study underpowered to detect a statistically significant difference [18]. A recent meta-analysis based on 9 studies highlights the efficacy and safety of TDF to prevent HBV MTCT [19]. Furthermore, HBIg remains unavailable and/or unaffordable in many LMICs, including Cambodia, and TDF represents an interesting alternative option as this drug is available through the human immunodeficiency virus (HIV) national programs.
HBV DNA quantification remains the reference method to determine eligibility for alternative preventive strategies. Recently, a point-of-care (POC) GeneXpert (Cepheid) HBV DNA viral load was marketed and 1 article reported good performance compared with the Aptima Quant HBV assay [20]. Further in-field evaluation of this POC assay as a predictive tool for MTCT risk is necessary, and logistical issues remain in terms of costs, availability of tests, and testing platforms. Furthermore, the optimal HBV DNA threshold predicting the risk of MTCT and for deciding preventive therapy needs to be confirmed. A large prospective cohort in China of 1177 mother–infant dyads reported a 7 log10 IU/mL maternal HBV DNA threshold associated with perinatal transmission of HBV [21].
The WHO global health sector strategy on viral hepatitis 2016–2021 emphasizes the necessity to develop and update global guidance on a comprehensive package of interventions to eliminate HBV MTCT, including the possible role of perinatal usage of antiviral drugs [3]. While HBV DNA quantification remains the gold standard to identify pregnant women eligible for antiviral drugs, it is crucial to identify alternative strategies in many countries facing challenges in term of access to HBV DNA quantification, including Cambodia, where this study was conducted. In many of those countries, ANC and deliveries are conducted by midwives at the decentralized level (primary health centers, district hospitals) with no access to HBV DNA viral load measurement. On the contrary, liver function tests and HBeAg RDTs could be performed in all district hospitals and these 2 tests could easily be scaled-up in all decentralized areas, resulting in improved availability of testing and subsequent access to TDF prophylaxis. A simple algorithm selecting HBeAg-positive women and HBeAg-negative women with ALT levels of 40 IU/L and higher as eligible for antiviral preventive treatment by TDF in conjunction with immediate HBV vaccination of newborns at birth could be an effective strategy to prevent HBV MTCT, as evaluated in the ANRS 12345-TA PROHM clinical trial.
Utilizing our novel algorithm, 30 women were not identified as being at risk of transmission at an HBV DNA level above 5.3 log10 IU/mL. The majority of these women had an HBV DNA level between 5.3 and 7.3 log10 IU/mL. In true-positive women, HBV DNA was mainly higher than 7.3 log10 IU/mL. Therefore, the risk of perinatal transmission could be lower for these false-negative women but, as stated above, exact thresholds need to be confirmed.
Compared with using the HBeAg RDT alone, this novel algorithm reduces the specificity to predict an HBV DNA level greater than 5.3 log10 IU/mL from 99% to 93%. This reduction would lead to prescribing TDF to 25 pregnant women of 371 with HBV DNA levels of 5.3 log10 IU/mL or less, so with limited risk of HBV MTCT. The safety of prenatal exposure to TDF has been well documented in association with HIV infection [22, 23], and data on HBV infection point in a similar direction [19, 24]. Therefore, the strategy to decrease specificity but increase sensitivity could be an acceptable trade-off for a public health approach.
Recently, the performance of a simple score (Treat-B score) based on HBeAg and ALT for selecting patients for HBV treatment in Africa was reported [15]. The score of 2 and above (HBeAg-positive and ALT ≥20 IU/L or HBeAg-negative and ALT ≥40 IU/L) had a sensitivity and specificity for treatment eligibility of 85% and 77%, respectively. This algorithm identified patients eligible for treatment in the general population in Africa, thus combining fibrosis, HBV-DNA, and ALT as a gold standard. In our study, adding an ALT measurement for HBeAg-negative women was included because of the lowered sensitivity of the current assay potentially related to the presence of BPC/PC mutations [14]. Indeed, according to the new nomenclature, HBeAg-negative chronic hepatitis B (phase 4) is characterized by the lack of serum HBeAg, persistent or fluctuating moderate to high levels of serum HBV DNA, and fluctuating or persistently elevated ALT values [25]. Most of these subjects harbor HBV variants in the PC and/or the BCP regions that impair or abolish HBeAg expression. Patients carrying these mutations, and more specifically, the precore G1896A mutation, could not express HBeAg and still suffer from severe liver disease with high HBV DNA levels and persistently elevated ALT values [26]. Therefore, adding an ALT measurement for HBeAg-negative pregnant women could represent an option to identify pregnant women with HBeAg-negative chronic hepatitis B. This information is particularly relevant as these women should probably continue treatment after delivery regardless of the degree of fibrosis [25].
This study does have some limitations. First, we are not able to rule out all the possible reasons for elevated ALT. Coinfections with HIV and hepatitis C virus were excluded as noneligible criteria in the TA PROHM study. However, hepatitis A and hepatitis E virus infections were not screened systematically, and the possibility of traditional or conventional medicine consumption could not be excluded. Second, analyses of BCP/PC mutations were not conducted on samples from the 55 women with negative HBeAg RDT and HBV DNA viral load levels greater than 5.3 log10 IU/mL. Finally, no direct comparison was performed with a strategy using the HBV DNA GeneXpert POC, which could represent an attractive alternative to scale-up access to HBV DNA quantification.
In conclusion, a comprehensive, affordable package of interventions to eliminate HBV MTCT needs to be scaled-up at the decentralized level in order to reach the WHO 2030 target of less than 0.1% prevalence of HBsAg among children. In the absence of increased access to HBV DNA testing, implementation of this novel algorithm could be a timely and highly feasible strategy to improve detection of pregnant women eligible for alternative preventive strategies, specifically in remote areas where many ANC visits take place. The effectiveness and cost-effectiveness of this free-HBIg strategy to reduce HBV MTCT need to be confirmed through the currently ongoing ANRS 12345-TA PROHM study.
Notes
Acknowledgments. The authors thank all of the pregnant women and their families for their participation in the study; all the medical, laboratory, and methodology staff for their contribution to the study; all the members of the Scientific Committee and Data and Safety Monitoring Board for their advice; Professor Laurence Meyer for her support and advice; Claire Rekacewicz, Isabelle Fournier, Nicolas Rouveau, Maria-Camila Calvo Cortes, and Laura Fernandez from the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) for their support; and Erik Karlsson for grammatical and textual corrections.
Financial support. This work was supported by the ANRS.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Schweitzer A, Horn J, Mikolajczyk RT, Krause G, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015; 386:1546–55. [DOI] [PubMed] [Google Scholar]
- 2. Ott JJ, Horn J, Krause G, Mikolajczyk RT. Time trends of chronic HBV infection over prior decades—a global analysis. J Hepatol 2017; 66:48–54. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Global health sector strategy on viral hepatitis 2016–2021. Available at: http://www.who.int/hepatitis/strategy2016-2021/ghss-hep/en/. Accessed 9 October 2019. [Google Scholar]
- 4. Shao ZJ, Zhang L, Xu JQ, et al. Mother-to-infant transmission of hepatitis B virus: a Chinese experience. J Med Virol 2011; 83:791–5. [DOI] [PubMed] [Google Scholar]
- 5. Zou H, Chen Y, Duan Z, Zhang H, Pan C. Virologic factors associated with failure to passive-active immunoprophylaxis in infants born to HBsAg-positive mothers. J Viral Hepat 2012; 19:e18–25. [DOI] [PubMed] [Google Scholar]
- 6. Wen WH, Chang MH, Zhao LL, et al. Mother-to-infant transmission of hepatitis B virus infection: significance of maternal viral load and strategies for intervention. J Hepatol 2013; 59:24–30. [DOI] [PubMed] [Google Scholar]
- 7. Sellier P, Maylin S, Amarsy R, et al. Untreated highly viraemic pregnant women from Asia or sub-Saharan Africa often transmit hepatitis B virus despite serovaccination to newborns. Liver Int 2015; 35:409–16. [DOI] [PubMed] [Google Scholar]
- 8. Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH; American Association for the Study of Liver Diseases AASLD guidelines for treatment of chronic hepatitis B. Hepatology 2016; 63:261–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int 2016; 10:1–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. European Association for the Study of the Liver. EASL clinical practice guidelines: management of chronic hepatitis B virus infection. J Hepatol 2012; 57:167–85. [DOI] [PubMed] [Google Scholar]
- 11. Xu DZ, Yan YP, Choi BC, et al. Risk factors and mechanism of transplacental transmission of hepatitis B virus: a case-control study. J Med Virol 2002; 67:20–6. [DOI] [PubMed] [Google Scholar]
- 12. Yamada H, Fujimoto M, Svay S, et al. Seroprevalence, genotypic distribution and potential risk factors of hepatitis B and C virus infections among adults in Siem Reap, Cambodia. Hepatol Res 2015; 45:480–7. [DOI] [PubMed] [Google Scholar]
- 13. Sa-Nguanmoo P, Tangkijvanich P, Thawornsuk N, et al. Molecular epidemiological study of hepatitis B virus among migrant workers from Cambodia, Laos, and Myanmar to Thailand. J Med Virol 2010; 82:1341–9. [DOI] [PubMed] [Google Scholar]
- 14. Ségéral O, S N’Diaye D, Prak S, et al. Usefulness of a serial algorithm of HBsAg and HBeAg rapid diagnosis tests to detect pregnant women at risk of HBV mother-to-child transmission in Cambodia, the ANRS 12328 pilot study. J Clin Virol 2018; 109:29–34. [DOI] [PubMed] [Google Scholar]
- 15. Shimakawa Y, Njie R, Ndow G, et al. Development of a simple score based on HBeAg and ALT for selecting patients for HBV treatment in Africa. J Hepatol 2018; 69:776–84. [DOI] [PubMed] [Google Scholar]
- 16. Pan CQ, Duan Z, Dai E, et al. ; China Study Group for the Mother-to-Child Transmission of Hepatitis B Tenofovir to prevent hepatitis B transmission in mothers with high viral load. N Engl J Med 2016; 374:2324–34. [DOI] [PubMed] [Google Scholar]
- 17. Jourdain G, Ngo-Giang-Huong N, Harrison L, et al. Tenofovir versus placebo to prevent perinatal transmission of hepatitis B. N Engl J Med 2018; 378:911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Terrault N, Brown R, Lok A, et al. Preventing perinatal transmission of hepatitis B necessitates consideration of antiviral treatment in pregnancy. Hepatology 2018; 68:1658–60. [Google Scholar]
- 19. Li W, Jia L, Zhao X, Wu X, Tang H. Efficacy and safety of tenofovir in preventing mother-to-infant transmission of hepatitis B virus: a meta-analysis based on 6 studies from China and 3 studies from other countries. BMC Gastroenterol 2018; 18:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abravanel F, Lhomme S, Trémeaux P, et al. Performance of the Xpert HBV viral load assay versus the Aptima Quant assay for quantifying hepatitis B virus DNA. Diagn Microbiol Infect Dis 2020; 96:114946. [DOI] [PubMed] [Google Scholar]
- 21. Lu Y, Zhu FC, Liu JX, et al. The maternal viral threshold for antiviral prophylaxis of perinatal hepatitis B virus transmission in settings with limited resources: a large prospective cohort study in China. Vaccine 2017; 35:6627–33. [DOI] [PubMed] [Google Scholar]
- 22. Sibiude J, Mandelbrot L, Blanche S, et al. Association between prenatal exposure to antiretroviral therapy and birth defects: an analysis of the French perinatal cohort study (ANRS CO1/CO11). PLoS Med 2014; 11:e1001635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brogly SB, Abzug MJ, Watts DH, et al. Birth defects among children born to human immunodeficiency virus-infected women: pediatric AIDS clinical trials protocols 219 and 219C. Pediatr Infect Dis J 2010; 29:721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greenup AJ, Tan PK, Nguyen V, et al. Efficacy and safety of tenofovir disoproxil fumarate in pregnancy to prevent perinatal transmission of hepatitis B virus. J Hepatol 2014; 61:502–7. [DOI] [PubMed] [Google Scholar]
- 25. European Association for the Study of the Liver. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 2017; 67:370–98. [DOI] [PubMed] [Google Scholar]
- 26. Lapalus M, Laouenan C, Cardoso AC, et al. Precore/Core promoter variants to predict significant fibrosis in both HBeAg positive and negative chronic hepatitis B. Liver Int 2015; 35:2082–9. [DOI] [PubMed] [Google Scholar]