Abstract
We report 2 independent patients from whom carbapenem and ceftazidime-avibactam–resistant Enterobacter cloacae complex strains were identified. The ceftazidime-avibactam resistance was attributed to a 2–amino acid deletion in the R2 loop of AmpC β-lactamase, which concurrently caused resistance to cefepime and reduced susceptibility to cefiderocol, a novel siderophore cephalosporin.
Keywords: R2 loop, deletion, β-lactamase, cefepime
(See the Editorial Commentary by Gallagher on pages 2717–8.)
Avibactam is a potent inhibitor of class A, class C, and some class D β-lactamases [1]. Since its clinical introduction in combination with ceftazidime, β-lactamase–mediated resistance to the combination of ceftazidime and avibactam has been reported against clinical isolates producing class A β-lactamases, including Klebsiella pneumoniae carbapenemases (KPCs) and CTX-M-14 group extended-spectrum β-lactamases (ESBLs) [2–4]. For class C enzymes, ceftazidime-avibactam resistance has been demonstrated in vitro [5], but not against clinical isolates. Here, we report the clinical evolution of ceftazidime-avibactam resistance in Enterobacter hormaechei, the major genomospecies of Enterobacter cloacae complex, likely due to a 2–amino acid deletion in AmpC that also conferred resistance to expanded-spectrum cephalosporins and reduced susceptibility to cefiderocol.
A 50-year-old morbidly obese woman presented to our hospital in septic shock 2 months after an open cholecystectomy. She was empirically treated with piperacillin-tazobactam and vancomycin. A computed tomographic scan showed multiple intra-abdominal fluid collections and concern for necrotizing pancreatitis, for which an exploratory laparotomy was performed with extensive debridement and drainage of pancreatic and peripancreatic necrosis. Intraoperative cultures of the necrotic tissue grew 2 E. hormaechei strains (Ent634 and Ent635; Supplementary Table), Serratia marcescens, and Enterococcus species, resulting in a change of her antibiotic regimen to meropenem and daptomycin. The patient remained critically ill, requiring mechanical ventilation and vasopressor support. On hospital day 23, she developed ventilator-associated pneumonia due to carbapenem-resistant Pseudomonas aeruginosa, and her antibiotic regimen was changed to cefepime (2 g intravenously every 8 hours over a 3-hour infusion). Despite active treatment, the patient required increasing ventilator support and frequent suctioning for mucus plugging. Cultures from a bronchoscopy performed on hospital day 34 (day 11 of cefepime treatment) grew P. aeruginosa and E. hormaechei (Ent630); both isolates were resistant to carbapenems and cefepime. Her antibiotic regimen was changed to ceftazidime-avibactam and ciprofloxacin on hospital day 37; however, a decision was made to transition care to comfort measures in the setting of multisystem organ failure, and she died the following day. On the day of her death, susceptibility testing of Ent630 demonstrated resistance to ceftazidime-avibactam that prompted an investigation into potential mechanisms of resistance.
The initial E. hormaechei strains Ent634 and Ent635 belonged to sequence type (ST) 133 and ST145, respectively. By broth microdilution, both isolates were susceptible to cefepime (minimum inhibitory concentrations [MICs], 2 and ≤ 0.25 µg/mL), ceftazidime-avibactam (MICs, 0.5 and 0.25 µg/mL) and meropenem (MICs, 0.12 and ≤0.06 µg/mL; Supplementary Table). Following consecutive exposures to meropenem (23 days) and cefepime (11 days), the subsequent E. hormaechei strain Ent630 was resistant to cefepime (MIC, >256 µg/mL), meropenem (MIC, 8 µg/mL), and ceftazidime-avibactam (MIC, 64 µg/mL). Like Ent634, Ent630 belonged to ST133. Whole genome sequencing (WGS), performed on Illumina NextSeq 500 (coverage, 59× to 105×; total length of contigs, 4 619 975 to 4 950 959), showed 2 core genome single-nucleotide polymorphisms (SNPs) present in Ent630 that were not present in Ent634. The core SNPs included nonsynonymous mutations in wzc (A404V), which encodes a tyrosine-protein kinase, and in rpoE (T160A), which encodes an RNA polymerase sigma factor. Neither strain harbored genes encoding ESBL or carbapenemase enzymes; however, Ent630 harbored IS903-like insertions in both ompC and ompF that were not present in Ent634, which likely contributed to carbapenem resistance [6]. Most importantly, the deduced Ent630 AmpC sequence contained a 2–amino acid deletion of alanine and leucine at positions 292 and 293, respectively (A292_L293del), not present in Ent634, its direct ancestor. This led us to hypothesize that the A292_L293del in AmpC was the cause of cefepime and ceftazidime-avibactam resistance. Draft genomes of Ent630, Ent634, Ent635, and Surv196 are deposited in the National Center for Biotechnology Information database under BioSample numbers SAMN13042297, SAMN13042298, SAMN13042299, and SAMN13245837, respectively.
To test the impact of the deletion, we obtained a 1.9-kb genomic fragment containing Ent630 ampC by partial digestion using restriction enzyme Sau3AI, ligation into pBK-CMV digested with BamHI and cloned into Escherichia coli TOP10 [7]. The resulting E. coli transformant demonstrated cefepime and ceftazidime-avibactam MICs of 64 µg/mL and 16 µg/mL, respectively, as compared to E. coli transformant carrying pBK-CMV alone, which demonstrated corresponding MICs of ≤ 0.25 µg/mL and 0.25 µg/mL, respectively. The results strongly suggest that the variant AmpC containing an A292_L293del is responsible for co-resistance to cefepime and ceftazidime-avibactam, a deletion that likely evolved under selective pressure from cefepime treatment. In support of this hypothesis, we encountered a second patient from whom E. hormaechei strain Surv196, distinct from Ent630 with > 38 000 pairwise SNPs between their genomes, was isolated by perirectal surveillance during empiric cefepime treatment of cholangitis (isolated on day 13 of therapy). Strain Surv196 also demonstrated resistance to cefepime (MIC, 256 µg/mL) and ceftazidime-avibactam (MIC, 64 µg/mL). By WGS analysis, the isolate harbored an identical A292_L293del in AmpC, representing convergent evolution.
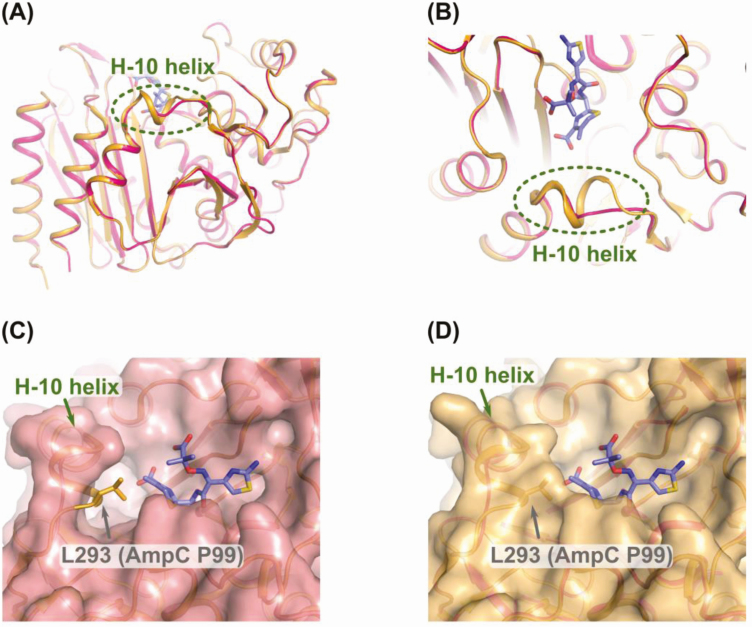
Next, we tested the susceptibility of E. coli TOP10 harboring the A292_L293del AmpC variant against cefiderocol, a novel siderophore cephalosporin with broad-spectrum activity against gram-negative bacteria. Compared to E. coli TOP10 with pBK-CMV alone, the transformant demonstrated a > 32-fold increase in the cefiderocol MIC (from 0.06 to 2 µg/mL). Similarly, clinical strains Ent630 and Surv196 showed reduced susceptibility to cefiderocol (MICs, ≥16 µg/mL and 4 µg/mL, respectively). Taken together, the data suggest that the A292_L293del in AmpC exhibits a broad impact on the cephalosporin class rather than any specific agent. Indeed, residues 292 and 293 in AmpC are located in the R2 loop of the enzyme that spans positions 289–307 and contains the H-10 helix. The R2 loop is in close proximity with the R2 side chain of cephalosporins [8]. Various amino acid substitutions, insertions, and deletions in AmpC have been reported to cause more efficient hydrolysis of ceftazidime and extend the substrate profile to include cefepime [9]. Given this association, we generated a homology model of AmpC of Ent630 using the MODELLER program with a published structure of AmpC in E. cloacae P99 (Protein Data Bank [PDB] accession number 5XHR) as a template [10, 11]. The ceftazidime molecule bound to AmpC of Ent630 was modeled by superposition of the homology model of AmpC of Ent630 on the structure of ceftazidime-bound AmpC of E. coli K12 (PDB accession number 1IEL) [9]. The model showed a significant structural change in and around the R2 loop, while the positions of key residues involved in the recognition and catalysis of ceftazidime appeared to be conserved overall. As seen in Figure 1, A292_L293del present in AmpC of Ent630 likely causes partial disappearance of the H-10 helix in the R2 loop, thereby expanding the substrate binding site to better accommodate cephalosporins with a bulkier R2 side chain, including ceftazidime, cefepime, and cefiderocol.
Figure 1.
A, Modeled structure of AmpC of Enterobacter hormaechei Ent630. AmpC of Enterobacter cloacae P99 is shown in orange, upon which the structure of AmpC of Ent630 is imposed in magenta. Bound ceftazidime is shown in purple. B–D, Enlarged view of the area containing A292_L293del. Molecular surfaces are shown in magenta (AmpC of Ent630, C) and orange (AmpC of P99, D), and L293 in AmpC of P99 is shown in orange sticks.
The A292_L293del AmpC variant did not impact MICs of imipenem or meropenem (≤ 0.25 µg/mL and ≤ 0.06 µg/mL, respectively; Supplementary Table) when cloned into E. coli TOP10, supporting our hypothesis that carbapenem resistance in Ent630 was mediated by porin loss due to the insertion of an IS903-like element in both ompC and ompF. Carbapenem resistance mediated by nonenzymatic mechanisms poses an important threat to the utility of newly developed carbapenem–β-lactamase inhibitor combination agents that were designed to inhibit serine carbapenemases. Indeed, Ent630 was nonsusceptible to both meropenem-vaborbactam (MIC, 8 µg/mL) and imipenem-relebactam (MIC, 8 µg/mL), suggesting that treatment with any of the available β-lactam antibiotics would have been ineffective for this patient. This case serves as an important warning for the emerging threat of carbapenem-resistant Enterobacter species in the United States [12], for which underlying mechanisms of resistance are diverse [13]. Our data attest to the continued need to employ cephalosporins and carbapenems judiciously to avoid selection of resistant pathogens, including E. cloacae complex.
To our knowledge, this is the first case to demonstrate that resistance to ceftazidime-avibactam may be conferred by exposure to cefepime, a preferred treatment option for AmpC-producing Enterobacteriaceae [14]. Selection of resistance to cefepime following treatment of infections caused by Enterobacter species remains rare [15]. An AmpC variant harboring a single amino acid substitution from valine to glycine inside the H-10 helix (V291G) was previously identified following cefepime treatment in a patient with pneumonia due to Klebsiella (formerly Enterobacter) aerogenes [16]; however, the impact of this substitution on ceftazidime-avibactam susceptibility is unknown. In contrast, reduced ceftazidime-avibactam susceptibility was conferred by an E. cloacae isolate harboring a 6–amino acid deletion in the H-10 helix at residues 289–294 (SKVALA), likely resulting in a widened R2 binding pocket to better accommodate later-generation cephalosporins [8]. This strain was identified from a neutropenic pediatric patient on long-term cefepime treatment. In vitro selection of ceftazidime-avibactam resistance against E. cloacae identified arginine at position 168 and glycine at position 176 as hot spots for mutations in AmpC, but less commonly within the H-10 helix [5]. Taken together with the new data reported here, resistance to ceftazidime-avibactam due to deletions within the H-10 helix appears to derive primarily from improved recognition and inactivation of ceftazidime, suggesting that avibactam in combination with an alternative β-lactam may still be effective. To underscore this point, we tested the combination of aztreonam-avibactam and found that, indeed, the addition of avibactam restored the activity of aztreonam against Ent630 and Surv196 (MIC, 2 µg/mL for each; Supplementary Table). Against E. coli TOP10 harboring the A292_L293del AmpC variant, aztreonam MICs were reduced by 32-fold with the addition of avibactam.
In conclusion, mutational resistance to ceftazidime-avibactam is a concern following treatment of KPC-producing Enterobacteriaceae [4] and, as evidenced by this report, may also be a concern for Enterobacteriaceae producing class C β-lactamases. Selection of co-resistance to cefepime, ceftazidime-avibactam, and cefiderocol following cefepime treatment highlight the potential impact of mutational resistance in AmpC, and more specifically deletions within the H-10 helix region. Further surveillance studies are needed to understand the prevalence and clinical impact of such extended-spectrum AmpC enzymes. As clinical experience with ceftazidime-avibactam grows, it will be equally important to compare and validate potential mechanisms of resistance following treatment of noncarbapenemase-producing carbapenem-resistant Enterobacterales, for which ceftazidime-avibactam may offer an advantage over carbapenem–β-lactamase inhibitor combinations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Marissa Griffith for her help in genome assembly and single-nucleotide polymorphism analysis, and Shionogi for the provision of frozen plates for minimum inhibitory concentration (MIC) testing of cefiderocol. Shionogi provided the MIC testing plates but otherwise was not involved in the design, conduct, or reporting of this work.
Financial support. A. I. is supported by the Physician-Scientist Institutional Award from the Burroughs Wellcome Fund awarded to the University of Pittsburgh. R. K. S. was supported by the National Institutes of Health (NIH) (grant numbers K08AI114883 and R03AI144636). Y. D. was supported by the NIH (grant numbers R01AI104895 and R21AI135522).
Potential conflicts of interest. R. K. S. has received grant support from Accelerate Diagnostics, Achaogen, Allergan, Merck, Melinta, Roche, Shionogi, Tetraphase, VenatoRx, and the NIH; has served on advisory boards for Accelerate Diagnostics, Achaogen, Allergan, Entasis, Merck, Nabriva, Shionogi, and VenatoRx; and has received speaking honoraria from Allergan, Menarini, Pfizer, and T2Biosystems. Y. D. has received grant support from Pfizer, Merck, Shionogi, Astellas, Kanto Chemical, the NIH, the Japan Society for the Promotion of Science, and Japan Agency for Medical Research and Development; has served on advisory boards for Roche, Pfizer, Tetraphase, Recida, Fedora, VenatoRx, and Entasis; and has received speaking honoraria from Pfizer, Merck, and Shionogi. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Falcone M, Paterson D. Spotlight on ceftazidime/avibactam: a new option for MDR gram-negative infections. J Antimicrob Chemother 2016; 71:2713–22. [DOI] [PubMed] [Google Scholar]
- 2. Both A, Büttner H, Huang J, et al. . Emergence of ceftazidime/avibactam non-susceptibility in an MDR Klebsiella pneumoniae isolate. J Antimicrob Chemother 2017; 72:2483–8. [DOI] [PubMed] [Google Scholar]
- 3. Shields RK, Chen L, Cheng S, et al. . Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 2017; 61:e02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother 2018; 62:e02497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Livermore DM, Mushtaq S, Doumith M, Jamrozy D, Nichols WW, Woodford N. Selection of mutants with resistance or diminished susceptibility to ceftazidime/avibactam from ESBL- and AmpC-producing Enterobacteriaceae. J Antimicrob Chemother 2018; 73:3336–45. [DOI] [PubMed] [Google Scholar]
- 6. Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 2009; 63:659–67. [DOI] [PubMed] [Google Scholar]
- 7. Doi Y, Poirel L, Paterson DL, Nordmann P. Characterization of a naturally occurring class D beta-lactamase from Achromobacter xylosoxidans. Antimicrob Agents Chemother 2008; 52:1952–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lahiri SD, Giacobbe RA, Johnstone MR, Alm RA. Activity of avibactam against Enterobacter cloacae producing an extended-spectrum class C β-lactamase enzyme. J Antimicrob Chemother 2014; 69:2942–6. [DOI] [PubMed] [Google Scholar]
- 9. Powers RA. Structural and functional aspects of extended-spectrum AmpC cephalosporinases. Curr Drug Targets 2016; 17:1051–60. [DOI] [PubMed] [Google Scholar]
- 10. Pan X, He Y, Chen T, Chan KF, Zhao Y. Modified penicillin molecule with carbapenem-like stereochemistry specifically inhibits class C β-lactamases. Antimicrob Agents Chemother 2017; 61:e01288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 2016; 54:Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilson BM, El Chakhtoura NG, Patel S, et al. . Carbapenem-resistant Enterobacter cloacae in patients from the US Veterans Health Administration, 2006–2015. Emerg Infect Dis 2017; 23:878–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol 2019; 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamma PD, Doi Y, Bonomo RA, Johnson JK, Simner PJ; Antibacterial Resistance Leadership Group A primer on AmpC β-lactamases: necessary knowledge for an increasingly multidrug-resistant world. Clin Infect Dis 2019; 69:1446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tamma PD, Girdwood SC, Gopaul R, et al. . The use of cefepime for treating AmpC β-lactamase-producing Enterobacteriaceae. Clin Infect Dis 2013; 57:781–8. [DOI] [PubMed] [Google Scholar]
- 16. Rodríguez-Martínez JM, Fernández-Echauri P, Fernández-Cuenca F, Diaz de Alba P, Briales A, Pascual A. Genetic characterization of an extended-spectrum AmpC cephalosporinase with hydrolysing activity against fourth-generation cephalosporins in a clinical isolate of Enterobacter aerogenes selected in vivo. J Antimicrob Chemother 2012; 67:64–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.