Abstract
Since accelerated metabolism produces much higher levels of reactive oxygen species (ROS) in cancer cells compared to ROS levels found in normal cells, human MutT homolog 1 (MTH1), which sanitizes oxidized nucleotide pools, was recently demonstrated to be crucial for the survival of cancer cells, but not required for the proliferation of normal cells. Therefore, dozens of MTH1 inhibitors have been developed with the aim of suppressing cancer growth by accumulating oxidative damage in cancer cells. While several inhibitors were indeed confirmed to be effective, some inhibitors failed to kill cancer cells, complicating MTH1 as a viable target for cancer eradication. In this review, we summarize the current status of developing MTH1 inhibitors as drug candidates, classify the MTH1 inhibitors based on their structures, and offer our perspectives toward the therapeutic potential against cancer through the targeting of MTH1.
Key words: Oxidized nucleotide, MTH1, Inhibitor, Anticancer, DNA repair
Abbreviations: AI, 7-azaindole; AID, 7-azaindazole; AP, aminopyrimidine; AQ, amidoquinolines; AZ, 2-aminoquinazoline; CETSA, cellular thermal shift assay; CR, cyclometalated ruthenium; DDR, DNA damage response; F, fragment; FP, farnesyl phenolic; IC50, half-maximal inhibitory concentrations; MMR, DNA mismatch repair; MTH1, human MutT homolog 1; NSCLC, non-small cell lung cancer; ROS, reactive oxygen species; TLR7, Toll-like receptor 7; P, purinone; PM, purinone macrocycle; Pu, purine; PDT, photodynamic therapy; TS-FITGE, thermal stability shift-based fluorescence difference in two-dimensional gel electrophoresis; TPP, thermal proteome profiling
Graphical abstract
As an essential enzyme for the survival of cancer cells, human MutT homolog 1 (MTH1), was recently demonstrated as a promising target for cancer eradication. This review summarizes the current progress of developing MTH1 inhibitors with various structures as drug candidates and presents our perspectives toward the therapeutic potential.
1. Introduction
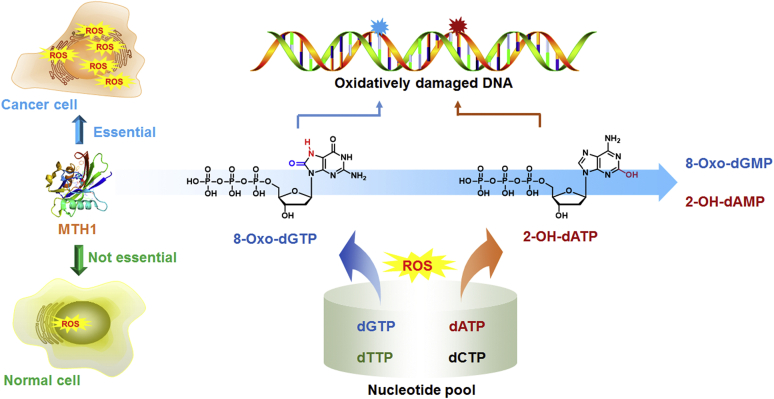
Cellular DNA strands and nucleotide pool continuously suffer from oxidative damage caused by reactive oxygen species (ROS) arising from endogenous oxygen metabolism and detrimental environmental exposure1,2. In particular, the nucleotide pool is much more susceptible to ROS than DNA strands3, resulting in various types of oxidized nucleotides, such as 8-oxo-dGTP, 8-oxo-dATP, 2-OH-dATP and 2-OH-ATP, etc. These oxidized nucleotides tend to be incorporated into DNA or RNA during replication and transcription steps, and consequently lead to severe genome instability and mutations, abundance of repair-associated single-strand breaks, and subsequent cytotoxic double-strand breaks4, 5, 6, 7. For instance, 8-oxo-dGTP not only forms Watson–Crick base pair with dCTP but also forms a Hoogsteen base pair with dATP, thereby introducing GC to TA transversion mutation during the replication process6,7.
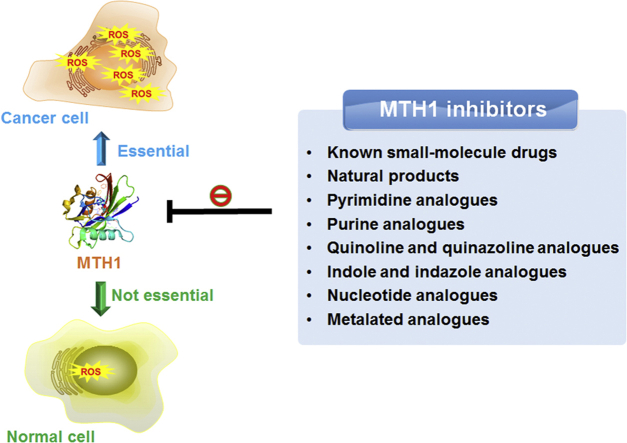
To prevent the mis-incorporation of oxidized nucleotides and maintain genome integrity, various repair enzymes, such as human MutT homolog 1 (MTH1), have evolved to counteract the potentially deleterious effects by hydrolyzing the oxidized nucleotides to their corresponding monophosphates8,9. Surprisingly, due to the accelerated metabolism that produces much higher level of ROS in cancer cells compared to ROS levels found in normal cells, MTH1 was recently demonstrated to be essential for the survival of cancer cells, but not required for the proliferation of normal cells (Fig. 1)10, 11, 12. As such, selective inhibition of MTH1 by small molecules could suppress cancer growth by accumulating oxidative damage11,12. Although dozens of inhibitors have been reported, MTH1 was regarded as a controversial therapeutic target because several inhibitors were proved to be effective while some other inhibitors were unable to kill cancer cells13,14. Since 2014, great advances of MTH1 inhibitors have been witnessed for the past five years, and therefore, we briefly introduce MTH1, summarize the current status of development of the MTH1 inhibitors as drug candidates, classify the MTH1 inhibitors based on their structures, and offer our perspectives toward the therapeutic potential against cancer by targeting MTH1 from the viewpoint of medicinal chemistry.
Figure 1.
MTH1 as a therapeutic target for cancer eradication.
2. MTH1
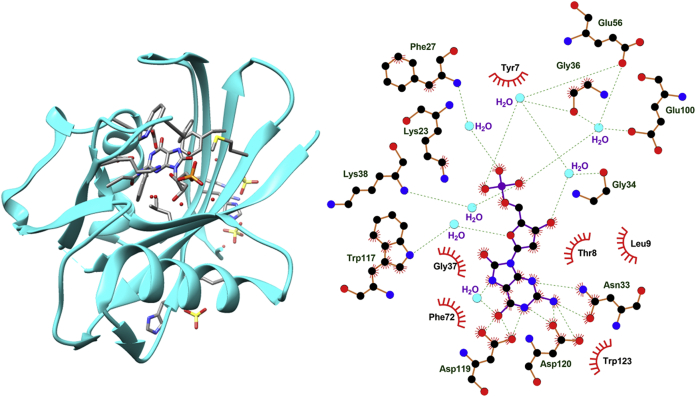
MTH1 harnesses broad substrate specificity toward hydrolysis of the oxidized nucleotides including 8-oxo-dGTP, 8-oxo-dATP, 2-OH-dATP, 8-oxo-rGTP, 8-oxo-rATP, 2-OH-rATP, 8-Cl-dGTP and 8-Br-dGTP, etc15, 16, 17, 18, 19. Aiming to investigate the mechanisms of recognition and hydrolysis by MTH1, researchers solved the X-ray crystal structure for the complex of 8-oxo-dGMP and MTH1, but unfortunately, there is no available analogous structure bearing 8-oxo-dGTP owing to the fast hydrolysis by MTH1 (Fig. 2)20,21. In the active site of MTH1, 8-oxo-dGMP adopts an anti-conformation and forms π-stacking interaction with Trp117 and Phe72, and its Watson–Crick face is recognized by Asp119 and Asp120 through hydrogen bonds to 6-O, 1-NH and 2-NH2 groups of 8-oxo-dGMP21. Asn33 is located at the bottom of the purine base in 8-oxo-dGMP and forms two hydrogen bonds with 2-NH2 and 3-N (Fig. 2). It is worthy to note that Trp117Ala mutant abolishes the ability of MTH1 to hydrolyze either 8-oxo-dGTP or 2-OH-dATP, suggesting an essential role for Trp11715. Asn33 also appears to be critical, as an Asn33Glu mutation does not allow for 8-oxo-dGTP hydrolysis at all, while the Asn33Ala mutation only retains 14% activity20. Taken together, Asp119, Asp120, Asn33, and Trp117 play the crucial roles for the recognition and excision of substrates by MTH1, and also represent the key residues for inhibiting MTH1 by inhibitors.
Figure 2.
MTH1 in complex with 8-oxo-dGMP. Left: 3D structure of MTH1 in complex with 8-oxo-dGMP (PDB file: 3ZR0); Right: 2D structure of MTH1 residues interacting with 8-oxo-dGMP.
3. Inhibitors against MTH1
Starting from 2014, a series of MTH1 inhibitors have been developed from some known small-molecule drugs, natural products, screening and rational drug design, etc. In this review, we summarize these inhibitors as described below, and the half-maximal inhibitory concentrations (IC50s) of inhibitors against 8-oxo-dGTP hydrolysis by MTH1 are consistently provided in order to easily display and compare the inhibitory activities of each inhibitor, except for some specially labeled cases using dGTP as a substrate. Additionally, partial inhibitors were renamed to discriminate each other as described in this review.
3.1. Known small-molecule drugs as MTH1 inhibitors
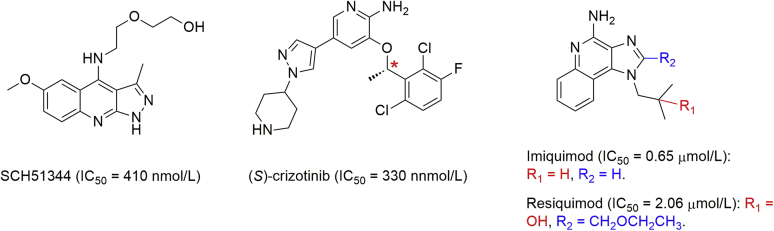
Huber et al.12 utilized a chemical proteomic approach and identified MTH1 as a main cellular target of SCH5 1344 (Fig. 3, IC50 = 410 nmol/L), which was previously discovered to suppress the growth of RAS-overexpressing fibroblasts. Moreover, crizotinib, a clinically approved anticancer agent in 2011 for the treatment of EML4-ALK-positive non-small cell lung cancer (NSCLC) by serving as a dual c-MET/ALK inhibitor, was also found able to inhibit MTH1 at nanomolar concentrations (Fig. 3)12. Surprisingly, (S)-crizotinib rather than (R)-crizotinib significantly inhibits MTH1 activity with the value of IC50 at 330 nmol/L. As a consequence, (S)-crizotinib efficiently suppressed colony formation of SW480 cells and KRAS-mutated PANC1 cells. Furthermore, (S)-crizotinib was demonstrated to display high selectivity—only a few kinases among 456 different recombinant kinases could bind to (S)-crizotinib. Encouraged by the above results, MTH1 inhibition with (S)-crizotinib proved to result in significantly more DNA single-strand breaks and activated DNA repair in human colon carcinoma cells followed by effectively inhibition of tumor progression in mouse xenograft studies utilizing SW480 cells. Kettle and colleagues22 fortuitously found that several agonists of Toll-like receptor 7 (TLR7) could be utilized as MTH1 inhibitors. For instance, imiquimod, the only approved small-molecule agonist of TLR7, showed reasonable submicromolar potency against MTH1 (IC50 = 0.65 μmol/L), while another TLR7 agonist (resiquimod) had 3-fold reduced potency (Fig. 3).
Figure 3.
Several known small-molecule drugs as MTH1 inhibitors.
3.2. Natural products as MTH1 inhibitors
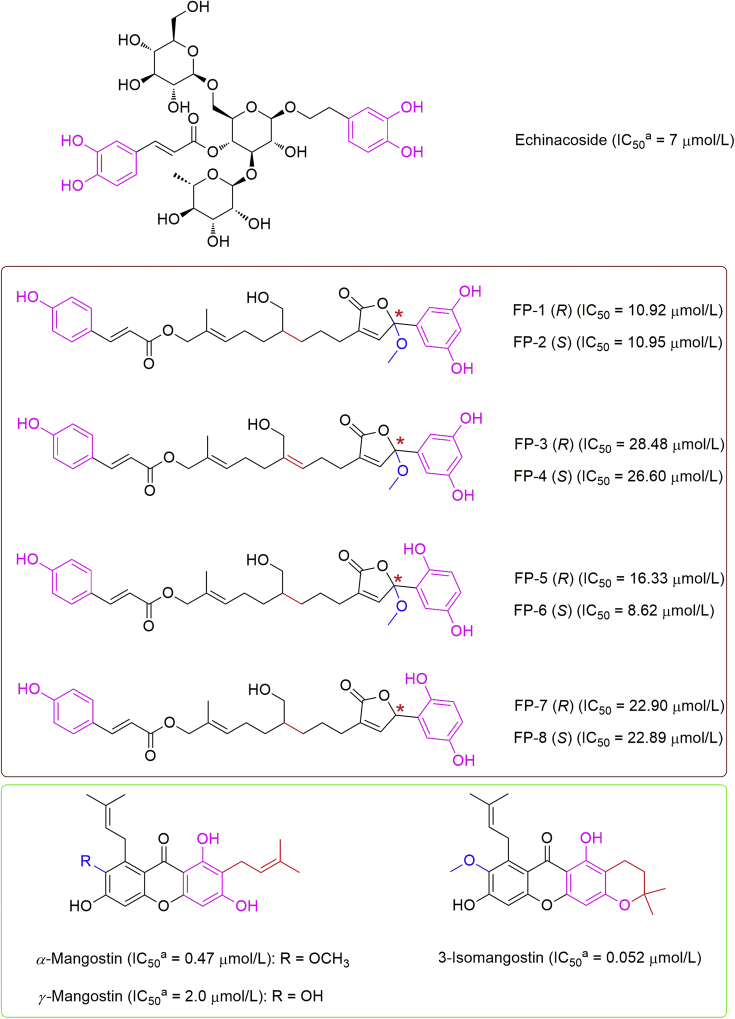
Natural products have been considered to be a rich source of drugs, and MTH1 inhibitors have likewise been isolated from natural products. Dong et al.23 discovered a natural product containing phenol groups, echinacoside, to be an effective inhibitor of MTH1 with an IC50 value of about 7 μmol/L from the medicinal plants Cistanche and Echinacea (Fig. 4). Treatment with echinacoside was further demonstrated to increase the cellular level of oxidative damage in various human cancer cell lines. As such, an immediate and dramatic increase in DNA damage markers and upregulation of P21 was induced followed by the remarkable apoptotic cell death and cell cycle arrest, but not in normal cells, implying the selectivity of echinacoside against cancer cells.
Figure 4.
Several natural products as MTH1 inhibitors. adGTP was used for determining the IC50 values.
Gao et al.24 isolated eight farnesyl phenolic (FP) compounds with a coumaroyl moiety, FP-1 to 8, from the fruiting bodies of Ganoderma sinense, and these compounds subsequently proved to actively inhibit MTH1 with the IC50 values ranging from 8.6 to 28.5 μmol/L (Fig. 4). Subsequently, it was confirmed that these farnesyl phenolic enantiomers could specifically bind to MTH1 within intact cells, and further demonstrated cell cytotoxicity and selectivity against several carcinoma cell lines.
Aiming to discover novel MTH1 inhibitors, Yokoyama et al.25 performed an X-ray crystallographic screening by soaking MTH1–(R)-crizotinib complex crystals into cocktails containing 62 natural products or 33 synthetic compounds, and first identified phenol-bearing α-mangostin as a hit followed by further determination of its inhibitory potency (IC50 = 0.47 μmol/L). Interestingly, the inhibitory potency of γ-mangostin was much lower than that of α-mangostin, showing the IC50 value at 2.0 μmol/L. Next, an inhibition assay against the additional nine natural xanthone derivatives revealed that 3-isomangostin as the most potent inhibitor with the IC50 value of 52 nmol/L (Fig. 4). It was speculated that mangostins might exhibit their antitumor activities against a broad spectrum of cancer cells by inhibiting the MTH1 activities25.
3.3. Development of a novel series of MTH1 inhibitors
3.3.1. Pyrimidine analogues as MTH1 inhibitors
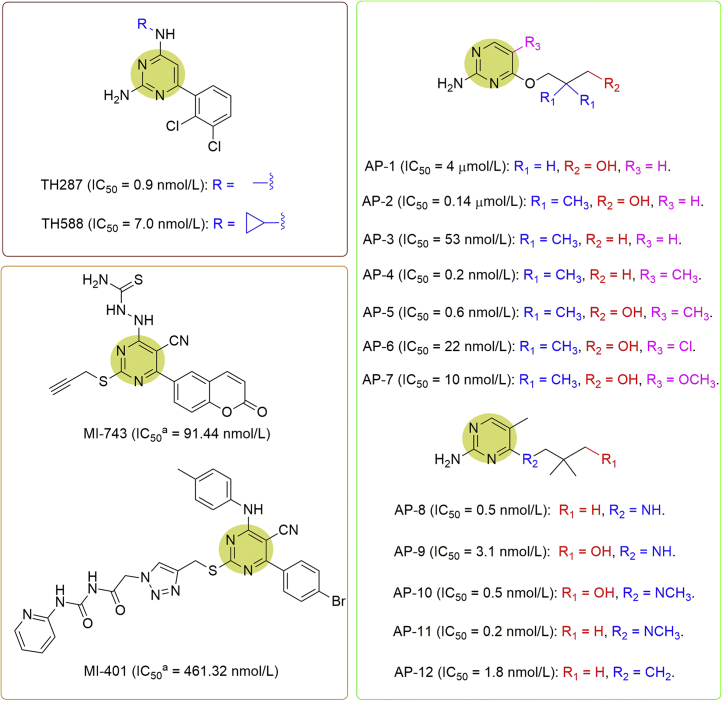
Helleday and co-workers11 identified TH287 as a potent MTH1 inhibitor with an IC50 value of approximately 0.9 nmol/L (Fig. 5). Nevertheless, TH287 was rapidly degraded in human and mouse liver microsomes via N-dealkylation of the aminomethyl substituent, limiting its potential application. TH588, however, having a cyclopropyl substituent instead of the methyl group resulted in improved metabolic stability both in vitro and in vivo, with only slightly reduced MTH1 inhibitory potency (IC50 = 7.0 nmol/L, Fig. 5). Both TH287 and TH588 were further demonstrated to cause the accumulation of oxidized dNTPs and cytotoxicity in cancer cells, and consequently exhibited notable selectivity and high potency to eliminate U2OS and other cancer cell lines with little toxicity to several primary or immortalized cells. Intriguingly, it was found in this study that the cytotoxic effect of MTH1 inhibitors was still not altered with differential expression levels or overexpression of base excision repair enzymes (hOGG1 or MUTYH) that are responsible for repairing 8-oxo-dG and 2-OH-dA lesions in DNA. This might be explained by the fact that the nucleotide pool is particularly susceptible to oxidation mediated by ROS, thereby producing an excessive amount of oxidized bases that may overwhelm the base excision repair system. Based on the fact that TH287 and TH588 were selectively cytotoxic to SV40-large-T-cells and Ras-expressing BJ cells, it was further surmised that MTH1 might be critical early in the transformation process having increased expression for MTH1. Furthermore, the possibility of inhibition by TH287 and TH588 towards other nudix proteins (MTH2, NUDT5, NUDT12, NUDT14 and NUDT16), other proteins with known nucleoside triphosphate pyrophosphatase activity (dCTPase, dUTPase and ITPA), and a much larger panel of 87 enzymes, GPCRs, kinases, ion channels and transporters were also tested, and it turned out that both molecules exhibited reasonable selectivity towards MTH1. Next, TH588 was applied for in vivo treatment using mice bearing BRAFV600E-mutated melanoma, SW480 colorectal, or MCF7 breast tumor xenografts, and responded with an observable reduced tumor growth rate, suggesting that MTH1 is indeed a promising target and potential for a range of different tumors11. In the further studies, TH588 was optimized to give TH1579 (karonudib), which showed more favorable pharmacokinetic and orally available properties and has been approved for phase I clinical testing in cancer patients with advanced solid malignancies (NCT03036228)26. Similarly, TH1579 was also proved to be highly selective and able to cause accumulation of oxidative lesions into DNA in an MTH1-dependent manner. In addition, it was demonstrated to be effective in both the in vitro studies and in vivo models including human colon cancer and chemotherapy-resistant patient-derived malignant melanoma mouse xenograft models, validating TH1579 as a promising inhibitor for cancer treatment26.
Figure 5.
Pyrimidine analogues as MTH1 inhibitors. adGTP was used for determining the IC50 values.
On the basis of the structures of TH287 and TH588, Petrocchi et al.27 first designed an aminopyrimidine (AP) analogue, AP-1, as an inhibitor for MTH1 (IC50 = 4 μmol/L) with the aid of molecular modeling using Schrödinger GLIDE XP docking (Fig. 5). To alleviate desolvation penalty of the ligand and engage with the lipophilic cavity, a dimethyl group was added in the middle of the chain in AP-1 to provide the compound AP-2 having 26-fold improved inhibitory potency against MTH1 (IC50 = 0.14 μmol/L). The potency was further improved by removing the terminal hydroxyl group in AP-2, resulting into AP-3 with an IC50 value of 53 nmol/L. It was amazing that the substitution of the aminopyrimidine ring with a methyl group at the 5-position caused a significant boost in potency, such as AP-4 (IACS-4619, IC50 = 0.2 nmol/L) versus AP-3, and AP-5 (IACS-4759, IC50 = 0.6 nmol/L) versus AP-2. Additional exploration with chloride or the methoxyl substituents at the 5-position of AP-2 gave AP-6 (IC50 = 22 nmol/L) or AP-7 (IC50 = 10 nmol/L), respectively, as relatively weaker inhibitors. Further replacement of the alkoxy chain at the 4-position with an alkyl amine in compounds AP-4 and AP-5 respectively afforded AP-8 (IC50 = 0.5 nmol/L) and AP-9 (IC50 = 3.1 nmol/L), which exhibited lower, but comparable potencies. AP-10 and AP-11 having the tertiary amines showed almost identical potency with IC50 values at 0.5 and 0.2 nmol/L, respectively. Similarly, a carbon-linked chain at the 4-position gave AP-12 which also showed comparable potency (IC50 = 1.8 nmol/L). Given their high enzymatic potency, AP-4 and AP-5 were further evaluated, and AP-5 showed excellent cell permeability, solubility, and stability in rat and human plasma and liver microsomes, while compound AP-4 showed good permeability but high turnover in liver microsomes. Furthermore, AP-5 was profiled and confirmed to be highly selective towards MTH1 by testing a panel of 97 kinases, with no off-target kinase activity. In comparison with TH287 and TH588, the anti-proliferative effects of AP-4 and AP-5 were also tested by using a broad range of human cancer and normal cell lines, but unfortunately displayed little or no expected anticancer responses even at compound concentrations up to 50 μmol/L27.
Zhou et al.28 recently screened approximately 1000 compounds followed by further optimization of the identified lead compound, and ultimately obtained two potent compounds containing 5-cyano-6-phenylpyrimidine structure, MI-743 (IC50 = 91.44 nmol/L) and MI-401 (IC50 = 461.32 nmol/L), as potential inhibitors against MTH1 (Fig. 5). Further evaluation by the cellular thermal shift assay (CETSA) demonstrated that MI-743 had specifically engagement to MTH1 at a cellular level. Moreover, MI-743 was able to cause cytotoxicity and induce accumulation of 8-oxo-dG lesions, DNA damage, and apoptosis in gastric cancer cell lines MGC-803 and HGC-27 cells, therefore leading to cytotoxicity and anti-proliferation effects on MGC-803 (IC50 = 2.91 μmol/L) and HGC-27 cells (IC50 = 1.14 μmol/L) along with little or no effect on four normal cell lines. Apart from the significantly increased 8-oxo-dG level in MGC-803 and HGC-27 cells, it was also confirmed that MI-743 could markedly induce cellular DNA damage response (DDR) and apoptosis, which may be related to its specific inhibition towards MTH1 activity. Furthermore, MI-743 exhibited notable antitumor effect on the MGC-803 cells-derived xenograft model in BALB/c nude mice without obvious global toxicity, suggesting that MI-743 may serve as a promising lead compound for gastric cancer treatment through the targeting of MTH128.
3.3.2. Purine analogues as MTH1 inhibitors
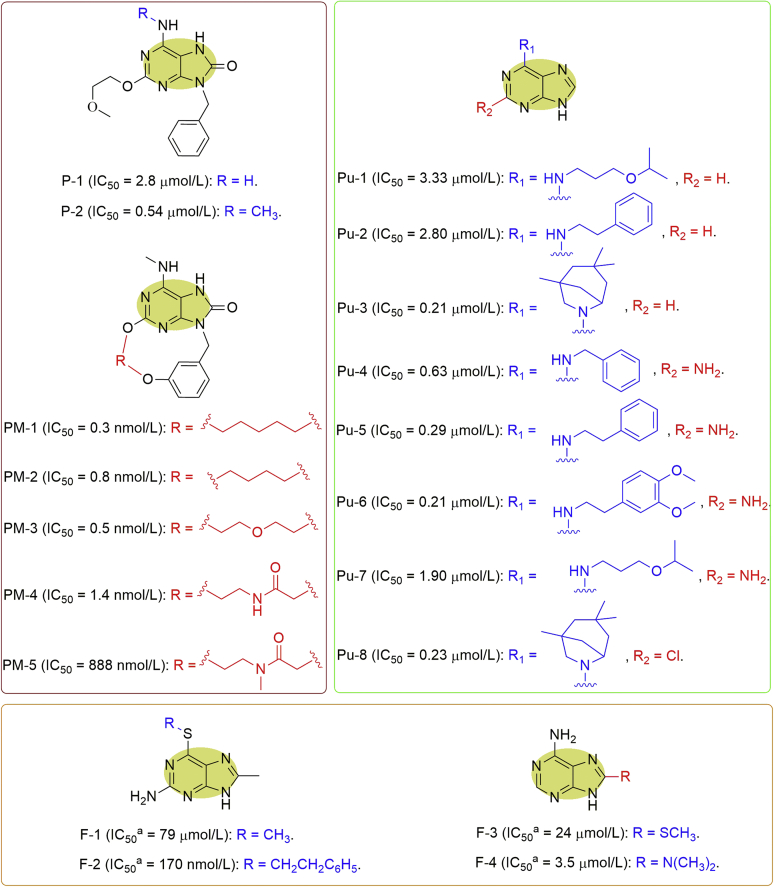
In addition to the known TLR-7 agonists (imiquimod and resiquimod) as described above, Kettle and colleagues22 also realized a series of purinone (P) analogues as MTH1 inhibitors (Fig. 6). While a purinone analogue, P-1, was demonstrated to inhibit MTH1 with IC50 value at 2.8 μmol/L, the N-methylation of P-1 gave P-2 having a modest increased affinity for MTH1. Amazingly, macrocyclization of P-2 via an n-pentyl bis-ether linker provided a purinone macrocycle (PM) analogue, PM-1, whose active binding conformation was locked, and thereby resulting in 1700-fold increase in potency against MTH1 (IC50 = 0.3 nmol/L, Fig. 6). However, this macrocyclization also increased lipophilicity, which led to an increase in in vitro clearance, although in vitro membrane permeability is also increased. By shortening the n-pentyl bis-ether in PM-1 to a n-butyl bis-ether linker, PM-2 was obtained with reduced lipophilicity but slight loss in potency (IC50 = 0.8 nmol/L). Introducing a polar ether linker into the all carbon side chain of PM-2 resulted in PM-3 having comparable inhibitory activity against MTH1 (IC50 = 0.5 nmol/L) along with significantly lowered lipophilicity. Further increasing the polarity by the introduction of an amide group afforded PM-4 (IC50 = 1.4 nmol/L) having much lower logD (1.8), but subsequently proved to be unstable in human plasma. The tertiary amide analogue PM-5 was markedly less active, and similarly unstable in human plasma. Importantly, three compounds of P-2, PM-1 and PM-3 were all tested to exhibit excellent correlation between MTH1 inhibitory potencies and target engagement in K562 cells by utilizing a whole CETSA. Furthermore, a representative inhibitor, PM-3, was profiled against 267 kinases and 153 secondary pharmacology targets, and observed with no notable off-target activities, suggesting their high selectivity targeting MTH1. Subsequently, a wide range of tumor cell lines were applied to assess the antiproliferative effects of the representative inhibitors. PM-1 showed some activity at the high concentrations only for a fraction of the cell lines, while macrocycle PM-3 had almost no impact on cell viability22.
Figure 6.
Purine analogues as MTH1 inhibitors. adGTP was used for determining the IC50 values.
By an alternative approach, Rudling et al.29 pursued structure-based virtual screening by means of molecular docking of 0.3 million fragments to the MTH1 binding site, and identified 22 commercially available fragment ligands. Further experimental evaluation discovered five fragments (F) including purine analogues, which could inhibit MTH1 activities with IC50 values within the range of 6–79 μmol/L. These fragments were subsequently optimized based on the predicted binding modes and led to several potential inhibitors. For instance, F-1 was first identified with the IC50 value at 79 μmol/L and optimized to F-2 having the IC50 value at 170 nmol/L, while F-3 (IC50 = 24 μmol/L) was optimized to F-4 (IC50 = 3.5 μmol/L, Fig. 6). It is intriguing that one of the identified fragments is a component for the compound discovered by Kettle et al.22, indicating the reliability of this strategy.
By means of chemical array platforms, Kumar et al.30 identified several purine (Pu) derivatives as MTH1 inhibitors from a chemical library of the RIKEN Natural Products Depository (NPDepo, Fig. 6). Pu-1 (NPD15095) exerted MTH1 inhibitory activity with IC50 value at 3.3 μmol/L. Further exploration of 131 structurally related compounds with a purine moiety provided seven compounds (Pu-2, Pu-3, Pu-4, Pu-5, Pu-6, Pu-7, and Pu-8) which displayed higher activities than NPD15095. In particular, Pu-3 (NPD7155), Pu-5 (NPD9948), Pu-6, and Pu-8 were found to be the strongest inhibitors with IC50 values of 0.21, 0.29, 0.21, and 0.23 μmol/L, respectively (Fig. 6). Moreover, Pu-3 and Pu-5 were further assessed and demonstrated to considerably stabilize cellular MTH1 rather than many other proteins in HeLa cells by the target engagement assay, suggesting their specificity towards MTH1 in HeLa cells. In spite of acceptable potency against MTH1, Pu-3 and Pu-5 displayed only weak cytotoxicity against HeLa cells (IC50 = 65 and 35 μmol/L, respectively) as well as the other cancer cell lines. This might be reasonable because it was further found that Pu-3 and Pu-5 induced DNA damage and increased the sub-G1 cell population only at the higher concentrations31.
3.3.3. Quinoline and quinazoline analogues as MTH1 inhibitors
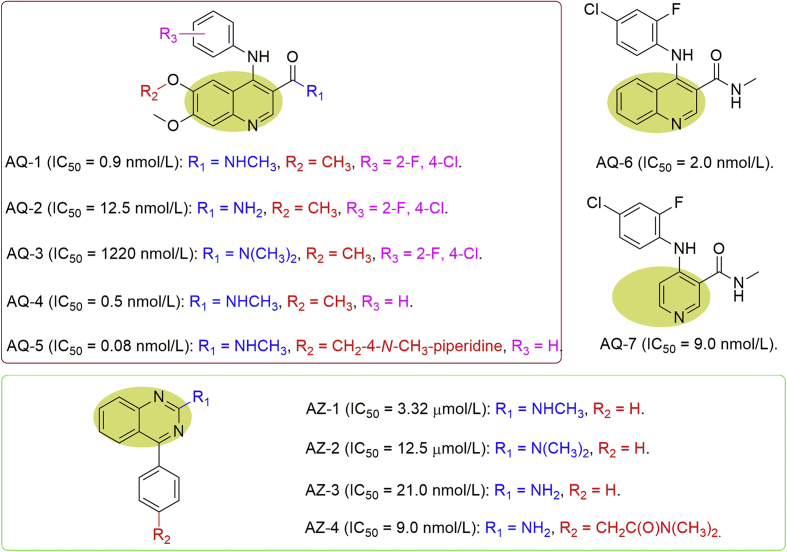
Kettle et al.22 also performed high-throughput screening and obtained a series of 3-amidoquinolines (AQ) as another chemical entity against MTH1 with high potency, such as AQ-1 (IC50 = 0.9 nmol/L) and AQ-2 (IC50 = 12.5 nmol/L, Fig. 7). But the dimethyl analogue AQ-3 showed over 130-fold lower activity relative to AQ-1. While deletion of both halogens of AQ-1 afforded AQ-4 which was consequently both more potent (IC50 = 0.5 nmol/L) and less lipophilic, introduction of a basic group to AQ-4 further improved the IC50 value to 80 pmol/L (AQ-5) together with improvements in solubility. Of interest, while removing both methoxy groups in AQ-1 provided AQ-6 showing slightly reduced potency with IC50 value at 2 nmol/L, further truncation of the quinoline produced the pyridyl AQ-7 with somewhat retained potency (IC50 = 9 nmol/L, Fig. 7)22.
Figure 7.
Quinoline and quinazoline analogues as MTH1 inhibitors.
Similarly, 2-aminoquinazoline (AZ) structure-based molecules were also identified by high-throughput screening as one class of inhibitors against MTH1. The hit compound, AZ-1, had some potency with IC50 value at 3.32 μmol/L while dimethylation of the amine to give AZ-2 reduced the potency by 4-fold. Intriguingly, deletion of the methyl in AZ-1 to give AZ-3 resulted in significantly improved activity against MTH1 (IC50 = 21 nmol/L). Moreover, the dimethylamide AZ-4 showed further enhanced MTH1 potency (IC50 = 9 nmol/L) and acceptable physicochemical properties (Fig. 7)22.
Furthermore, three representative inhibitors (AQ-1, AQ-4, and AZ-4) were assessed and exhibited superior MTH1 engagement in K562 cells and high selectivity against MTH1. However, while AQ-1 showed somewhat antiproliferative effect only at the high concentrations for a fraction of tumor cell lines, AQ-4 was almost completely inactive. In the further studies, it was demonstrated that compound AQ-1 failed to induce the increasing of DDR signaling markers, which might explain the weak effect of AQ-122.
3.3.4. Indole and indazole analogues as MTH1 inhibitors
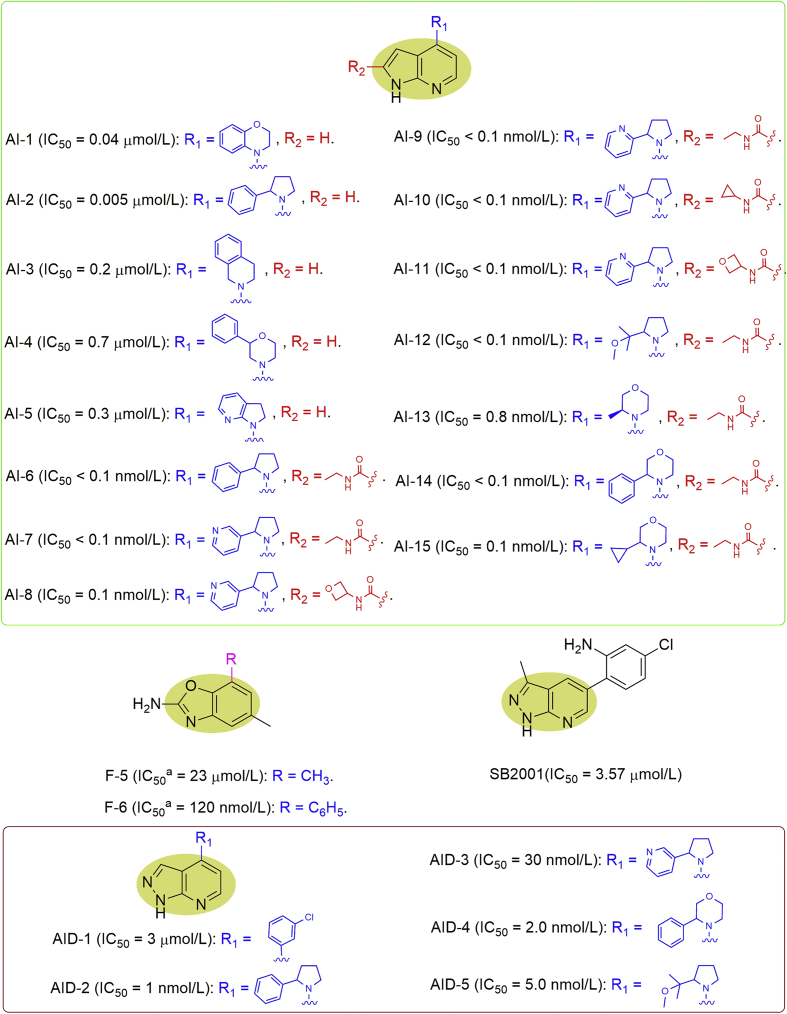
By performing a fragment-based screening, Rahm et al.32 identified a 7-azaindole (AI) fragment hit followed by introduction of cyclic amines in the 4-position via structure-based drug design and rational medicinal chemistry approaches to provide compounds AI-1 to 5, of which AI-2 having a 2-phenylpyrrolidine was demonstrated to be highly potent (IC50 = 5 nmol/L, Fig. 8). Further adding a secondary amide substituent to AI-2 gave compound AI-6 showing the notable potency with IC50 value below 0.1 nmol/L. Replacement of the phenyl group in AI-6 with a pyridine afforded compound AI-7 with increased hydrophilicity and microsomal stability. It was further surveyed by combining a few pyrrolidine-based substituents in the 4-position and a small number of secondary amide substituents to give compounds AI-8 to 11, especially for compounds AI-10 and AI-11, which were the most promising representatives with high potency, moderate stability in microsomes and medium permeability. Moreover, AI-10 was demonstrated to be 1000 times more selective against other tested targets including 97-membered kinase as well as 215-membered ATPase. The three most potent compounds (AI-6, AI-10, and AI-11) also exerted high cellular on-target engagement. Aiming to improve the cell permeability and metabolic stability, four new compounds, AI-12 to 15, were obtained displaying picomolar potency, high solubility, high permeability, and acceptable metabolic stability32. In particular, AI-13 (BAY-707) was chosen for further in vivo studies (Fig. 8). However, inhibition of MTH1 with AI-13 did not exhibit in vitro or in vivo anticancer efficacy either in mono- or in combination therapies unfortunately33. In addition to the above purine fragments, Rudling et al.29 also identified another fragment (F-5) which could inhibit MTH1 with IC50 value at 23 μmol/L via structure-based virtual screening followed by optimization to give F-6 with IC50 value at 120 nmol/L (Fig. 8).
Figure 8.
Indole and indazole analogues as MTH1 inhibitors. adGTP was used for determining the IC50 values.
Some 7-azaindazole (AID) analogues have also been verified as MTH1 inhibitors. Using two label-free target identification methods, including thermal stability shift-based fluorescence difference in two-dimensional gel electrophoresis (TS-FITGE) and thermal proteome profiling (TPP), Park et al.34 found that an indazole analogue, SB2001, could bind and inhibit MTH1 (IC50 = 3.57 μmol/L) and LTA4H (Fig. 8). Subsequently, SB2001 was confirmed to induce the accumulation of oxidative damages in DNA mismatch repair (MMR) pathway defective HeLa cells, thereby resulting in the cell cytotoxicity. Rahm et al.32 created several 7-azaindazole analogues, of which AID-1 showed comparable inhibitory activity (IC50 = 3.0 μmol/L) to SB2001, while AID-2 to 5 exerted much stronger inhibitory activities than SB2001 with the IC50 values ranging from 1 to 30 nmol/L (Fig. 8).
3.3.5. Nucleotide analogues as MTH1 inhibitors
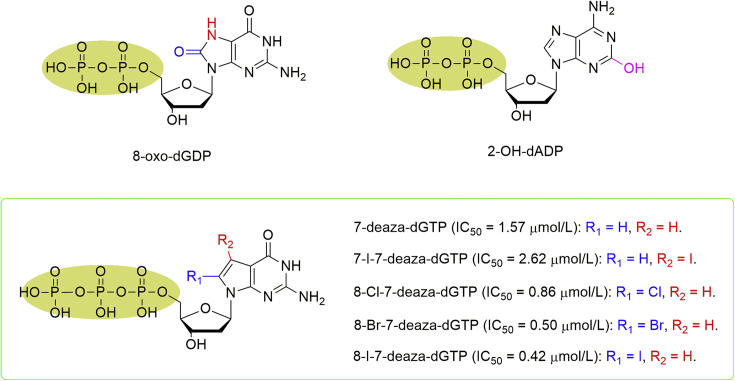
Despite the fact that MTH1 can degrade 8-oxo-dGTP and 2-OH-dATP to their corresponding monophosphates, 8-oxo-dGDP and 2-OH-dADP having the identical oxidized nucleosides but in diphosphate manner were demonstrated not to be hydrolyzed by MTH1, whereas these diphosphate derivatives could bind and inhibit MTH1 activities at low-micromolar range (Fig. 9)35,36. However, the other human Nudix proteins, such as MTH2 and MTH3, can hydrolyze 8-oxo-dGDP and 2-OH-dADP to their monophosphates, and therefore limiting their potential as antitumorigenic agents37.
Figure 9.
Nucleotide analogues as MTH1 inhibitors.
Taniguchi and co-workers38,39 have developed 8-halogenated 7-deazadGTP analogues as 8-oxo-dGTP mimics, and it was further found that these 7-deazadGTP analogues were seldom hydrolyzed but obviously inhibited MTH1 activity even though the minor difference at 7- and 8-postion. While IC50 value of 7-deazadGTP was 1.57 μmol/L, 7-I-7-deazadGTP exhibited inhibitory effect against MTH1 showing IC50 value at 2.62 μmol/L. Interestingly, introduction of halogen at 8-position could further enhance the inhibitory activities with the IC50 values of 8-Cl-7-deazadGTP, 8-Br-7-deazadGTP, and 8-I-7-deazadGTP at 0.857, 0.496, and 0.415 μmol/L, respectively (Fig. 9). This high inhibitory activity of 7-deazadGTP analogues might attribute to the increased π−π interaction with Trp117 and Phe72 in the active site of MTH1 due to 7-CH replacement. Further anticancer effects would be expected after addressing the activation of 8-halogenated 7-deazadG analogues by nucleoside kinases or efficient delivery of 8-halogenated 7-deazadGTP analogues.
3.3.6. Metalated analogues as MTH1 inhibitors
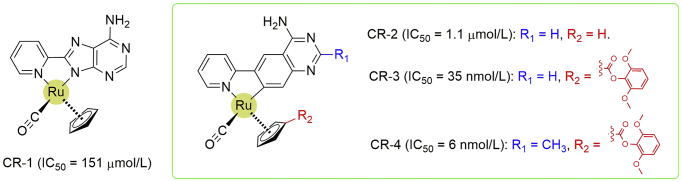
Some transition metal ions, such as Cd(II) and Cu(II), were also found as inhibitors against MTH1 (IC50 values for Cd(II) and Cu(II) are 30 and 17 μmol/L, respectively)40. In another study, Streib et al.41 developed several unconventional cyclometalated ruthenium (CR) half-sandwich complexes, which were subsequently demonstrated as strong inhibitors against MTH1 with IC50 values at low-nanomolar levels (Fig. 10). The striking specificity towards MTH1 was further confirmed by testing with a large panel of protein kinases and other ATP binding proteins, indicating their potentials as bio-probes and antitumor agents. In this study, the complex CR-1 was first identified as a binder to MTH1 but with the IC50 value at 151 μmol/L, and the inhibitory activity against MTH1 was improved by more than 100-fold through replacing the 8-(pyridin-2-yl)adenine ligand of CR-1 with a cyclometalated 4-amino-6-(pyridin-2-yl)quinazoline, resulting in the complex CR-2 (IC50 = 1.1 μmol/L). The following derivatization of cyclopentadienyl moiety afforded the complex CR-3, which exhibited a dramatically improved activity with IC50 value at 35 nmol/L. The further introduction of a methyl group at 2-position of the quinazoline moiety in complex CR-3 provided the complex CR-4 as a single-digit nanomolar inhibitor for MTH1 (IC50 = 6 nmol/L, Fig. 10)41.
Figure 10.
Metalated analogues as MTH1 inhibitors.
4. Conclusions and perspectives
In conclusion, a growing number of MTH1 inhibitors have been developed since 2014 by means of various strategies involving with drug repositioning, natural product component extraction, high-throughput screening, structure-based virtual screening, fragment-based screening, structure-based drug design and rational medicinal chemistry approaches after screening, substrate-based drug design, etc. A series of structures including known small-molecule drugs, phenolic natural products, pyrimidine analogues, purine analogues, quinoline and quinazoline analogues, indole and indazole analogues, nucleotide analogues, metalated analogues have been reported as MTH1 inhibitors.
While a wide range of inhibitors of MTH1 as described above were indeed demonstrated to be effective for cancer eradication, several studies also suggested that inhibition of MTH1 failed to obtain the desired anticancer activity, leading to a controversial question whether MTH1 is a valuable target13,14. Accordingly, the viability of this anticancer strategy, as well as the detailed mechanisms through the inhibition of MTH1, needs to be rigorously addressed, and there might be multiple critical factors that could affect the cellular response to MTH1 inhibition. Importantly, the indispensability of MTH1 for cancer cell growth or survival under oxidative conditions should be inspected. Some provoking factors to elevate ROS levels in certain cell lines, such as oncogenic RAS, could emphasize the importance of MTH1 to cellular viability, and thus, as a therapeutic target, to some degree42, 43, 44. In contrast, some resistance mechanisms, such as the highly efficient DNA maintenance and repair pathways involving base excision repair glycosylases hOGG1 and MUTYH, as well as the other enhanced antioxidant cellular pathways including the peroxiredoxin 1 (PRDX1) that, cooperating with MTH1, might arise to undermine the role of MTH145,46. Besides, the discrepancy between in vitro and in vivo tumorigenic models was also found to affect the cellular response to MTH1 inhibitors45. Kawamura et al.26,31,47,48 have suggested that TH287 and TH588 might display their antitumor activities through off-target effect by suppressing tubulin polymerization, whereas these inhibitors were subsequently proved to activate in a different manner as compared to anti-microtubule agent. Indeed, it was recently demonstrated that MTH1 could bind tubulin and promote the progression of mitosis in cancer cells. Some MTH1 inhibitors, such as TH588 and TH1579, could block the interaction of MTH1–tubulin, therefore inducing the mitotic arrest of cancer cells49. Taken together, the redox balance of the cellular processes involving oxidative metabolism and elimination, the tumorigenic models, off-target possibility and so on, might directly or indirectly determine the outcome of MTH1 inhibitors, complicating it as a viable therapeutic approach for cancer eradication by inhibiting MTH1. As it was found that the loss of anticancer effect for some reported MTH1 inhibitors was derived from the failure to introduce oxidized damages into DNA strands as confirmed by immunofluorescence, it might be worthy of visualizing the accumulation of oxidized damages prior to the next assessment stage26.
Despite its complexity, MTH1 is still regarded as a promising therapeutic target. The overexpression and crucial role of MTH1 have been verified for a broad spectrum of tumors, such as renal-cell carcinoma, brain tumors, primary non-small cell lung tumors, colorectal cancer, non-small cell lung cancer, gliomas, breast cancer, myeloma, squamous cell carcinoma, etc.28,50. It was also proposed that MTH1 might be a therapeutic target for metastasis of cancer, as MTH1 was observed to stimulate migration and invasion potential of thyroid cancer cells51. Wei and co-workers52 recently demonstrated that depletion of MTH1 could significantly inhibit the ovarian tumor growth by delivering a CRISPR-Cas9 system targeting MTH1 gene using a multifunctional nucleus-targeting “core-shell” artificial virus (RRPHC), validating MTH1 as a therapeutic target for ovarian cancer. Moreover, MTH1 suppression leads to elevated 8-oxo-dG which can introduce 8-oxo-dG associated structural alterations in promoter G-quadruplexes residing in the KRAS promoter, thereby impairing the proliferation of KRAS-mutant NSCLC cells and xenograft tumor formation44. Inhibitions of PRDX1 and MTH1 could introduce 8-oxo-dG at the 3′-ends of telomeric substrates, which could result in efficient inhibition of telomere extension by telomerase46. Additionally, photodynamic therapy (PDT) has been applied to achieve effective cancer treatment through the highly toxic ROS53,54, and accordingly MTH1 suppression was recently demonstrated to play a synergistic role for PDT on the basis of the improvement of cellular sensitivity to ROS in cancer cells55,56. These studies open new avenues for targeting oncogenic RAS and telomerase that are notoriously difficult anticancer targets by inhibiting MTH1, and also suggest that MTH1 inhibition might be suitable for polytherapy due to its synergistic effect. In the future, it will be anticipated that the increasing quantity of MTH1 inhibitors with novel structures will be discovered for cancer eradication and validation of MTH1 as a potential anticancer target on one hand; on the other hand, further shedding light on the mechanisms of MTH1 in cancer and normal cells in depth may open some new avenues, such as polytherapy involving MTH1 as a target, for cancer therapy. It is also worth noting in the future that MTH1 deficiency elevates oxidative stress, which can result in vulnerability to neurodegeneration13, and thus, the possible detrimental effects on normal cells from MTH1 inhibitors should also be tracked.
Acknowledgments
This work was funded by the grant from the National Natural Science Foundation of China (81903425, to Yizhen Yin). We thank Dr. Christopher John Hipolito (University of Tsukuba, Tsukuba, Japan) for his kind proofreading.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Author contributions
Yizhen Yin generated the manuscript draft. Fener Chen edited and revised the manuscript. All authors reviewed and approved the final version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Lindahi T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Evans M.D., Dizdaroglu M., Cooke M.S. Oxidative DNA damage and disease: induction, repair and significance. Mutation Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Topal M.D., Baker M.S. DNA precursor pool: a significant target for N-methyl-N-nitrosourea in C3H/10T1/2 clone 8 cells. Proc Natl Acad Sci U S A. 1982;79:2211–2215. doi: 10.1073/pnas.79.7.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrows C.J., Muller J.G. Oxidative nucleobase modifications leading to strand scission. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 5.Maynard S., Schurman S.H., Harboe C., Souza-Pinto N.C., Bohr V.A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibutani S., Takeshita M., Grollaman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 7.Hsu G.W., Ober M., Carell T., Beese L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 8.David S.S., O'Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuzuki T., Nakatsu Y., Nakabeppu Y. Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci. 2007;98:465–470. doi: 10.1111/j.1349-7006.2007.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nature Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 11.Gad H., Koolmeister T., Jemth A.S., Eshtad S., Jacques S.A., Ström C.E. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 12.Huber K.V., Salah E., Radic B., Gridling M., Elkins J.M., Stukalov A. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakabeppu Y., Ohta E., Abolhassani N. MTH1 as a nucleotide pool sanitizing enzyme: friend or foe? Free Radical Biol Med. 2017;107:151–158. doi: 10.1016/j.freeradbiomed.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Papeo G. MutT Homolog 1 (MTH1): the silencing of a target. J Med Chem. 2016;59:2343–2345. doi: 10.1021/acs.jmedchem.6b00283. [DOI] [PubMed] [Google Scholar]
- 15.Sakai Y., Furuichi M., Takahashi M., Mishima M., Iwai S., Shirakawa M., Nakabeppu Y. A molecular basis for the selective recognition of 2-hydroxy-dATP and 8-oxo-dGTP by human MTH1. J Biol Chem. 2002;277:8579–8587. doi: 10.1074/jbc.M110566200. [DOI] [PubMed] [Google Scholar]
- 16.Fujikawa K., Kamiya H., Yakushiji H., Fujii Y., Nakabeppu Y., Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J Biol Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 17.Fujikawa K., Kamiya H., Yakushiji H., Nakabeppu Y., Kasai H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001;29:449–454. doi: 10.1093/nar/29.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamiya H., Yakushiji H., Dugué L., Tanimoto M., Pochet S., Nakabeppu Y., Harashima H. Probing the substrate recognition mechanism of the human MTH1 protein by nucleotide analogs. J Mol Biol. 2004;336:843–850. doi: 10.1016/j.jmb.2003.12.060. [DOI] [PubMed] [Google Scholar]
- 19.Fujikawa K., Yakushiji H., Nakabeppu Y., Suzuki T., Matsuda M., Ohshima H. 8-Chloro-dGTP, a hypochlorous acid-modified nucleotide, is hydrolyzed by hMTH1, the human MutT homolog. FEBS Lett. 2002;512:149–151. doi: 10.1016/s0014-5793(02)02240-8. [DOI] [PubMed] [Google Scholar]
- 20.Mishima M., Sakai Y., Itoh N., Kamiya H., Furuichi M., Takahashi M. Structure of human MTH1, a nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates. J Biol Chem. 2004;279:33806–33815. doi: 10.1074/jbc.M402393200. [DOI] [PubMed] [Google Scholar]
- 21.Svensson L.M., Jemth A.S., Desroses M., Loseva O., Helleday T., Högbom M. Crystal structure of human MTH1 and the 8-oxo-dGMP product complex. FEBS Lett. 2011;585:2617–2621. doi: 10.1016/j.febslet.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 22.Kettle J.G., Alwan H., Bista M., Breed J., Davies N.L., Eckersley K. Potent and selective inhibitors of MTH1 probe its role in cancer cell survival. J Med Chem. 2016;59:2346–2361. doi: 10.1021/acs.jmedchem.5b01760. [DOI] [PubMed] [Google Scholar]
- 23.Dong L., Wang H., Niu J., Zou M., Wu N., Yu D. Echinacoside induces apoptotic cancer cell death by inhibiting the nucleotide pool sanitizing enzyme MTH1. OncoTargets Ther. 2015;8:3649–3664. doi: 10.2147/OTT.S94513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y., Zhu L., Guo J., Yuan T., Wang L., Li H. Farnesyl phenolic enantiomers as natural MTH1 inhibitors from Ganoderma sinense. Oncotarget. 2017;8:95865–95879. doi: 10.18632/oncotarget.21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama T., Kitakami R., Mizuguchi M. Discovery of a new class of MTH1 inhibitor by X-ray crystallographic screening. Eur J Med Chem. 2019;167:153–160. doi: 10.1016/j.ejmech.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Berglund U.W., Sanjiv K., Gad H., Kalderen C., Koolmeister T., Pham T. Validation and development of MTH1 inhibitors for treatment of cancer. Ann Oncol. 2016;27:2275–2283. doi: 10.1093/annonc/mdw429. [DOI] [PubMed] [Google Scholar]
- 27.Petrocchi A., Leoy E., Reyna N.J., Hamilton M.M., Shi X., Parker C.A. Identification of potent and selective MTH1 inhibitors. Bioorg Med Chem Lett. 2016;26:1503–1507. doi: 10.1016/j.bmcl.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W., Ma L., Yang J., Qiao H., Li L., Guo Q. Potent and specific MTH1 inhibitors targeting gastric cancer. Cell Death Dis. 2019;10:434. doi: 10.1038/s41419-019-1665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudling A., Gustafsson R., Almlöf I., Homan E., Scobie M., Berglund U.W. Fragment-based discovery and optimization of enzyme inhibitors by docking of commercial chemical space. J Med Chem. 2017;60:8160–8169. doi: 10.1021/acs.jmedchem.7b01006. [DOI] [PubMed] [Google Scholar]
- 30.Kumar A., Kawamura T., Kawatani M., Osada H., Zhang K.Y.J. Identification and structure–activity relationship of purine derivatives as novel MTH1 inhibitors. Chem Biol Drug Des. 2017;89:862–869. doi: 10.1111/cbdd.12909. [DOI] [PubMed] [Google Scholar]
- 31.Kawamura T., Kawatani M., Muroi M., Kondoh Y., Futamura Y., Aono H. Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival. Sci Rep. 2016;6:26521. doi: 10.1038/srep26521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahm F., Viklund J., Trésaugues L., Ellermann M., Giese A., Ericsson U. Creation of a novel class of potent and selective MutT homologue 1 (MTH1) inhibitors using fragment-based screening and structure-based drug design. J Med Chem. 2018;61:2533–2551. doi: 10.1021/acs.jmedchem.7b01884. [DOI] [PubMed] [Google Scholar]
- 33.Ellermann M., Eheim A., Rahm F., Viklund J., Guenther J., Andersson M. Novel class of potent and cellularly active inhibitors devalidates MTH1 as broad-spectrum cancer target. ACS Chem Biol. 2017;12:1986–1992. doi: 10.1021/acschembio.7b00370. [DOI] [PubMed] [Google Scholar]
- 34.Park H., Park S.B. Label-free target identification reveals oxidative DNA damage as the mechanism of a selective cytotoxic agent. Chem Sci. 2019;10:3449–3458. doi: 10.1039/c8sc05465g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bialkowski K., Kasprzak K.S. A novel assay of 8-oxo-2′-deoxyguanosine 5′-triphosphate pyrophosphohydrolase (8-oxo-dGTPase) activity in cultured cells and its use for evaluation of cadmium(II) inhibition of this activity. Nucleic Acids Res. 1998;26:3194–3201. doi: 10.1093/nar/26.13.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bialkowski K., Kasprzak K.S. Inhibition of 8-oxo-2ʹ-deoxyguanosine 5ʹ-triphosphate pyrophosphohydrolase (8-oxo-dGTPase) activity of the antimutagenic human MTH1 protein by nucleoside 5ʹ-diphosphates. Free Radic Biol Med. 2003;35:595–602. doi: 10.1016/s0891-5849(03)00362-9. [DOI] [PubMed] [Google Scholar]
- 37.Takagi Y., Setoyama D., Ito R., Kamiya H., Yamagata Y., Sekiguchi M. Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates. J Biol Chem. 2012;287:21541–21549. doi: 10.1074/jbc.M112.363010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Y., Sasaki S., Taniguchi Y. Inhibitory effect of 8-halogenated-7-deaza-2ʹ-deoxyguanosine triphosphates on human 8-oxo-2ʹ-deoxyguanosine triphosphatase, hMTH1, activities. ChemBioChem. 2016;17:566–569. doi: 10.1002/cbic.201500589. [DOI] [PubMed] [Google Scholar]
- 39.Yin Y., Sasaki S., Taniguchi Y. Effects of 8-halo-7-deaza-2ʹ-deoxyguanosine triphosphate on DNA synthesis by DNA polymerases and cell proliferation. Bioorg Med Chem. 2016;24:3856–3861. doi: 10.1016/j.bmc.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 40.Kasprzak K.S., Bialkowski K. Inhibition of antimutagenic enzymes, 8-oxo-dGTPases, by carcinogenic metals. Recent developments. J Inorg Biochem. 2000;79:231–236. doi: 10.1016/s0162-0134(99)00240-8. [DOI] [PubMed] [Google Scholar]
- 41.Streib M., Kräling K., Richter K., Xie X., Steuber H., Meggers E. An organometallic inhibitor for the human repair enzyme 7,8-dihydro-8-oxoguanosine triphosphatase. Angew Chem Int Ed. 2014;53:305–309. doi: 10.1002/anie.201307849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rai P., Young J.J., Burton D.G., Giribaldi M.G., Onder T.T., Weinberg R.A. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene. 2011;30:1489–1496. doi: 10.1038/onc.2010.520. [DOI] [PubMed] [Google Scholar]
- 43.Giribaldi M.G., Munoz A., Halvorsen K., Patel A., Rai P. MTH1 expression is required for effective transformation by oncogenic HRAS. Oncotarget. 2015;6:11519–11529. doi: 10.18632/oncotarget.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel A., Burton D.G., Halvorsen K., Balkan W., Reiner T., Perez-Stable C. MutT homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways. Oncogene. 2015;34:2586–2596. doi: 10.1038/onc.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samaranayake G.J., Huynh M., Rai P. MTH1 as a chemotherapeutic target: the elephant in the room. Cancers. 2017;9:47. doi: 10.3390/cancers9050047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed W., Lingner J. PRDX1 and MTH1 cooperate to prevent ROS-mediated inhibition of telomerase. Gene Dev. 2018;32:658–669. doi: 10.1101/gad.313460.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Waals L.M., Laoukili J., Jongen J.M.J., Raats D.A., Borel Rinkes I.H.M., Kranenburg O. Differential anti-tumour effects of MTH1 inhibitors in patient-derived 3D colorectal cancer cultures. Sci Rep. 2019;9:819. doi: 10.1038/s41598-018-37316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gul N., Karlsson J., Tängemo C., Linsefors S., Tuyizere S., Perkins R. The MTH1 inhibitor TH588 is a microtubule-modulating agent that eliminates cancer cells by activating the mitotic surveillance pathway. Sci Rep. 2019;9:14667. doi: 10.1038/s41598-019-51205-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gad H., Mortusewicz O., Rudd S.G., Stolz A., Amaral N., Brautigham L. MTH1 promotes mitotic progression to avoid oxidative DNA damage in cancer cells. BioRxiv. 2019 doi: 10.1101/575290. Available from: [DOI] [Google Scholar]
- 50.McPherson L.A., Troccoli C.I., Ji D., Bowles A.E., Gardiner M.L., Mohsen M.G. Increased MTH1-specific 8-oxodGTPase activity is a hallmark of cancer in colon, lung and pancreatic tissue. DNA Repair. 2019;83:102644. doi: 10.1016/j.dnarep.2019.102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arczewska K.D., Stachurska A., Wojewódzka M., Karpińska K., Kruszewski M., Nilsen H. hMTH1 is required for maintaining migration and invasion potential of human thyroid cancer cells. DNA Repair. 2018;69:53–62. doi: 10.1016/j.dnarep.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 52.Li L., Song L., Liu X., Yang X., Li X., He T. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J., Jiang C., Figueiró Longo J.P., Azevedo R.B., Zhang H., Muehlmann L.A. An updated overview on the development of new photosensitizers for anticancer photodynamic therapy. Acta Pharm Sin B. 2018;8:137–146. doi: 10.1016/j.apsb.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y., Zhao D., Wang G., Wang Y., Cao L., Sun J. Recent progress of hypoxia-modulated multifunctional nanomedicines to enhance photodynamic therapy: opportunities, challenges, and future development. Acta Pharm Sin B. 2020;10:1382–1396. doi: 10.1016/j.apsb.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan H., Zhang L., Hu X., Zhao Z., Bai H., Fu X. MTH1-targeted nanosystem for enhanced PDT via improving cellular sensitivity to reactive oxygen species. Chem Commun. 2018;54:4310–4313. doi: 10.1039/c8cc01841c. [DOI] [PubMed] [Google Scholar]
- 56.Zhao L., Li J., Su Y., Yang L., Chen L., Qiang L. MTH1 inhibitor amplifies the lethality of reactive oxygen species to tumor in photodynamic therapy. Sci Adv. 2020;6:eaaz0575. doi: 10.1126/sciadv.aaz0575. [DOI] [PMC free article] [PubMed] [Google Scholar]