Abstract
The human gut microbiota metabolizes the Parkinson’s disease medication Levodopa (L-dopa), potentially reducing drug availability and causing side effects. However, the organisms, genes, and enzymes responsible for this activity in patients and their susceptibility to inhibition by host-targeted drugs are unknown. Here, we describe an interspecies pathway for gut bacterial L-dopa metabolism. Conversion of L-dopa to dopamine by a pyridoxal phosphate-dependent tyrosine decarboxylase from Enterococcus faecalis is followed by transformation of dopamine to m-tyramine by a molybdenum-dependent dehydroxylase from Eggerthella lenta. These enzymes predict drug metabolism in complex human gut microbiotas. Although a drug that targets host aromatic amino acid decarboxylase does not prevent gut microbial L-dopa decarboxylation, we identified a compound that inhibits this activity in Parkinson’s patient microbiotas and increases L-dopa bioavailability in mice.
INTRODUCTION
Parkinson’s disease is a debilitating neurological condition affecting more than 1% of the global population aged 60 and above. The primary medication used to treat Parkinson’s disease is levodopa (L-dopa). To be effective, L-dopa must enter the brain and be converted to the neurotransmitter dopamine by the human enzyme aromatic amino acid decarboxylase (AADC). However, the gastro-intestinal tract is also a major site for L-dopa decarboxylation, and this metabolism is problematic because dopamine generated in the periphery cannot cross the blood-brain barrier and causes unwanted side effects. Thus, L-dopa is coadministered with drugs that block pe-ripheral metabolism, including the AADC in-hibitor carbidopa. Even with these drugs, up to 56% of L-dopa fails to reach the brain. Moreover, the efficacy and side effects of L-dopa treatment are extremely heterogeneous across Parkinson’s patients, and this variability cannot be completely explained by differences in host metabolism
RATIONALE
Previous studies in humans and animal models have demonstrated that the gut microbiota can metabolize L-dopa. The major proposed pathway involves an initial decarboxylation of L-dopa to dopamine, followed by conversion of dopamine to m-tyramine by means of a distinctly microbial dehydroxylation reaction. Although these metabolic activities were shown to occur in complex gut microbiota samples, the specific organisms, gene, and enzymes responsible were unknown. The effects of host-targeted inhibitors such as carbidopa on gut microbial L-dopa metabolism were also unclear. As a first step toward understanding the gut microbiota’s effect on Parkinson’s disease therapy, we sought to elucidate the molecular basis for gut microbial L-dopa and dopamine metabolism.
RESULTS
Hypothesizing that L-dopa decarboxylation would require a pyridoxal phosphate (PLP)–dependent enzyme, we searched gut bacterial genomes for candidates and identified a conserved tyrosine decarboxylase (TyrDC) in Enterococcus faecalis. Genetic and biochemical experiments revealed that TyrDC simultaneously decarboxylates both L-dopa and its preferred substrate, tyrosine. Next, we used enrichment culturing to isolate a dopamine dehydroxylating strain of Eggerthella lenta, a species previously implicated in drug metabolism. Transcriptomics linked this activity to a molybdenum cofactor–dependent dopamine dehydroxylase (Dadh) enzyme. Unexpectedly, the presence of this enzyme in gut bacterial genomes did not correlate with dopamine metabolism; instead, we identified a single-nucleotide polymorphism (SNP) in the dadh gene that predicts activity. The abundance of E. faecalis, tyrDC, and the individual SNPs of dadh correlated with L-dopa and dopamine metabolism in complex gut microbiotas from Parkinson’s patients, indicating that these organisms, genes, enzymes, and even nucleotides are relevant in this setting.
We then tested whether the host-targeted AADC inhibitor carbidopa affected L-dopa decarboxylation by E. faecalis TyrDC. Carbidopa displayed greatly reduced potency toward bacteria and was completely ineffective in complex gut microbiotas from Parkinson’s patients, suggesting that this drug likely does not prevent microbial L-dopa metabolism in vivo. To identify a selective inhibitor of gut bacterial L-dopa decarboxylation, we leveraged our molecular understanding of gut microbial L-dopa metabolism. Given TyrDC’s preference for tyrosine, we examined tyrosine mimics and found that (S)-α-fluoromethyltyrosine (AFMT) prevented L-dopa decarboxylation by TyrDC and E. faecalis as well as complex gut microbiota samples from Parkinson’s patients. Coadministering AFMT with L-dopa and carbidopa to mice colonized with E. faecalis also increased the peak serum concentration of L-dopa. This observation is consistent with inhibition of gut microbial L-dopa metabolism in vivo.
CONCLUSION
We have characterized an interspecies pathway for gut bacterial L-dopa metabolism and demonstrated its relevance in human gut microbiotas. Variations in these microbial activities could possibly contribute to the heterogeneous responses to L-dopa observed among patients, including decreased efficacy and harmful side effects. Our findings will enable efforts to elucidate the gut microbiota’s contribution to treatment outcomes and highlight the promise of developing therapies that target both host and gut microbial drug metabolism.
Gut microbes metabolize the Parkinson’s drug L-dopa. Decarboxylation of L-dopa by E. faecalis TyrDC and human AADC likely limits drug availability and contributes to side effects. E. lenta dehydroxylates dopamine produced from L-dopa using a molybdenum-dependent enzyme. Although the host-targeted drug carbidopa did not affect gut bacterial L-dopa decarboxylation, AFMT inhibited this activity in complex human gut microbiotas.
A growing body of evidence links the trillions of microbes that inhabit the human gastrointestinal tract (the human gut microbiota) to neurological conditions, including the debilitating neurodegenerative disorder Parkinson’s disease (1, 2). Gut microbes from Parkinson’s patients exacerbate motor deficits when transplanted into germ-free mouse models of disease (2). This effect is reversed with antibiotic treatment, suggesting a causal role for gut microbes in neurodegeneration. Multiple studies have revealed differences in gut microbiota composition in Parkinson’s disease patients compared with healthy controls that may correlate with disease severity (3-9). However, the influence of the human gut microbiota on the treatment of Parkinson’s and other neurodegenerative diseases remains poorly understood.
The primary treatment for Parkinson’s disease is Levodopa (L-dopa) (10), which is prescribed to manage motor symptoms that result from dopaminergic neuron loss in the substantia nigra. After crossing the blood-brain barrier, L-dopa is decarboxylated by aromatic amino acid decarboxylase (AADC) to give dopamine, the active therapeutic agent. However, dopamine generated in the periphery by AADC cannot cross the blood-brain barrier, and only 1 to 5% of L-dopa reaches the brain, owing to extensive presystemic metabolism in the gut by enzymes such as AADC (11-13). Peripheral production of dopamine also causes gastrointestinal side effects, can lead to orthostatic hypotension through activation of vascular dopamine receptors, and may induce cardiac arrhythmias (14,15). To decrease peripheral metabolism, L-dopa is coadministered with AADC inhibitors such as carbidopa. Despite this, 56% of L-dopa is metabolized peripherally (16), and patients display highly variable responses to the drug, including loss of efficacy over time (17).
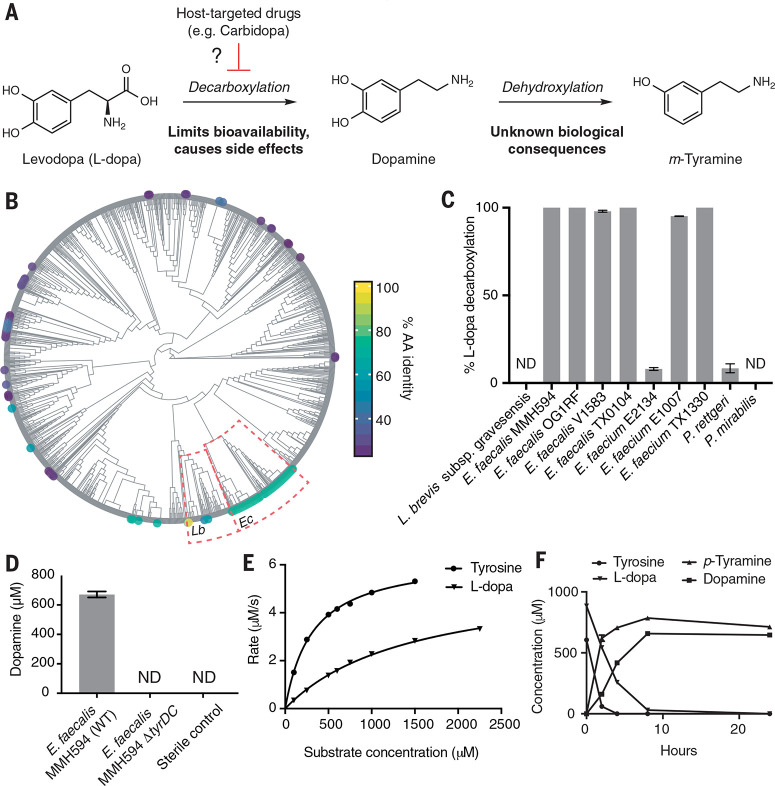
Multiple lines of evidence suggest that gut microbial interactions with L-dopa influence treatment outcomes (18). Administering broad-spectrum antibiotics improves L-dopa therapy, suggesting that gut bacteria interfere with drug efficacy (19, 20). The gut microbiota can also metabolize L-dopa, potentially reducing its bioavailability and leading to side effects (21-24). The major proposed pathway involves an initial decarboxylation of L-dopa to dopamine followed by a distinctly microbial dehydroxylation reaction that converts this neurotransmitter to m-tyramine by selectively removing the para hydroxyl group of the catechol ring (Fig. 1A) (25,26). When we began our work, the gut microbial species, genes, and enzymes involved in these transformations were unknown because previous studies examined undefined and uncharacterized consortia. The clinical relevance of this pathway was also unclear given the potential effects of coadministered inhibitors of host peripheral L-dopa metabolism on these gut microbial activities.
Fig. 1.
E. faecalis metabolizes L-dopa using a PLP-dependent tyrosine decarboxylase. (A) Proposed major pathway for L-dopa metabolism by the human gut microbiota and potential for interaction with host-targeted drugs. (B) Phylogenetic distribution of TyrDC in the human microbiota. Human Microbiome Project reference genomes were queried by means of BLASTP for homologs of the L. brevis TyrDC, and the results are visualized on a cladogram of phylogeny [based on 16S ribosomal RNA (rRNA) alignment]. TyrDC homologs found sporadically within Lactobacillus spp. (Lb) are widely distributed among Enterococcus (Ec; average amino acid identity 67.8% over 97.6% query length). (C) Testing representative gut microbial strains encoding TyrDC reveals that E. faecalis strains reproducibly convert L-dopa to dopamine. Strains were cultured for 48 hours anaerobically. Bar graphs represent the mean ± SEM of three biological replicates. (D) Deletion of tyrDC abolishes L-dopa decarboxylation by E. faecalis. Dopamine was detected in culture supernatants after 48 hours of anaerobic growth with 0.5 mM L-dopa. Bar graphs represent the mean ± SEM of three biological replicates. (E) Kinetic analysis of E. faecalis TyrDC reveals a preference for tyrosine. Error bars represent the mean ± SEM of three biological replicates. ND, not detected. (F) L-dopa and tyrosine are simultaneously decarboxylated in anaerobic cultures of E. faecalis MMH594 grown at pH 5 with 1 mM L-dopa and 0.5 mM tyrosine. Bar graphs represent the mean ± SEM of three biological replicates.
The human gut bacterium Enterococcus faecalis decarboxylates L-dopa
We sought to elucidate the genetic and biochemical bases for gut microbial L-dopa metabolism and understand how coadministered AADC inhibitors affect this pathway. Using a genome-mining approach, we first identified strains that encode candidate L-dopa decarboxylating enzymes. Aromatic amino acid decarboxylation is typically performed by enzymes using pyridoxal-5'-phosphate (PLP), an organic cofactor that provides an electron sink (27). Recently, a PLP-dependent tyrosine decarboxylase (TyrDC) from the food-associated strain Lactobacillus brevis CGMCC 1.2028 was shown to have promiscuous activity toward L-dopa in vitro (28). To locate TyrDC homologs in human gut bacteria, we performed a BLASTP (Protein Basic Local Alignment Search Tool) search against the complete set of Human Microbiome Project (HMP) reference genomes available through the National Center for Biotechnology Information (NCBI). The majority of hits were found in the neighboring genus Enterococcus, with some hits within lactobacilli and Proteobacteria (Fig. 1B, fig. S1, and data file S1). We selected 10 representative gut strains that contain TyrDC homologs (29 to 100% amino acid ID) and examined their ability to decarbox-ylate L-dopa in anaerobic culture. Although both Enterococcus faecalis and Enterococcus faecium displayed activity, only E.faecalis showed complete decarboxylation across all strains tested (Fig. 1C). All E. faecalis strains tested share the highly conserved four-gene tyrDC operon (fig. S2), and we found tyrDC in 98.4% of the E. faecalis assemblies deposited in NCBI with a median amino acid identity of 99.8 (range 97.0 to 100). This high degree of sequence conservation and prevalence is consistent with tyrosine decarboxylation being a common phenotypic trait of E. faecalis (29). We therefore chose this prevalent, genetically tractable gut organism as a model for characterizing L-dopa decarboxylation (30).
Although lyophilized E. faecalis cells decarbox-ylate L-dopa (31) and the tyrDC operon’s role in tyrosine decarboxylation in E. faecalis is well-characterized (32), the connection between tyrDC and L-dopa decarboxylation was unknown. We used genetics and in vitro biochemistry experiments to confirm that TyrDC is necessary and sufficient for L-dopa decarboxylation by E.faecalis. E. faecalis MMH594 mutants carrying a 2-kb Tet-cassette disrupting tyrDC could not decarboxylate L-dopa (Fig. 1D and fig. S3) and displayed no growth defects compared with wild type (fig. S4). In vitro characterization of TyrDC revealed a fivefold higher catalytic efficiency toward L-tyrosine compared with L-dopa, suggesting that drug metabolism arises from promiscuous enzyme activity (Fig. 1E, fig. S5, and table S1). This selectivity contrasts sharply with that of AADC, which displays very low activity toward L-tyrosine (33). Although TyrDC from E. faecalis was previously shown to decarboxylate tyrosine and phenylalanine (34-37), its ability to accept L-dopa had not been demonstrated. A recent, independent report also corroborates this finding (38).
We next tested whether tyrosine, which is the preferred substrate for TyrDC and is present in the small intestine, could interfere with L-dopa decarboxylation by E.faecalis (39, 40). In competition experiments, purified TyrDC (fig. S6) and anaerobic E.faecalis cultures decarboxylated L-dopa and tyrosine simultaneously (500 mM tyrosine, approximating the resting small intestinal concentration) (Fig. 1F and fig. S7) (40). This observation sharply contrasts with previous investigations of phenylalanine, which is metabolized by E.faecalis only when tyrosine is completely consumed (36). Simultaneous decarboxylation of L-dopa and tyrosine also occurred in E. faecalis MMH594 cultures that contained higher tyrosine concentrations (1.5 mM, approximating the small intestinal post-meal concentration) (fig. S8) and in three human fecal suspensions (fig. S9). As observed previously for tyrosine, L-dopa decarboxylation occurred more rapidly at lower pH across all strains tested (figs S7 and S8), suggesting that this metabolism is likely accelerated at the lower pH of the upper small intestine (41,42). Because the Michaelis constant (Km) of TyrDC for L-dopa (1.5 mM) is below the estimated maximum in vivo small intestinal L-dopa concentration even at its lowest clinically administered dose (5 mM), these data strongly suggest that peripheral decarboxylation is performed by both host and gut bacterial enzymes.
Eggerthella lenta dehydroxylates dopamine using a molybdenum-dependent enzyme
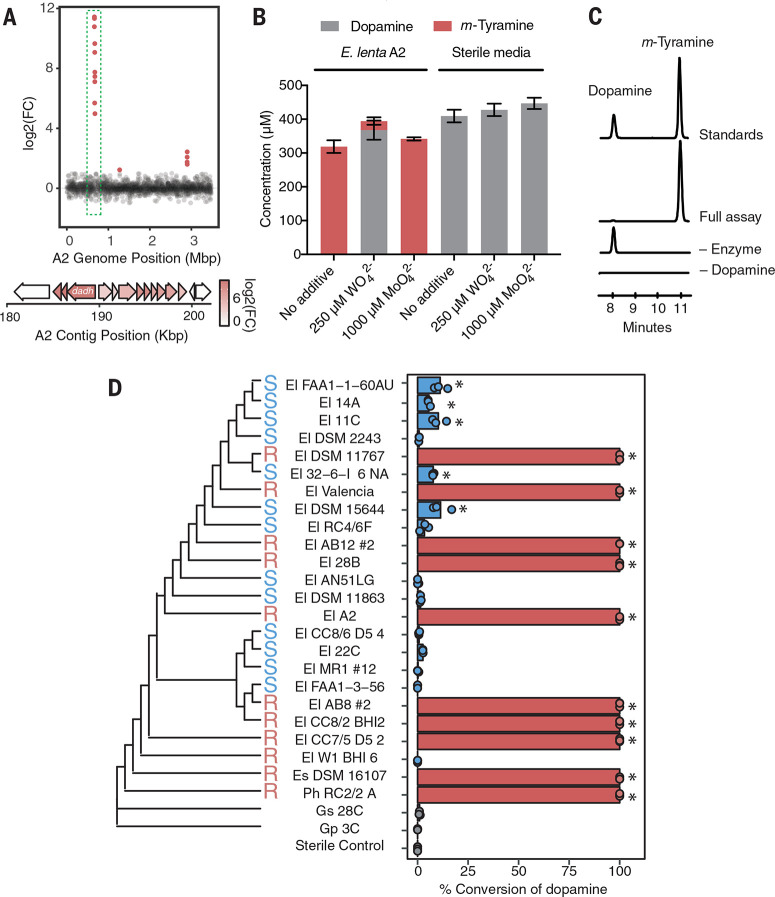
Having identified a gut bacterial L-dopa decarboxylase, we next examined the conversion of dopamine to m-tyramine because this activity may influence the side effects associated with peripheral L-dopa decarboxylation. E. faecalis did not further metabolize dopamine, indicating that this step was performed by a different microorganism. Dehydroxylation of dopamine has not been reported for any bacterial isolate, and a screen of 18 human gut strains failed to uncover metabolizers. Therefore, we used enrichment culturing to obtain a dopamine-dehydroxylating organism. Recognizing the chemical parallels between this reductive dehydroxylation and reductive dehalogenation of chlorinated aromatics, which enables anaerobic respiration in certain bacteria (43), we inoculated a stool sample from a human donor into a minimal growth medium containing 0.5 mM dopamine as the sole electron acceptor (figs. S10 and S11). Passaging over multiple generations enriched for active strains, as assessed by means of a colorimetric assay for catechol dehydroxylation (fig. S11). This effort identified a strain of the gut Actinobacterium Eggerthella lenta (referred to herein as strain A2) that is capable of selectively removing the para hydroxyl group of dopamine to give m-tyramine (fig. S12). Because E. lenta also inactivates the cardiac drug digoxin, our finding suggests a wider role for this gut organism in drug metabolism (44, 45).
Catechol dehydroxylation is a chemically challenging reaction that has no equivalent in synthetic chemistry and likely involves unusual enzymology. To identify the dopamine-dehydroxylating enzyme, we first searched the E. lenta A2 genome for genes that encode homologs of the only characterized aromatic para-dehydroxylase, 4-hydroxybenzoyl-CoA reductase (46), but found no hits. Assays with E. lenta A2 cell lysates showed dopamine dehydroxylation required anaerobic conditions and was induced by dopamine (fig. S13). We therefore used RNA-sequencing of E. lenta A2 to identify the de hydroxylase. This experiment revealed >2500-fold up-regulation of three colocalized genes in response to dopamine (Fig. 2A and table S2). These genes encode a predicted bis-molybdopterin guanine dinucleotide cofactor (moco)-containing enzyme belonging to the dimethyl sulfoxide reductase family. Moco-dependent enzymes catalyze a wide variety of oxygen-transfer reactions but have not been demonstrated to catalyze catechol dehydroxylation in vitro (47). We therefore hypothesized that this enzyme was a dopamine dehydroxylase (Dadh).
Fig. 2.
E. lenta dehydroxylates dopamine using a molybdenum-dependent enzyme. (A) RNAsequencing identifies a putative molybdenum (moco)–dependent dopamine dehydroxylase (Dadh) in E. lenta A2. Differentially expressed candidate genes (false discovery rate < 0.1 and fold change > j2j) are plotted as a function of genome position, revealing three discrete loci of differentially expressed genes. (Inset) Analysis of the largest cluster of differentially expressed genes at 0.665 Mbp in the scaffolded assembly (190 kg base pairs in the reference contig) revealed that a putative dadh was up-regulated by 2568-fold in response to dopamine. (B) Tungstate treatment inhibits dehydroxylation of dopamine by E. lenta A2. Cultures were grown anaerobically with tungstate (WO42–) or molybdate (MoO42–) for 48 hours with 0.5 mM dopamine. Bar graphs represent the mean ± SEM of three biological replicates. (C) In vitro activity of Dadh-containing fractions purified from E. lenta A2. Extracted LC-MS/MS ion chromatograms for simultaneous detection of dopamine and m-tyramine after 12 hours of anaerobic incubation of enzyme preparation with 500 mM dopamine and artificial electron donors at room temperature. Peak heights show the relative intensity of each mass, and all chromatograms are shown on the same scale. (D) A single amino acid variant predicts dopamine metabolism by E. lenta and related strains (P = 0.013 Fisher’s exact test) and does not correlate with phylogeny. Strains were cultured anaerobically with 500 mM dopamine for 48 hours (El, E. lenta; Es, Eggerthella sinensis; Gs, Gordonibacter sp.; and Gp, Gordonibacter pamelaeae; Ph, Paraeggerthella hongkongensis). High (100% conversion) and low (<11% conversion) metabolizers are denoted in red and blue. For each strain, data points represent biological replicates (P *< 0.05 analysis of variance with Dunnett’s test versus sterile controls).
To assess Dadh’s role in dopamine dehydroxylation, we first explored whether this activity was molybdenum-dependent by culturing E. lenta A2 in the presence of tungstate. Substitution of molybdate with tungstate during moco biosyn thesis generates an inactive metallocofactor (fig. S14) (48). Treating cultures of E. lenta A2 with tungstate inhibited dopamine dehydroxylation without affecting growth (Fig. 2B and fig. S15), whereas incubating cell lysates with tungstate had no effect, which is consistent with inhibition requiring active moco biosynthesis (fig. S16). We next confirmed the activity of Dadh in vitro. Heterologous expression of >20 constructs in multiple hosts failed to provide active enzyme, prompting us to pursue native purification. Anaerobic activity-guided fractionation of E. lenta A2 cell lysates yielded a dopamine-dehydroxylating fraction containing four proteins as assessed by means of SDS-polyacrylamide gel electrophoresis (Fig. 2C, fig. S17, and table S3). Dehydroxylation activity correlated with a 115-kDa band that was confirmed with mass spectrometry (MS) to be Dadh. Dadh was the only isolated protein up-regulated in the presence of dopamine (tables S2 and S3). Together, these data strongly support the assignment of this enzyme.
We next assessed whether the presence of dadh in microbial genomes correlated with dopamine dehydroxylation. A BLASTP search revealed that this enzyme is restricted to E. lenta and its close Actinobacterial relatives (table S4), prompting us to screen a collection of 26 gut Actinobacterial isolates (49) for their ability to dehydroxylate dopamine in anaerobic culture. Although Dadh appeared to be encoded by 24 of the 26 strains (92 to 100% amino acid ID) (fig. S18 and table S5), only 10 Eggerthella strains quantitatively converted dopamine to m-tyramine, with low (<11%) or no metabolism in the others (Fig. 2D). This strain-level variability in dopamine metabolism reinforces that gut microbial species identity is often not predictive of metabolic functions (49, 50).
To better understand this variation, we first performed RNA-sequencing experiments with metabolizing (E. lenta 28B) and nonmetabolizing (E. lenta DSM2243) strains in the presence and absence of dopamine. Surprisingly, dadh was up-regulated in response to dopamine in both strains, indicating that lack of activity in E. lenta DSM2243 did not arise from differences in transcription (tables S6 and S7). Aligning the Dadh protein sequences, we instead found a single amino acid substitution that almost perfectly predicted metabolizer status: Position 506 is an arginine in metabolizing strains and a serine in inactive strains (Fig. 2D and fig. S19). This change arises from a single-nucleotide polymorphism (SNP) in dadh. The only exception, E. lenta W1BHI6, has the Arg506 variant and an additional substitution nearby (Cys500) (fig. S19). Thus, specific amino acid residues in the Dadh enzyme, rather than presence or transcription of dadh, predict dopamine dehydroxylation among gut bacterial strains. The Dadh variants do not correlate with E. lenta phylogeny (Fig. 2D), suggesting that this activity has been gained and/or lost multiple times.
E. faecalis and E. lenta metabolize L-dopa in human gut microbiotas
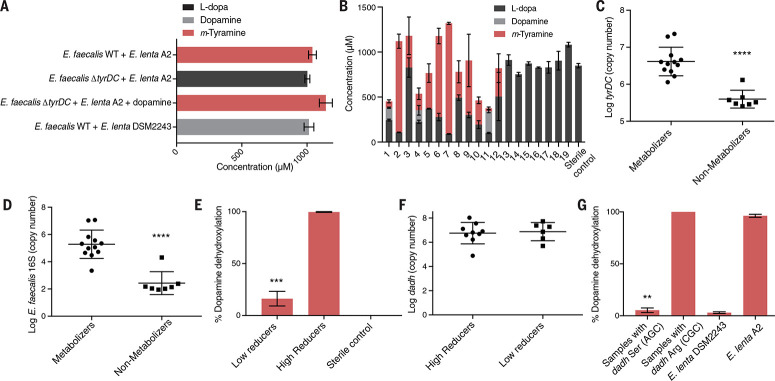
Having identified organisms and enzymes that perform the individual steps in the L-dopa pathway, we next tested whether E. faecalis and E. lenta generated m-tyramine in coculture. Wild-type E. faecalis grown with E. lenta A2 (Arg506) fully converted L-dopa to m-tyramine (Fig. 3A). Although a coculture containing the E. faecalis tyrDC mutant could not consume L-dopa, m-tyramine was produced when exogenous dopamine was added to this culture, revealing that E. lenta A2 was still metabolically active. Incubating wild-type E.feacalis with the nonmetabolizing E. lenta DSM2243 (Ser506) strain produced only dopamine, indicating that this Dadh variant is also inactive in a coculture setting (Fig. 3A).
Fig. 3.
E. faecalis and E. lenta Dadh predict L-dopa metabolism in complex human gut microbiotas. (A) Metabolismof L-dopa by cocultures of E. faecalis and E. lenta strains cocultured for 48 hours with 1mMd3-phenyl-L-dopa or 1 mM dopamine. Results are mean ± SEM (n = 3 replicates). (B) Metabolism of d3-phenyl-L-dopa by 19 unrelated human gut microbiota samples ex vivo. Samples were cultured anaerobically with d3-phenyl-L-dopa (1 mM) for 72 hours. Results are mean concentration ± SEM (n = 3 replicates). (C) The abundance of tyrDC predicts L-dopa decarboxylation in human gut microbiota samples. Data represent the average tyrDC abundance (as assessed with qPCR) across the three replicates for samples in (B). Results are mean ± SEM (****P < 0.0001, one tailed Mann-Whitney test). (D) The abundance of E. faecalis (as assessed with qPCR) predicts L-dopa decarboxylation in human gut microbiota samples. Each data point is the average abundance across three biological replicates for each sample shown in (B). Results are mean ± SEM (****P < 0.0001, one-tailed Mann-Whitney test). (E) Dopamine dehydroxylation by gut microbiota samples of 15 unrelated individuals. Samples were cultured for 48 hours with 0.5 mM dopamine. Bars are mean ± SEM of n = 6 for low reducers (<50%) and n = 9 for high reducers (>50%) (*** P = 0.0002, one tailed Mann-Whitney test). (F) Dadh abundance does not correlate with dehydroxylation by human gut microbiotas. Data represent qPCR with Dadh-specific primers. Each data point is the dadh abundance in each sample shown in (E). Bars represent the mean and SE. (G) Dadh sequence variants predict dopamine dehydroxylation ex vivo. Full-length dadh from each culture in (E) was sequenced by using primers specific for the region containing position 506. Samples in which a mix of variants were present (n = 5) were removed. Bars represent the mean and SEM [n = 3 for samples encoding the Arg506 Dadh variant, n = 7 for samples encoding the Ser506 Dadh variant., n = 3 for DSM2243, and n = 3 for A2] (** P = 0.0083, one-tailed Mann-Whitney test, CGC samples versus AGC samples).
To investigate whether E. faecalis and E. lenta transform L-dopa in the human gut microbiota, we assessed the metabolism of deuterated L-dopa by fecal suspensions ex vivo. Whereas 7 of 19 samples did not show detectable depletion of L-dopa, the remaining samples displayed substantial variability in metabolism, ranging from partial (25%) to almost full conversion (98%) of L-dopa to m-tyramine (Fig. 3B). We next asked whether the abundance of tyrDC predicted metabolism in these samples. Quantitative polymerase chain reaction (qPCR) enumeration of tyrDC (51) and E. faecalis discriminated metabolizing and nonmetabolizing samples (P < 0.0001, onetailed Mann-Whitney test) (Fig. 3, C and D). By contrast, E. lenta abundance showed no association with L-dopa metabolism (fig. S20). We found a strong linear correlation between tyrDC abundance and E. faecalis abundance [coefficient of determination (R2) = 0.99, P < 0.0001] (fig. S21), which likely reflects the high conservation of tyrDC in E. faecalis genomes. These data also suggest that E. faecalis is the dominant microorganism responsible for L-dopa decarboxylation in these complex human gut microbial communities. Consistent with this, E. faecalis abundance significantly correlated with tyrDC abundance in 1870 human gut microbiomes (R2 > 0.812, P < 2.2 x 10-16, Pearson’s correlation) (fig. S22).
To confirm that E. faecalis could decarboxylate L-dopa in complex gut microbiotas, we added this organism to nonmetabolizing samples. Although introducing the tyrDC-deficient strain did not change L-dopa levels, including the wild-type strain led to complete depletion of L-dopa (fig. S23, B to E). In some samples, addition of wild-type E. faecalis was sufficient to yield quantitative production of m-tyramine, indicating the presence of dopamine-dehydroxylating organisms in these communities (fig. S23, B and D). Last, addition of both the wild-type E. faecalis and the metabolizing strain E. lenta A2 to nonmetabolizing samples or the addition of E. lenta A2 alone to a decarboxylating sample generated m-tyramine (fig. S23, A and C to E). Taken together, these data indicate that the abundance of E. faecalis and its encoded tyrDC predicts the considerable interindividual variation in L-dopa metabolism observed in complex human gut microbiota samples.
As expected from our previous experiments, neither the abundance of E. lenta nor dadh predicted dopamine dehydroxylation in complex gut microbial communities (Fig. 3, E and F, and fig. S24). However, when we amplified dadh from these cultures and determined the SNP status at position 506, we found samples that contained the Arg506 variant quantitatively metabolized dopamine, whereas the activity of samples that carried the Ser506 variant was indistinguishable from the nonmetabolizing E. lenta DSM2243 strain (Fig. 3G). These findings indicate that a single amino acid residue in a gut microbial enzyme predicts dopamine metabolism in complex communities. Given that dadh is highly prevalent (>70%) in gut microbiomes from human subjects and the two dadh variants are present among this population (figs. S22 and S25), we speculate that SNPs may influence xenobiotic metabolism in the context of both the host genome (52) and the human gut microbiome (53).
To further explore the clinical relevance of our findings, we assessed themetabolismof dopamine and L-dopa by fecal suspensions from Parkinson’s disease patients ex vivo. Similar to control subjects, these individuals displayed seubstantial variability in metabolism of L-dopa (fig. S26A). qPCR assays revealed that tyrDC abundance and E. faecalis abundance discriminated between L-dopa decarboxylating and nondecarboxylating samples (P < 0.005, one-tailed Mann-Whitney test) (fig. S26, C and D).We also observed depletion of L-dopa without corresponding production of dopamine or m-tyramine in three samples (fig. S26A). Instead, L-dopa was converted to hydroxyphenylpropionic acid (fig. S26B), a pathway thought to make a minor contribution to drug metabolism in vivo (22, 25, 26). Last, we found that the dadh SNP predicted dopamine dehydroxylation in these samples (fig. S27). Overall, these data support a role for gut bacteria in the extensive interindividual variability in L-dopa decarboxylation observed in Parkinson’s patients (13). A recent study reported that stool tyrDC abundance is positively correlated with L-dopa dosage in patients (38) but did not demonstrate a connection between tyrDC and L-dopa decarboxylation in these samples. Our findings indicate this metabolic activity may indeed affect L-dopa therapeutic efficacy.
(S)-α-Fluoromethyltyrosine (AFMT) inhibits gut microbial L-dopa metabolism
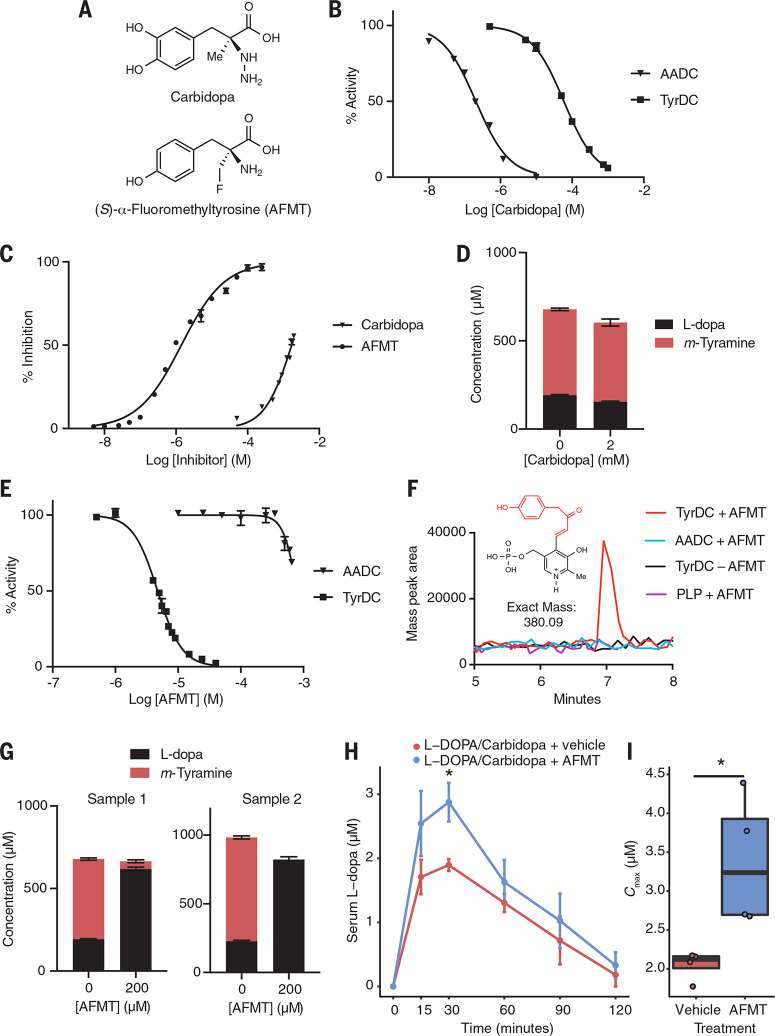
Having shown that E. faecalis and E. lenta enzymes predict L-dopa metabolism by complex patient gut microbiotas, we next investigated whether this interspecies pathway was susceptible to inhibition by drugs that target peripheral L-dopa decarboxylation. In the United States, Parkinson’s patients are coprescribed carbidopa (Fig. 4A), an L-dopa mimic that inhibits AADC by forming a stable, covalent hydrazone linkage with its PLP cofactor (54). We found carbidopa was 200 times less active towardpurified E. faecalis TyrDC [half maximal inhibitory concentration (IC50) = 57 μM] relative to H. sapiens AADC (IC50 = 0.21 μM) and showed only ~50% inhibition of L-dopa decarboxylation by E. faecalis cultures at the solubility limit of 2 mM (Fig. 4, B and C, and table S8),which is consistent with recently reported findings (38). Additionally, carbidopa did not affect growth of E. faecalis or metabolism or growth of E. lenta (figs. S28 to S30). Given the maximum predicted gastrointestinal concentration of car-bidopa (0.4 to 9 mM), these data suggest that this drug does not fully inhibit gut bacterial L-dopa decarboxylation in Parkinson’s patients. We found that 2 mM carbidopa did not alter the kinetics of L-dopa degradation (fig. S31) or endpoint m-tyramine production in stool samples from both Parkinson’s patients and neurologically healthy controls (Fig. 4D and fig. S32). These observations support previous findings that carbidopa administration does not affect m-tyramine production in patients (55).
Fig. 4.
L-dopa decarboxylation by E. faecalis is inhibited by AFMT but not the host-targeted drug carbidopa. (A) Carbidopa and AFMT. (B) Carbidopa preferentially inhibits human AADC over TyrDC. AADC or TyrDC were incubated with inhibitor, and reaction rates were measured with LC-MS/MS. “% Activity” represents the rate relative to a no inhibitor (vehicle) control. Results are mean ± SEM (n = 3 replicates). (C) Activity of carbidopa and AFMT in cultures of E. faecalis grown for 16 hours anaerobically with 0.5 mM L-dopa. Error bars represent the mean ± SEM for three biological replicates. (D) Activity of carbidopa in a human fecal microbiota from a Parkinson’s patient. The sample was cultured anaerobically with carbidopa and 1 mM d3-phenyl-L-dopa for 72 hours. Error bars represent the mean ± SEM for three biological replicates. (E) AFMT preferentially inhibits TyrDC over AADC in vitro. AADC or TyrDC were incubated with inhibitor, and reaction rates were measured with LC-MS/MS. “% Activity” represents the rate relative to a no inhibitor (vehicle) control. Error bars represent the mean ± SEM for three biological replicates. (F) Detection of an AFMT-PLP covalent adduct after incubation of TyrDC or AADC with AFMT for 1 hour. The data shown is the extracted ion chromatogram of the mass of the predicted covalent adduct. (G) Action of AFMT in human fecal microbiotas from Parkinson’s patients incubated anaerobically with AFMT and 1 mM d3-phenyl-L-dopa for 72 hours. Error bars represent the mean ± SEM for three biological replicates. (H) Pharmacokinetic analysis in gnotobiotic mice colonized with E. faecalis and given L-dopa + carbidopa + AFMT demonstrates higher serum L-dopa relative to vehicle controls. Error bars represent the mean ± SEM. (I) The maximum serum concentration (Cmax) of L-dopa is significantly higher with AFMT relative to vehicle controls. In (H) and (I), *P < 0.05, Mann-Whitney U test; n = 4 to 5 mice per group.
Our results also highlight the possibility of therapeutically targeting gut microbial L-dopa decarboxylation to increase L-dopa efficacy. To selectively manipulate gut bacterial TyrDC in complex microbiotas, we turned to α-fluoromethyl amino acids, which are known mechanism-based inhibitors of PLP-dependent decarboxylases (33).
A survey of potential amino acid substrates revealed that TyrDC requires a p-hydroxyl group for robust activity, whereas AADC prefers a m-hydroxyl substituent (fig. S33), leading us to hypothesize that the L-tyrosine analog (S)-α-fluoromethyltyrosine (AFMT) (Fig. 4A) might selectively inhibit the microbial enzyme. In vitro, AFMT strongly inhibited L-dopa decarboxylation by TyrDC (IC50 = 4.7 mM) but not AADC (~20% inhibition at solubility limit of 650 mM) (Fig. 4E and table S8). Consistent with this selectivity, AFMT formed a covalent PLP adduct only in the presence of TyrDC (Fig. 4F). AFMT was also effective in E.faecalis cultures (EC50 = 1.4 mM) (Fig. 4C), outperforming carbidopa by 1000-fold without affecting growth (table S8 and fig. S29).
It also reduced L-dopa decarboxylation by cocultures of E.faecalis and E. lenta without affecting growth or metabolism of E. lenta (figs. S29, S30, and S34). Last, AFMT completely inhibited L-dopa decarboxylation in gut microbiota samples from Parkinson’s disease patients and neurologically healthy control subjects (Fig. 4G and fig. S35) and was nontoxic to eukaryotic cells (fig. S36).
To investigate AFMT activity in vivo, we administered either AFMT (25 mg/kg) or a vehicle control in combination with L-dopa (10 mg/kg) and carbidopa (30 mg/kg) to gnotobiotic mice colonized with E.faecalis MMH594 (Fig. 4H). We found that AFMT significantly increased the peak serum concentration (Cmax) of L-dopa compared with vehicle (P < 0.05, two-tailed Mann Whitney test) (Fig. 4I), which is consistent with inhibition of first-pass gut microbial metabolism in the intestine. Although we cannot rule out the possibility that AFMT modulates additional, uncharacterized targets, this observation is consistent with our in vitro inhibition data. This result also aligns with a recent report that small intestinal tyrDC abundance negatively correlates with plasma L-dopa levels in conventional rats receiving L-dopa and carbidopa (38). Overall, these data suggest that AFMT could be a promising tool compound for the study of bacterial L-dopa metabolism (56) and highlight the promise of developing L-dopa-based combination therapies containing drugs that target both host and gut microbial decarboxylation.
Conclusions
We have used chemical knowledge and interdisciplinary tools to decipher the molecular mechanisms by which gut bacteria interfere with the treatment of Parkinson’s disease. The decarboxylation of L-dopa by E.faecalis mirrors host drug metabolism and, together with human AADC, likely limits drug availability and contributes to interindividual variation in efficacy. Together with recent work dissecting host and gut microbial contributions to the antiviral drug brivudine (57), our findings show that gut bacterial metabolism need not be chemically distinct from host activities to alter drug efficacy and suggest that such interactions may be underappreciated. Moreover, carbidopa’s failure to prevent L-dopa decarboxylation by E. faecalis implies that additional host-targeted drugs may lack efficacy toward activities also present in the gut microbiota. Although a recent, independent study also characterized E. faecalis TyrDC’s role in L-dopa decarboxylation and its lack of susceptibility to carbidopa (38), it did not show that this activity occurs in human gut microbiotas or identify strategies for inhibiting the bacterial enzyme. By contrast, we demonstrate that TyrDC predicts drug metabolism in Parkinson’s patient microbiotas and use an understanding of its substrate specificity to identify a small molecule that prevents L-dopa decarboxylation in patient samples and increases L-dopa bioavailability in vivo. Through discovery of predictive biomarkers for L-dopa metabolism and identification of an inhibitor of this activity, this work will enable efforts to elucidate the contribution of the gut microbiota to drug availability, patient drug response, and treatment outcomes.
We also show that E. lenta further metabolizes the dopamine produced by L-dopa decarboxylation using a distinctly microbial reaction, catechol dehydroxylation. It is possible that this transformation influences the multiple side effects of L-dopa administration linked to dopamine production. This discovery also raises questions about the biological consequences of gut microbial metabolism of endogenous dopamine, which is present in the gastrointestinal tract and has been linked to phenotypes ranging from gut motility to pathogen colonization (58-60). The biological activity of the gut microbial metabolite m-tyramine in the host and the benefits of this metabolism for E. lenta are also poorly understood. Our findings will enable further study of these phenomena. Given that gut microbes de-hydroxylate catechol groups found in numerous aromatic drugs and dietary compounds (18,61-63), the discovery of Dadh will enable identification of additional catechol dehydroxylases and help to elucidate the biological role of this enigmatic transformation. Uncovering the unexpected effect of SNPs on gut microbial dopamine metabolism suggests that simply detecting functional genes may not accurately predict the activities encoded by the human gut microbiome and underscores the importance of studying enzymes from this community.
Materials and methods summary
Our methods for the identification and biochemical characterization of E.faecalis TyrDC; characterization of anaerobic L-dopa metabolism by E.faecalis and gut microbiota samples; enrichment culturing for dopamine dehydrox-ylating organisms; RNA-sequencing; culture-based assays; purification of Dadh; assays of anaerobic dopamine metabolism by Actinobacteria and complex gut microbiota samples; PCR and qPCR assays; liquid chromatography-MS (LC-MS) methods; and assays for evaluating inhibitors in vitro, ex vivo, and in vivo are provided in the supplementary materials. Additional information about our protocols, including references to the supplementary materials, can be found throughout the main text.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Broad Institute Microbial Omics Core (MOC) for assistance with RNA and 16S rRNA gene sequencing analysis and experimental design, the Harvard Bauer Core Proteomics facility for assistance with proteomics, M. Wilson (Harvard University) for helpful discussions and input, M. Gilmore and Selleck (Massachusetts Eye and Ear, Harvard Medical School, and Broad Institute) for supplying the E. faecalis tyrDC mutant and helpful discussions, F. Lebreton (Massachusetts Eye and Ear and Harvard Medical School) for supplying E. faecalis and E. faecium strains, R. Nayak [University of California, San Francisco (UCSF)] and M. Krueger (UCSF) for help with sample collection from healthy control subjects, and the Biocollective for collection and provision of stool samples from Parkinson’s patients. We thank Merck for the gift of AFMT.
Funding
This work was supported by the Packard Fellowship for Science and Engineering (2013-39267) (E.P.B), the Howard Hughes Medical Institute (HHMI)-Gates Faculty Scholars Program (OPP1158186) (E.P.B), the National Institutes of Health (R01HL122593) (P.J.T.), the Searle Scholars Program (SSP-2016-1352) (P.J.T), the UCSF-Stanford Arthritis Center of Excellence (supported in part by the Arthritis Foundation) (P.J.T), and the Rheumatology Research Foundation (P.J.T.). V.M.R. is the recipient of a National Science Foundation Graduate Research Fellowship, a Gilliam Fellowship from HHMI, and the Ardis and Robert James Graduate Research Fellowship from Harvard University and acknowledges support from a National Institutes of Health Training Grant (5T32GM007598-38). E.N.B. is a Howard Hughes Medical Institute fellow of the Life Sciences Research Foundation. J.E.B. has fellowship support from the Natural Sciences and Engineering Research Council of Canada. P.J.T. is a Chan Zuckerberg Biohub investigator and a Nadia’s Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42-16). This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visitto figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Author contributions
V.M.R. and E.P.B. conceived of the project. V.M.R. purified and characterized all proteins biochemically with and without inhibitors, performed assays for L-dopa and dopamine metabolism in pure cultures and complex microbiotas in the presence and absence of inhibitors, performed qPCR experiments and analysis, and performed RNA-sequencing experiments in E. lenta A2 and E. lenta 28B. E.N.B. performed RNA-sequencing experiments in E. lenta DSM2243, performed assays for dopamine metabolism across the Actinobacterial library, and contributed to the design of the ex vivo experiments and other culture-based assays. J.E.B. performed RNA sequencing analysis, comparative genomics, and metagenomic analysis and contributed to the design of and conducted in vivo AFMT experiments. V.M.R., E.P.B., E.N.B., J.E.B., and P.J.T. provided critical feedback on experiments. V.M.R., J.E.B., P.J.T., and E.P.B. wrote the manuscript. Competing interests: E.P.B. has consulted for Merck, Novartis, and Kintai Therapeutics; is on the Scientific Advisory Boards of Kintai Therapeutics and Caribou Biosciences; and is an Associate Member of the Broad Institute of Harvard and MIT. P.J.T. is on the scientific advisory board for Kaleido, Seres, uBiome, and WholeBiome. Data and materials availability: The E. lenta A2 genome has been deposited into GenBank (PRJNA412637). RNA-sequencing data has been deposited into the Sequence Read Archive available by way of BioProjec PRJNA507796. The small-molecule AFMT was obtained under a materials transfer agreement with Merck.
References And Notes
- 1.Sharon G., Sampson T. R., Geschwind D. H., Mazmanian S. K., The central nervous system and the gut microbiome.Cell 167, 915-932 (2016). doi: 10.1016/j.cell.2016.10.027;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sampson T. R., et al. , Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167,1469—1480.e12 (2016). doi: 10.1016/j.cell.2016.11.018;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheperjans F., et al. , Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 30, 350—358. doi: 10.1002/mds.26069;pmid: [DOI] [PubMed] [Google Scholar]
- 4.Unger M. M., et al. , Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Parkinsonism Relat. Disord. 32, 66—72. doi: 10.1016/j.parkreldis.2016.08.019;pmid: [DOI] [PubMed] [Google Scholar]
- 5.Bedarf J. R., et al. , Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson’s disease patients. Genome Med. 9, 39 (2017). doi: 10.1186/s13073-017-0428-y;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill-Burns E. M., et al. , Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 32, 739—749 (2017). doi: 10.1002/mds.26942;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minato T. ,et al., Progression of Parkinson’s disease is associated with gut dysbiosis: Two-year follow-up study. PLOS ONE 12, e0187307 (2017). doi: 10.1371/journal.pone.0187307;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrov V. A., et al. , Analysis of gut microbiota in patients with Parkinson’s disease. Bull. Exp. Biol. Med. 162, 734—737 (2017). doi: 10.1007/s10517-017-3700-7;pmid: [DOI] [PubMed] [Google Scholar]
- 9.Heintz-Buschart A., et al. , The nasal and gut microbiome in Parkinson’s disease and idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 33, 88—98 (2018). doi: 10.1002/mds.27105;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornykiew O.icz, L-DOPA. J. Parkinsons Dis. 7 (s1), S3—S10. doi: 10.3233/JPD-179004;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papavasiliou P. S., et al. , Levodopa in Parkinsonism: Potentiation of central effects with a peripheral inhibitor. N. Engl. J. Med. 286, 8—14 (1972). doi: 10.1056/NEJM197201062860102;pmid: [DOI] [PubMed] [Google Scholar]
- 12.Gey K. F., Pletscher A., Distribution and metabolism of DL-3,4-dihydroxy[2-14C]-phenylalanine in rat tissues. Biochem. J. 92, 300—308 (1964). doi: 10.1042/bj0920300;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann S., et al. , The absorption and metabolism of a standard oral dose of levodopa in patients with Parkinsonism. Br. J. Clin. Pharmacol. 1, 417—424 (1974). doi: 10.1111/j.1365-2125.1974.tb00280.x;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldenberg M. M., Medical management of Parkinson’s disease. P&T 33, 590—606 (2008). [PMC free article] [PubMed] [Google Scholar]
- 15.Whitfield A. C., Moore B. T., Daniels R. N., Classics in chemical neuroscience: Levodopa. ACS Chem. Neurosci. 5, 1192—1197 (2014). doi: 10.1021/cn5001759;pmid: [DOI] [PubMed] [Google Scholar]
- 16.SINEMET CR, (carbidopa-levodopa) [package insert]. Princeton: Bristol-Myers Squibb Company, NJ; 2008. [Google Scholar]
- 17.Gray R., et al. , Long-term effectiveness of dopamine agonists and monoamine oxidase B inhibitors compared with levodopa as initial treatment for Parkinson’s disease (PD MED): A large, open-label, pragmatic randomised trial. Lancet 384, 1196-1205 (2014). doi: 10.1016/S0140-6736(14)60683-8;pmid: [DOI] [PubMed] [Google Scholar]
- 18.Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J., The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273-287 (2016). doi: 10.1038/nrmicro.2016.17;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashim H., et al. , Eradication of Helicobacter pylori infection improves levodopa action, clinical symptoms and quality of life in patients with Parkinson’s disease. PLOS ONE 9, e112330 (2014). doi: 10.1371/journal.pone.0112330;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasano A., et al. , The role of small intestinal bacterial overgrowth in Parkinson’s disease. Mov. Disord. 28, 1241-1249 (2013). doi: 10.1002/mds.25522;pmid: [DOI] [PubMed] [Google Scholar]
- 21.Deeds F., Booth A. N., Jones F. T., Methylation and dehydroxylation of phenolic compounds by rats and rabbits. J. Biol. Chem. 225, 615-621 (1957). [PubMed] [Google Scholar]
- 22.Goldin B. R., Peppercorn M. A., Goldman P., Contributions of host and intestinal microflora in the metabolism of L-dopa by the rat. J. Pharmacol. Exp. Ther. 186, 160-166 (1973). [PubMed] [Google Scholar]
- 23.Sandler M., Goodwin B. L., Ruthven C. R., Calne D. B., Therapeutic implications in Parkinsonism of m-tyramine formation from L-dopa in man. Nature 229, 414-415 (1971). doi: 10.1038/229414a0;pmid: [DOI] [PubMed] [Google Scholar]
- 24.Sandler M., Ruthven C. R., Goodwin B. L., Hunter K. R., Stern G. M., Variation of levodopa metabolism with gastrointestinal absorption site. Lancet 1, 238-240 (1974). doi: 10.1016/S0140-6736(74)92547-1;pmid: [DOI] [PubMed] [Google Scholar]
- 25.Goldman P., Peppercorn M. A., Goldin B. R., Metabolism of drugs by microorganisms in the intestine. Am. J. Clin. Nutr. 27, 1348-1355 (1974). doi: 10.1093/ajcn/27.11.1348;pmid: [DOI] [PubMed] [Google Scholar]
- 26.Goodwin B. L., Ruthven C. R. J., King G. S., Sandler M., Leask B. G. S., Metabolism of 3, 4-dihydroxyphenylalanine, its metabolites and analogues in vivo in the rat: Urinary excretion pattern. Xenobiotica 8, 629-651 (1978). doi: 10.3109/00498257809069575;pmid: [DOI] [PubMed] [Google Scholar]
- 27.Eliot A. C., Kirsch J. F., Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383-415 (2004). doi: 10.1146/annurev.biochem.73.011303.074021;pmid: [DOI] [PubMed] [Google Scholar]
- 28.Zhu H. ,et al., Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding. Sci. Rep. 6, 27779 (2016). doi: 10.1038/srep27779;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladero V., et al., Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 30, 132-138 (2012). doi: 10.1016/j.fm.2011.12.016;pmid: [DOI] [PubMed] [Google Scholar]
- 30.Lebreton F., et al., Tracing the Enterococci from Paleozoic origins to the hospital. Cell 169, 849-861.e13 (2017). doi: 10.1016/j.cell.2017.04.027;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blashcko H., Substrate specificity of amino acid decarboxylases. Biochim. Biophys. Acta 4, 130-137 (1950). doi: 10.1016/0006-3002(50)90016-310.1016/0006-3002(50)90016-3;pmid: [DOI] [PubMed] [Google Scholar]
- 32.Connil N., et al., Identification of the Enterococcus faecalis tyrosine decarboxylase operon involved in tyramine production. Appl. Environ. Microbiol. 68, 3537-3544 (2002). doi: 10.1128/AEM.68.7.3537-3544.2002;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kollonitsch J., et al., Selective inhibitors of biosynthesis of aminergic neurotransmitters. Nature 274, 906-908 (1978). doi: 10.1038/274906a0;pmid: [DOI] [PubMed] [Google Scholar]
- 34.B0rresen T., Klausen N. K., Larsen L. M., S0rensen H., Purification and characterisation of tyrosine decarboxylase and aromatic-L-amino-acid decarboxylase. Biochim. Biophys. Acta 993, 108-115 (1989). doi: 10.1016/0304-4165(89)90149-9;pmid: [DOI] [PubMed] [Google Scholar]
- 35.Liu F., et al., Heterologous expression and characterization of tyrosine decarboxylase from Enterococcus faecalis R612Z1 and Enterococcus faecium R615Z1. J. Food Prot. 77, 592-598 (2014). doi: 10.4315/0362-028X.JFP-13-326;pmid: [DOI] [PubMed] [Google Scholar]
- 36.Pessione E., et al., First evidence of a membrane-bound, tyramine and beta-phenylethylamine producing, tyrosine decarboxylase in Enterococcus faecalis: A two-dimensional electrophoresis proteomic study. Proteomics 9, 2695-2710 (2009). doi: 10.1002/pmic.200800780;pmid: [DOI] [PubMed] [Google Scholar]
- 37.Allenmark S., Servenius B., Characterization of bacterial L-(-)-tyrosine decarboxylase by isoelectric focusing and gel chromatography. J. Chromatogr. 153, 238-245 (1978). doi: 10.1016/S0021-9673(00)89878-7;pmid: [DOI] [PubMed] [Google Scholar]
- 38.van Kessel S. P., et al., Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat. Commun. 10, 310 (2019). doi: 10.1038/s41467-019-08294-y;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abrams W. B., Coutinho C. B., Leon A. S., Spiegel H. E., Absorption and metabolism of levodopa. JAMA 218,1912-1914 (1971). doi: 10.1001/jama.1971.03190260028007;pmid: [DOI] [PubMed] [Google Scholar]
- 40.Adibi S. A., Mercer D. W., Protein digestion in human intestine as reflected in luminal, mucosal, and plasma amino acid concentrations after meals. J. Clin. Invest. 52, 1586–1594 (1973). doi: 10.1172/JCI107335;pmid:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez M., et al., Tyramine biosynthesis is transcriptionally induced at low pH and improves the fitness of Enterococcus faecalis in acidic environments. Appl. Microbiol. Biotechnol. 99, 3547–3558 (2015). doi: 10.1007/s00253-014-6301-7;pmid: [DOI] [PubMed] [Google Scholar]
- 42.Fallingborg J., Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 46, 183–196 (1999). [PubMed] [Google Scholar]
- 43.Holliger C., Wohlfarth G., Diekert G., Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22, 383–398 (1998). doi: 10.1111/j.1574-6976.1998.tb00377.x [DOI] [Google Scholar]
- 44.Koppel N., Bisanz J. E., Pandelia M. E., Turnbaugh P. J., Balskus E. P., Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. eLife 7, e33953 (2018). doi: 10.7554/eLife.33953;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haiser H. J., et al., Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013). doi: 10.1126/science.1235872;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boll M., Schink B., Messerschmidt A., Kroneck P. M. H., Novel bacterial molybdenum and tungsten enzymes: Three-dimensional structure, spectroscopy, and reaction mechanism. Biol. Chem. 386, 999–1006 (2005). doi: 10.1515/BC.2005.116;pmid: [DOI] [PubMed] [Google Scholar]
- 47.Rothery R. A., Weiner J. H., Shifting the metallocentric molybdoenzyme paradigm: The importance of pyranopterin coordination. J. Biol. Inorg. Chem. 20, 349–372 (2015). doi: 10.1007/s00775-014-1194-6;pmid: [DOI] [PubMed] [Google Scholar]
- 48.Zhu W., et al., Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018). doi: 10.1038/nature25172;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bisanz J. E., et al. , Illuminating the microbiome’s dark matter: A functional genomic toolkit for the study of human gut Actinobacteria. bioRxiv 10.1101/304840 (2018); accessed 18 April 2019]. doi: 10.1101/304840 [DOI] [Google Scholar]
- 50.Martinez-del Campo A., et al., Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6, e00042–e15 (2015). doi: 10.1128/mBio.00042-15;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torriani S., et al., Rapid detection and quantification of tyrosine decarboxylase gene (tdc) and its expression in gram-positive bacteria associated with fermented foods using PCR-based methods. J. Food Prot. 71, 93–101 (2008). doi: 10.4315/0362-028X-71.1.930362-028X-71.1.93;pmid: [DOI] [PubMed] [Google Scholar]
- 52.Preissner S. C., et al., Polymorphic cytochrome P450 enzymes (CYPs) and their role in personalized therapy. PLOS ONE 8, e82562 (2013). doi: 10.1371/journal.pone.0082562;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins J., et al., Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553, 291–294 (2018). doi: 10.1038/nature25178;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burkhard P., Dominici P., Borri-Voltattorni C., Jansonius J. N., Malashkevich V. N., Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol. 8, 963–967 (2001). doi: 10.1038/nsb1101-963;pmid: [DOI] [PubMed] [Google Scholar]
- 55.Sandler M., Johnson R. D., Ruthven C. R. J., Reid J. L., Calne B., Transamination is a major pathway of L-dopa metabolism following peripheral decarboxylase inhibition. Nature 247, 364–366 (1974). doi: 10.1038/247364b0;pmid: [DOI] [PubMed] [Google Scholar]
- 56.Wallace B. D., et al., Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). doi: 10.1126/science.1191175;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A. L., Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 363, eaat9931 (2019). doi: 10.1126/science.aat9931;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eisenhofer G., et al., Substantial production of dopamine in the human gastrointestinal tract. J. Clin. Endocrinol. Metab. 82, 3864–3871 (1997). doi: 10.1210/jcem.82.11.4339;pmid: [DOI] [PubMed] [Google Scholar]
- 59.Li Z. S., Schmauss C., Cuenca A., Ratcliffe E., Gershon M. D., Physiological modulation of intestinal motility by enteric dopaminergic neurons and the D2 receptor: Analysis of dopamine receptor expression, location, development, and function in wild-type and knock-out mice. J. Neurosci. 26, 2798–2807 (2006). doi: 10.1523/JNEUR0SCI.4720-05.2006;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dichtl S., et al., Dopamine is a siderophore-like iron chelator that promotes Salmonella enterica serovar typhimurium virulence in mice. mBio 10, e02624–e18 (2019). doi: 10.1128/mBio.02624-18;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheline R. R., Williams R. T., Wit J. G., Biological dehydroxylation. Nature 188, 849–850 (1960). doi: 10.1038/188849a0;pmid: [DOI] [PubMed] [Google Scholar]
- 62.Sweeny D. J., et al., Metabolism of fostamatinib, the oral methylene phosphate prodrug of the spleen tyrosine kinase inhibitor R406 in humans: Contribution of hepatic and gut bacterial processes to the overall biotransformation. Drug Metab. Dispos. 38, 1166–1176 (2010). doi: 10.1124/dmd.110.032151;pmid: [DOI] [PubMed] [Google Scholar]
- 63.Koppel N., Maini Rekdal V., Balskus E. P., Chemical transformation of xenobiotics by the human gut microbiota. Science 356, eaag2770 (2017). doi: 10.1126/science.aag2770;pmid: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.