Abstract
The Coronavirus disease 2019 (COVID-19) outbreak, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), had emerged as a pandemic affecting almost all countries in the world in a short span after it was first reported in December. Clinical laboratory have a crucial role in mitigating this new pandemic. Timely and accurate diagnosis of COVID-19 is of paramount importance for detecting cases early and to prevent transmission. Clinical Laboratories have adopted different test modalities and processes to tackle this unprecedented situation with directives from regulatory bodies such as the WHO. The varying presentations, as well as complications attributed to comorbidities in COVID-19, have created hurdles in the management of these patients. Various clinical laboratory parameters have been investigated for their potential for diagnosis and prognosis of the disease, prediction of complications and monitoring of treatment response. Different routine and uncommon parameters have been shown to have the diagnostic and prognostic capacity. This update discusses the role of the laboratory in diagnosis, prognosis and monitoring of treatment response. Different methodologies for diagnostic testing as well as various clinical laboratory parameters having diagnostic and predictive powers have been discussed. This compilation organises relevant available information on various clinical laboratory parameters and their role in COVID-19 mitigating pandemic.
Key words: COVID-19, SARS-CoV-2, laboratory, clinical parameter, diagnosis, prognosis
1. INTRODUCTION:
Novel Coronavirus induced pneumonia, which was given the name of coronavirus disease 2019 (COVID-19) by the WHO on the 11th of February 2020, has rapidly amplified to the full scale of a pandemic since it was first reported in Wuhan, China, back in December 2019 (1,2). COVID-19 is the clinical syndrome associated with SARS-CoV-2 infection. The disease signifies a respiratory syndrome starting from mild upper respiratory illness to severe pneumonia and acute respiratory distress syndrome (ARDS). SARS-CoV-2 belongs to the beta coronavirus genus of the coronaviruses. Although Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) also belongs to the same genus, SARS-CoV-2 leads to milder infections.
However, SARS-CoV-2 have a broader community transmission when compared with SARS and MERS. Hence, laboratory testing is of paramount importance to distinguish between COVID-19 and other respiratory diseases. Moreover, extensive testing will help in COVID diagnosis and a better understanding of disease prevalence in asymptomatic infections. As of November 10, 2020, there have been over 50 million confirmed cases of COVID-19 and over 1.2 million deaths across the world. Contribution of Laboratory medicine in diagnosis, prognosis, risk prediction and management is indispensable in most of the human pathologies, and COVID-19 is not an exception. The current COVID-19 pandemic has reconfirmed that laboratory diagnostics will remain the core of every clinical decision made. This review covers recent laboratory modalities available for diagnosis, prognosis and monitoring of treatment response in COVID-19.
2. LABORATORY TESTING IN COVID-19
Clinical Laboratories are of paramount importance in mitigating the COVID-19 pandemic. From early diagnosis, Clinical Laboratories play a crucial role in monitoring comorbidities, diagnosing complications, assessment of treatment responses and in assessing the prevalence of diseases in the community. Timely and accurate diagnosis of the disease is essential for early initiation of treatment as well as to prevent the transmission to contacts.
Different counties had followed and implemented different testing strategies targeting different genes based on the availability of diagnostic methods and consumables (Table 1). Further, the WHO has meanwhile taken strict steps and created the diagnostics available with the mission to “detect, protect and treat” to break the chain of transmission of SARS-CoV-2(3). Early diagnosis and immediate treatment will significantly decrease future COVID-19 cases. Therefore, early laboratory diagnosis of SARS-CoV-2 plays a vital role in controlling the COVID-19 pandemic.
Table 1.
Currently targeting different genes by the different country protocol as per WHO
| Country | Institute | Targeting gene | References |
|---|---|---|---|
| China | China CDC | ORF 1ab and N genes | (4) |
| Hong Kong SAR | Hong Kong University | ORF 1b-nsp14, N genes | (5) |
| Germany | Charitè | RdRp, E, N genes | (6) |
| Japan | National Institute of Infectious Diseases | N gene | (7) |
| Thailand | National Institute of Health | N gene | (8) |
| USA | US CDC | Three targets in N gene (N1, N2, and N3) RP-RNase | (9) |
3.1 DIAGNOSTIC METHODS FOR COVID-19
Compared to symptomatic testing and CT scan method, the molecular techniques are more appropriate in accurate diagnosis since they target the identification of pathogens (Figure 1). Despite this, the Real-time reverse transcriptase-polymerase chain reaction (rRT–PCR) serves as a gold standard method for nucleic acid screening of SARS–CoV-2. Since this a time consuming and sophisticated method, rRT-PCR serves better as a diagnostic tool than a screening tool(10). Considering the current stage of the pandemic, a large number of patient screening is needed using novel screening methods which require lesser equipment and materials.
Figure 1.
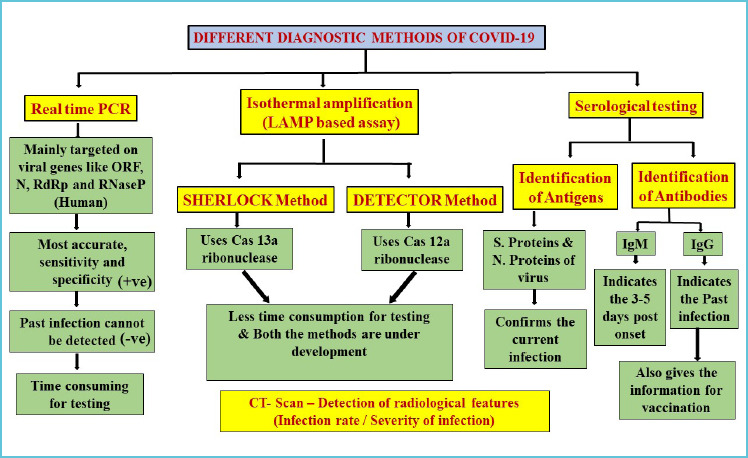
Different diagnostic methods of COVID-19
The advancement of molecular techniques is mainly reliant on understanding the genomic and proteomic composition of the pathogen. Similarly, the changes in the host gene or protein expressions induced by the pathogen after infection (11). World Health Organization (WHO) and China jointly described genome sequence of SARS-CoV-2 and its genetic characterisation (12,13). This genome sequencing has given a road map to researchers for designing primers and probe sequences for rRT-PCR and some other nucleic acid amplification tests.
3.2 SPECIMEN COLLECTION, SAMPLE STORAGE AND SAMPLE STABILITY
The World Health Organization (WHO), Centre for Disease Control and Prevention (CDC) and Indian Council of Medical Research (ICMR) have recommended a set of guidelines to collect the samples from patients affected or suspected by COVID-19 (14–16). For the safety of clinicians and researchers, it is highly necessary and suggested that the specimens should collected in a BSL-2 laboratory. It is compulsory by law for the individuals concerned or suspected to cooperate appropriately with health departments for collection, storage and shipment of the specimens. In the case of unavailability of immediate testing, store the specimens at 2-8°C for up to 72 hours after collection. If a delay in testing or shipping is expected, store specimens at -70°C or below. Rodino et al demonstrated the stability and reliable detection of SARS-CoV-2 RNA in stored swabs in viral transport medium, saline, PBS and minimal essential medium after seven days at 2-8°C and frozen at –20°C using an in-house Emergency Use Authorization (EUA) assay as well as the Roche Cobas EUA assay (17). In another study Perchetti GA et al. shown that SARS-CoV-2 stability can be retained at 4°C for up to a month if the storage of -80°C is not available (18).
The sample should be isolated from two main sites, the lower respiratory or upper respiratory tract, depending on WHO’s suggested guidelines. The nasopharyngeal or oropharyngeal swab specimens should be collected from the upper respiratory tract, while tracheal aspirate, bronchoalveolar lavage, and sputum should be collected from the lower respiratory tract. Bronchoalveolar lavage fluid specimens remain the ideal sample for detection of COVID-19. Sputum, nasal swabs, fibre bronchoscope brush biopsy, pharyngeal swabs and faeces demonstrated different rates of positivity for COVID-19 virus. Urine was not found to be a suitable sample for detection of COVID-19 (19).
3.3 NUCLEIC ACID AMPLIFICATION TEST (NAAT)
Since the outbreak of COVID-19, nucleic acid amplification testing is the primary method of diagnosis. Multiple real-time reverse transcription-polymerase chain reaction (rRT-PCR) kits have been invented to detect SARS-CoV-2. Corman et al. aligned and scrutinised SARS-CoV-2 related viral genome sequences to construct a set of oligo primers and probe sequences(6). Among these mainly three conserved sequences have been revealed. 1) In open reading frame ORF1ab region the RdRP gene (RNA-dependent RNA polymerase gene), 2) Envelope protein gene (E gene), 3) Nucleocapsid protein gene (N gene) (6). Different countries submitted their primary probe designs to the WHO. As an example, the rRT-PCR can be designed as two genes target system or three genes target systems, where one primer set detects family of coronaviruses, the second set detects specifically SARS-CoV-2 and third is human RNase P as the internal control (Table 1). Similarly, ICMR also released some recommendations for COVID-19 diagnosis. The ICMR has recommended the use of US-based RT-PCR probes distributed to national laboratories (16).
The United States Centers for Disease Control and Prevention (CDC) set up a panel of genes through RT-PCR for the specific finding of SARS-CoV-2 and overall detection of SARS-like beta coronaviruses (9). Primarily designed by targeting three different sets of primers to the N gene among these two primers sets are specific to SARS-CoV-2 and one primer set is specific to all beta coronaviruses. If all three genes are positive, then it specifies the COVID-19 confirmation. Similarly, in Germany Charite (6) developed two sets of nucleic acid tests for detection of SARS-CoV-2, SARS-CoV and bat-like beta-CoVs by targeting the RdRp and E genes, if both tests were positive then COVID-19 confirmation through SARS-CoV-2 specific RdRp gene. The results of the Chu et al. study suggested targeting the N gene as primary screening and ORF1ab as a confirmative target. Studies targeted at two or more genes, thus had a stronger outcome performance compared to single genes alone (20). Now, molecular testing was developed as the gold standard for the diagnosis of SARS-CoV-2, hence the E and RdRb genes suggesting better analytical sensitivity compared to the N and ORF1ab genes combination.
While different institutions have developed various SARS-CoV-2 research protocols, it remains uncertain if the findings of nucleic acid tests based on multiple targets are comparable. In a recent study compared the analytical sensitivities of the United States, Germany, Hong Kong and China qRT-PCR assays by using RNA transcripts isolated from a COVID-19 patient (21). They found that all primer-probe sets used in the qRT-PCR tests could detect SARS-CoV-2, but the significant difference was observed in the limit of detection (LOD) and the ability to distinguish the positives and negatives while the viral load is at lower levels. The highest sensitivity of primer-probe sets was found E-gene (Germany), N1 gene (US CDC), ORF1 (Hongkong) but RdRp gene (Germany) showed the lowest sensitivity. In another study from Germany Konrad et al. found that by using a single-step qRT-PCR method, the E gene target was more sensitive than the RdRp target (22).
3.4 DIRECT RT-PCR
The positive controls (2019-nCoV pseudovirus) provide a nucleic acid extraction and a reverse transcription control to validate the entire procedure and reagent integrity. Similarly, the RNAse P internal control provides an RNA extraction of practical control and secondary negative control. However, RNA extraction from clinical samples creates a major bottleneck in the diagnostic process, as it either runs manually and thus is laborious or automated and expensive. To overcome this, recently, some research groups developed direct RT-PCR by omitting RNA extraction procedure (23-25). In this method, after the collection of patient material and deposition of potential SARS-CoV-2 viral particles in transport medium followed by the inactivation of the virus through detergent/chaotropic reagents or heating process step. Then, transfer the lysate to single-step RT-PCR format in which cDNA synthesis by RT and detection by qPCR may take place. Wee Sk et al. showed that direct RT-PCR has a high sensitivity of 6 RNA copies per reaction and is quantitative over a dynamic range of 7 orders of magnitude (25). Direct amplification of SARS-CoV-2 viral RNA from samples without RNA purification allows the reducing hands-on-time, time-to-results, and costs.
As per WHO guidelines, one of the following conditions should be met for considering a case as a NAAT-confirmed laboratory in areas with no circulation of SARS-CoV-2(18).
1) A positive NAAT result for at least two different targets on the SARS-CoV-2 virus genome, of which at least one target is preferably specific for SARS-CoV-2 virus using a validated assay;
2) One positive NAAT result for the presence of beta coronavirus, and SARS-CoV-2 virus further identified by sequencing partial or whole genome of the virus as long as the sequence target is larger or different from the amplicon probed in the NAAT assay used.
At the moment, it’s important to identify that a negative result may not eliminate the possibility of COVID-19, it might be due to the poor-quality specimens, early or late collection, inadequate sample, and incorrect test procedures. When a patient with a high level of suspicion obtains a negative result for SARS-CoV-2 virus infection, especially when only upper respiratory tract specimens have been collected, additional specimens should be collected and tested, including, where possible, from the lower respiratory tract (26).
3.5 LOOP-MEDIATED ISOTHERMAL AMPLIFICATION
Isothermal amplification depended nucleic acid tests are currently under progression for SARS-CoV-2. Recently a few studies reported the development of reverse transcription LAMP (RT-LAMP) tests(27–29) and some are clinically tested for SARS-CoV-2(30,31).
Primarily RT-LAMP is based on the DNA polymerase and 4-6 primers to bind at distinct regions on the target genome. RT-LAMP is a highly specific method since it uses a greater number of primers, like two inner primers and two outer primers on different regions on the genome. In LAMP diagnostic tests, SARS-CoV-2 family genes such as ORF1ab, spike (S), envelope (E) or/and N gene can be targeted, and the procedure will be done in a single step at 63 °C isothermal conditions, and within 15-40 minutes the results will be obtained (27,28,30,31). For the POCT of SARS-CoV-2, many institutes are keen to implement isothermal nucleic acid amplification technology, eliminating the need for a highly costly thermal cycler. The most promising alternative to PCR may be loop-mediated isothermal amplification (LAMP) because it provides many advantages in terms of precision, sensitivity, reaction efficiency and product yield. Recently, a reverse transcription (RT)-LAMP assay targeting non-structural protein 3 (Nsp3) for SARS-CoV-2 detection was developed by Park et al., whose LOD was 100 copies per reaction (32). Similarly, RT-LAMP assay within 60 min targeting an ORF1ab and the S gene, whereby the LOD was 20 copies/reaction and 200 copies/reaction, was prepared by Yan et al. (33).
3.6 CRISPR BASED METHODS
Along with isothermal amplification, another category of nucleic acid tests that could be used for SARS-CoV-2 detection based on dyes employing inherent by-products of comprehensive DNA synthesis, such as calcein, malachite orange, and hydroxynaphthol blue can be utilised for performing visual detection methods. Clustered regularly interspaced short palindromic repeats (CRISPR) based diagnostic tests have been developed for point-of-care nucleic acid detection (34), such as Specific High-sensitivity Enzymatic Reporter unlocking (SHERLOCK) or DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR).
CRISPR based method nucleic acid tests mainly in a combination of Recombinase Polymerase Amplification with CRISPR–Cas enzymology for specific recognition of targeted RNA or DNA sequences (34). In SHERLOCK testing strategy is based on Cas13a ribonuclease for RNA sensing (35). Recently, studies have reported the development and evaluation of a CRISPR based Diagnostic For 2019-Novel Coronavirus (36). Similar to the SHERLOCK method, CRISPR–Cas12-based assay was developed termed as DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter). Broughton et al. reported the development and initial validation of a CRISPR–Cas12-based assay for detection of SARS-CoV-2 from extracted patient sample RNA (37). In addition to that, Broughton et al. compared the detection strategies of DETECTR, and the RT-qPCR which is recommended by CDC/WHO for SARS-CoV-2 detection, however, they found that the limit of detection these methods is ten copies/μL, 1 or 3.2 copies/μL input sample, respectively. Also, the assays turnaround time is 45 min and four hours, respectively (37). Since less time consumption and equipment requirement, these methods can be set up in emergency departments and local community hospitals. Recently, Hou et al. exploited polymerase mediated amplification by the combination of recombinase polymerase amplification (RPA) and CRISPR-Cas13-mediated enzymatic signal amplification for detection of SARS-CoV-2 with high sensitivity and 7.5 copies/reaction within 40 min. The CRISPR-Cas13-based assay has a higher detection potential than the RT-PCR assay, according to a comparative clinical study. (38). In another study, Ding at al developed the protocol by integrating RT-RPA and CRISPR-based detection in a one-pot reaction and incubating at a single temperature (39). This “All-In-One Dual CRISPR-Cas12a” (AIOD-CRISPR) assay detected as little as 4.6 SARS-CoV-2 RNA copies per μL input at 40 minutes per μL input.
3.7 SEROLOGY TESTING
It is emphasised that nucleic acid-based testing methods need to extract nucleic acid in advance, the requirement of trained technicians, complex operation, expensive equipment; it is complicated to do in epidemiological and surveillance purposes. With the aid of viral protein antigen and antibodies which are produced in response to a SARS-CoV-2 infection can be used for diagnosis. Since variations in the viral load throughout infection, it may difficult to detect the viral proteins. In contrast to this, the detection of antibodies which are generated to viral proteins may enable the indirect ways to detect SARS-CoV-2.
Serology testing involves the screening test by qualitative assays and measurement of different classes of immunoglobulins (IgA, IgM, IgG) against SARS-CoV-2 by using quantitative assays for establishing whether a person has been infected by SARS-CoV-2. Zhang et al. detected immunoglobulin G and M (IgG and IgM) from the human serum of COVID-19 patients using an enzyme-linked immunosorbent assay (40). Although recent reports suggesting that detection of antibody-based methods targeted to IgM and IgG by using recombinant N and S proteins of SARS-CoV-2are consistent with the results obtained by real-time RT-PCR (41–43). In addition to this, the receptor-binding domain (RBD) of the viral S protein presented a better antigenicity than viral N protein in the diagnosis of SARS-CoV-2 infection (44). Also, IgA levels in patient serum have positively correlated with the severity of SARS-CoV-2 infection, signifying that serum IgA can be used as a biological marker (44). In clinical diagnosis, the IgA and IgM antibodies against viral proteins can be detected seven days after SARS-CoV-2 infection or within 3-4 days after symptoms appear, as well as for IgG antibodies appears in 7-10 days later SARS-CoV-2 infection.
Serology testing has some advantages over other techniques, apart from being inexpensive. The primary application of serology testing is to identify individuals who previously had SARS-CoV-2 infections. This knowledge can be used to guide studies of epidemiology and seroprevalence, and to facilitate contact tracing. Serology tests can also be used to determine possible convalescent donors of plasma and to assess the immune response to candidate vaccines. Finally, serology tests can also aid in diagnosing Covid-19 in patients with clinical suspicion but having repeated RT-PCR-negative results (45,46). Serology testing has its limitations too. The serology test cannot be used to diagnose acute or recent COVID-19 cases. Antibody tests for COVID-19 may also interact with other pathogens, including other human coronaviruses and leads to false-positive results. Based on current data, the WHO does not recommend the use of antibody-detecting rapid diagnostic tests for patient care but encourages the continuation of work to establish their usefulness in disease surveillance and epidemiologic research (14).
Serology testing helps in the assessment of seroprevalence of COVID-19 disease in the community. Nationwide serology testing would help in tailoring the public health measures to control and avoid renewed COVID-19 epidemic wave (47). Serologic surveillance also can help in anticipate and modify treatment modalities as in perinatal clinical practices pregnant women (48). Seroprevalence surveys can also help in understand the geographical profile of the COVID-19 disease and help in creating a regional level approach in controlling the pandemic (49). However, serology testing cannot be used to determine the infectivity status or the susceptibility to reinfection for the patient. The presence of antibodies does not render the patient non-infectious, as the antibodies can be of non-neutralising in nature (50,51). Virus neutralisation tests have to be performed to assess the neutralising capability of antibodies generated by the body against the SARS-CoV-2 virus. Hence clinical laboratories are recommended not to promote so-called “immunity passports” due to a lack of evidence for the neutralising capability of antibodies (52).
3.8 VIRAL SEQUENCING
Sequencing does not play a part in the initial SARS-CoV-2 laboratory diagnosis but can be beneficial in the following circumstances; 1) Provides evidence of virus existence; 2) Monitoring for viral genome mutations that could affect medical countermeasure performance, including diagnostic testing; 3) Virus sequencing of entire genomes can also inform studies on molecular epidemiology. Virus isolation, currently, is not recommended as part of the routine diagnostic methodology.
4.1 CLINICAL LABORATORY PARAMETERS IN COVID-19
Biochemical and haematological parameters have been investigated to assess their role in diagnosis and prognosis. Further, the role of laboratory testing in assessing severity and selecting treatment modalities and monitoring the effectiveness of treatment has been elucidated through multiple studies. Figure 2 depicts the important parameters that can be used for determining diagnosis, prognosis and treatment response.
Figure 2.
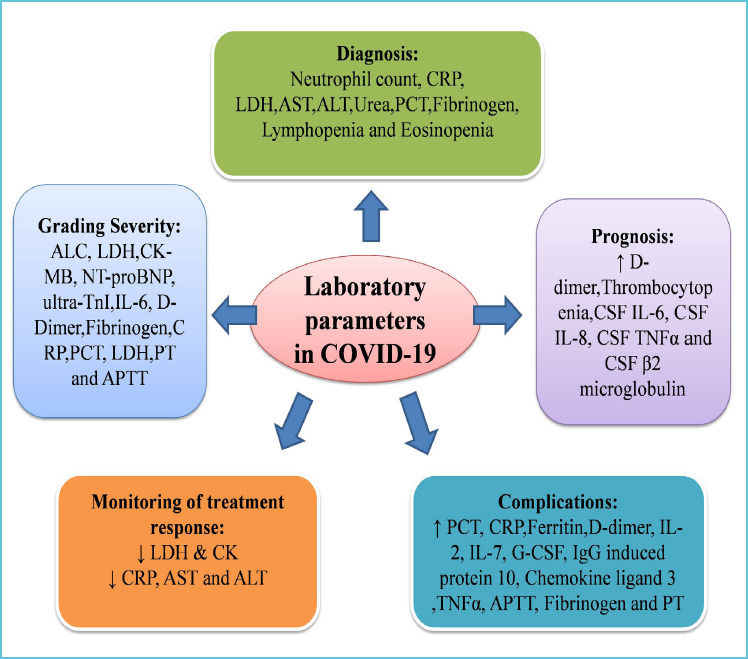
Important parameters used for determining diagnosis, prognosis and treatment response
4.2 LABORATORY PARAMETERS AIDING IN DIAGNOSIS OF COVID-19
Several Laboratory Parameters are significantly increased in COVID-19 positive patients when compared with others. RT-PCR diagnosed COVID-19 patients had significantly higher neutrophil (NEU) count, and C-reactive protein (CRP), aspartate aminotransferase, alanine aminotransferase (ALT), lactate dehydrogenase and Urea levels in serum(53). Serum albumin levels and White blood cell (WBC) count are decreased in COVID-19 positive patients when compared to others (53). More number of control patients had a higher procalcitonin (PCT) level of more than 0.5 ng/ml than that of COVID-19 patients (54). At admission, the COVID-19 patients showed elevated levels of fibrinogen than the control group. Further, a greater percentage of COVID-19 patients had fibrinogen levels >400 mg/dL compared to the control group (55). Normal or decreased number of leukocytes, lymphopenia, eosinopenia, and elevated hs-CRP were presented in COVID-19 patients when compared with controls. It has been found that the use of eosinopenia alone or the combination of eosinopenia and elevated hs-CRP improves the predictive capacity for the detection of COVID-19 patients (56). Table 2 depicts the Laboratory parameters assessed to distinguish COVID-19 Positive patients from Negative patients.
Table 2.
Laboratory parameters in COVID-19 positive and negative patients
| Study | Sample size | Parameter | Remarks |
|---|---|---|---|
| Mardani et al. (53) | Two hundred cases RT-PCR for COVID-19 was positive in 70 | Neutrophil (NEU) count, C-reactive protein (CRP), Aspartate aminotransferase, alanine aminotransferase, Lactate dehydrogenase, Urea, Lower white blood cell (WBC) count, Lower serum albumin level |
ALT (AUC = 0.879), NEU (AUC = 0.858), CRP (AUC = 0.870), LDH (AUC = 0.835), Urea (AUC = 0.835) |
| Chen et al (54) | 78 COVID-19 patients 26 control patients | PCT | - |
| Di Micco et al. (55) | 67 COVID-19 patients and 67 patients with non-COVID-19 acute respiratory illness |
Increased levels of fibrinogen | - |
| Li et al. (56) | 458 | Normal or decreased number of leukocytes, lymphopenia, eosinopenia and elevated hs-CRP | Eosinopenia the sensitivity of 74.7% and specificity of 68.7% Combination of eosinopenia and elevated hs-CRP showed a sensitivity of 67.9% and specificity of 78.2% (AUC=0.730). |
| Ferrari et al. (57) | 207 | WBC, AST, ALT,CRP, and LDH | For LDH cut off 210 U/L: positive or negative with PPV: 83.3% and NPV: 90.6%. |
| Liu et al. (58) | 119 | Presence of urine occult blood and proteinuria. Lower urine specific gravity |
- |
4.3 LABORATORY PARAMETERS HELPING IN ASSESSING THE SEVERITY OF COVID-19
Multiple parameters are useful in assessing the severity of the disease. The parameters that were found to have a significant difference between mild and severe disease include interleukin-6 (IL-6), d-dimer (d-D), glucose, fibrinogen, thrombin time, and C-reactive protein (59). Fibrinogen was found to be higher in COVID-19 patients with SARS compared to those without SARS (55).
The role of laboratory parameters indicating inflammation have been discussed elsewhere (60). IL-6, an inflammatory cytokine, was found to have a potential value for monitoring the process of severe cases (61). The increased concentration of ultra-TnI, MYO, and NT-proBNP was also found to be associated with the severity of COVID-19(62). The dysregulated activity of CD3+ CD8+ T lymphocytes, CD16+ CD56+ NK cells and altered C1q and IL-6 have been found to accentuate the severity of disease and death (63). Further, on correlation analysis between multiple cytokines and coagulation indicators in critically ill COVID-19 patients, a high correlation was observed between IL-6 and the International normalised ratio (INR) (64).
The severity of lung abnormalities is quantified by chest imaging. Different laboratory parameters are associated with stages of lung diseases in COVID-19 patients as quantified on chest CT. Early-stage as per CT scoring was found to be correlated with the neutrophil count. The progressive stage was correlated with the neutrophil count, white blood cell count, C-reactive protein, procalcitonin, and lactate dehydrogenase. Contrastingly, peak and absorption stages were not correlated with any parameter (65). The paradoxical increase in D-dimer levels despite decreased fibrinolytic capacity had prompted the researchers to hypothesise that the major source of D-dimer could be the lungs (66).
Apart from altered coagulation profile, low activities of natural anticoagulants, increased factor VIII level and antiphospholipid antibodies presence have also been found to accentuate the severity of the disease in COVID-19 patients (67). In severe and critically ill patients, the specific immunoglobulin G antibodies to the SARS-CoV-2 were found to be significantly low when compared with patients with mild disease (68).
Different parameters have been assessed for their dynamic trend in different stages as well as the severity of the disease. Lymphocytes in the severe COVID-19 were found to be progressively decreasing at the progression and the peak stages. C-reactive protein (CRP) was higher in the severe group at the initial and progression stages than those in the mild group (64). Table 3 depicts the laboratory parameters associated with severity of the disease in COVID -19 patients.
Table 3.
Laboratory parameters associated with severity of the disease in COVID-19 patients
| Study | Sample size | Parameter | Category |
|---|---|---|---|
| Fan et al. (53) | Between the ICU (n=9) and non-ICU (n=58) patients | ALC and LDH, ALC and Absolute Monocyte Count (AMC) nadir | ICU vs Non-ICU |
| Han et al. (62) | mild (198 cases), severe (60 cases) and critical (15 cases) | CK-MB, MYO, ultra-TnI and NT-proBNP | Severity and case fatality |
| Gao et al. (59) | 43 COVID-19 patients mild group (28 patients) and severe group (15 patients). |
Interleukin-6 (IL-6),d-dimer (d-D), glucose, thrombin time, fibrinogen, and C-reactive protein IL 6 (AUC=0.795) D-Dimer (AUC=0.75) Glu, TT, CRP and FIB (AUC<0.75) IL 6 + D-Dimer (AUC=0.84) |
Mild vs severe |
| Zhang et al. (63) | 84 COVID-19 patients | Early stage: neutrophil count. Progressive stage: neutrophil count, white blood cell count, C-reactive protein, procalcitonin, lactate dehydrogenase. |
early and progressive stages of lung abnormalities (CT Finding) |
| Tan et al. (64) | 27 COVID-19 and 75 Flu patients | Progression and the peak stages: lymphocytes decreased Initial and progression stages: C-reactive protein (CRP) higher. Correlation analysis showed that CRP, erythrocyte sedimentation rate and granulocyte/lymphocyte ratio were positively associated with the CT severity scores. CRP (AUC=0.87) at 20.42 mg/L cut-off, with sensitivity and specificity 83% and 91%, respectively. |
Mild vs Severe |
| Di Micco et al. (55) | SARS: 24 Without SARS: 43 | Fibrinogen | Mild vs Severe |
| Fu et al. (65) | 75 | WBC, NLR,D-dimer, and fibrinogen levels Incresed. Lymphocyte level Decreased. AUC isNLR (0.88), AUC of D-dimer and fibrinogen was 0.74, and AUC of lymphocyte and PCT were 0.72 and 0.67 respectively. |
mild/moderate COVID-19 group |
| Zhu et al. (61) | 127 16 severe cases | High level of interleukin-6 (IL-6), C-reaction protein (CRP). The area under the ROC curve was 0.835 for IL-6, sensitivity was 87.50%, specificity was 74.77%. |
severity of COVID-19 |
| Liu et al. (58) | 119 | The positive rates of urine glucose (GLU-U) and PRO in the severe and critical groups were higher | mild/moderate COVID-19 group |
| Tang et al. (71) | 183 | Higher D-dimer and FDP levels, longer PT and APTT | Between survivors and non-survivors |
4.4 LABORATORY PARAMETERS INDICATING PROGNOSIS OF THE DISEASE IN COVID-19 PATIENTS
Laboratory parameters at admission have been investigated for their prognostic power for the severity of the disease. Logistic regression analysis showed that IL-6 and D-Dimer could be important predictors in the severity of COVID-19. Further, it had also been found that combined detection using IL-6 and D-Dimer was more efficient than independent detection (59). Various parameters have also been used to predict admission to ICU. ALC and LDH stood out as parameters that can, with the levels at admission, reliably predict the admission of the patient to ICU (53). The change of neutrophil to lymphocyte ratio (NLR) and D-dimer level has been found to help in discriminating severe COVID-19 cases from mild/moderate ones on consequent days after admission (65). The early increase in Fibrinogen in COVID-19 patients makes it a good risk stratification marker for the early detection of a subgroup of COVID-19 patient at increased risk to develop SARS (55).
Non-survivors mainly presented with laboratory abnormalities of serious inflammation response and multiple organ failure, manifesting as high levels of cytokines and deranged coagulation parameters. Neutrophil count, hypersensitivity C-reactive protein, creatine kinase, and blood urea nitrogen were identified to help in early detection of COVID-19 severe patients with poor outcomes on admission (72). Further, the non-survivors of COVID-19 disease revealed significantly higher D-dimer and FDP levels compared to survivors on admission (71). Hence, the use of Sepsis-induced coagulopathy scoring system for early assessment and management have been advised in patients with the critical disease (73). mRNA clearance rates indicate the resolution of the disease. It has been found that the decline of serum creatine kinase (CK) and lactate dehydrogenase (LDH) levels significantly correlated with mRNA clearance rates (74). CSF analyses revealed relatively slightly increased levels of interleukin 6 (IL-6), interleukin 8, tumour necrosis factor-alpha and (β2-microglobulin. Ten days after the admission, CSF IL-8 and TNF-α decreased, whereas IL-6 and (β2-microglobulin values were stable (76). Table 4 depicts the important laboratory parameters that can be used to determine the prognosis in COVID-19 patients.
Table 4.
Laboratory parameters determining prognosis in COVID-19 patients
| Study | Sample size | Parameter |
|---|---|---|
| Li et al. (75) | 279 | The higher D-dimer levels on admission progressively improved only in the mild disease group. |
| Liu et al. (77) | 383 | Thrombocytopenia. An increment of per 50 × 109/L in platelets was associated with a 40% decrease in mortality (hazard ratio: 0.60, 95% CI: 0.43, 0.84). |
4.5 LABORATORY PARAMETERS IN COMORBIDITIES AND TREATMENT IN COVID-19 PATIENTS
Various laboratory parameters have been assessed for its role in complications in COVID-19 patients. Patients with abnormal liver function had higher levels of procalcitonin and C-reactive protein(78). Various inflammatory markers are elevated in patients with COVID-19 related cardiac injury. They include C- reactive protein (CRP), procalcitonin, ferritin, D- dimer, Interleukin - 2 (IL-2) interleukin – 7 (IL-7), granulocyte – colony-stimulating factor, IgG- induced protein 10, chemokine ligand three and tumour necrosis alpha(79). Lymphocyte counts, activated partial thromboplastin time (APTT) and D-dimer was found to be different in patients with venous thromboembolism when compared with the non-VTE group. The significant increase of D-dimer observed in severe patients makes it a good index for identifying high-risk groups of VTE(80). On comparison of COVID-19 patients with and without HBV co-infection, although the level of liver function parameters showed no differences, prealbumin levels were found to be lower in HBsAg+ patients(81). In solid organ transplant recipients with COVID-19, a biphasic pattern was observed with initial increases in inflammatory markers, followed by an increase in WBC, CRP, ferritin and D-dimer (82).
To assess the efficacy of treatment, the primary tool for analysis have been the trend shown by Laboratory parameters. Lymphocytopenia improved after Convalescent Plasma transfusion. C-reactive protein (CRP), alanine aminotransferase, and aspartate aminotransferase decreased after treatment (83). Table 5 depicts the laboratory parameters which are associated with complications and monitoring of response to treatment in COVID-19 patients.
Table 5.
Laboratory parameters associated with complications and response to treatment in COVID-19 patients
| Study | Sample size | Parameter | Remarks | Category |
|---|---|---|---|---|
| Yuan et al. (74) | 94 COVID-19 patients | Decline in Serum LDH or CK | - | Response to treatment |
| Fan et al. (78) | 148 | Increased levels of procalcitonin and C-reactive protein | - | Abnormal liver function |
| Han et al. (79) | 273 | Increased levels of CK-MB, MYO, ultra-TnI, and NT-proBNP | - | COVID-19 related cardiac injury |
| Duan et al. (83) | 10 | Lymphocytopenia tended to be improved after CP transfusion. Decreased in C-reactive protein (CRP), alanine amino-transferase and aspartate aminotransferase. |
- | Convalescent plasma therapy |
| Cui et al. (80) | 81 | Lymphocyte counts, activated partial thromboplastin time (APTT), D-dimer | D-dimer cut-off 1.5 μg/mLfor predicting VTE had a sensitivity of 85.0%, the specificity of 88.5%, and the negative predictive value (NPV) of 94.7%. | VTE/NonVTE group |
| Wright et al (84) | 44 | Elevated D-dimer, fibrinogen, PT, and PTT | A D-Dimer cutoff of 2600 ng/ml predicted need for dialysis with an AUROC of .779 | Predict thromboembolic outcomes and new-onset renal failure |
| Lin et al. (85) | 137 | CD8+ T cells | HR=2.376 | Duration of SARS-CoV-2 viral positivity |
4.6 LABORATORY PARAMETERS AND BODY FLUIDS IN COVID-19
Different fluids have also been assessed in COVID-19 patients for different parameters. The presence of urine occult blood and proteinuria were found to be higher in COVID-19 patients than in healthy controls, whereas urine specific gravity was found to be lower in patients than in healthy controls. The presence of urine glucose and proteinuria were higher in the severe and critical groups when compared with that of the moderate group (58). CSF analyses have revealed relatively slightly increased levels of interleukin 6 (IL-6), interleukin 8, tumour necrosis factor-alpha, and β2-microglobulin in a single patient (76).
5. POOLED AND META-ANALYSIS OF LABORATORY PARAMETERS INCOVID-19
Multiple meta-analyses had been undertaken to find the significance of various laboratory parameters in COVID-19. Soraya et al. had concluded thrombocyte count to have a crucial role in the diagnosis and prognosis of COVID-19. Further, lymphocyte count, D-dimer and CRP levels helped to assess the severity of the disease (86). Henry et al. had observed that markers of inflammation, coagulation markers and organ damage to be significantly elevated severe and fatal COVID-19 patients. In patients with severe disease, interleukins 6 (IL-6) and 10 (IL-10) and serum ferritin were found to be predominantly elevated (87). Interestingly, in a pooled analysis of Laboratory Parameters paediatric COVID-19 patients, contrary to adult patients, leukocyte indices showed inconsistent trends (88). The elevated levels of the neutrophil count, D-dimer, prothrombin time (PT), fibrinogen erythrocyte sedimentation rate, procalcitonin, IL-6, and IL-10 were found to be better predictors for severe COVID-19 disease (89, 90). Further, high IL-6, CRP, D-dimer and neutrophils were found to be better predictors of mortality in COVID-19(89). A similar meta-analysis also observed severe or critical COVID-19 to be associated with innate immune response and tissue damage (91).
6. CONCLUSION
In summary, the crucial role that clinical laboratory plays in the management of diseases has never been more evident than today. Validating various assays for diagnosing COVID-19 helps in early diagnosis and initiation of treatment as well as prevent transmission. The assessment of the clinical utility of tests in different scenarios in COVID-19 and ensuring its accuracy adds to the efforts to treatment of the disease as well as predicting complications. This review has emphasised the importance of laboratory in the COVID-19 crisis. The emergence of diagnostic assays with better sensitivity and specificity equips the laboratories with an enhanced ability to identify COVID-19 cases early and prevents transmission (92). The routine laboratory parameters have been shown to be able to distinguish between positive and negative patients, have the capacity to predict prognosis & complications and have usefulness in monitoring treatment response. Further studies in this arena would lead to validation of better assays for precise diagnosis and newer biomarkers for monitoring treatment and disease progression. Decision-makers should not underestimate the role of the laboratory as it plays a pivotal role in patient-centred and sustainable future of health care.
Footnotes
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020. March;579(7798): 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020. February;395(10225):689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Director-General’s opening remarks at the media briefing on COVID-19. [Google Scholar]
- 4.Specific Primers and Probes for Detection 2019 Novel Coronavirus; China National Institute For Viral Disease Control and Prevention: Beijing, 2020. [Google Scholar]
- 5.Detection of 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases by RT-PCR; School of Public Health, Hong Kong University: Hong Kong, 2020. [Google Scholar]
- 6.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance [Internet]. 2020. Jan 23 [cited 2020 May 31];25(3). Available from: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naganori N., Shirato K., et al. Detection of Second Case of 2019-nCoV Infection in Japan; Department of Virology III, National Institute of Infectious Diseases, Japan, 2020. [Google Scholar]
- 8.Diagnostic Detection of Novel Coronavirus 2019 by Real Time RT-PCR; Department of Medical Sciences, Ministry of Public Health, Thailand, 2020. [Google Scholar]
- 9.CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel; Division of Viral Diseases, U.S. Centers for Disease Control and Prevention: Atlanta, GA, 2020. [Google Scholar]
- 10.Coronavirus Disease 2019 (COVID-19); Centers for Disease Control and Prevention. [Google Scholar]
- 11.Miller S, Chiu C, Rodino KG, Miller MB. Point-Counter-point: Should We Be Performing Metagenomic Next-Generation Sequencing for Infectious Disease Diagnosis in the Clinical Laboratory? Ledeboer N, editor. J Clin Microbiol [Internet]. 2019. Oct 16 [cited 2020 May 31];58(3). Available from: https://jcm.asm.org/content/58/3/e01739-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19); WHO: Geneva, Switzerland, 2020. [Google Scholar]
- 13.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel Coronavirus: implications for virus origins and receptor binding. The Lancet. 2020. February;395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization (WHO) (2020) Advice on the use of point-of-care immunodiagnostic tests for COVID-19 https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiag-nostic-tests-forcovid-19.
- 15.Centers for Disease Prevention and Control (CDC) (2020) Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19). https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html.
- 16.Indian Council of Medical Research (ICMR) (2020) https://icmr.nic.in/sites/default/files/upload_documents/Validation_of_Commercial_Kits_02042020.pdf.
- 17.Rodino KE, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E, Boerger A, Schuetz AN, Yao JD, Binnicker MJ. 30 March 2020. Evaluation of saline, phosphate-buffered saline, and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol doi:10.1128/JCM.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perchetti GA, Huang ML, Peddu V, Jerome KR, Greninger AL.J. Stability of SARS-CoV-2 in Phosphate-Buffered Saline for Molecular Detection. Clin Microbiol. 2020. Jul 23;58(8):e01094-20 doi: 10.1128/JCM.01094-20. Print 2020 Jul 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020. May 12;323(18):1843-1844. doi: 10.1001/jama.2020.3786. PMID: 32159775; PMCID: PMC7066521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, Peiris M, Poon LLM. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020. Apr 1;66(4):549-555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chantal B F, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020. October;5(10):1299-1305. doi: 10.1038/s41564-020-0761-6. Epub 2020 Jul 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konrad R., Eberle U., Dangel A., Treis B., Berger A., Bengs K., Fingerle V., Liebl B., Ackermann N., Sing A, Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February. 2020, Euro Surveill 25(9) (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyrlaki I, et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nature Communications volume 11, Article number: 4812 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morehouse Z.P., Proctor C.M., Ryan G.L., et al. A novel two-step, direct-to-PCR method for virus detection off swabs using human coronavirus 229E. Virol J 17, 129 (2020). 10.1186/s12985-020-01405-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wee SK, Sivalingam SP, Yap EPH. Rapid Direct Nucleic Acid Amplification Test without RNA Extraction for SARS-CoV-2 Using a Portable PCR Thermocycler. Genes. 2020. June;11(6). DOI: 10.3390/genes11060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: WHO Interim guidance 19 March 2020. [Google Scholar]
- 27.Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid Detection of Novel Coronavirus (COVID-19) by Reverse Transcription- Loop-Mediated Isothermal Amplification.:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, et al. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin Chem [Internet]. 2020. May 12 [cited 2020 May 31]; Available from: https://academic.oup.com/clinchem/advance-article/doi/10.1093/clinchem/hvaa102/5823294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP [Internet]. Infectious Diseases (except HIV/AIDS); 2020. Feb [cited 2020 May 31]. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.02.26.20028373 [Google Scholar]
- 30.Yan C, Cui J, Huang L, Du B, Chen L, Xue G, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020. June;26(6):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int J Mol Sci. 2020. April 18;21(8):2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting SARS-CoV-2. J. Mol. Diagn.: J. Mod. Dynam. 2020;22(6):729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26(6):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017. April 28;356(6336):438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019. October;14(10):2986–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, et al. Development and Evaluation of A CRISPR-based Diagnostic For 2019-novel Coronavirus [Internet]. Infectious Diseases (except HIV/AIDS); 2020. Feb [cited 2020 May 31]. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.02.22.20025460 [Google Scholar]
- 37.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol [Internet]. 2020. Apr 16 [cited 2020 May 31]; Available from: http://www.nature.com/articles/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, Wu J, Liao Y, Gou X, Li Y, Wang X, Su H, Gu B, Wang J, Xu T. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020. August 27;16(8):e1008705 doi: 10.1371/journal.ppat.1008705. eCollection 2020 Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Ding, 1, Kun Yin, 1, Ziyue Li, 1, Rajesh V Lalla, 2, Enrique Ballesteros, 3, Maroun M Sfeir, 3, Changchun Liu., 4 Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020. September 18;11(1):4711 doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Du R-H, Li B, Zheng X-S, Yang X-L, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020. January 1;9(1):386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong L, Chuan J, Gong B, Shuai P, Zhou Y, Zhang Y, et al. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci China Life Sci. 2020. May;63(5):777–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia X, Zhang P, Tian Y, Wang J, Zeng H, Wang J, et al. Clinical significance of IgM and IgG test for diagnosis of highly suspected COVID-19 infection [Internet]. Infectious Diseases (except HIV/AIDS); 2020. Mar [cited 2020 May 31]. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.02.28.20029025 [Google Scholar]
- 43.Evaluations of serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak.:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma H, Zeng W, He H, Zhao D, Yang Y, Jiang D, et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by chemiluminescence immunoanalysis [Internet]. Infectious Diseases (except HIV/AIDS); 2020. Apr [cited 2020 May 31]. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.04.17.20064907 [Google Scholar]
- 45.Guo L., et al. , Profiling early humoral response to diagnose novel coronavirus dis-ease (COVID-19), Clin. Infect. Dis. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao R., et al. , Early detection of SARS-CoV-2 antibodies in COVID-19 patients as a serologic marker of infection, Clin. Infect. Dis. (2020). [Google Scholar]
- 47.Pollán M, Pérez-Gómez B, Pastor-Barriuso R, Oteo J, Hernán MA, Pérez-Olmeda M, et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. The Lancet [Internet]. 2020. Jul [cited 2020 Jul 21]; Available from: https://link-inehub.elsevier.com/retrieve/pii/S0140673620314835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flannery DD, Gouma S, Dhudasia MB, et al. SARS-CoV-2 Seroprevalence Among Parturient Women. Pre-print. medRxiv. 2020;2020.07.08.20149179. Published 2020 Jul 10. doi:10.1101/2020.07.08.20149179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takita M, Matsumura T, Yamamoto K, Yamashita E, Hosoda K, Hamaki T, et al. Geographical Profiles of COVID-19 Outbreak in Tokyo: An Analysis of the Primary Care Clinic–Based Point-of-Care Antibody Testing. J Prim Care Community Health. 2020. January;11:215013272094269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wölfel, et al. , Virological assessment of hospitalised patients with COVID-2019, Nature 581 (2020) 465–469. [DOI] [PubMed] [Google Scholar]
- 51.Atkinson B., Petersen E, SARS-CoV-2 shedding and infectivity, Lancet 395 (2020) 1339–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey D, Konforte D, Barakauskas VE, Yip PM, Kulasingam V, Abou El Hassan M, Beach LA, Blasutig IM, Catomeris P, Dooley KC, Gong Y, Kavsak P, Randell EW, Robinson JL, Shaw J, Taher J, White-Al Habeeb N. Canadian society of clinical chemists (CSCC) interim consensus guidance for testing and reporting of SARS-CoV-2 serology. Clin Biochem. 2020. October 6:S0009-9120(20)30844-30844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan BE, Chong VCL, Chan SSW, Lim GH, Lim KGE, Tan GB, et al. Hematologic parameters in patients with COVID-19 infection. Am J Hematol [Internet]. 2020. Jun [cited 2020 May 25];95(6). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/ajh.25774 [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Yang Y, Huang M, Liu L, Zhang X, Xu J, et al. Differences between COVID-19 and suspected then confirmed SARS-CoV-2-negative pneumonia: a retrospective study from a single center. J Med Virol [Internet]. 2020. Apr 1 [cited 2020 May 25]; Available from: http://doi.wiley.com/10.1002/jmv.25810 [DOI] [PubMed] [Google Scholar]
- 55.Di Micco P, Russo V, Carannante N, Imparato M, Rodolfi S, Cardillo G, et al. Clotting Factors in COVID-19: Epidemiological Association and Prognostic Values in Different Clinical Presentations in an Italian Cohort. J Clin Med. 2020. May 7;9(5):1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q, Ding X, Xia G, Chen H-G, Chen F, Geng Z, et al. Eosinopenia and elevated C-reactive protein facilitate triage of COVID-19 patients in fever clinic: a retrospective case-control study. EClinicalMedicine. 2020. May;100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferrari D, Motta A, Strollo M, Banfi G, Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med CCLM [Internet]. 2020. Apr 16 [cited 2020 May 25];0(0). Available from: https://www.deeruyter.com/view/journals/cclm/ahead-of-print/article-10.1515-cclm-2020-0398/article-10.1515-cclm-2020-0398.xml [DOI] [PubMed] [Google Scholar]
- 58.Liu R, Ma Q, Han H, Su H, Liu F, Wu K, et al. The value of urine biochemical parameters in the prediction of the severity of coronavirus disease 2019. Clin Chem Lab Med CCLM [Internet]. 2020. Apr 14 [cited 2020 May 25];0(0). Available from: https://www.deeruyter.com/view/journals/cclm/ahead-of-print/article-10.1515-cclm-2020-0220/article-10.1515-cclm-2020-0220.xml [DOI] [PubMed] [Google Scholar]
- 59.Gao Y, Li T, Han M, Li X, Wu D, Xu Y, et al. Diagnostic Utility of Clinical Laboratory Data Determinations for Patients with the Severe COVID-19. J Med Virol [Internet]. 2020. Mar 17 [cited 2020 May 25]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lingeswaran M, Goyal T, Ghosh R, Suri S, Mitra P, Misra S, et al. Inflammation, Immunity and Immunogenetics in COVID-19: A Narrative Review. Indian J Clin Biochem [Internet]. 2020. Jun 6 [cited 2020 Jun 10]; Available from: http://link.springer.com/10.1007/s12291-020-00897-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu Z, Cai T, Fan L, Lou K, Hua X, Huang Z, et al. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int J Infect Dis. 2020. June;95:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han H, Xie L, Liu R, Yang J, Liu F, Wu K, et al. Analysis of heart injury laboratory parameters in 273 COVID-19 patients in one hospital in Wuhan, China. J Med Virol [Internet]. 2020. Apr 15 [cited 2020 May 25]; Available from: http://doi.wiley.com/10.1002/jmv.25809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y, Huang X, Sun J, Xie T, Lei Y, Muhammad J, et al. Clinical Characteristics and Immune Injury Mechanisms in 71 Patients with COVID-19. Rosenberg HF, editor. mSphere [Internet]. 2020. Jul 15 [cited 2020 Jul 21];5(4). Available from: https://msphere.asm.org/content/5/4/e00362-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang D, Zhou X, Yan S, Tian R, Su L, Ding X, et al. Correlation between cytokines and coagulation-related parameters in patients with coronavirus disease 2019 admitted to ICU. Clin Chim Acta. 2020. November;510:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang B, Zhang J, Chen H, Chen L, Chen Q, Li M, et al. Novel coronavirus disease 2019 (COVID-19): relationship between chest CT scores and laboratory parameters. Eur J Nucl Med Mol Imaging [Internet]. 2020. May 12 [cited 2020 May 25]; Available from: http://link.springer.com/10.1007/s00259-020-04854-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ibañez C, Perdomo J, Calvo A, Ferrando C, Reverter JC, Tassies D, et al. High D dimers and low global fibrinolysis coexist in COVID19 patients: what is going on in there? J Thromb Thrombolysis [Internet]. 2020. Jul 15 [cited 2020 Jul 21]; Available from: http://link.springer.com/10.1007/s11239-020-02226-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Cao W, Jiang W, Xiao M, Li Y, Tang N, et al. Profile of natural anticoagulant, coagulant factor and antiphospholipid antibody in critically ill COVID-19 patients. J Thromb Thrombolysis [Internet]. 2020. Jul 9 [cited 2020 Jul 21]; Available from: http://link.springer.com/10.1007/s11239-020-02182-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan X, Huang W, Ye B, Chen C, Huang R, Wu F, et al. Changes of hematological and immunological parameters in COVID-19 patients. Int J Hematol [Internet]. 2020. Jul 12 [cited 2020 Jul 21];Availablefrom: http://link.springer.com/10.1007/s12185-020-02930-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan C, Huang Y, Shi F, Tan K, Ma Q, Chen Y, et al. C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J Med Virol [Internet]. 2020. Apr 25 [cited 2020 May 25]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.25871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu J, Kong J, Wang W, Wu M, Yao L, Wang Z, et al. The clinical implication of dynamic neutrophil to lymphocyte ratio and D-dimer in COVID-19: A retrospective study in Suzhou China. Thromb Res. 2020. August;192:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020. April; 18(4):844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X, Yan L, Fei Y, Zhang C. Laboratory abnormalities and risk factors associated with in-hospital death in patients with severe COVID-19. J Clin Lab Anal [Internet]. 2020. Jul 12 [cited 2020 Jul 21]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jcla.23467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao D, Zhou F, Luo L, Xu M, Wang H, Xia J, et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: a retrospective cohort study. Lancet Haematol [Internet]. 2020. Jul [cited 2020 Jul 21]; Available from: https://linkinghub.elsevier.com/retrieve/pii/S2352302620302179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan J, Zou R, Zeng L, Kou S, Lan J, Li X, et al. The correlation between viral clearance and biochemical outcomes of 94 COVID-19 infected discharged patients. Inflamm Res. 2020. June;69(6):599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Zhao K, Wei H, Chen W, Wang W, Jia L, et al. Dynamic relationship between D-dimer and COVID-19 severity. Br J Haematol [Internet]. 2020. May 18 [cited 2020 May 25]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/bjh.16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pilotto A, Odolini S, Stefano Masciocchi S, Comelli A, Volonghi I, Gazzina S, et al. Steroid-responsive encephalitis in Covid-19 disease. Ann Neurol [Internet]. 2020. May 17 [cited 2020 May 25]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/ana.25783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Sun W, Guo Y, Chen L, Zhang L, Zhao S, et al. Association between platelet parameters and mortality in coronavirus disease 2019: Retrospective cohort study. Platelets. 2020. May 18;31(4):490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, et al. Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin Gastroenterol Hepatol. 2020. June;18(7):1561–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Han H, Xie L, Liu R, Yang J, Liu F, Wu K, Chen L, Hou W, Feng Y, Zhu C. Analysis of heart injury laboratory parameters in 273 COVID-19 patients in one hospital in Wuhan, China. J Med Virol. 2020. July;92(7):819-823. doi: 10.1002/jmv.25809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cui S, Chen S, Li X, Liu S, Wang F. Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J Thromb Haemost [Internet]. 2020. May 6 [cited 2020 May 25]; Available from: https://on-linelibrary.wiley.com/doi/abs/10.1111/jth.14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L, Huang S, Yang J, Cheng X, Shang Z, Lu H, et al. Clinical characteristics in patients with SARS-CoV-2/HBV co-infection. J Viral Hepat [Internet]. 2020. Jul 15 [cited 2020 Jul 21]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/jvh.13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roberts MB, Izzy S, Tahir Z, Al Jarrah A, Fishman JA, El Khoury J. COVID-19 in solid organ transplant recipients: dynamics of disease progression and inflammatory markers in ICU and non-ICU admitted patients. Transpl Infect Dis [Internet]. 2020. Jul 12 [cited 2020 Jul 21]; Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/tid.13407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci. 2020. April 28;117(17):9490–9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright FL, Vogler TO, Moore EE, Moore HB, Wohlauer MV, Urban S, et al. Fibrinolysis Shutdown Correlates to Thromboembolic Events in Severe COVID-19 Infection. J Am Coll Surg [Internet]. 2020. May [cited 2020 May 25]; Available from: https://linkinghub.elsevier.com/retrieve/pii/S1072751520304002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin A, He Z-B, Zhang S, Zhang J-G, Zhang X, Yan W-H. Early risk factors for the duration of SARS-CoV-2 viral positivity in COVID-19 patients. Clin Infect Dis [Internet]. 2020. Apr 27 [cited 2020 May 25]; Available from: https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa490/5825508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soraya GV, Ulhaq ZS. Crucial laboratory parameters in COVID-19 diagnosis and prognosis: An updated metaanalysis. Med Clin (Barc). 2020. August 28;155(4):143-151. doi: 10.1016/j.medcli.2020.05.017. Epub 2020 Jun 5. PMID: 32586670; PMCID: PMC7274591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med. 2020. June 25;58(7):1021-1028. doi: 10.1515/cclm-2020-0369. PMID: 32286245. [DOI] [PubMed] [Google Scholar]
- 88.Henry BM, Benoit SW, de Oliveira MHS, Hsieh WC, Benoit J, Ballout RA, Plebani M, Lippi G. Laboratory abnormalities in children with mild and severe coronavirus disease 2019 (COVID-19): A pooled analysis and review. Clin Biochem. 2020. July;81:1-8. doi: 10.1016/j.clinbiochem.2020.05.012. Epub 2020 May 27. PMID: 32473151; PMCID: PMC7251358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elshazli RM, Toraih EA, Elgaml A, El-Mowafy M, El-Mesery M, Amin MN, Hussein MH, Killackey MT, Fawzy MS, Kandil E. Diagnostic and prognostic value of hematological and immunological markers in COVID-19 infection: A meta-analysis of 6320 patients. PLoS One. 2020. August 21;15(8):e0238160 doi: 10.1371/journal.pone.0238160. PMID: 32822430; PMCID: PMC7446892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henry B, Cheruiyot I, Vikse J, Mutua V, Kipkorir V, Benoit J, Plebani M, Bragazzi N, Lippi G. Lymphopenia and neutrophilia at admission predicts severity and mortality in patients with COVID-19: a meta-analysis. Acta Biomed. 2020. September 7;91(3):e2020008 doi: 10.23750/abm. v91i3.10217. PMID: 32921706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moutchia J, Pokharel P, Kerri A, McGaw K, Uchai S, Nji M, Goodman M. Clinical laboratory parameters associated with severe or critical novel coronavirus disease 2019 (COVID-19): A systematic review and metaanalysis. PLoS One. 2020. October 1;15(10):e0239802 doi: 10.1371/journal.pone.0239802. PMID: 33002041; PMCID: PMC7529271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mitra P, Misra S, Sharma P. COVID-19 Pandemic in India: What Lies Ahead. Indian J Clin Biochem [Internet].2020. Apr 20 [cited 2020 Jun 10]; Available from: http://link.springer.com/10.1007/s12291-020-00886-6 [DOI] [PMC free article] [PubMed] [Google Scholar]


