Abstract
P97/VCP, an endoplasmic reticulum associated protein, belongs to AAA ATPase family, ubiquitous ATPases associated with various cellular activities. Recent research has elucidated the roles of p97/VCP and evaluated its potential as a therapeutic target for some kinds of cancer diseases. We screened the small molecule compounds from a previously established library and found promise in the compound 2-[3-(2-aminoethyl)-1H-indol-1-yl]-N-benzylquinazolin-4-amine (FQ393). Data from docking simulation indicates FQ393 acts as an ATP competitor, and ATPase activity assays showed FQ393 was an inhibitor of p97/VCP. Furthermore, in vitro FQ393 is able to promote apoptosis and prohibit proliferation in a variety of cancer cell lines. Using comparative proteomic profiling of HCT-116 cells, we found significantly different canonical KEGG pathways, which revealed that the protein changes in FQ393 groups were associated with p97/VCP or tumor-related pathways. The present data suggests that FQ393 exerts antitumor activity, at least in part through p97/VCP inhibition.
1. Introduction
An evolutionarily conserved type II AAA ATPase, valosin-containing protein p97/VCP, acts as a segregase to extract target protein organelle membranes and protein or DNA complexes, thereby promoting their recycling, refolding, relocation, or degradation.1−3 A VCP forms a double-ring-shaped homohexameric complex to perform its ATPase activity. In this complex, the C terminal and N terminal are responsible for interacting with the cofactors, the D1 domain is in charge of oligomerization, and the D2 domain catalyzes the ATP hydrolysis.4,5 By combining with more than 40 cofactors, the VCP is recruited to various subcellular fractions to participate in different cellular processes, such as, autophagy, endocytosis, membrane fusion, apoptosis, DNA repair, cell cycle progression, and endoplasmic reticulum associated protein degradation (ERAD).6,7 The VCP regulates these processes through the ubiquitin–proteasome system, which regulates intracellular levels of all proteins by tagging the proteins with ubiquitin. Then, these tagged proteins are transported to the proteasome and degraded.8−10
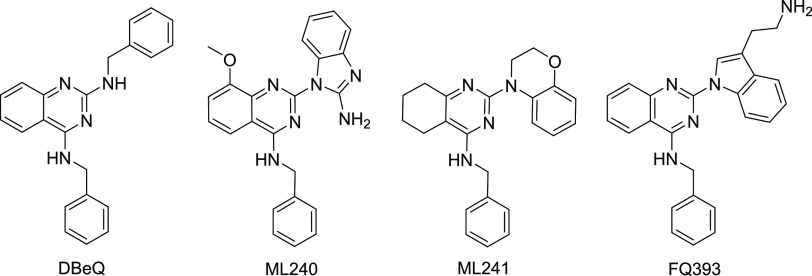
Because of its important role in regulating various physiological responses, p97/VCP has gradually become a potential therapeutic target.11,12 Autosomal dominant mutations in p97/VCP lead to a multisystem degenerative disease called IBMPFD/ALS that can manifest in patients with any combination of the following phenotypes: frontotemporal dementia (FTD), Paget’s disease of bone (PDB), inclusion body myopathy (IBM), and amyotrophic lateral sclerosis (ALS).1,13−15 Elevated p97/VCP expression has been found in several different tumors and is associated with poor clinical outcomes.16−19 Some labs have previously shown that increased levels of p97 /VCP in cancer cells allow the cancer cells to proliferate and metastasize.8−10,20 Inhibiting the function of this protein has shown to reduce cancerous cellular growth by inducing apoptosis, as well as inhibiting the migration and arresting cell cycle.8,9,20−23 Since p97/VCP plays an significant role in the malignant progress of tumor cells, some research groups have been looking for p97/VCP inhibitors with a view to eventually developing drugs for cancer treatment. DBeQ (Figure 1) is a selective, reversible ATP-competitive small molecule inhibitor of p97/VCP that inhibits the proliferation of cancer cells by impairing both autophagic protein clearance pathways and ubiquitin-independent degradation.22 By modifying the structure of DBeQ, researchers found more potent p97 /VCP inhibitors, such as ML240 and ML241 (Figure 1).12 From these molecules, the most advanced p97 /VCP inhibitor CB-5083 was subsequently developed by Cleave Biosciences.24 Due to retention of endoplasmic reticulum-associated degradation (ERAD) substrates, CB-5083 induces apoptosis of cancer cells,25 which has been indicated to be a novel approach of cancer treatment. CB-5083 entered two phase I clinical trials in 2015. However, when toxicities due to off-target effects of the compound were found, the trials were halted.26 Recently, a crystal structure combining CB-5083 with D1-D2 p97/VCP (lacking a N domain) was solved, which revealed the molecular basis of selective inhibition of CB-5083 on the D2 domain of p97/VCP.27
Figure 1.
Structures of DBeQ, ML240, ML241, and FQ393.
Since p97/VCP is identified as a potential target in cancer cells and there is no inhibitor of p97/VCP clinically available for this target, finding new p97/VCP inhibitors is worth investigating. We screened small molecule compounds from the compound library in our laboratory to exploit novel small compounds with anticancer cell proliferation activity for discovering p97/VCP inhibitors. Among those compounds, FQ393 showed a relative activity. We found that FQ393 has similar structural units with DBeQ, both of which contain quinazoline and phenylmethanamine. The difference between the two compounds is that FQ393 replaces benzylamine with tryptamine. Perhaps because of their structural similarity, FQ393 has similar p97-inhibiting activity. In the present study, we show inhibitory activity of FQ393 through p97/VCP inhibition in an ATP-competitive manner and scores on the docking with the p97/VCP protein. FQ393 inhibited hydrolysis of ATP in a dose-dependent method, and it also has a favorable binding score of −7.9 kJ/mol, which is similar to ATP (a natural ligand of p97/VCP) with a binding score of −8.3 kJ/mol. These encouraged the present study to synthesize FQ393 and evaluate its in vitro activities.
2. Results and Discussion
2.1. Synthesis of FQ393
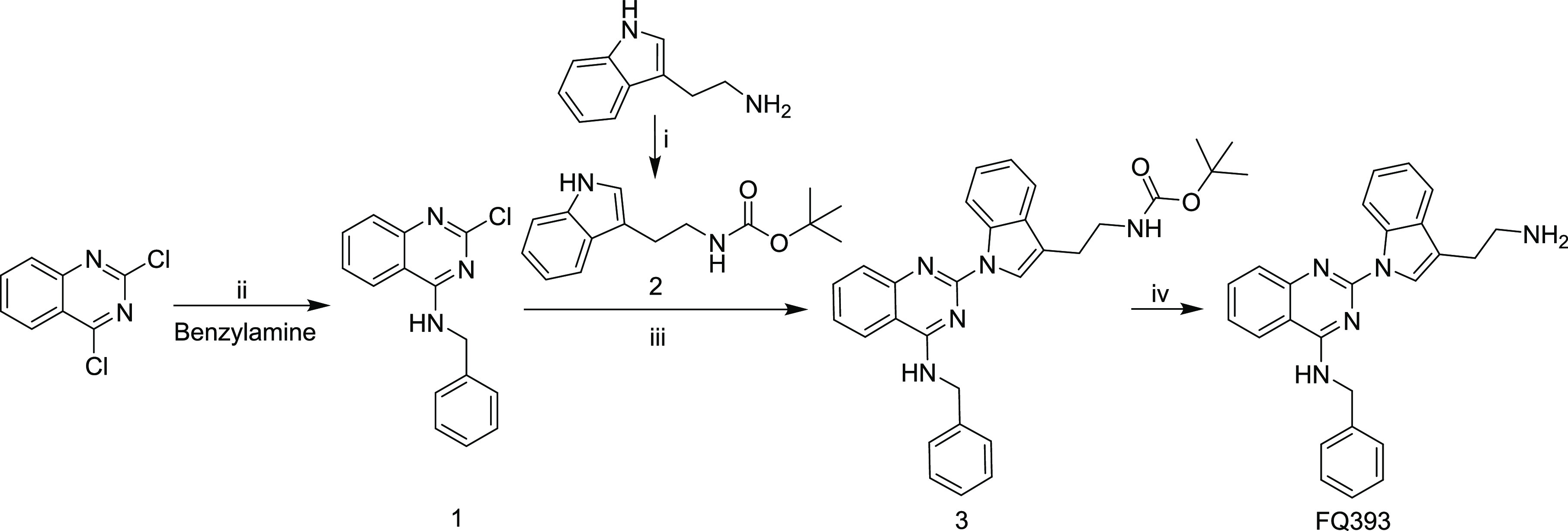
FQ393 was prepared via a four-step reaction based on Scheme 1. The yields of the four-step reaction were 92.6, 83.9, 55.6, and 92.8%, respectively. The details and related data are provided in the Experimental Section. The synthetic route is suitable for preparation of FQ393 with appropriate conditions and a simple procedure.
Scheme 1. Synthetic Route of FQ393: (i) (Boc)2O, 1,4-Dioxane, and NaOH (2 M); (ii) MeCN at RT; (iii) Cs2CO3, Pd2(dba)3, X-Phos, and MeCN at 100 °C; (iv) Hydrogen Chloride in Ethyl Acetate (4 M).
2.2. FQ393 Inhibits ATPase Activity of p97/VCP
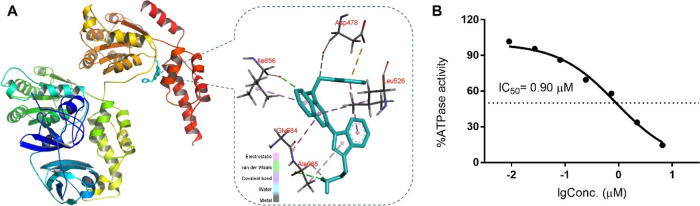
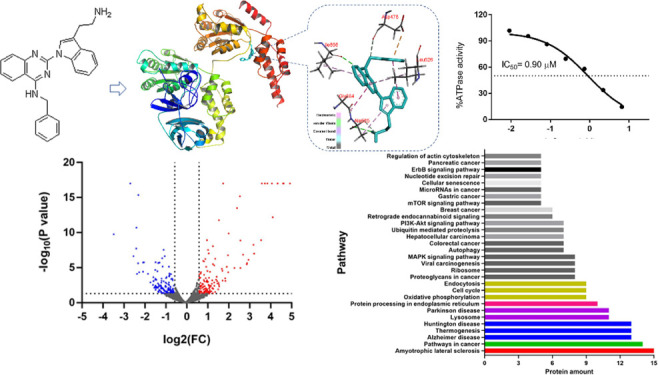
Since FQ393 has a similar structure to DBeQ, a known p97/VCP inhibitor, we investigated whether FQ393 has inhibitory activity against p97/VCP. Using structural data of active sites in the p97/VCP catalytic domain, in silico flexible docking simulations were conducted. The total binding scores and 3D docking poses between FQ393 and p97/VCP are shown in Figure 2A. The intermolecular interaction between the active pocket of p97/VCP and FQ393 mainly consists of van der Waals and electrostatic interactions in the force field representation. Van der Waals were formed by the quinazoline of FQ393 and the side chain of Ile656 of the active pocket, the phenylmethanamine of FQ393 and the side chain of Asp478 of the active pocket, and the amidogen of FQ393 and the side chain of Ala685 of the active pocket. Electrostatic interactions were formed by the quinazoline of FQ393 and the side chain of Ile656, Leu526, and Gly684 of the active pocket; and the indole of FQ393 and the side chain of Ala685 and Leu526. FQ393 has a favorable binding score of −7.9 kJ/mol, which is similar to ATP (a natural ligand of p97/VCP) with a binding score of −8.3 kJ/mol. The results indicate that FQ393 may have inhibitory activity against p97/VCP in cancer cells.
Figure 2.
(A) Docking simulation results of FQ393 with the active site of the catalytic domain of p97/VCP (PDB ID: 3CF1). Residue interactions between the compounds and the molecular surface of p97/VCP are indicated by dashed lines. The docking simulations were carried out with the molecular docking program AutoDock Vina. (B) ATPase inhibition assay in vitro shows that FQ393 suppressed ATPase activity of p97/VCP in a dose-dependent manner.
Then, we tested the performance of FQ393 as a viable inhibitor of p97/VCP using a cell-free assay. A purified p97/VCP protein was used for p97/VCP inhibition assays. FQ393 inhibited ATP hydrolysis in a dose-dependent method (Figure 2B, IC50 = 0.90 ± 0.11 μmol/L). These data encouraged us to evaluate its other in vitro antitumor activities.
2.3. FQ393 against Proliferation of Various Carcinoma Cells
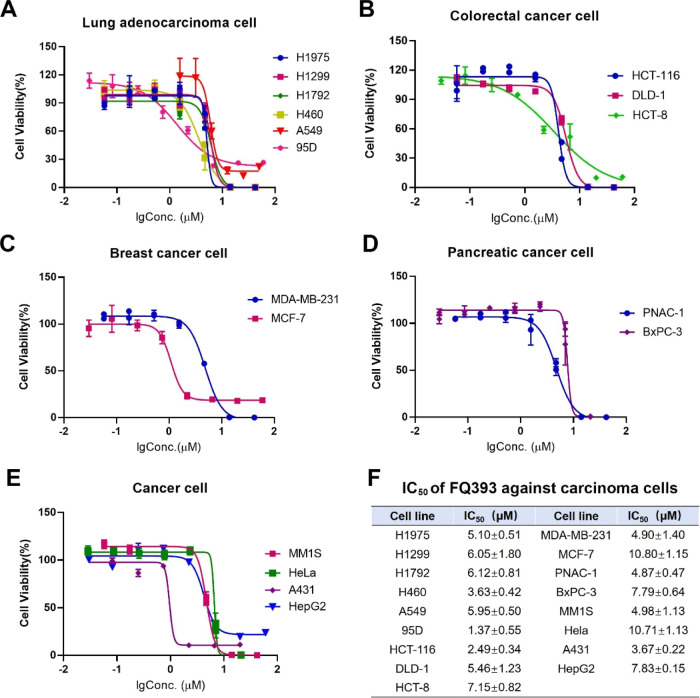
The in vitro antiproliferation of FQ393 against lung adenocarcinoma cells (H1975, H1299, H1792, H460, A549, and 95D), colorectal carcinoma cells (HCT-116, DLD-1, and HCT-8), breast carcinoma cells (MCF-7 and MDA-MB-231), pancreatic carcinoma cells (PNAC-1 and BxPC-3), and other cancer cells (human myeloma MM1S, human cervical carcinoma HeLa, human skin cancer cell line A431, and human hepatoma cell line HepG2) were evaluated by the MTT method and represented with an IC50 value. Figure 3 shows that when the cancer cells are exposed to serial concentrations (1.6–50 μM) of FQ393, the IC50 values range from 1.37 to 10.80 μM; therefore, FQ393 efficiently inhibits the proliferation of 17 carcinoma cell lines. Figure 3 also shows that when exposed to FQ393, cell lines 95D, HCT-116, H460, and A549 are more sensitive than MCF-7 and HeLa cells to apoptosis.
Figure 3.
IC50 of FQ393 against (A) lung adenocarcinoma cells, (B) colorectal cancer cells, (C) breast cancer cells, (D) pancreatic cancer cells, and (E) other cancer cells, n = 4. The exact mean and SD are shown in (F).
2.4. Apoptosis Activity of FQ393 In Vitro
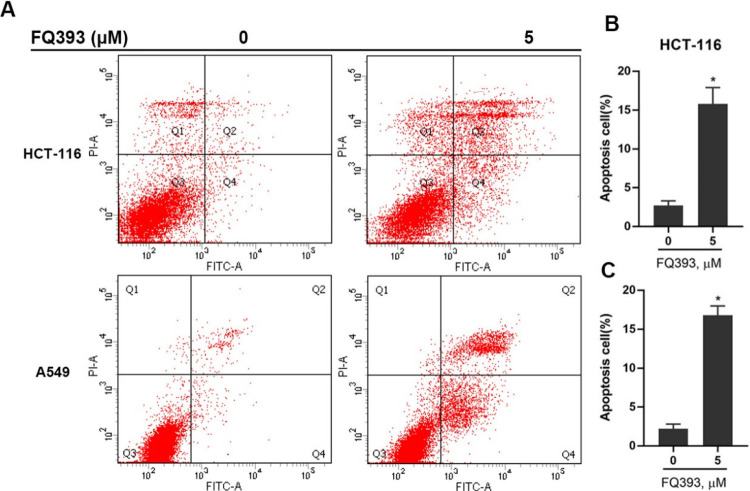
An inhibitor lead of p97/VCP, FQ393, was confirmed as an apoptosis inducer by the flow cytometry assay, and suspensions of HCT-116 and A549 cells were used in this assay. The cells were exposed to 5 μM FQ393, and the resulting data is presented in Figure 4. The data revealed that at 12 h, the total percentage of apoptosis for HCT-116 an A549 cells were 15.8 and 16.8%, respectively, indicating that FQ393 is an apoptosis inducer. We discovered that the p97/VCP inhibitor lead FQ393 can inhibit carcinoma cells proliferation and effectively induce apoptosis, and these findings provide a potential avenue for the development of tumor therapies.
Figure 4.
(A) Apoptosis assay of HCT-116 and A549 cells treated with FQ393. The cells were treated for 12 h, and cell apoptosis was detected by PI/annexin V staining and flow cytometry. Percentages of apoptotic (B) HCT-116 and (C) A549 cells after FQ393 treatment are shown. *p < 0.01 compared with the control group.
2.5. Mass Spectrometric Analysis of Proteomic Profiling after FQ393 Treatment
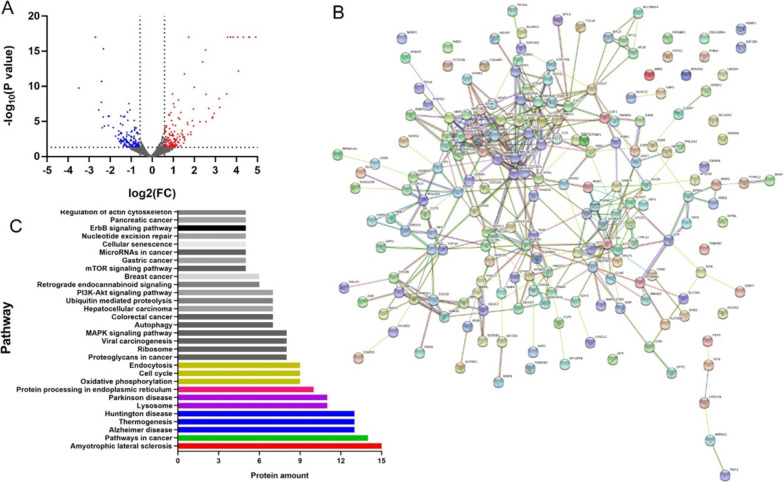
Using proteomic profiling of HCT-116 cells from DMSO and FQ393 groups, 4960 proteins were detected with at least one unique peptide and a 1% false discovery rate (FDR). Figure 5A shows a volcanic plot of all the proteins. Compared with the DMSO group, proteins with p values < 0.05 and 1.5-fold changes (>1.50 or < 0.66) for the proteomic analysis were finally considered to be differentially expressed and chosen for further analysis. Based on the criteria, 301 differentially expressed proteins were identified and selected for further analysis. Among them, 149 proteins showed increased expression, while the remaining 152 experienced a decrease. A total of 252 altered proteins were annotated by the “String” database. Figure 5B shows the protein–protein interaction network of these proteins. The significantly different canonical Kyoto Encyclopedia of Genes and Genomics (KEGG) pathways are displayed in Figure 5C. Based on differentially expressed proteins, the KEGG pathway analysis revealed ALS signaling, protein processing in endoplasmic reticulum signaling, cell cycle signaling, endocytosis signaling, ribosome signaling, autophagy signaling, ubiquitin-mediated proteolysis signaling, and nucleotide excision repair signaling, which have previously been associated with p97/VCP.6−10 Also, the KEGG pathway analysis revealed other signaling, such as pathways in cancer, proteoglycans in cancer signaling, viral carcinogenesis signaling, MAPK signaling pathway, colorectal cancer signaling, hepatocellular carcinoma signaling, PI3K-Akt signaling pathway, breast cancer signaling, mTOR signaling pathway, gastric cancer signaling, microRNAs in cancer signaling, and pancreatic cancer signaling, which are recognized tumor-related pathways.28 The data above indicate that FQ393, as a potential inhibitor of p97/VCP, may play a role in inhibiting cancer cells by inhibiting the main function of p97/VCP.
Figure 5.
Proteomic analysis of HCT-116 cells incubated with FQ393. (A) Volcano plot of all proteins (red and blue points represent increased and decreased expression respectively). (B) PPI network of 252 differently expressed proteins that were performed with String database. Nodes represent proteins. Lines represent protein–protein associations. (C) Significantly enriched pathways identified by KEGG pathway analysis (p < 0.05).
P97/VCP plays a significant role in protein homeostasis and cancer cell-dependent protein quality control mechanisms, which makes its modulation an attractive target.29 Evidence is accumulating that the expression level of p97/VCP is markedly elevated in non-small-cell lung carcinoma, colorectal carcinomas, multiple myeloma, breast carcinoma, hepatocellular carcinoma, pancreatic endocrine neoplasms, gastric carcinoma, esophageal squamous cell carcinoma, gingival squamous cell carcinoma, follicular thyroid cancer, and prostate cancer and has been found to be associated with poor prognosis.8,20,30−32 Clearly, targeting p97/VCP has potential as a therapeutic strategy for the treatment of cancer. The current study showed that FQ393 was active against many kinds of cancer cells, in which p97/VCP was overexpressed in cancer cells. The results are consistent with those reported in the literature.
Traditional heterocyclic compounds represented by DBeQ, ML-240, ML-241, NMS-873, CB-5083, and UPCDC30245 show potent effects targeting p97/VCP.29,33 In 2015, CB-5083 entered phase I clinical trials for multiple myeloma and advanced solid tumors. The first drugs targeting p97 are expected to play a role in oncology, with the potential for future trials in solid cancers and acute myeloid leukemia.29 However, clinical development was terminated due to visual loss, a consequence of off-target inhibition of phosphodiesterase-6.34 As a potent and selective, second-generation, oral small molecule inhibitor of p97/VCP, CB-5339 have entered phase I clinical trials for acute myeloid leukemia or myelodysplastic syndrome reported in the ClinicalTrails.gov (NCT04402541). Despite p97/VCP offering therapeutic opportunities, the development of effective and selective therapeutic agents for p97/VCP remains challenging. There is still an urgent need for more chemotypes of p97/VCP inhibitors. Based on unmet clinical needs, we screened the compound library, the internal library of our laboratory, including heterocyclic small molecules, small molecule peptides, and their conjugates, totaling about 3000 compounds. Interestingly, FQ393 is the most active compound, and the structure of FQ393 is similar to DBeQ. We preliminarily verified the antitumor activity of FQ393 in vitro, and FQ393 showed a relative activity. At present, the activity of FQ393 is not satisfactory, and FQ393 will be used as the lead compound for structural modification in the future, hoping to obtain candidate compounds with high activity, good selectivity, and low off-target effect.
3. Conclusions
In summary, p97/VCP is a ubiquitous protein involved in many biological processes, and p97/VCP inhibition is a promising strategy to treat cancer patients. We screened the small molecule compounds in our compound library to exploit a novel small compound, FQ393. The data from in silico docking simulations and ATPase activity assays indicate that FQ393 acts as both an ATP competitor and inhibitor of p97/VCP. Furthermore, in vitro FQ393 is able to promote the apoptosis of carcinoma cells and inhibit the proliferation of carcinoma cells. Using proteomic profiling of HCT-116 cells from DMSO and FQ393 groups, the significantly different canonical KEGG pathways revealed that the protein changes in FQ393 groups were associated with p97/VCP or a tumor-related pathway. FQ393 is thus considered a promising lead compound as an inhibitor of p97/VCP for chemotherapy.
4. Experimental Section
4.1. General
Organic solvents were dried and purified when necessary by standard methods. All commercially available reagents were purchased from Sigma (St Louis, MO, U.S.). Silica gel of 200–300 mesh was used for column chromatography. The purities of the intermediates were identified with thin-layer chromatography (TLC). The purities of products were identified with both TLC and high-performance liquid chromatography (HPLC, Waters Corporation, U.S.A., C18 column 4.6 × 150 mm) and were higher than 97%. Reactions were monitored by TLC (silica gel coated with a fluorescent indicator F 254). The melting point of FQ393 was measured on an XT5 hot stage apparatus (uncorrected, Beijing Keyi Electro-Optic Factory, China). With tetramethylsilane as the internal standard, proton nuclear magnetic resonance (1H NMR) and 13C nuclear magnetic resonance (13C NMR) spectra were recorded on a Bruker Avance 300 (300 and 75 MHz, respectively). Electrospray ionization mass spectrometry (ESI-MS) was measured on a ZQ 2000 (Waters Corp).
The data was analyzed by GraphPad Prism (version 7.0), and the results of assays are presented with mean ± standard deviation. A p value less than 0.05 was considered to have statistical significance.
4.2. Synthesis of FQ393
The preparation of FQ393 was carried out according to Scheme 1. After the four-step reaction, the target compound FQ393 is obtained in a 40.1% overall yield. Processes for the preparation and the physicochemical data of FQ393 and intermediates are given as follows.
4.2.1. Preparing N-Benzyl-2-chloroquinazolin-4-amine (1)
At 0 °C, to the solution of 2.00 g (10 mmol) of 2,4-dichloroquinazoline in 30 mL of acetonitrile (ACN), 30 mL of benzylamine was added. Then, the reaction mixture was stirred for 12 h at room temperature. The result of TLC (CH2Cl2:MeOH, 10:1) indicated the complete disappearance of 2,4-dichloroquinazoline. The reaction mixture was filtered, and the residue was washed with ACN. The obtained residue was dried to give the title compound as a colorless powder (yield = 2.51 g, 92.6%). ESI-MS (m/z): 292.21 [M + Na]+. 1H NMR (300 MHz, DMSO-d6, δ/ppm): 9.31 (t, J = 5.4 Hz, 1H), 8.32 (d, J = 8.1 Hz, 1H), 7.33 (d, J = 7.8 Hz, 1H), 7.82 (t, J = 7.5 Hz, 1H), 7.64 (d, J = 8.1 Hz, 1H), 7.55 (t, J = 7.5 Hz, 1H), 7.34–7.26 (m, 5H), 4.76 (d, J = 5.4 Hz, 2H). 13C NMR (75 MHz, DMSO-d6, δ/ppm): 161.58, 157.37, 150.84, 138.97, 134.20, 128.86, 127.88, 127.50, 127.15, 126.68, 123.57, 113.96, 44.35.
4.2.2. Preparing tert-Butyl [2-(1H-indol-3-yl)ethyl]carbamate (2)
To the solution of 2.00 g (10 mmol) of tryptamine in 5 mL of distilled water ,10 mL of aqueous NaOH (2 N) was added dropwise. In 15 mL of anhydrous 1,4-dioxane, 2.66 g (12 mmol) of (Boc)2O was dissolved and added to the reaction solution dropwise. The reaction mixture was stirred for 0.5 h and bubble removal was performed. The reaction mixture was stirred for 12 h at room temperature, and its pH is kept at 8–9 by adding 2 N aqueous NaOH. TLC detection (CH2Cl2:MeOH, 20:1) indicated raw material tryptamine was disappeared. The pH of the solution was adjusted to 7 by adding saturated aqueous KHSO4. Reaction solution removes the solvent under a reduced pressure, and the residue was dissolved in 25 mL of water, adjusted to pH 2 by adding saturated aqueous KHSO4, transferred into a separating funnel and extracted with 40 mL of ethyl acetate (3x). The combined organic phase was washed with saturated aqueous NaCl and dried over anhydrous Na2SO4 for 2 h. The solution was filtered and evaporated under vacuum. The residue was crystallized in petroleum ether to give colorless powder (2.22 g), yield = 83.9%. ESI-MS (m/z): 283.30 [M + Na]+. 1H NMR (300 MHz, DMSO-d6, δ/ppm): 10.79 (s, 1H), 7.51 (d, J = 7.8 Hz, 1H), 7.33 (d, J = 7.8 Hz, 1H), 7.13–6.89 (m, 4H), 3.20 (q, J = 6.6 Hz, 2H), 2.79 (t, J = 7.8 Hz, 2H), 1.39 (s, 9H).
4.2.3. Preparing tert-Butyl[2-(1-(4-(benzylamino)quinazolin-2-yl)-1H-indol-3-yl)ethyl]carbamate (3)
A mixture of 0.35 g (1.3 mmol) of compound 2 and 1.19 g (3.7 mmol) of Cs2CO3 in 10 mL of ACN was refluxed in a 100 °C oil bath for 20 min to form solution A. A mixture of 0.33 g (1.2 mmol) of compound 1, 0.17 g (0.4 mmol) of X-Phos, and 0.09 g (0.4 mmol) of Pd2(dba)3 in 25 mL of ACN were dissolved to form solution B. Solution B was added to solution A dropwise, and the reaction mixture was refluxed for 6 h in a 100 °C oil bath. TLC (CH2Cl2:MeOH, 20:1) detection indicated the raw material disappeared. The reaction mixture was filtered to remove the precipitate immediately. After purification on a silica gel column, the title compound was provided (0.33 g, 55.6%) as a light yellow powder. ESI-MS (m/z): 494.48 [M + H]+. 1H NMR (300 MHz, DMSO-d6, δ/ppm): 9.18 (t, J = 4.5 Hz, 1H), 8.77 (d, J = 7.8 Hz, 1H), 8.21 (d, J = 7.8 Hz, 1H), 8.13 (s, 1H), 7.80–7.70 (m, 2H), 7.57 (d, J = 6.9 Hz, 1H), 7.50–7.13 (m, 8H), 6.96 (m, 1H), 4.92 (d, J = 4.5 Hz, 2H), 3.29 (q, J = 6.0 Hz, 2H), 2.84 (t, J = 6.9 Hz, 2H), 1.31 (s, 9H). 13C NMR (75 MHz, DMSO-d6, δ/ppm): 161.05, 156.10, 154.28, 150.80, 139.76, 135.58, 133.76, 131.05, 128.85, 127.60, 127.31, 127.13, 124.48, 124.00, 123.48, 121.52, 119.02, 116.82, 116.42, 113.22, 77.95, 44.81, 28.73, 25.57.
4.2.4. Preparing 2-[3-(2-Aminoethyl)-1H-indol-1-yl]-N-benzylquinazolin-4-amine (FQ393)
The solution of 0.15 g (0.3 mmol) of compound 3 in 7 mL of ethyl acetate containing hydrogen chloride (4 M) was stirred at 0 °C for 2 h; then, the reaction solution was evaporated under vacuum. The residue was dissolved in ethyl acetate and evaporated under vacuum. This procedure was repeated for three times so as to remove the excess hydrogen chloride thoroughly. The residue is reconstituted with ethyl acetate and washed with saturated NaHCO3 to neutrality; then, the combined organic layers were washed with saturated aqueous NaCl and dried over anhydrous Na2SO4 for 2 h. The solution was filtered and evaporated under a reduced pressure to give the title compound (0.13 g, 92.8%) as a colorless powder. ESI-MS (m/z): 394.42 [M + H]+. Mp: 248–250 °C. Infrared (IR): 3085.26, 3050.18, 2872.35, 2163.74, 1632.87, 1566.54, 1462.17, 1380.66, 1253.61, 1205.00, 745.33, 694.70, 678.97 cm–1. 1H NMR (300 MHz, DMSO-d6, δ/ppm): 9.66 (m, 1H), 8.66 (d, J = 5.7 Hz, 1H), 8.44 (d, J = 5.1 Hz, 1H), 8.31 (s, 1H), 8.05 (m, 3H), 7.84 (m, 2H), 7.64 (t, J = 7.5 Hz, 2H), 7.28–7.22 (m, 3H), 4.95 (d, J = 4.8 Hz, 2H), 3.14–3.07 (m, 4H). 13C NMR (75 MHz, DMSO-d6, δ/ppm): 161.17, 139.37, 135.70, 134.26, 130.49, 128.90, 127.55, 127.40, 125.85, 125.08, 124.74, 124.01, 123.86, 122.11, 119.08, 116.92, 114.98, 113.05, 44.98, 23.24.
4.3. Determining IC50 Values of FQ393 in ATPase Assays
The detailed method has been reported in the literature.23 Briefly, inhibition of human p97/VCP (25 nM monomer) was carried out in the assay buffer (50 mM Tris, pH 7.4; 1 mM EDTA; 20 mM MgCl2; 0.5 mM TCEP) containing 200 μM ATP and 0.01% Triton X-100. The IC50 value of FQ393 in blocking ATPase activity was determined by eight-dose titration through the addition of Biomol Green reagent (Enzo Life Sciences, Farmingdale, NY, U.S.A.).
4.4. In Vitro Anti-Proliferation Assay
A549, DLD-1, MM1S, NCI-H1975, NCI-H1299, NCI-H1792, H460, HCT-8, BxPC-3, 95D, and HCT-116 cells were cultured in an RPMI 1640 medium supplemented with 10% fetal calf serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin. MDA-MB-231, PANC, HeLa, A431, HepG2, and MCF-7 were maintained in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. The tumor cells were maintained in a 37 °C, 5% CO2-humidified incubator. The medium was changed every 2 days. The proliferation of carcinoma cells above was determined by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. In brief, A549, DLD-1, MM1S, NCI-H1975, NCI-H1299, NCI-H1792, H460, HCT-8, HCT-116, MDA-MB-231, PANC, HeLa, A431, HepG2, and MCF-7 cells in the logarithmic growth phase were plated into 96-well plates (4 × 103 cells per well) and incubated overnight in an RPMI 1640 medium or DMEM supplemented with 10% FBS.
The cells were treated with FQ393 (final concentrations: 1.6, 3.1, 6.3, 12.5, 25, and 50 μM) for 48 h; MTT reagent (25 μL, 5 mg/mL) was added to each well. After a 4 h incubation at 37 °C, the supernatant was replaced by 100 μL DMSO. The optical density (O.D) was measured at 492 nm using a Spectra Max M3 microplate reader (BioTek, Winooski, VT, U.S.). The proliferation of A549, DLD-1, MM1S, NCI-H1975, NCI-H1299, NCI-H1792, H460, HCT-8, BxPC-3, 95D, HCT-116, MDA-MB-231, PANC, HeLa, A431, HepG2, and MCF-7 cells was measured in the O.D value. Each measurement was performed in triplicate.
4.5. Flow Cytometry Assay
To explore the apoptotic activity of FQ393, flow cytometry assay was performed. HCT-116 and A549 cells (106 cells/mL) were incubated in an RPMI 1640 medium with 10% FBS, 60 μg/mL penicillin, and 100 μg/mL streptomycin at 37 °C under a humidified atmosphere containing 5% CO2 for 4 h. A solution of FQ393 (final concentration = 5 μM) was added into the 6-well plates, and cells were incubated at 37 °C for another 12 h. After removing the medium, the cells were washed twice with a fresh medium (1 mL), followed by staining with Annexin V-FITC (KeyGEN Biological Technology Co., Ltd., Nanjing, P.R. China) and propidium iodide (PI) for 10 min. FITC and PI fluorescence for cells were analyzed for ∼10,000 events (counts) per sample aliquot by flow cytometry.
4.6. Mass Spectrometric Analyses
HCT-116 cell lysis was proteolyzed and labeled with TMT 10-plex (Thermo Fisher Scientific) following the manufacturer’s protocol. For the proteome analysis, HPLC-MS/MS analysis was performed using an Orbitrap Fusion Lumos mass spectrometer equipped with an EASY-nLC 1000 liquid chromatography system and a nanoelectrospray ionization source (Thermo Scientific, U.S.A.). The specific method of the experiment was the same as that reported in the literature.35 Compared with the DMSO group, proteins quantified with a p value ≤ 0.05 and a fold change of >1.50 or <0.66 were considered to be significantly expressed and used for further proteomic analysis. Differentially expressed protein interaction data was identified by the STRING online tool (version 9.0).
Acknowledgments
We are grateful to Dr. Tsui-fen Chou for her suggestions on the revision of the article. This work was supported by Funding for Training Excellent Talents in Beijing and Scientific research, Cultivation Fund of the Capital Medical University (PYZ19145).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c04478.
1H NMR for compound 2-[3-(2-aminoethyl)-1H-indol-1-yl]-N-benzylquinazolin-4-amine (FQ393), 13C NMR for compound FQ393, and MS for compound FQ393 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- van den Boom J.; Meyer H. VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol. Cell 2018, 69, 182–194. 10.1016/j.molcel.2017.10.028. [DOI] [PubMed] [Google Scholar]
- Stach L.; Freemont P. S. The AAA+ ATPase p97, a cellular multitool. Biochem. J. 2017, 474, 2953–2976. 10.1042/BCJ20160783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekaria P. H.; Home T.; Weir S.; Schoenen F. J.; Rao R. Targeting p97 to Disrupt Protein Homeostasis in Cancer. Front. Oncol. 2016, 6, 181. 10.3389/fonc.2016.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B.; Brunger A. T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003, 10, 856–863. 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- Xia D.; Tang W. K.; Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene 2016, 583, 64–77. 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.; Bug M.; Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012, 14, 117–123. 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Buchberger A.; Schindelin H.; Hänzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015, 589, 2578–2589. 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Valle C. W.; Min T.; Bodas M.; Mazur S.; Begum S.; Tang D.; Vij N. Critical role of VCP/p97 in the pathogenesis and progression of non-small cell lung carcinoma. PLoS One 2011, 6, e29073 10.1371/journal.pone.0029073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij N. AAA ATPase p97/VCP: cellular functions, disease and therapeutic potential. J. Cell Mol. Med. 2008, 12, 2511–2518. 10.1111/j.1582-4934.2008.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parzych K.; Chinn T. M.; Chen Z.; Loaiza S.; Porsch F.; Valbuena G. N.; Kleijnen M. F.; Karadimitris A.; Gentleman E.; Keun H. C.; Auner H. W. Inadequate fine-tuning of protein synthesis and failure of amino acid homeostasis following inhibition of the ATPase VCP/p97. Cell Death Dis. 2015, 6, e2031 10.1038/cddis.2015.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P.; D’alessio R.; Valsasina B.; Avanzi N.; Rizzi S.; Asa D.; Gasparri F.; Cozzi L.; Cucchi U.; Orrenius C.; Polucci P. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013, 9, 548–556. 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Chou T. F.; Li K.; Frankowski K. J.; Schoenen F. J.; Deshaies R. J. Structure–Activity Relationship Study Reveals ML240 and ML241 as Potent and Selective Inhibitors of p97 ATPase. ChemMedChem 2013, 8, 297–312. 10.1002/cmdc.201200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halawani D.; LeBlanc A. C.; Rouiller I.; Michnick S. W.; Servant M. J.; Latterich M. Hereditary Inclusion Body Myopathy-Linked p97/VCP Mutations in the NH2 Domain and the D1 Ring Modulate p97/VCP ATPase Activity and D2 Ring Conformation. Mol. Cell. Biol. 2009, 29, 4484–4494. 10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. K.; Xia D. Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants. J. Biol. Chem. 2013, 288, 36624–36635. 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. O.; Mandrioli J.; Benatar M.; Abramzon Y.; Van Deerlin V. M.; Trojanowski J. Q.; Gibbs J. R.; Brunetti M.; Gronka S.; Wuu J.; Ding J.; McCluskey L.; Martinez-Lage M.; Falcone D.; Hernandez D. G.; Arepalli S.; Chong S.; Schymick J. C.; Rothstein J.; Landi F.; Wang Y. D.; Calvo A.; Mora G.; Sabatelli M.; Monsurrò M.; Battistini S.; Salvi F.; Spataro R.; Sola P.; Borghero G.; Galassi G.; Scholz S.; Taylor J.; Restagno G.; Chiò A.; Traynor B. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010, 68, 857–864. 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.; Fry A. N.; Kang M. J. The complexities of p97 function in health and disease. Mol. BioSyst. 2011, 7, 700–710. 10.1039/C0MB00176G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai R. M.; Li C. C. H. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 2001, 3, 740–744. 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Asai T.; Tomita Y.; Nakatsuka S.; Hoshida Y.; Myoui A.; Yoshikawa H.; Aozasa K. VCP (p97) regulates NFkappaB signaling pathway, which is important for metastasis of osteosarcoma cell line. Jpn. J. Cancer Res. 2002, 93, 296–304. 10.1111/j.1349-7006.2002.tb02172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G.; Graumann J.; Smith G. T.; Kolawa N. J.; Fang R.; Deshaies R. J. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell 2008, 134, 804–816. 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessart D.; Marza E.; Taouji S.; Delom F.; Chevet E. P97/CDC-48: proteostasis control in tumor cell biology. Cancer Lett. 2013, 337, 26–34. 10.1016/j.canlet.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Fang C. J.; Gui L.; Zhang X.; Moen D. R.; Li K.; Frankowski K. J.; Lin H. J.; Schoenen F. J.; Chou T. F. Evaluating p97 inhibitor analogues for their domain selectivity and potency against the p97-p47 complex. ChemMedChem 2015, 10, 52–56. 10.1002/cmdc.201402420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. F.; Brown S. J.; Minond D.; Nordin B. E.; Li K.; Jones A. C.; Chase P.; Porubsky P. R.; Stoltz B. M.; Schoenen F. J.; Patricelli M. P.; Hodder P.; Rosen H.; Deshaies R. J. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 4834–4839. 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. F.; Bulfer S. L.; Weihl C. C.; Li K.; Lis L. G.; Walters M. A.; Schoenen F. J.; Lin H. J.; Deshaies R. J.; Arkin M. R. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J. Mol. Biol. 2014, 426, 2886–2899. 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J.; Le Moigne R.; Djakovic S.; Kumar B.; Rice J.; Wong S.; Wang J.; Yao B.; Valle E.; Kiss von Soly S.; Madriaga A.; Soriano F.; Menon M. K.; Wu Z. Y.; Kampmann M.; Chen Y.; Weissman J. S.; Aftab B. T.; Yakes F. M.; Shawver L.; Zhou H. J.; Wustrow D.; Rolfe M. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 2015, 28, 653–665. 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. F.; Deshaies R. J. Development of p97 AAA ATPase inhibitors. Autophagy 2011, 7, 1091–1092. 10.4161/auto.7.9.16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S.; Bartesaghi A.; Merk A.; Rao P.; Bulfer S. L.; Yan Y.; Green N.; Mroczkowski B.; Neitz R. J.; Wipf P.; Falconieri V.; Deshaies R. J.; Milne J. L. S.; Huryn D.; Arkin M.; Subramaniam S. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 2016, 351, 871–875. 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. K.; Odzorig T.; Jin W.; Xia D. Structural Basis of p97 Inhibition by the Site-Selective Anticancer Compound CB-5083. Mol. Pharmacol. 2019, 95, 286–293. 10.1124/mol.118.114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling S.; Xie H.; Yang F.; Shan Q.; Dai H.; Zhuo J.; Wei X.; Song P.; Zhou L.; Xu X.; Zheng S. Metformin potentiates the effect of arsenic trioxide suppressing intrahepatic cholangiocarcinoma: roles of p38 MAPK, ERK3, and mTORC1. J. Hematol. Oncol. 2017, 10, 59. 10.1186/s13045-017-0424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huryn D. M.; Kornfilt D. J.; Wipf P. p97: An Emerging Target for Cancer, Neurodegenerative Diseases, and Viral Infections. J. Med. Chem. 2020, 63, 1892–1907. 10.1021/acs.jmedchem.9b01318. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y.; Tomita Y.; Hoshida Y.; Kono T.; Oka T.; Yamamoto S.; Nonomura N.; Okuyama A.; Aozasa K. Elevated expression of valosin-containing protein (p97) is associated with poor prognosis of prostate cancer. Clin. Cancer Res. 2004, 10, 3007–3012. 10.1158/1078-0432.CCR-03-0191. [DOI] [PubMed] [Google Scholar]
- Le Moigne R.; Aftab B. T.; Djakovic S.; Dhimolea E.; Valle E.; Murnane M.; King E. M.; Soriano F.; Menon M. K.; Wu Z. Y.; Wong S. T.; Lee G. J.; Yao B.; Wiita A. P.; Lam C.; Rice J.; Wang J.; Chesi M.; Bergsagel P. L.; Kraus M.; Driessen C.; Kiss von Soly S.; Yakes F. M.; Wustrow D.; Shawver L.; Zhou H. J.; Martin T. G. III; Wolf J. L.; Mitsiades C. S.; Anderson D. J.; Rolfe M. The p97 Inhibitor CB-5083 Is a Unique Disrupter of Protein Homeostasis in Models of Multiple Myeloma. Mol. Cancer Ther. 2017, 16, 2375–2386. 10.1158/1535-7163.MCT-17-0233. [DOI] [PubMed] [Google Scholar]
- Meyer H.; Weihl C. C. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 2014, 127, 3877–3883. 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Li S.; Gan T.; Stott G. M.; Flint A.; Chou T. F. Allosteric p97 Inhibitors Can Overcome Resistance to ATP-Competitive p97 Inhibitors for Potential Anticancer Therapy. ChemMedChem 2020, 15, 685–694. 10.1002/cmdc.201900722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshow J. H.; Parchment R.; Moscow J.. NCI Experimental Therapeutics (NExT) Program, FMLAC Meeting May 8, 2018. https://deainfo.nci.nih.gov/advisory/fac/0518/Doroshow.pdf.
- Zhang Y.; Jing H.; Wen T.; Wang Y.; Zhao Y.; Wang X.; Qian X.; Ying W. Phenylboronic acid functionalized C3N4 facultative hydrophilic materials for enhanced enrichment of glycopeptides. Talanta 2019, 191, 509–518. 10.1016/j.talanta.2018.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.