Abstract
Colorectal cancer (CRC) is one of the most frequently diagnosed cancers worldwide. Lifestyle-related factors, such as diet, are associated with the development of CRC. Cumulating evidence indicates noticeable chemopreventive effects of phytochemicals on CRC, suggesting that drinking herbal tea potentially reduces the risk of distal colon cancer via its antiproliferative and anti-angiogenic activities. We examine the antitumor effects of nine components frequently found in herbal tea and uncover the underlying molecular mechanism. Among them, the hot water extract of Melissa officinalis (MO) exhibited the highest anticancer activity on CRC cells. We revealed that MO reduced cell proliferation, induced cell cycle arrest at the G2/M phase, triggered caspase-dependent apoptotic cell death, and inhibited cell migration ability by modulating the epithelial–mesenchymal transition in HCT116 CRC cells. To examine the metabolite composition in the MO hot water extract, we applied mass spectrometry-based analysis and identified 67 compounds. Among them, the phenolic compounds, including lignans, phenylpropanoids, and polyketides, are widely found in natural products and possess various bioactivities such as anti-inflammatory, antioxidation, and anticancer effects. The results indicate that herbal tea consumption benefits CRC prevention and management.
1. Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers worldwide and the third leading cause of cancer death in the United States.1 Most of the CRC cases are sporadic, and about 25% of cases are from family inheritance, implying both genetic background and environment play roles in CRC occurrence.2,3 Among the CRC associated risk factors, diet strongly affects the risk of CRC. For example, intaking high dietary fat and meats can promote the occurrence of CRC. Conversely, a proper diet like fish, fiber, vitamin D, and calcium benefits in preventing CRC.4−6 Additionally, a previous study showed that people who consume one or more cups of herbal tea per week were related to a reduced risk of distal colon cancer,7 suggesting a potential role of drinking herbal tea in CRC prevention.
Noticeable chemopreventive effects on CRC from numerous phytochemicals, such as curcumin, polysaccharides, resveratrol, quercetin, flavonoids, rosmarinic acid (RA), and phenolic acids have been reported.8−10 These phytochemicals derived from natural products exhibit anticarcinogenic properties by regulating multiple cellular signaling molecules.11 For example, curcumin inhibits transcription factors, NF-κB, AP-1, and STAT3, and downregulates growth- and metastasis-promoting genes, such as cyclin D, matrix metalloproteinase-9 (MMP-9), and MMP-2, to prevent cancer cell growth and induce apoptosis;12 polyphenols like RA reduce cancer metastasis of CRC by activating the AMPK pathway.13 Furthermore, strong evidence of in vitro, in vivo, and clinical trials indicates that a plant-based diet can reduce the risk of CRC.14
Herbal tea has been widely consumed as beverages for health in the world. Because of rich bioactive ingredients such as polyphenols, herbal tea possessing various biological effects such as antioxidation, anti-inflammatory, and anti-carcinogenicity.15−17 Along with the popularity of drinking tea, numerous scientists investigated the health benefits of the consumption of herbal tea. However, detailed molecular mechanisms of herbal tea in preventing CRC remained limited. To reveal the major bioactive components of herbal tea and its effects on CRC, we measured the antitumor activity of hot water extracts of nine frequently applied herbs including Verbena officinalis, Melissa officinalis (MO), Hyssopus officinalis, Salvia officinalis, Urtica dioica, Hemerocallis fulva, Citrus maxima, Citrus limon, and Ficus formosana. We demonstrated that MO is the most active extract to inhibit the proliferation of CRC cells.
MO, also known as lemon balm, is one of the commonly used herbs in Europe, Iran, the Mediterranean region, and central Asia. It has been used for cooking and relieving mental diseases, cardiovascular diseases, respiratory problems, and several types of cancer.18 MO extracts in ways of water, ethanol, methanol, and essential oil differ in pharmacological activities, including antioxidant, antimicrobial, antiviral, anticancer, cytotoxic effects, anti-anxiety, antidepressant, antinociceptive, and anti-inflammatory effects.18−21 Although the antitumor potential of the MO extract has been revealed in CRC cells,19,22 the cellular effects and molecular mechanisms induced by the MO water extract are still poorly understood. In this study, we investigate how MO inhibits CRC progression by accessing cell physiological functions and analyzing the major ingredients of the hot water extract that potentially benefit humans. We demonstrate that MO prevented cell proliferation by hindering the entry of the mitotic phase of the cell cycle and triggering apoptotic cell death in a caspase-dependent manner and inhibited the cell migration ability by modulating epithelial–mesenchymal transition (EMT) in HCT116 CRC cells.
2. Materials and Methods
2.1. Sample Extraction
Although the optimal brewing temperature of MO tea is about 95 °C and steeping for 5 min, we used the method of 100 °C for 30 min to improve the extraction efficiency. Fifty grams of the dried samples were boiled twice at 100 °C for 30 min in 500 mL of deionized water and filtered with filter paper. The extraction step was repeated. After extraction, all of the filtrates were collected and freeze-dried to powder for storage.
(V. officinalis: 22.3%; MO: 20.9%; H. officinalis: 15.1%; S. officinalis; 21.9%, U. dioica: 13.9%; H. fulva: 19.8%; C. maxima: 19.2%; C. limon: 26.9%; F. formosana: 4.6%)
The lyophilized water extract was dissolved in deionized water at indicated concentrations and sterile with 0.22 μm filter for functional assays.
2.2. Cell Culture
Human colon cancer cell lines HCT116, HT-29, SW620, and DLD-1 were purchased from Bioresource Collection and Research Center (BCRC, Taiwan) and were cultured at 37 °C in a humidified atmosphere of 5% CO2 in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with the 10% fetal bovine serum (FBS; Gibco). Cells were treated with the water extracts of herbs at indicated concentrations for 48 h before harvest.
2.3. Cell Viability
In MTS assay, 2000 cells per well were seeded in 96-well plates overnight and treated as indicated. After treatment, cell viability was determined by coulometric measurement with CellTiter 96 AQueous non-radioactive cell proliferation assay (Promega, Madison, WI, USA) and measured at 490 nm using an Epoch Microplate Spectrophotometer (BioTek Instruments, Winooski, VT, USA) according to the user manual. In trypan blue staining assay, a total of 1 × 106 cells were seeded in 10 cm dish overnight and treated as indicated. Cell viability was determined by cell counting with trypan blue dye exclusion method (Invitrogen, Carlsbad, CA, USA).
2.4. Colony Formation
Cells were pretreated with MO (water extract from MO) or deionized water (control) for 48 h. After treatment, the cells washed with PBS, trypsinized, seeded in 6-well plates at a density of 250 cells/well, and then cultured for eight days to form colonies. The colonies were fixed with methanol and stained with Giemsa (Sigma-Aldrich, St. Louis, MO, USA). The statistical significance of the difference was compared with the control. All experiments were performed in triplicate (n = 3).
2.5. Western Blot Analysis
Cells were washed with cold PBS and harvested in iced RIPA lysis buffer (Bioman, Taipei, Taiwan) containing a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA). The cells lysate was homogenized on ice using a homogenizer (UP50H with sonotrode 3; Hielscher, Ultrasound Technology) and clarified by centrifugation at 17,000g for 20 min at 4 °C. The protein concentration was determined by the T-pro BCA Protein Assay (T-Probiotechnology, Taipei, Taiwan). Protein samples were boiled at 95 °C for 5 min, separated by SDS-PAGE, and then transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The blots were blocked with BlockPROTM Blocking buffer (Visual PROTEIN Biotechnology Corporation, Taipei, Taiwan), incubated with primary antibodies at 4 °C overnight, and subsequently probed with corresponding horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h. The hybridization signals were developed by the enhanced chemiluminescence (Advansta, Menlo Park, CA, USA) and detected by an Amersham Imager 600 (GE Healthcare Life Sciences, MA, USA). Western blots signal intensities were quantified by ImageJ software.
2.6. Antibodies
Antibodies probing Cyclin B1, CDC25C, p-CDC2 (Tyr 15), CDC2, Caspase 3, N-cadherin, and E-cadherin were purchased from GeneTex (San Antonio, TX, USA). Antibodies probing Cleaved PARP, Caspase 7, and anti-rabbit/mouse IgG-horseradish peroxidase were obtained from Cell Signaling Technology (Danvers, MA, USA). Anti-β-actin from Millipore (Billerica, MA, USA) was used as an internal control.
2.7. Cell Cycle Analysis
Cells were treated with MO at concentrations of 0, 250, 375, and 500 μg/mL for 48 h. After treatment, cells were washed with PBS three times and fixed with 70% ethanol at −20 °C overnight. The fixed cells were washed with PBS, incubated with RNase A at 37 °C for 1 h, and stained with propidium iodide (PI) for 15 min at room temperature. The cell cycle phase distribution was examined by an Attune NxT acoustic focusing flow cytometer (Thermo Fisher Scientific Inc.) and analyzed using ModFit LT software (Verity Software House).
2.8. Apoptosis Assay of Annexin V/PI Staining
Cells were treated with MO at concentrations of 0, 250, 375, and 500 μg/mL for 48 h. After treatment, cells were harvested and stained with a FITC Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, USA) for cell apoptosis analysis. Cells were stained with Annexin V-FITC and PI for 15 min in the dark at room temperature. Then, the stained cells were analyzed by an Attune NxT acoustic focusing flow cytometer.
2.9. Migration Assay
After treatment, cells were trypsinized and seeded onto the insert of 8 μm pore size transwell plates (Corning, MA, USA) in medium containing 1% FBS. The inserts containing cells were assembled above the lower compartments containing medium with 10% FBS and incubated for 18 h. After incubation, the cells were fixed with methanol and stained with Giemsa (Sigma-Aldrich).
2.10. UPLC System
We acquired the metabolite profiling at the mass spectrometry core of the Taipei Medical University Core Facility. The untargeted metabolite profiling of the MO extract was performed by UPLC-QTOF under the data-dependent analysis (DDA) and data-independent analysis (DIA) modes. Each mode was conducted in the negative and positive ionization modes. The MO water extract was injected into a Waters Acquity UPLC system coupled a SYNAPT G2 Q-ToF (Waters Co., Manchester, UK). Separation was achieved on a UPLC HSS T3 C18 guard column and an analytical column (2.1 × 100 mm, 1.8 μm, Waters Co., Milford, MA, USA) thermostatted at 40 °C using the following binary gradient composed of solvent A (water with 0.1% formic acid) to solvent B (acetonitrile with 0.1% formic acid): 0 to 1 min, isocratic 5% B; 1 to 7 min, gradient phase to 20% B; 7 to 14 min, gradient phase to 40% B; 14 to 15 min, isocratic 40% B; 15 to 16 min, gradient phase to 100% B; 16 to 18 min, isocratic 100% B; 18.1 to 20 min, isocratic 5% B. The flow rate was set at 0.3 mL/min.
2.11. Q-TOF MS Analysis
Data were collected in the electrospray ionization (ESI) mode (both ESI+ and ESI–), in the range of m/z 150–1000 every 1.2 s during the chromatographic separation. Capillary voltages were optimized separately for positive (2.5 kV) and negative (2 kV) ion modes. MSE parameters were as listed: sampling cone, 30.0 V; extraction cone, 4.0 V; desolvation gas flow, 900 L h–1; source temperature, 120 °C; and desolvation temperature, 450 °C; trap CE, off (low CE collection), and trap CE ramp 8–40 V (high CE collection). Lockspray configuration used the average of three m/z measurements every 10 s of protonated leucine-enkephalin (m/z 556.2771) formed from the infusion of a 1 μg/mL solution. This configuration typically yields mass accuracies <2 ppm. All the acquisition was controlled by Waters MassLynx v4.1 software (Waters Co., Manchester, UK).
2.12. Metabolome Data Analysis
Data processing was conducted by the Progenesis QI (Version 2.4, Waters Co., Milford, MA, USA) with default settings for peak alignment, normalization, signal integration, and initial compound assignments. Metabolites were identified by comparing accurate masses, MS/MS fragmentation patterns, and isotope patterns with authentic standards against METLIN mass spectral database.23 To obtain high-confident results, we only accepted the identified metabolites with the highest score and evidential MS2 spectrum. The identified compounds were classified using ClassyFire, a chemical classification web tool (http://classyfire.wishartlab.com).24
2.13. Statistics
All experiments were repeated three times. The data were analyzed by using the 2-tailed Student’s t-test. In Figure 1, the statistically significant p-value of the comparison between the water control and each extract in different concentrations is are denoted: *p < 0.01, **p < 0.001, ***p < 0.0001 for 100 μg/mL treatments; #p < 0.01, ##p < 0.001, ###p < 0.0001 for 500 μg/mL treatments; and +p < 0.01, ++p < 0.001, +++p < 0.0001 for 1000 μg/mL treatments. In other figures, statistically significant p-value is denoted: *p < 0.05, **p < 0.01, ***p < 0.001.
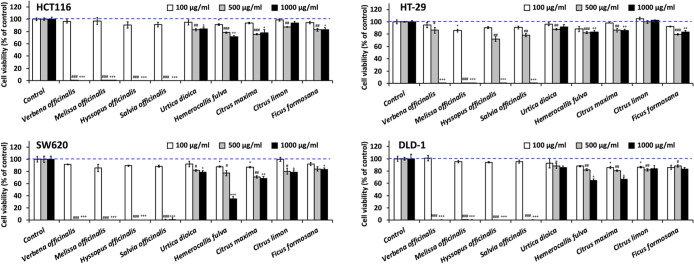
Figure 1.
Effects on cell viability of hot water extracts of herbs on CRC cell lines. HCT116, HT-29, SW620, or DLD-1 cells were seeded in 96-well plates at a density of 2000 cells/well and incubated overnight. Cells were then treated with hot water extracts of herbal tea with a concentration of 100, 500, and 1000 μg/mL for 48 h. After 48 h treatment, cell viability was measured by the MTS assay at 490 nm. The results were normalized to water control and shown in mean (% of control) ± SD of three sampled replicates. Dash lines indicate 100% viability. *, p < 0.01; **, p < 0.001; ***, and p < 0.0001 compares the 100 μg/mL of extracts to the swater control. #, p < 0.01; ##, p < 0.001; and ###, p < 0.0001 compares the 500 μg/mL of extracts to the water control. +, p < 0.01; ++, p < 0.001; and +++, p < 0.0001 compares the 1000 μg/mL of extracts to water control.
3. Results
3.1. Effects of Herbal Tea Extracts on CRC Cell Viability
To evaluate the antitumor activity of nine kinds of herbal tea extracts against CRC cells, we performed MTS cell proliferation assay on four CRC cell lines, HCT116, HT-29, SW620, and DLD-1. For initial screening, we treated cells with herbal tea extracts of concentrations at 100, 500, and 1000 μg/mL for 48 h and measured the cell viability. Among nine herbal tea extracts, we revealed that the extracts of V. officinalis, MO, H. officinalis, and S. officinalis caused marked growth inhibition on all CRC cells (Figure 1). These four herbal tea extracts effectively repressed the growth of HCT116, SW620, and DLD-1 cells at a concentration of 500 μg/mL and inhibited HT-29 cells at a concentration of 1000 μg/mL. Of note, the most active extract was MO, which significantly inhibited all CRC cells at a concentration of 500 μg/mL.
3.2. MO Caused Irreversible Growth Inhibition in HCT116 Cells
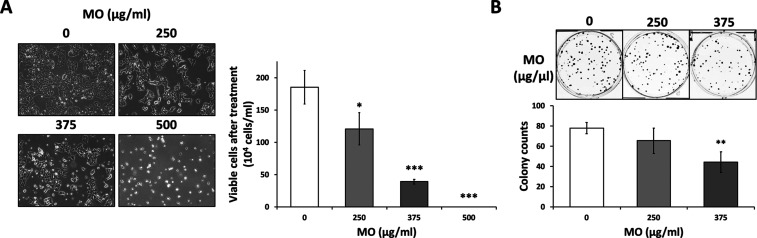
To further investigate the antiproliferation effect of the MO extract on CRC cells, we selected HCT116, which is a commonly used cell line in CRC studies, as our experimental model. We treated HCT116 at concentrations of 0, 250, 375, and 500 μg/mL for 48 h. After treatment, we measured cell viability by the trypan blue assay. The cell viabilities of HCT116 were 65, 21, and 0% at the concentrations of 250, 375, and 500 μg/mL, respectively. We found that MO significantly decreased the cell proliferation in a dose-dependent manner, and the inhibitory concentration (IC50) of MO in HCT116 was 264.4 μg/mL (Figure 2A).
Figure 2.
MO extract inhibited cell growth in HCT116 cells. (A) HCT116 cells were seeded in 10 cm dish overnight and then treated with control (water) or MO for 48 h with the concentration ranging from 0 to 500 μg/mL for HCT116 cells. After 48 h treatment, the cellular morphology was observed by optical microscopy before cell counting by trypan blue exclusion staining. (B) MO-pre-treated-48 h cells were seeded in 6-well plates (250 cells/well) with MO-free medium and incubated for eight days for HCT116. Formed colonies were fixed, stained, and counted. All experiments were performed in three independent replicates. Significant difference indicated as ***p < 0.001, **p < 0.01, *p < 0.05 compared to 0 μg/mL control.
Moreover, we performed the colony formation assay to determine the long-term effect of the MO extract on cell survival. The 48 h-treated cells were seeded in 6-well plates and further cultured for eight days in MO-free RPMI medium. The colony formation ability of CRC cells was diminished by 48 h MO treatment (Figure 2B), suggesting that MO treatment generated an irreversible inhibition on cell growth, and the MO extract could be a potential antitumor compound.
3.3. MO Induced Cell Cycle Arrest at S and G2/M Phases in HCT116 Cells
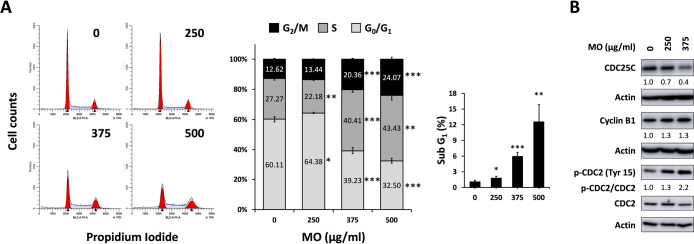
To investigate whether the MO hot water extract affected the cell cycle progression, we treated the HCT116 at concentrations of 0, 250, 375, and 500 μg/mL for 48 h and performed flow cytometry analysis to measure the DNA content. MO treatment increased the cell populations of the S and G2/M phases in a dose-dependent manner, accompanied by decreasing cell numbers in the G1 phase (Figure 3A). Consistent with the previously reported effects of ethanolic extracts of MO,19 the water extract of the MO induced G2/M cell cycle arrest in HCT116 (Figure 3A), indicating that the active anticancer components from MO might be hydrophilic.
Figure 3.
MO extract induced G2/M phase cell cycle arrest in HCT116 cells. (A) HCT116 cells were treated with MO extract for 48 h with indicated concentrations. After treatment, cells were fixed with 70% ethanol, treated with RNase A, stained with PI, and analyzed cell cycle by flow cytometry. Cell cycle distributions were obtained from three independent experiments and represented the mean ± SD. ***p < 0.001, **p < 0.01, *p < 0.05. (B) Western blot showed expression levels of the G2/M arrest associated protein with 48 h MO treatment at the indicated concentration. The protein levels were normalized with actin and phospho-CDC2 levels were normalized with total CDC2.
In the cell cycle progression, cyclin B1 steadily increases during the G2 phase and formed the cyclin B1/CDC2 complex.25 Phosphatase CDC25C activates the cyclin B1/CDC2 complex by dephosphorylating Tyr15 on CDC2, and thus triggers mitosis.26 To further understand the molecular mechanism by the MO extract-induced G2/M phase arrest, we measured the protein expression levels of the G2/M associated proteins. We revealed that the level of CDC25C was decreased, accompanied by increasing CDC2 phosphorylation at Tyr15 (inactive form) in HCT116 cell lines (Figure 3B). The levels of Cyclin B1 was increased in HCT116 cells, indicating that MO inhibited the late G2/M phase in HCT116 cells. These results demonstrated that MO attenuated cell cycle progression through the inhibition of cyclin B/CDC2 activity by decreasing the CDC25C phosphatase expression and induced cell cycle arrest at the G2/M phase.
3.4. MO Induced Caspase-Dependent Apoptotic Cell Death in HCT116 Cells
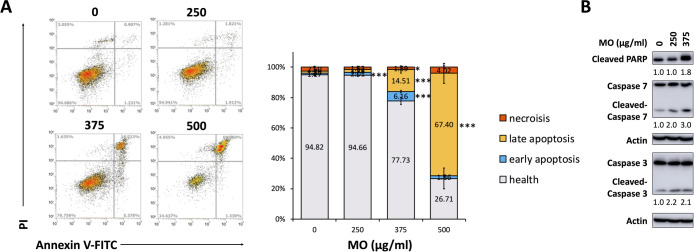
In addition to the G2/M arrest, we found that the MO extract increased the cell population in the sub-G1 phase in a dose-dependent manner (Figure 3A), prompting us to explore whether MO triggered apoptosis in HCT116 cells. When the apoptosis signal is triggered, phosphatidylserine (PS) translocates to the outer leaflets of the plasma membrane because of the inactivation of flippases and activation of scramblases by caspases.27 To detect the apoptosis population based on the exposure of PS to the cell surface, we treated HCT116 at concentrations of 0, 250, 375, and 500 μg/mL for 48 h and applied fluorescent-labeled Annexin V, which readily binds to PS. The MO-treated cells were stained with Annexin V-FITC coupled with PI and detected using flow cytometry. We observed that the FITC+ population (the early and late apoptosis) was increased by MO treatment in a dose-dependent manner (Figure 4A).
Figure 4.
MO extract induced apoptosis cell death in HCT116 cells. (A) HCT116 cells were treated with the MO extract for 48 h with indicated concentrations. After treatment, cells were harvested and stained with Annexin V-FITC and PI, then analyzed by flow cytometry. All measurements were obtained from three independent experiments and represented the mean ± SD. ***p < 0.001, **p < 0.01, and *p < 0.05. (B) Western blot showed expression levels of apoptosis-associated proteins with 48 h MO treatment at the indicated concentration. The protein levels were examined and normalized with actin.
During apoptosis, the effector caspases, such as caspase 3 and caspase 7, can be activated by cleaving and, subsequently, caused the cleavage of PARP, which is a well-known cellular substrate for caspases and considered as a hallmark of apoptosis.28,29 To further understand the molecular mechanism of the MO extract-induced apoptosis, we measured these protein markers of apoptotic cell death. The results showed that the MO extract significantly augmented the cleavage of PARP and increased the cleavage of caspase 3 and caspase 7 (Figure 4B). These data indicated that MO induced caspase-dependent apoptotic cell death in HCT116 CRC cells.
3.5. MO Suppressed Cell Migration in HCT116 Cells
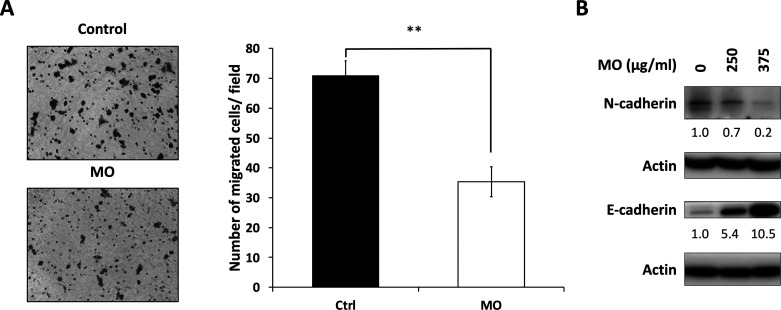
To understand whether the MO extract reduced cell migration, we treated HCT116 cells with 375 μg/mL of MO or vehicle control for 48 h and determined the migratory ability by the transwell assay. The results showed that the number of migrated cells was markedly reduced by MO treatment (Figure 5A). Furthermore, we measured the expression levels of EMT cadherins30 and found that the expression of mesenchymal hallmark N-cadherin was decreased (Figure 5B) and that of epithelial marker E-cadherin was significantly increased by MO treatment. This switch from N-cadherin to E-cadherin indicated the reduced migration ability and reversed EMT in HCT116 cells, suggesting that the MO extract might prevent metastasis by reducing the EMT process.
Figure 5.
MO extract inhibited cell migration in HCT116 cells. HCT116 cells were treated with 0 or 375 μg/mL of MO for 48 h. (A) Cell migration was measured by the Transwell migration assay **p < 0.01, Student’s t-test. (B) EMT markers were analyzed by the western blot. The expression level of proteins was normalized to that of actin.
3.6. Metabolomic Analysis of the MO Extract
To further investigate the main constituents of the MO extract that are responsible for the antitumor activities, we performed untargeted metabolomic analysis to identify the metabolites in the MO hot water extract. We used data-dependent acquisition (DDA in MSE mode) or data independent acquisition (DIA in survey mode) in both positive and negative ion modes. To obtain high qualified results, we accepted the compounds with the highest scores for each matched spectrum and only kept the identified metabolites with evidence at both MS and MS/MS levels. The identified compounds were listed in Tables S1–S4 accompanied by their retention time, m/z, molecular formula, score, and mass error. We identified 10, 30, 17, and 20 compounds in MSE-negative mode, MSE-positive mode, survey-negative mode, and survey-positive mode, respectively. In total, 67 different compounds were identified in the MO extracts and can be classified into nine different chemical classes by ClassyFire, including alkaloids, benzenoids, lipids, nucleotides, organic acids, sugars, organoheterocyclic compounds, lignans, phenylpropanoids, and polyketides (Table S5). Among the nine superclasses, compounds belonging to superclasses of lignans, phenylpropanoids, and polyketides are phenolic compounds, which are widely found in natural products and possess various bioactivities such as anti-inflammatory, antioxidation, and anticancer effects.31,32 Twenty-eight phenolic compounds were identified in the MO extract and can be further classified into the class of cinnamic acids, coumarins, flavonoids, isoflavonoids, and lignan glycosides (Table 1). We suggest that the compounds such as melitric acid B,33 RA,34−41 sagecoumarin,37 and sagerinic acid,34,36−40 might be the major bioactive ingredients in the MO extract and play roles in anticancer activities on HCT116 CRC cells.
Table 1. Classification of Phenolic Compounds in MO Extracts.
| identified compound | superclass | class |
|---|---|---|
| 2,3-dehydrosilychristin | phenylpropanoids and polyketides | 2-arylbenzofuran flavonoids |
| maritimetin | aurone flavonoids | |
| 3-(3,5-dihydroxyphenyl)-2-propenoic acid | cinnamic acids and derivatives | |
| melitric acid B | ||
| rosmarinate (rosmarinic acid) | ||
| 3-hydroxycoumarin | coumarins and derivatives | |
| 4-hydroxycoumarin | ||
| 7,7′-dihydroxy-6,8′-bicoumarin | ||
| sagecoumarin | ||
| umbelliferone | ||
| gangleoidin acetate | depsides and depsidones | |
| psoromic acid | ||
| epicatechin pentaacetate | flavonoids | |
| epigallocatechin | ||
| phyllocoumarin | ||
| theaflavin monogallates | ||
| centaurein | ||
| chrysanthemin | ||
| kaempferol 3-glucuronide | ||
| kaempferol 3-glucuronide-7-glucoside | ||
| luteolinidin 3-O-glucoside | ||
| petunidin-3-O-arabinoside | ||
| eupatorin | ||
| thevetiaflavone | ||
| maximaisoflavone A | isoflavonoids | |
| koparin 2′-methyl ether | ||
| sagerinic acid | lignans, neolignans, and related compounds | cyclobutane lignans |
| occidentoside | lignan glycosides |
4. Discussion
Drinking herbal tea is relevant to a decreased risk of distal colon cancer,7 prompting us to investigate whether herbal tea possesses antitumor activities to prevent CRC. We demonstrated that the hot water extract of V. officinalis, MO, H. officinalis, and S. officinalis diminished cell proliferation in several CRC cell lines (Figure 1). MO was the most effective one and has been studied for its bioactivity in many types of diseases, including cancer.18 Weidner and colleagues demonstrated that the hydroethanolic MO extract induced apoptosis and inhibited cell proliferation through the formation of reactive oxygen species in human colon cancer cells HT-29 and T84.19 Encalada and colleagues showed the antiproliferative effect of the aqueous and ethanolic MO extract in HCT116 cells,22 yet the molecular mechanism induced by MO in CRC cells remains unclear. Considering the ways to extract natural products vary the constitution of the bioactive compounds,42 we focused on the effects of hot water extracts of herbs in CRC cells to mimic the outcome of drinking herbal tea. Consistent with previous studies, we observed that MO significantly decreased cell proliferation in HCT116 (IC50 = 264.4 μg/mL) (Figure 2A) with comparable IC50 of the hydroethanolic MO extract in HT-29 cells (IC50 = 346 μg/mL).19 Furthermore, the prolonged inhibition of MO on the anchorage-dependent colony formation ability was observed even we remove MO after 2-day incubation (Figure 2B), suggesting an irreversible signal of cell growth inhibition was achieved.
Prior studies showed that natural compounds derived from plants possess antiproliferation activity by modulating the cell cycle arrest. The extraction of natural products was shown to inhibit the activities of cyclins and cyclin-dependent kinases (CDKs) and upregulate the expression of CDK inhibitor proteins p53, p21, and p27 to block cell cycle progression.43 Similar activities were found in MO extracts. Hydroethanolic MO extract induced cell population which accumulated at the G2/M phase in HT-29 cells;19 dichloromethane fraction of the MO induced G2/M phase arrest in Jurkat cells;44 and essential oil citral (3,7-dimethyl-2,6-octadienal) extract from MO suppressed cell cycle at the G2/M phase in MCF-7 cells.45 Similar to previous studies, we observed the MO hot water extract-induced cell cycle arrest at the G2/M phase in CRC cells HCT116. We confirmed that the regulation was associated with the blockade of cyclin B1/CDC2 complex activity through the decreased dephosphorylation from a dropped level of CDC25C (Figure 3). Following the cell cycle arrest, MO induced apoptosis in several types of cancer cell lines in the forms of hydroethanolic, ethanolic, and essential oil extracts.19,34,46 The same property was observed in the hot water extract that the MO treatment triggered apoptosis in CRC cells and caused PARP cleavage in a caspase-dependent fashion (Figure 4). Although MO ethanolic extracts have been reported to inhibit cell migration in MDA-MB-231 breast cancer cells,47 the anti-metastasis effect of the MO water extract remains unclear. In this study, we showed that 375 μg/mL of the MO hot water extract significantly reduced the cell migratory ability to 35.4% of control through modulating the EMT events in HCT116 cells (Figure 5).
To explore the potent anticancer compounds in MO, we applied untargeted metabolomics analysis using UPLC-Q-TOF mass spectrometry. In total, 67 compounds were identified in the MO water extract and can be classified into nine superclasses (Table S5). Among these superclasses, compounds belonging to lignans, phenylpropanoids, and polyketides are notable because of the presence of phenols. Phenolic and polyphenolic compounds can be categorized into many classes, mainly known as phenolic acids, flavonoids, tannins, lignans, lignins, coumarins, and stilbenes.31,48−50 We identified numerous compounds that belong to flavonoids, phenolic acid, coumarins, and lignans (Table 1). Phenolic acids and their derivatives have been demonstrated to act as antioxidants and exhibit a broad spectrum of bioactivities, including anticancer, anti-inflammation, anti-atherogenesis, anti-thrombosis, and analgesic activities.51 In line with previous findings,33,34,36−40 we identified RA and its derivatives, melitric acid, and sagerinic acid, assuming their contribution to the anticancer effects of MO.
Among the phenolic acids, RA was the major identified metabolite in the MO hot water extract. This compound has been widely reported in previous studies34−41 as the major metabolite in total phenolic compounds of MO.34,35,41 RA has been documented to possess anticancer effects in many types of cancer, such as glioma, pancreatic cancer, CRC, and breast cancer.13,35,52−54 In contrast to the previous finding that RA significantly inhibited cell proliferation and induced cell cycle arrest at the G0/G1 phase by decreasing cyclin D1 and CDK4 expression in CRC cells,13 we revealed that the MO extract caused cell cycle arrest at the G2/M phase, implying that the complexity of the water extract compromises the effect from the single compound. Additionally, Han et al. showed that RA also inhibited cell migration activity in CRC cells and regulated EMT by increasing E-cadherin and decreasing N-cadherin,13 suggesting that RA serves as the major compound in the MO extract to inhibit cell mobility.
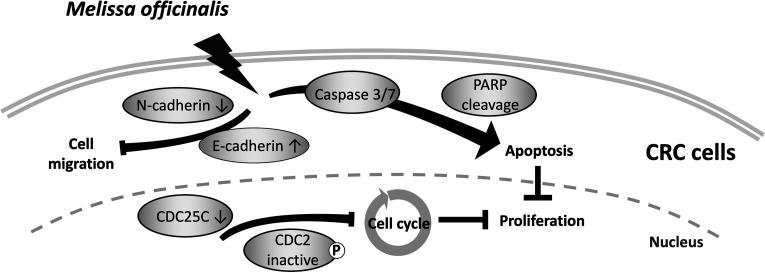
In conclusion, we demonstrated the anticancer effects of herbal tea, especially in the hot water extract of MO in CRC cells. We found that MO extract reduced cell proliferation, induced cell cycle arrest in theG2/M phase via blocking cyclin B1/CDC2 complex activation, triggered caspase-dependent apoptotic cell death, and inhibited cell migration ability via EMT modulation in CRC HCT116 cells (Figure 6), suggesting that the habit of drinking herbal tea prevents CRC progression.
Figure 6.
Summary of the anticancer effects of the MO extract in CRC cells. A mechanistic scheme for MO-mediated cell death.
Acknowledgments
We would like to the technical support provided by the TMU Core Facility.
Glossary
Abbreviations
- AMPK
5′ AMP-activated protein kinase
- AP-1
activator protein 1
- CDC2
cell division cycle 2
- CDC25C
cell division cycle 25C
- CDK
cyclin-dependent kinase
- CDK4
cyclin-dependent kinase 4
- CRC
colorectal cancer
- DDA
data-dependent analysis
- DIA
data-independent analysis
- EMT
epithelial–mesenchymal transition
- ESI
electrospray ionization
- MMP-2
matrix metalloproteinase-2
- MMP-9
matrix metalloproteinase-9
- MO
Melissa officinalis
- NF-κB
nuclear factor-κB
- PARP
poly (ADP-ribose) polymerase
- PI
propidium iodide
- RA
rosmarinic acid
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- STAT3
signal transducer and activator of transcription 3
- ZO1
zonula occludens-1
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c04489.
Compounds identified in the hot water extract of MO in MSE-negative ionization mode, compounds identified in the hot water extract of MO in MSE-positive ionization mode, compounds identified in the hot water extract of MO in survey-negative ionization mode, compounds identified in the hot water extract of MO in survey-positive ionization mode, and classification of MO-extract compounds (PDF)
Author Contributions
T.-T.K. and H.-Y.C. contributed equally to this work. T.-T.K. and H.-Y.C. designed and performed the experiments, analyzed and interpreted the data, and wrote the manuscript; T.-Y.C. performed the experiments and contributed to scientific discussion; B.-C.L. and Y.-C.H. performed the experiments; S.-M.H. contributed to scientific discussion; and C.-J.C. and T.-C.H. acquired funding, conceptualized the study, and reviewed and edited the manuscript.
This work was supported by the health and welfare surcharge of tobacco products of Taiwan (Wan-Fang Hospital, Chi-Mei Medical Center, and Hua-lien Tzu-Chi Hospital Joint Cancer Center Grant-Focus on Colon Cancer Research, grant number: MOHW108-TDU-B-212-124020; MOHW109-TDU-B-212-134020), the University System of Taipei Joint Research Program (USTP-NTOU-TMU-106-01; USTP-NTOU-TMU-106-03), Taipei Medical University (TMU107-AE1-B36), and the “TMU Research Center of Cancer Translational Medicine” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
The authors declare no competing financial interest.
Supplementary Material
References
- Siegel R. L.; Miller K. D.; Jemal A. Cancer statistics, 2018. Ca-Cancer J. Clin. 2018, 68, 7–30. 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- Migliore L.; Migheli F.; Spisni R.; Coppede F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J. Biomed. Biotechnol. 2011, 2011, 792362. 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggar F.; Boushey R. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009, 22, 191–197. 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzeh F. S.; Alshammari E. M.; Alazzeh A. Y.; Jazar A. S.; Dabbour I. R.; El-Taani H. A.; Obeidat A. A.; Kattan F. A.; Tashtoush S. H. Healthy dietary patterns decrease the risk of colorectal cancer in the Mecca Region, Saudi Arabia: a case-control study. BMC Public Health 2017, 17, 607. 10.1186/s12889-017-4520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena R.; Salinas P. Diet and colorectal cancer. Maturitas 2015, 80, 258–264. 10.1016/j.maturitas.2014.12.017. [DOI] [PubMed] [Google Scholar]
- Lipkin M.; Reddy B.; Newmark H.; Lamprecht S. A. Dietary factors in human colorectal cancer. Annu. Rev. Nutr. 1999, 19, 545–586. 10.1146/annurev.nutr.19.1.545. [DOI] [PubMed] [Google Scholar]
- Green C. J.; de Dauwe P.; Boyle T.; Tabatabaei S. M.; Fritschi L.; Heyworth J. S. Tea, coffee, and milk consumption and colorectal cancer risk. J. Epidemiol 2014, 24, 146–153. 10.2188/jea.je20130063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.-m.; Yang Z.-j.; Xie Q.; Zhang Z.-k.; Zhang H.; Ma J.-y. Natural products for treating colorectal cancer: A mechanistic review. Biomed. Pharmacother. 2019, 117, 109142. 10.1016/j.biopha.2019.109142. [DOI] [PubMed] [Google Scholar]
- Alam M. N.; Almoyad M.; Huq F. Polyphenols in Colorectal Cancer: Current State of Knowledge including Clinical Trials and Molecular Mechanism of Action. BioMed Res. Int. 2018, 2018, 4154185. 10.1155/2018/4154185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-H.; Niu Y. B.; Sun Y.; Zhang F.; Liu C. X.; Fan L.; Mei Q. B. Role of phytochemicals in colorectal cancer prevention. World J. Gastroenterol. 2015, 21, 9262–9272. 10.3748/wjg.v21.i31.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh Y.-J. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 2003, 3, 768–780. 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- Tomeh M. A.; Hadianamrei R.; Zhao X. A Review of Curcumin and Its Derivatives as Anticancer Agents. Int. J. Mol. Sci. 2019, 20, 1033. 10.3390/ijms20051033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-H.; Kee J. Y.; Hong S. H. Rosmarinic Acid Activates AMPK to Inhibit Metastasis of Colorectal Cancer. Front. Pharmacol. 2018, 9, 68. 10.3389/fphar.2018.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S.; Dixit M. Role of Polyphenols and Other Phytochemicals on Molecular Signaling. Oxid. Med. Cell. Longevity 2015, 2015, 504253. 10.1155/2015/504253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekara A.; Shahidi F. Herbal beverages: Bioactive compounds and their role in disease risk reduction - A review. J. Tradit. Complement. Med. 2018, 8, 451–458. 10.1016/j.jtcme.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini M.; Del Rio D.; Yao D. N.; Bettuzzi S.; Peluso I.. Health Benefits of Tea. In Herbal Medicine: Biomolecular and Clinical Aspects; Benzie I. F. F., Wachtel-Galor S., Eds.; CRC Press: Boca Raton, FL, 2011. [PubMed] [Google Scholar]
- McKay D. L.; Blumberg J. B. The role of tea in human health: an update. J. Am. Coll. Nutr. 2002, 21, 1–13. 10.1080/07315724.2002.10719187. [DOI] [PubMed] [Google Scholar]
- Shakeri A.; Sahebkar A.; Javadi B. Melissa officinalis L.—A review of its traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2016, 188, 204–228. 10.1016/j.jep.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Weidner C.; Rousseau M.; Plauth A.; Wowro S. J.; Fischer C.; Abdel-Aziz H.; Sauer S. Melissa officinalis extract induces apoptosis and inhibits proliferation in colon cancer cells through formation of reactive oxygen species. Phytomedicine 2015, 22, 262–270. 10.1016/j.phymed.2014.12.008. [DOI] [PubMed] [Google Scholar]
- López V.; Martín S.; Gómez-Serranillos M. P.; Carretero M. E.; Jäger A. K.; Calvo M. I. Neuroprotective and neurological properties of Melissa officinalis. Neurochem. Res. 2009, 34, 1955–1961. 10.1007/s11064-009-9981-0. [DOI] [PubMed] [Google Scholar]
- de Sousa A. C.; Gattass C. R.; Alviano D. S.; Alviano C. S.; Blank A. F.; Alves P. B. Melissa officinalis L. essential oil: antitumoral and antioxidant activities. J. Pharm. Pharmacol. 2004, 56, 677–681. 10.1211/0022357023321. [DOI] [PubMed] [Google Scholar]
- Encalada M. A.; Hoyos K. M.; Rehecho S.; Berasategi I.; de Ciriano M. G.-Í.; Ansorena D.; Astiasaran I.; Navarro-Blasco I.; Cavero R. Y.; Calvo M. I. Anti-proliferative effect of Melissa officinalis on human colon cancer cell line. Plant Foods Hum. Nutr. 2011, 66, 328–334. 10.1007/s11130-011-0256-y. [DOI] [PubMed] [Google Scholar]
- Guijas C.; Montenegro-Burke J. R.; Domingo-Almenara X.; Palermo A.; Warth B.; Hermann G.; Koellensperger G.; Huan T.; Uritboonthai W.; Aisporna A. E.; Wolan D. W.; Spilker M. E.; Benton H. P.; Siuzdak G. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018, 90, 3156–3164. 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou Feunang Y.; Eisner R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform 2016, 8, 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J.; Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 1989, 58, 833–846. 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- Berry L. D.; Gould K. L. Regulation of Cdc2 activity by phosphorylation at T14/Y15. Prog. Cell Cycle Res. 1996, 2, 99–105. 10.1007/978-1-4615-5873-6_10. [DOI] [PubMed] [Google Scholar]
- Segawa K.; Nagata S. An apoptotic “eat me” signal: phosphatidylserine exposure. Trends Cell Biol. 2015, 25, 639–650. 10.1016/j.tcb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- Kitazumi I.; Tsukahara M. Regulation of DNA fragmentation: the role of caspases and phosphorylation. FEBS J. 2011, 278, 427–441. 10.1111/j.1742-4658.2010.07975.x. [DOI] [PubMed] [Google Scholar]
- Shi Y. Caspase activation, inhibition, and reactivation: a mechanistic view. Protein Sci. 2004, 13, 1979–1987. 10.1110/ps.04789804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof A.; Berx G. Cadherins and epithelial-to-mesenchymal transition. Prog. Mol. Biol. Transl. Sci. 2013, 116, 317–336. 10.1016/b978-0-12-394311-8.00014-5. [DOI] [PubMed] [Google Scholar]
- Basli A.; Belkacem N.; Amrani I.. Health benefits of phenolic compounds against cancers. Phenolic Compounds-Biological Activity; InTechOpen: London, 2017; pp 193–210. [Google Scholar]
- Zhou Y.; Zheng J.; Li Y.; Xu D. P.; Li S.; Chen Y. M.; Li H. B. Natural Polyphenols for Prevention and Treatment of Cancer. Nutrients 2016, 8, 515. 10.3390/nu8080515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agata I.; Kusakabe H.; Hatano T.; Nishibe S.; Okuda T. Melitric acids A and B, new trimeric caffeic acid derivatives from Melissa officinalis. Chem. Pharm. Bull. 1993, 41, 1608–1611. 10.1248/cpb.41.1608. [DOI] [Google Scholar]
- Magalhães D. B.; Castro I.; Lopes-Rodrigues V.; Pereira J. M.; Barros L.; Ferreira I. C. F. R.; Xavier C. P. R.; Vasconcelos M. H. Melissa officinalis L. ethanolic extract inhibits the growth of a lung cancer cell line by interfering with the cell cycle and inducing apoptosis. Food Funct. 2018, 9, 3134–3142. 10.1039/c8fo00446c. [DOI] [PubMed] [Google Scholar]
- Ramanauskiene K.; Raudonis R.; Majiene D. Rosmarinic Acid and Melissa officinalis Extracts Differently Affect Glioblastoma Cells. Oxid. Med. Cell. Longevity 2016, 2016, 1564257. 10.1155/2016/1564257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sánchez A.; Barrajón-Catalán E.; Herranz-López M.; Castillo J.; Micol V. Lemon balm extract (Melissa officinalis, L.) promotes melanogenesis and prevents UVB-induced oxidative stress and DNA damage in a skin cell model. J. Dermatol. Sci. 2016, 84, 169–177. 10.1016/j.jdermsci.2016.08.004. [DOI] [PubMed] [Google Scholar]
- Ozarowski M.; Mikolajczak P. L.; Piasecka A.; Kachlicki P.; Kujawski R.; Bogacz A.; Bartkowiak-Wieczorek J.; Szulc M.; Kaminska E.; Kujawska M.; Jodynis-Liebert J.; Gryszczynska A.; Opala B.; Lowicki Z.; Seremak-Mrozikiewicz A.; Czerny B. Influence of the Melissa officinalis Leaf Extract on Long-Term Memory in Scopolamine Animal Model with Assessment of Mechanism of Action. J. Evidence-Based Complementary Altern. Med. 2016, 2016, 9729818. 10.1155/2016/9729818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carocho M.; Barros L.; Calhelha R. C.; Ćirić A.; Soković M.; Santos-Buelga C.; Morales P.; Ferreira I. C. F. R. Melissa officinalis L. decoctions as functional beverages: a bioactive approach and chemical characterization. Food Funct. 2015, 6, 2240–2248. 10.1039/c5fo00309a. [DOI] [PubMed] [Google Scholar]
- Miron T. L.; Herrero M.; Ibáñez E. Enrichment of antioxidant compounds from lemon balm (Melissa officinalis) by pressurized liquid extraction and enzyme-assisted extraction. J. Chromatogr. A 2013, 1288, 1–9. 10.1016/j.chroma.2013.02.075. [DOI] [PubMed] [Google Scholar]
- Barros L.; Dueñas M.; Dias M. I.; Sousa M. J.; Santos-Buelga C.; Ferreira I. C. F. R. Phenolic profiles of cultivated, in vitro cultured and commercial samples of Melissa officinalis L. infusions. Food Chem. 2013, 136, 1–8. 10.1016/j.foodchem.2012.07.107. [DOI] [PubMed] [Google Scholar]
- Lin J.-T.; Chen Y.-C.; Lee Y.-C.; Rolis Hou C.-W.; Chen F.-L.; Yang D.-J. Antioxidant, anti-proliferative and cyclooxygenase-2 inhibitory activities of ethanolic extracts from lemon balm (Melissa officinalis L.) leaves. LWT--Food Sci. Technol. 2012, 49, 1–7. 10.1016/j.lwt.2012.04.009. [DOI] [Google Scholar]
- Zhang Q. W.; Lin L. G.; Ye W. C. Techniques for extraction and isolation of natural products: a comprehensive review. Chin. Med. 2018, 13, 20. 10.1186/s13020-018-0177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailon-Moscoso N.; Cevallos-Solorzano G.; Romero-Benavides J.; Ramirez Orellana M. Natural Compounds as Modulators of Cell Cycle Arrest: Application for Anticancer Chemotherapies. Curr. Genomics 2017, 18, 106–131. 10.2174/1389202917666160808125645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimnezhad Darzi S.; Amirghofran Z. Dichloromethane fraction of Melissa officinalis induces apoptosis by activation of intrinsic and extrinsic pathways in human leukemia cell lines. Immunopharmacol. Immunotoxicol. 2013, 35, 313–320. 10.3109/08923973.2013.768268. [DOI] [PubMed] [Google Scholar]
- Chaouki W.; Leger D. Y.; Liagre B.; Beneytout J.-L.; Hmamouchi M. Citral inhibits cell proliferation and induces apoptosis and cell cycle arrest in MCF-7 cells. Fundam. Clin. Pharmacol. 2009, 23, 549–556. 10.1111/j.1472-8206.2009.00738.x. [DOI] [PubMed] [Google Scholar]
- Queiroz R. M. d.; Takiya C. M.; Guimarães L. P. T. P.; Rocha G. d. G.; Alviano D. S.; Blank A. F.; Alviano C. S.; Gattass C. R. Apoptosis-inducing effects of Melissa officinalis L. essential oil in glioblastoma multiforme cells. Cancer Invest. 2014, 32, 226–235. 10.3109/07357907.2014.905587. [DOI] [PubMed] [Google Scholar]
- Moaca E.-A.; Farcas C.; Ghitu A.; Coricovac D.; Popovici R.; Caraba-Meita N. L.; Ardelean F.; Antal D. S.; Dehelean C.; Avram S. A Comparative Study of Melissa officinalis Leaves and Stems Ethanolic Extracts in terms of Antioxidant, Cytotoxic, and Antiproliferative Potential. J. Evidence-Based Complementary Altern. Med. 2018, 2018, 7860456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahidi F.; Yeo J. Bioactivities of Phenolics by Focusing on Suppression of Chronic Diseases: A Review. Int. J. Mol. Sci. 2018, 19, 1573. 10.3390/ijms19061573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanakis C. M.Polyphenols: Properties, Recovery, and Applications; Woodhead Publishing, 2018. [Google Scholar]
- Abbas M.; Saeed F.; Anjum F. M.; Afzaal M.; Tufail T.; Bashir M. S.; Ishtiaq A.; Hussain S.; Suleria H. A. R. Natural polyphenols: An overview. Int. J. Food Prop. 2017, 20, 1689–1699. 10.1080/10942912.2016.1220393. [DOI] [Google Scholar]
- de Camargo A. C.; Regitano-d’Arce M. A. B.; Rasera G. B.; Canniatti-Brazaca S. G.; do Prado-Silva L.; Alvarenga V. O.; Sant’Ana A. S.; Shahidi F. Phenolic acids and flavonoids of peanut by-products: Antioxidant capacity and antimicrobial effects. Food Chem. 2017, 237, 538–544. 10.1016/j.foodchem.2017.05.046. [DOI] [PubMed] [Google Scholar]
- Han Y.; Ma L.; Zhao L.; Feng W.; Zheng X. Rosmarinic inhibits cell proliferation, invasion and migration via up-regulating miR-506 and suppressing MMP2/16 expression in pancreatic cancer. Biomed. Pharmacother. 2019, 115, 108878. 10.1016/j.biopha.2019.108878. [DOI] [PubMed] [Google Scholar]
- Swamy M. K.; Sinniah U. R.; Ghasemzadeh A. Anticancer potential of rosmarinic acid and its improved production through biotechnological interventions and functional genomics. Appl. Microbiol. Biotechnol. 2018, 102, 7775–7793. 10.1007/s00253-018-9223-y. [DOI] [PubMed] [Google Scholar]
- Juskowiak B.; Bogacz A.; Wolek M.; Kamiński A.; Uzar I.; Seremak-Mrozikiewicz A.; Czerny B. Expression profiling of genes modulated by rosmarinic acid (RA) in MCF-7 breast cancer cells. Ginekol. Pol. 2018, 89, 541–545. 10.5603/gp.a2018.0092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.