Abstract
Transposable elements (TEs) are mobile DNA and constitute approximately half of the human genome. LINE-1 (L1) is the only active autonomous TE in the mammalian genome and has been implicated in a number of diseases as well as aging. We have previously reported that skeletal muscle L1 expression is lower following acute and chronic exercise training in humans. Herein, we used a rodent model of voluntary wheel running to determine whether long-term exercise training affects markers of skeletal muscle L1 regulation. Selectively bred high-running female Wistar rats (n = 11 per group) were either given access to a running wheel (EX) or not (SED) at 5 wk of age, and these conditions were maintained until 27 wk of age. Thereafter, mixed gastrocnemius tissue was harvested and analyzed for L1 mRNA expression and DNA content along with other L1 regulation markers. We observed significantly (P < 0.05) lower L1 mRNA expression, higher L1 DNA methylation, and less L1 DNA in accessible chromatin regions in EX versus SED rats. We followed these experiments with 3-h in vitro drug treatments in L6 myotubes to mimic transient exercise-specific signaling events. The AMP-activated protein kinase (AMPK) agonist 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR; 4 mM) significantly decreased L1 mRNA expression in L6 myotubes. However, this effect was not facilitated through increased L1 DNA methylation. Collectively, these data suggest that long-term voluntary wheel running downregulates skeletal muscle L1 mRNA, and this may occur through chromatin modifications. Enhanced AMPK signaling with repetitive exercise bouts may also decrease L1 mRNA expression, although the mechanism of action remains unknown.
Keywords: exercise, LINE-1, L1, methylation, retrotransposons
INTRODUCTION
Transposable elements (TEs), often referred to as mobile DNA or “jumping genes,” are genetic elements that have the ability to change their position in a genome (4, 41). There are numerous types of TEs that exist based on their mechanism of transposition (or movement) and are categorized into two distinct classes: DNA transposons and retrotransposons (63). The former uses a DNA intermediate termed a “cut and paste” mechanism, while the latter utilizes a “copy and paste” mechanism, where an RNA intermediate is reverse transcribed into cDNA before integration into the genome, resulting in an amplification of the retrotransposon (63).
Long interspersed element-1 (LINE-1, or L1) is one of the most abundant TEs in humans and constitutes large portions of the genome of rats and mice, with estimations of 17, 18, and 21%, respectively (9, 20, 36, 54). L1 is the only autonomous retrotransposon active in mammalian genomes, meaning that it encodes for all the machinery needed for its retrotransposition (for review, see Refs. 4, 32, 54)). L1 is a 6-kb genetic element that contains an internal RNA polymerase II (Pol II) promoter within the 5′-untranslated region (UTR). L1 encodes for two different proteins: open reading frame 1 protein (ORF1p), an RNA binding protein, and open reading frame 2 protein (ORF2p), which has both endonuclease and reverse transcriptase activities. The first step of L1 retrotransposition is the recruitment of RNA Pol II to its internal promoter, followed by transcription of the L1 DNA into a bicistronic mRNA. L1 mRNA is then translated into its respective proteins, and the L1 proteins preferentially bind to their mRNA, forming an L1 ribonucleoprotein (L1RNP) complex. L1RNP then translocates to the nucleus, where ORF2p catalyzes the nicking of DNA at consensus TTAAAA target sites. Reverse transcription of L1 mRNA into cDNA occurs via ORF2p, and this ultimately leads to the de novo integration of L1 cDNA into the genome, resulting in target site duplications on both the 5′- and 3′-ends of the newly inserted element (10, 35, 49, 61). Recent elegant genetic studies have shed light on the amplification mechanisms and site selection of L1 (see Refs. 15 and 60). Although L1 ORF proteins show cis preference for L1 transcripts, other mRNAs and TEs can also bind with the ORF proteins, ultimately leading to amplification of those genes and TEs, respectively (54). In this regard, other TEs, such as short interspersed elements (SINEs), can “hijack” L1 machinery, evidenced by the number of SINEs in the human genome (~1,000,000 copies) (54). The relevance and potential physiological impact of these amplifications are detailed elsewhere (8, 51).
It has been widely speculated that L1, as well as other TEs, have had “positive” implications during evolution. For example, there is evidence that the DNA binding domain of the DNA transposon Tc1/mariner has been co-opted by animals in the PAX3 gene, which is important for skeletal muscle development (27, 50). Another example is that L1 may be involved in a quality control mechanism in oogenesis by which defective oocytes are removed (17, 39). However, TEs, and L1 specifically, have been implicated in a number of diseases (22, 32). These include both associative as well as causative examples, such as increased L1 expression in various cancers and direct insertions, causing specific cases of Duchenne’s muscular dystrophy (13, 23, 24, 29, 47).
With respect to TE and L1 research, skeletal muscle has been highlighted in only a handful of studies (2, 34, 38, 44). A seminal paper by De Cecco et al. (7) reported that skeletal muscle and liver L1 mRNA and DNA levels are significantly higher in older mice (36 mo of age) versus younger mice (5 mo of age), and the authors speculated that increased tissue L1 activity may accelerate the aging process. In line with these findings, Sirt6 has been shown to act as a repressor of L1 expression, but Sirt6-mediated L1 repression is lost in aged mice (43). Interestingly, the same laboratory demonstrated that muscle mass loss was attenuated in Sirt6-knockout mice that were treated with reverse transcriptase inhibitors or drugs that inhibit ORF2p enzyme activity (59).
Our laboratory has previously shown that skeletal muscle L1 mRNA expression was significantly decreased while L1 DNA methylation was increased in college-aged males after both acute and chronic resistance training (58). More recent data from our laboratory have also suggested that one bout of endurance exercise downregulates skeletal muscle L1 expression in younger and older humans (55). De Cecco et al. (7) also reported that skeletal muscle and liver L1 mRNA levels were significantly lower in lifelong calorically restricted mice compared with ad libitum fed counterparts. Although preliminary, these data highlight the possibility that exercise and caloric restriction may downregulate tissue L1 expression through a conserved mechanism. Given the data presented above, the goal of the present study was to examine how long-term voluntary wheel running affects skeletal muscle L1 markers in rats. Additionally, we sought to determine potential mechansims through which exercise may operate to regulate L1 gene expression. We hypothesized that long-term exercise would downregulate skeletal muscle L1 markers, although we had no a priori hypothesis regarding how this mechanistically occurs. Consistent with our previous results in humans, we found that long-term endurance training through voluntary wheel running downregulates skeletal muscle L1 expression and increases L1 DNA methylation in rats. Furthermore, we provide in vitro evidence in L6 myotubes that enhanced AMP-activated protein kinase (AMPK) signaling may also act to decrease L1 mRNA expression.
MATERIALS AND METHODS
Animals.
All live animal experiments and dissections occurred at the University of Missouri, and these experimental procedures were approved by the Institution’s Animal Care and Use Committee. For the present study, muscle was procured and analyzed by our research group at Auburn University.
Rats used were from a previous study in which high-voluntary wheel running (HVR) rats were selected and bred as described earlier (56, 57, 62). Females were chosen given that they have much higher voluntary running tendencies compared with males. Briefly, female Wistar rats that were selectively bred for high levels of wheel running were divided into two groups based on whether they did or did not have access to a voluntary running wheel [exercise (EX) and sedentary (SED), respectively]. Both groups were weaned at 28 days of age. EX rats (n = 11) were provided access to running wheels from 5 wk of age until the end of the experiment, when they were 27 wk of age, whereas SED rats (n = 11) were housed in standard cages without running wheels until 27 wk of age. Rats were provided drinking water and standard rodent chow ad libitum. On the day of dissections, animals were euthanized using CO2 asphyxiation in the afternoon hours during the middle of the light cycle (1400–1800); gastrocnemius muscles were rapidly removed, and muscles were flash-frozen in liquid N2 and stored at −80°C until they were shipped to Auburn University.
Tissue preparation for protein analyses.
Muscles were removed from −80°C storage; tissue was crushed on a liquid nitrogen-cooled ceramic mortar and pestle, and ~50 mg of tissue from each rodent was placed in 7 mL glass vials containing 500 μL of ice-cold cell lysis buffer [20 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM Na-EDTA, 1 mM EGTA, 1% Triton, 20 mM sodium pyrophosphate, 25 mM sodium fluoride, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 μg/mL leupeptin; Cell Signaling Technology; Danvers, MA]. Tissues were homogenized via glass dounce homogenization, and homogenates were centrifuged at 12,000 g for 10 min. Supernatants were transferred to 1.7-mL tubes, and protein concentration was determined using a BCA assay (Thermo Scientific, Waltham, MA). Supernatants were subsequently prepared for SDS-PAGE using 4× Laemmli buffer at 2 μg/μL for Western blot analysis, and 15 μL was loaded onto 4–15% SDS-polyacrylamide precasted gels obtained from (Bio-Rad Laboratories, Hercules, CA). 1× SDS-PAGE run buffer (Ameresco, Framingham, MA) was used for electrophoresis at 180 V for 60 min. Thereafter, proteins were transferred via 200 mA constant current for 120 min to polyvinylidene difluoride membranes (Bio-Rad Laboratories). Membranes were then stained with Ponceau S, and digital images were captured using a gel documentation system (UVP, Upland, CA) to ensure equal loading of samples among lanes. Membranes were then blocked at room temperature with 5% nonfat milk powder in Tris-buffered saline with 0.1% Tween-20 (TBST) for 60 min. All of the following primary antibodies were incubated overnight at 4°C in a solution of TBST containing 5% bovine serum albumin (BSA; Ameresco): rabbit anti-cytochrome c (1:1,000, cat. no. GTX108585; Genetex), mouse anti-ORF1p (1:1,000, cat. no. ab76726; Abcam), or rabbit anti-phosphorylated AMPKα (Thr172) (1:1,000, cat. no. 2531; Cell Signaling Technology). Notably, the mouse antiORF1p antibody has previously been validated by our group using HeLa cell lysate (55). The following day, membranes were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit IgG secondary antibodies (1:2,000, cat. no. 7076 and 7074, respectively; Cell Signaling Technology) in a solution of TBST containing 5% BSA at room temperature for 60 min. Thereafter, membranes were developed using an enhanced chemiluminescent reagent (Luminata Forte HRP substrate, EMD Milli-pore, Billerica, MA), with band densitometry subsequently assessed by use of a digital gel documentation system and associated densitometry software (UVP). Densitometry on white band values for each aforementioned target was normalized to a corresponding dark band using Ponceau densitometry values. Additionally, values were normalized to SED group to yield relative protein expression levels.
RNA isolation and quantitative PCR prep.
Approximately 20 mg of powdered frozen gastrocnemius muscle from each rodent were placed in 500 μL of Ribozol (Ameresco, Solon, OH) per the manufacturer’s recommendations. Thereafter, phase separation was achieved according to the manufacturer’s instructions for RNA isolation. Following RNA precipitation and pelleting, pellets were resuspended in 20 μL of RNase-free water, and RNA concentrations were determined in duplicate at an absorbance of 260 nm by using a NanoDrop Lite (Thermo Scientific, Waltham, MA). Isolated RNA (10 μg) was DNase treated (Turbo DNase, cat. no. AM2238; Invitrogen, Carlsbad, CA), and cDNA (2 μg) was synthesized using a commercial qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD) per the manufacturer’s recommendations. Quantitative PCR (qPCR) was performed with gene-specific primers and SYBR-green-based methods (Quanta Biosciences) in a real-time PCR thermal cycler (Bio-Rad Laboratories). The final volume of qPCR reactions was 20 μL, which contained a final concentation of 2 μM of forward and reverse primers and 25 ng of cDNA. All reactions were performed in duplicate. Primers for housekeeping genes (Table 1) were designed with primer designer software (Primer3Plus, Cambridge, MA), and melt curve analyses demonstrated that one PCR product was amplified per reaction for all reactions. For rats, two primer sets were designed to interrogate L1 mRNA expression based on previous data (33). The first primer set (L1–3) was designed to probe for the most active L1, while the second primer set (L1-Tot) was designed to encompass all full-length L1 elements, including those that contained a 5′-promoter, but did not have the ability to undergo retrotransposition based on mutations throughout the L1 gene. The L1–3 and L1-Tot primers were designed for the 5′-end of L1 elements based on a previous study characterizing functional elements (33). A diagram of general primer location is depicted in Fig. 1A. Fold change values from the SED rats were performed using the 2ΔΔCq method, where 2ΔCq = 2^ [house-keeping gene (HKG) Cq – gene of interest Cq], and 2ΔΔCq (or fold change) = [2ΔCq value/2ΔCq average of SED group]. The geometric mean of housekeeping genes (Fbl, Ppia, and Hprt) was used to normalize mRNA expression results for the in vivo data, while the geometric mean of B2m, Ppia, and Hprt were used for in vitro data. There were no between-group differences in the geometric mean of housekeeping genes.
Table 1.
Rat primer sequences used for real-time qPCR
| Gene | Accession No./L1 Family |
|---|---|
| (Rat) Fibrillarin (Fbl; HKG), forward (5′ → 3′): CTGCGGAATGGAGGACACTT; reverse (5′ → 3′): GATGCAAACACAGCCTCTGC | NM_001025643.1 |
| (Rat) 02-Microglobulin (B2m; HKG), forward (5′ → 3′): GGAAACTGAGGGGAGTAGGG; reverse (5′ → 3′): CCTGGGCTTTCATCCTAACA | NC_005102.4 |
| (Rat) Glyceraldehyde-3-phosphate dehydrogenase (Gapdh; HKG), forward (5′ → 3′): TGATGCCCCCATGTTTGTGA; reverse (5′ → 3′): GGCATGGACTGTGGTCATGA | NC_005103.4 |
| (Rat) Cyclophilin A (Ppia; HKG), forward (5′ → 3′): GCATACAGGTCCTGGCATCT; reverse (5′ → 3′): AGCCACTCAGTCTTGGCAGT | NM_017101.1 |
| (Rat) Hypoxanthine phosphoribosyltransferse 1 (Hprt; HKG), forward (5′ → 3′): AAGACAGCGGCAAGTTGAAT; reverse (5′ → 3′): GGGCCTGTGTCTTGAGTTCA | NM_012583.2 |
| (Rat) L1 (L1–3), forward (5′ → 3′): GACCATCTGGAACCCTGGTG; reverse (5′ → 3′): GGGCCTGTGTCTTGAGTTCA | DQ100473.1 |
| (Rat) L1 (L1-Tot), forward (5′ → 3′): GGAAGAGACCACCAACACTG; reverse (5′ → 3′): GAAGGTTTAGCTCTCCCTCC | DQ100473.1 |
| DQ100475.1 | |
| DQ100476.1 | |
| DQ100477.1 | |
| DQ100474.1 | |
| DQ100482.1 |
HKG, housekeeping gene; bp, base pairs; UTR, untranslated regions.
Fig. 1.
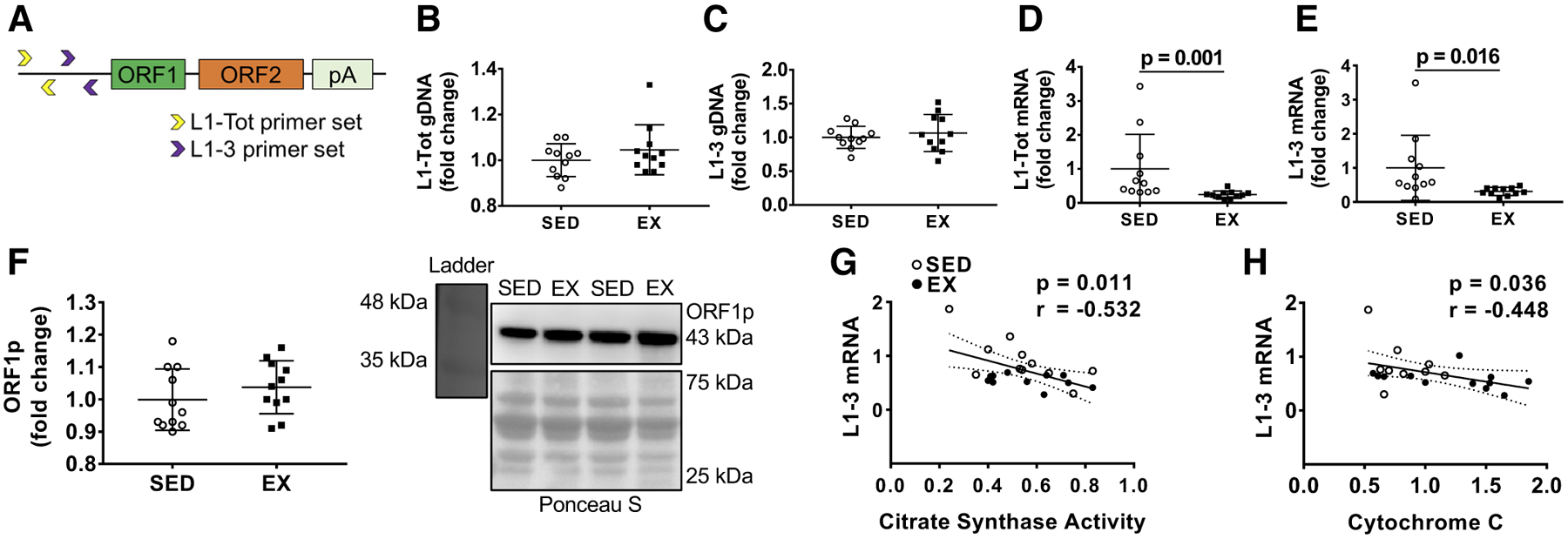
Long interspersed element-1 (L1) primer design, skeletal muscle L1 DNA content, L1 mRNA expression, and correlations with select mitochondrial markers. A: general location of L1 primers. B and C: L1 DNA content differences between the groups using the L1-Tot (B) and L1–3 (C) primer sets described in the text. D and E: mRNA expression for L1-Tot (D) and L1–3 (E). F: open reading frame protein 1 (ORF1p) protein expression with a Western blot representative image (right). All data in B–F are presented as individual values as well as fold change relative to the sedentary rat (SED) group ± SD, and group comparisons were performed using independent-samples t tests (n = 11 animals per group). G and H: associations between L1–3 mRNA expression and mitochondrial markers (citrate synthase activity and cytochrome c, respectively). EX, exercised rats.
DNA isolation and qPCR prep.
Powdered gastrocnemius muscle was removed from −80°C storage, and ~15 mg was processed using the commercially available DNeasy Blood & Tissue Kit (Qiagen, Venlo, The Netherlands) per the manufacturer’s recommendations, including RNase treatment. Following DNA precipitation and pelleting, DNA was eluted with 100 μL of elution buffer from the kit per the manufacturer’s recommendations, and DNA concentrations were determined in duplicate at an absorbance of 260 nm by using a NanoDrop Lite (Thermo Scientific). Isolated DNA was then used for the downstream assays described below as well as L1 DNA content using qPCR with the L1–3 and L1-Tot qPCR primers.
DNA methylation analysis.
L1 promoter methylation analysis was performed on isolated gastrocnemius DNA (described above) using a commercially available methylated DNA immunoprecipitation (MeDIP) kit (Abcam, Cambridge, MA). Prior to performing the MeDIP assay, 1.5 μg of gastrocnemius DNA was digested using MseI due to the fact that this enzyme did not digest DNA within our qPCR primer sequences (New England BioLabs, Ipswich, MA). Following digestion reactions, 1 μg of DNA was used for immunoprecipitation with an anti-5-methylcytosine antibody provided within the kit. qPCR was then performed on the methylated DNA-enriched sample using the L1–3 and L1-Tot primers listed above, given that both primer pairs span CpG-rich areas in the 5′-UTR. Additionally, 0.1 μg of residual input DNA from each sample was used as a control in a parallel reaction to normalize qPCR results. Both the experimental and control wells contained 25 ng of DNA for the reactions and were carried out using the same primer- and SYBR green-based methods as described above for qPCR. Fold change scores in L1 DNA methylation were calculated as follows: 1) 2ΔCq values were calculated whereby ΔCq = input DNA Cq – methylated DNA Cq, and 2) fold change values were then obtained by dividing each individual 2ΔCq value by the SED 2ΔCq group mean.
L1 chromatin accessibility analysis.
LINE-1 chromatin accessibility was assessed from each rodent using a commercially available kit (Chromatin Accessibility Assay Kit, product no. ab185901; Abcam) per the manufacturer’s recommendations. Because of sample limitations, only a subset of animals was analyzed in this experiment (SED, n = 10; EX, n = 9). Briefly, methods involved obtaining DNA, digesting the DNA using a proprietary nuclear digestion buffer, and performing qPCR on digested versus undigested samples. The premise of the assay is that a gene of interest localized to euchromatin regions is more susceptible to digestion and thus possesses a lower qPCR amplification signal relative to genes in heterochromatin regions. Gastrocnemius DNA was freshly isolated using propriety columned-based methods provided by the kit. qPCR was performed as described above on 25 ng of digested DNA using the L1–3 and L1-Tot primers to decipher fold change in genomic L1 DNA residing in euchromatin. Undigested DNA from each sample (25 ng) was used as a control to normalize RT-PCR results. Fold-change in L1–3 heterochromatin DNA was calculated using the 2ΔΔCq method, where 2 ΔCq = 2^ [undigested L1–3 DNA Cq – digested L1–3 DNA Cq] and 2 ΔΔCq (or fold change) = [2ΔCq value/2ΔCq average of SED group].
Nuclear DNMT activity assay.
Prior to assaying nuclear DNA methyltransferase (DNMT) activity, nuclear protein extraction was performed on frozen gastrocnemius muscle (~25 mg) using a commercially available kit (Nuclear Extraction Kit, cat. no. ab113474; Abcam) per the manufacturer’s recommendations. Global DNMT activity of nuclear isolates (10 μL) was assessed using a commercially available kit (DNMT Activity Assay Kit, cat. no. ab113467; Abcam) per the manufacturer’s recommendations. Briefly, gastrocnemius tissue was homogenized in an extraction buffer provided by the kit, followed by centrifugation at 12,000 rpm for 15 min. Supernatant was then discarded followed by the addition of a dithiothreitol solution and protease inhibitor cocktail to the aforementioned extraction buffer, all provided in the kit. The newly made solution was added to the pellet and incubated on ice (15 min) while vortexing every 3 min. The suspension was centrifuged at 14,000 rpm, the supernatant was transferred to a new tube, and protein content was determined using the BCA method described above. DNMT activity was measured on the nuclear extracts. Reactions were carried out on a 96-well plate that included Adomet substrate (provided in the kit) and 10 μg of nuclear extract followed by incubation at 37°C. Wells were then washed, and the capture antibody was added to the wells and incubated at room temperature for 60 min. Wells were washed again, and detection antibody was then added, followed by another incubation at room temperature for 30 min. Wells were washed again, followed by the addition of enhancer solution and a 30-min incubation time at room temperature. Wells were washed, a developer solution was added, and wells were incubated for 10 min away from light at room temperature, followed by the addition of a stop solution. Wells were then read in a microplate reader at 450 nm for 10 min. DNMT activity is expressed as relative expression units, which were normalized to input muscle weights.
Citrate synthase activity assay.
Citrate synthase (CS) activity was performed using frozen muscle obtained from rats following euthanasia, as previously described by our laboratory (25). The assay principle is based on the reduction of 5,50-dithiobis-(2-nitrobenzoic acid) (DTNB) at 412 nm (extinction coefficient 13.6 mmol·L−1·cm−1) coupled to the reduction of acetyl-CoA by the CS reaction in the presence of oxaloacetate. Briefly, 2 μg of skeletal muscle protein was added to a mixture composed of 0.125 mol/L Tris·HCl (pH 8.0), 0.03 mmol/L acetyl-CoA, and 0.1 mmol/L DTNB. The reaction was initiated by the addition of 5 μL of 50 mmol/L oxaloacetate, and the absorbance change was recorded for 1 min. Notably, the purpose of this assay was to associate CS activity levels (along with muscle cytochrome c protein levels) to skeletal muscle L1 mRNA expression levels.
mRNA microarray.
Total RNA was extracted from the gastrocnemius muscle as described above, and a subset of samples from each group (n = 8 per group) was shipped to a commercial service for transcriptome-wide analysis (Thermo Fisher Scientific). Briefly, RNA integrity was first assessed using microfluidic gel electrophoresis. Thereafter, RNA was subjected to first- and second-strand cDNA synthesis reactions. A series of reactions was then used to generate fragmented, single-stranded cDNA, which was labeled using the WT Terminal Labeling Kit. Labeled cDNA was then hybridized on Rat Clariom S array chips according to the manufacturer’s instructions. Differential gene expression was calculated using Transcriptome Analysis Console 4.0, summarization was performed using the SST-RMA algorithm, and results were provided as tab-delimited files. Log2 signal intensity values for mRNAs related to DNA methylation were then calculated and statistically compared via independent-samples t test between SED and EX rats. Dnmt and Tet genes were selectively chosen for analysis based on their ability to regulate L1 elements in prior literature (11, 45, 46, 53).
Cell culture.
For all in vitro experiments, rat L6 myoblasts (passage 2) were grown in growth medium (DMEM, 4.5 g/L glucose, 10% FBS, 1% penicillin-streptomycin, and 0.1% gentamycin) on 12-well plates at a seeding density of 2 × 105/mL under standard culture conditions (37°C in a 5% CO2 atmosphere). Once myoblast growth reached 80–90% confluency, differentiation was induced by removing growth medium and replacing it with differentiation medium [DM; DMEM, 2% (vol/vol) horse serum, 1% penicillin-streptomycin, and 0.1% gentamycin]. DM was then replaced every 24 h for 6 days to allow for myotube growth.
For drug screening experiments in L6 myotubes, cells were treated (n = 4 cell culture wells per condition) with one of six treatments, or vehicle, to investigate the possible exercise-associated mechanisms that regulate L1 gene expression (Table 2).
Table 2.
Drug treatments for in vitro screening
| Treatment | Mechanism of Action | Concentration | Ref. No. |
|---|---|---|---|
| DMSO (control) | Vehicle for all drugs (served as control) | 0.1% | 1 |
| AICAR | Increase AMPK activity | 1 mM | 1 |
| Caffeine | Increase intracellular calcium release | 5 mM | 1 |
| Rot | Increase intracellular NADH levels | 100 nM | 21 |
| Res | Increase SIRT activity | 10 μM | 12 |
| TA | Decrease HDAC activity | 100 nM | 26 |
| 5-AZA | Decrease DNMT activity | 10 μM | 31 |
AICAR; 5-aminoimidazole-4-carboxamide ribonucleotide; AMPK, AMP-activated protein kinase; 5-AZA, 5-azacytidine; DNMT, DNA methyltransferase; HDAC, histone deacetylase; Res, resveratrol; Rot, rotenone; SIRT, sirtuin; TA, trichostatin A.
Treatments included AICAR (1 mM, cat. no. BML-EI330; Enzo, Farmingdale, NY), caffeine (5 mM, cat. no. A10431; Alfa Aesar, Ward Hill, MA), rotenone (100 nM, cat. no. ALX350360G001; Enzo), resveratrol (10 μM, cat. no. R0071; TCI America, Portland, OR), the global HDAC inhibitor trichostatin A (100 nM, cat. no. G656A; Promega, Madison, WI), 5-azacytidine (10 μM, TCI America), or 0.1% DMSO (cat. no. 25–950-CQC; Corning Inc., Corning, NY) as a vehicle control. All conditions contained the same amount of DMSO in their final concentrations. Treatments were added to fresh DM when DM was replaced, and treatment DM was left on cells for a 3-h incubation. After treatments, cells were washed once with PBS and lysed with Ribozol for RNA isolation or DNA isolation, as described above. Following RNA isolation, qPCR was performed as described above to interrogate L1 mRNA expression using the SYBR green-based qPCR methods and the rat primers described above.
Follow-up AICAR-only experiments in L6 myotubes (n = 6 wells/dose) were performed as described above, with the exception of different doses of AICAR [1 (n = 6), 2 (n = 6), and 4 mM (n = 6)] being applied to cells over a 3-h treatment period along with their respective controls (n = 6 wells of DMSO). All wells contained the same percentage of DMSO.
Statistics.
Statistics were performed using SPSS version 23.0 (IBM, Armonk, NY). Prior to statistical analyses, assumptions testing was performed on all dependent variables. In cases where data were not normally distributed, we applied either log10 or square root transformations. Dependent variables were analyzed using independent-samples t tests. For data that were not normally distributed after transformation, we performed nonparametric statistics (Mann-Whitney U test). For in vitro data analysis, dependent variables were analyzed using independent-samples t tests relative to the vehicle condition. For data that violated Levene’s test for equality of variances, a Welch’s t test was used. Additionally, Pearson product correlations were also performed on L1 expression and select dependent variables. The magnitude of effects is expressed in the results, using Cohen’s d calculations, and effect sizes of 0.2, 0.5, and 0.8 were considered small, moderate, and large, respectively. Statistical significance for all null hypothesis testing was set at P < 0.05. All data are presented as means ± SD.
RESULTS
Animal characteristics.
SED and EX animal characteristics have previously been reported by Toedebusch et al. (62) and are only described here for convenience to the reader. Notably, of the 11 animals per group, only n = 8 per group were phenotyped. Body composition according to dual X-ray absorptiometry indicated that percent body fat was greater in SED versus EX rats (18.9 ± 2.2 versus 11.6 ± 1.4%, respectively, P < 0.001), while whole body lean mass was similar between groups (262 ± 8 vs. 257 ± 5 g, respectively, P = 0.156). Dissection analysis using an analytical scale revealed that gastrocnemius masses were greater in SED versus EX rats (1.67 ± 0.04 vs. 1.61 ± 0.06 g, respectively, P = 0.034). Indirect calorimetry during a maximal treadmill test during the week of euthanasia revealed that body mass-adjusted VO2peak was greater in EX versus SED rats (71 ± 3 ml O2·kg−1·min−1 vs. 57 ± 1 ml O2·kg−1·min−1, respectively, P < 0.001). Finally, EX animals (on average) ran >40 km per week during the first week when housed in running wheels and >60 km per week thereafter until they were euthanized.
L1 DNA content and mRNA expression.
One concern when analyzing L1 data is the fact that different animals can have differing amounts of L1 encoded in their genome. There were no significant between-group differences for either L1-Tot or L1–3 DNA [t(20) = 1.154, P = 0.262, d = 0.49 and t(20) = 0.663, P = 0.515, d = 0.28, respectively; Fig. 1, B and C]. L1-Tot and L1–3 mRNA expression, however, were significantly lower in the EX group [t(20) = 3.941, P = 0.001, d = 1.68 and t(12) = 2.831, P = 0.016 (Welch’s t test), d = 1.21, respectively; Fig. 1, D and E]. Thus, we posit that lower mRNA levels in EX animals were due to the exercise stimulus and not inherent L1 DNA content between groups. Despite L1 mRNA differences, there was no significant between-group difference for ORF1 protein levels [t(20) = 1.009, P = 0.325, d = 0.43; Fig. 1F]. We next wanted to test for potential associations between our markers of exercise training with the expression of the most active L1 gene, L1–3. We observed a significant negative correlation between L1–3 mRNA and CS activity (P = 0.011, r =−0.532; Fig. 1G). Moreover, we observed a similar effect with cytochrome c (P = 0.036, r =−0.448; Fig. 1H).
DNA methylation and chromatin dynamics.
With the microarray data, we focused on the mRNA expression of enzymes that regulate DNA methylation given that L1 expression is highly regulated by DNA methylation (Fig. 2A) (11). Interestingly, Dnmt1 mRNA expression was significantly greater [t(10) = 3.024, P = 0.012 (Welch’s t test), d = 1.51; Fig. 2A] in EX versus SED, and Tet2 mRNA expression was significantly lower in EX versus SED [t(14) = 3.544, P = 0.003, d = 1.77; Fig. 2A]. Neither Dnmt3a nor Dnmt3b were significantly different between groups [t(14) = 0.672, P = 0.512, d = 0.34 and t(14) = 0.232, P = 0.820, d = 0.12, respectively]. Tet1 and Tet3 mRNA was also not significantly different between groups [t(14) = 0.357, P = 0.726, d = 0.18 and t(14) = 0.623, P = 0.543, d = 0.32, respectively].
Fig. 2.
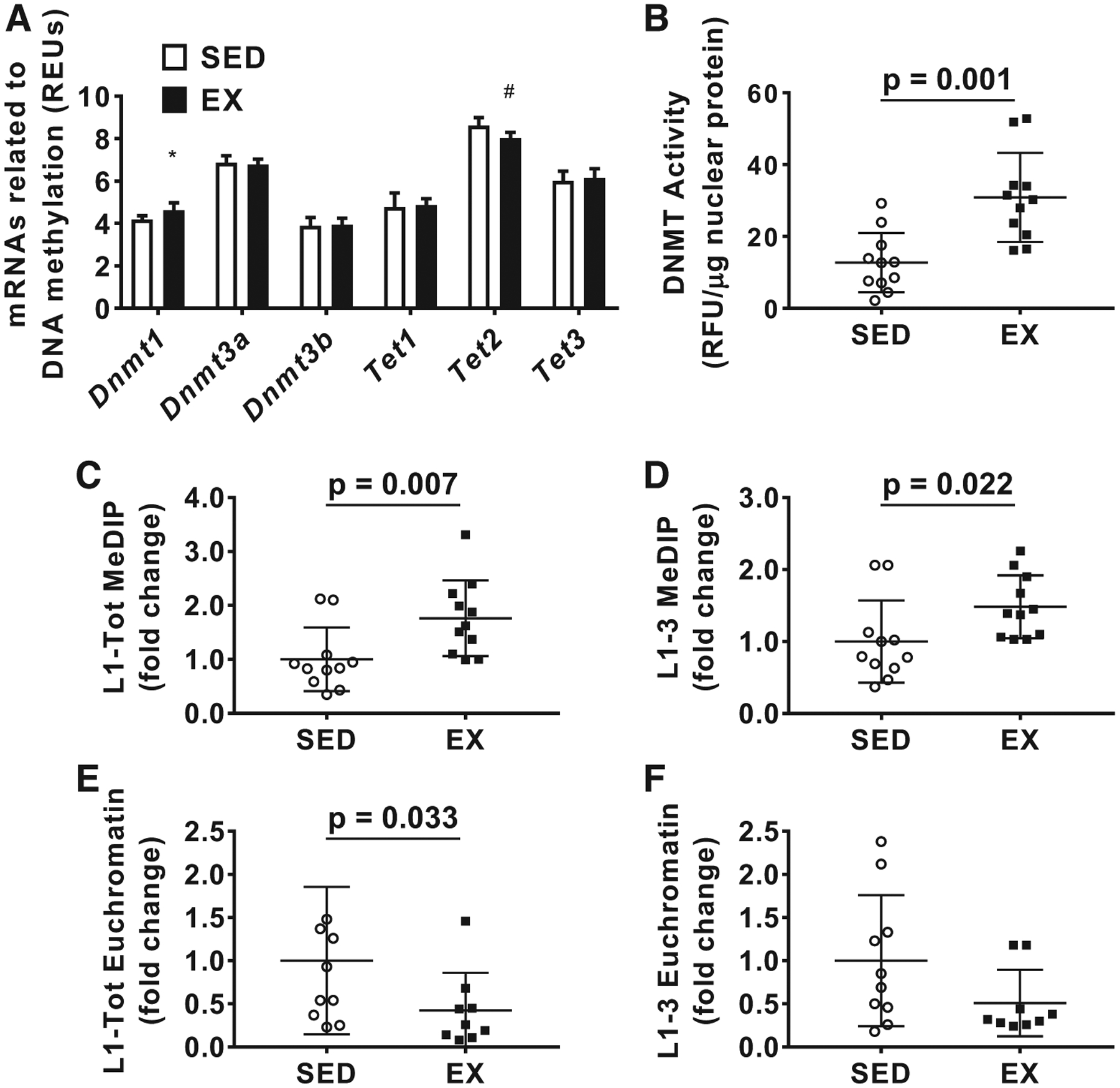
Skeletal muscle microarray results and long interspersed element-1 (L1) methylation changes. A: microarray results for mRNAs of genes involved with DNA methylation between groups; n = 8 animals per group. B: nuclear enzyme activity for DNA methyltransferases (DNMTs). C and D: methylated DNA immunoprecipitation (MeDIP) assay results for L1-Tot (C) and L1–3 (D). E and F: chromatin accessibility, represented as amount of L1-Tot (E) and L1–3 (F) in euchromatin. Data in B–F are presented as individual values; data in C–F are presented as fold change relative to the sedentary rat (SED) group ± SD, and group comparisons were performed using independent samples t tests (n = 11 animals per group). *Higher levels in exercised rats (EX) vs. SED (P < 0.05). #Lower levels in EX vs. SED (P < 0.05). REU, relative expression units; RFU, relative fluorescence units.
Next, we tested global DNMT activity and found that the EX group had significantly higher DNMT activity [t(20) = 4.039, P = 0.001, d = 1.72; Fig. 2B]. Interestingly, the L1–3 and L1-Tot primer sets also indicated that L1 DNA methylation was significantly higher in EX versus SED rats [t(20) = 2.994, P = 0.007, d = 1.28 and t(20) = 2.478, P = 0.022, d = 1.06, respectively; Fig. 2, C and D]. Based on our microarray as well as our DNA methylation data, we chose to investigate L1 chromatin accessibility using the L1–3 and L1-Tot primer sets. The EX group had significantly lower amounts of L1-Tot in accessible chromatin [t(17) = 2.323, P = 0.033, d = 1.06; Fig. 2E], while L1–3 was trending lower in the EX group as well [t(15) = 1.784, P = 0.093, d = 0.81; Fig. 2F].
Cell culture screen for exercise-specific pathways.
An independent in vitro screen experiment was used to examine whether one or multiple exercise-related signaling mediators could decrease L1 mRNA expression (Fig. 3, A and B). Relative to vehicle-only treatments, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) numerically decreased L1-Tot and L1–3 mRNA, although statistical significance was not reached [t(6) = 1.439, P = 0.200, d = 1.02, t(6) = 0.661, P = 0.533, d = 0.47, respectively]. L1-Tot and L1–3 mRNA trended upward with caffeine treatments [t(6) = 2.279, P = 0.063, d = 1.61 and t(6) = 2.110, P = 0.079, d = 1.49, respectively], while rotenone did not affect these mRNAs [t(6) = 0.625, P = 0.555, d = 0.44 and t(6) = 1.394, P = 0.213, d = 1.11, respectively], nor did resveratrol [t(6) = 0.199, P = 0.849, d = 0.14 and t(6) = 0.486, P = 0.644, d = 0.34, respectively]. L1-Tot and L1–3 mRNAs trended upward with trichostatin A treatments [t(6) = 2.238, P = 0.067, d = 1.58 and t(6) = 2.051, P = 0.086, d = 1.45, respectively], while 5-azacytidine did not affect these mRNAs [t(4) = 0.537, P = 0.620 (Welch’s t test), d = 0.38 and t(3) = 0.673, P = 0.547 (Welch’s t test), d = 0.48, respectively].
Fig. 3.
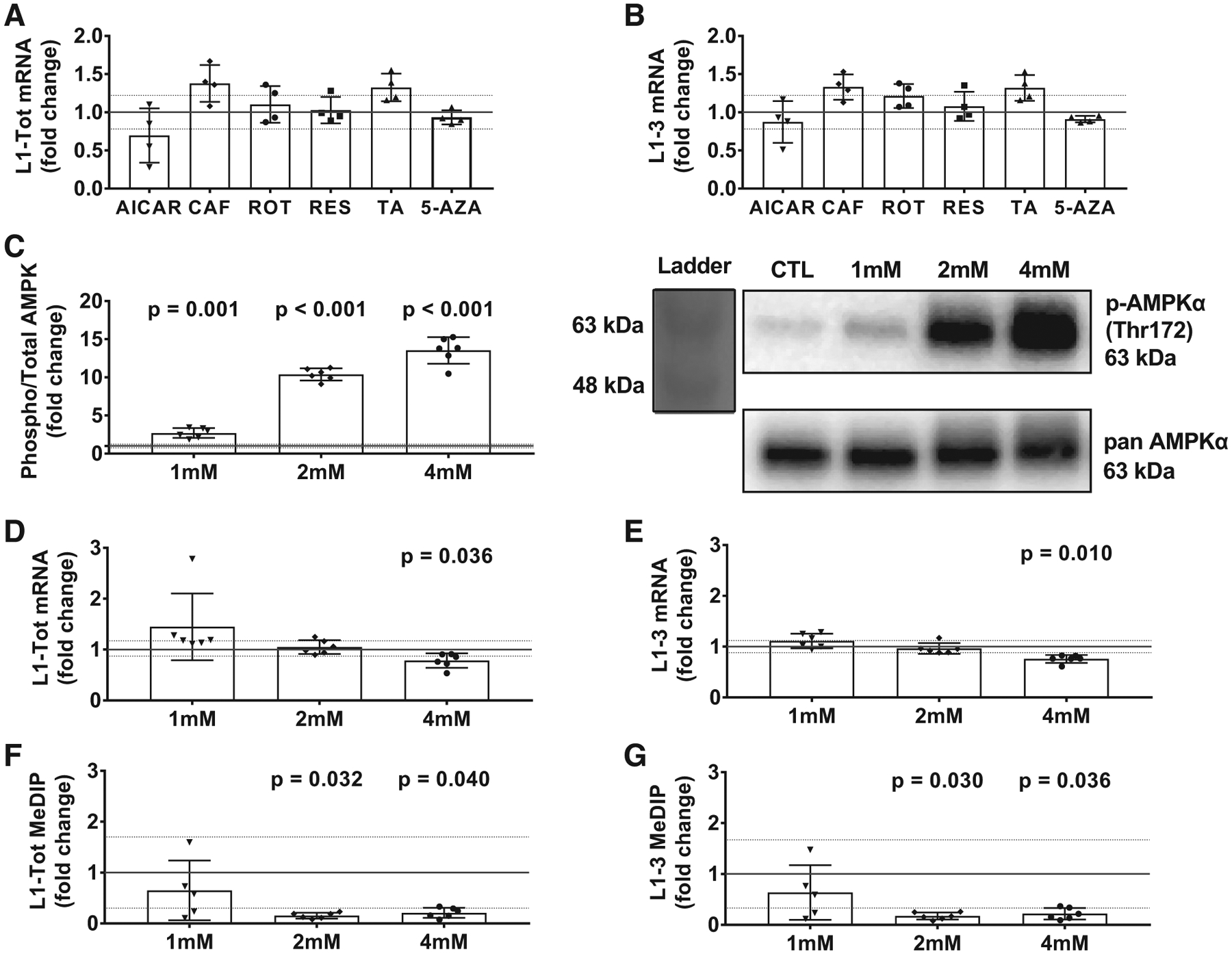
Cell culture experiments to determine possible exercise-specific pathways that regulate long interspersed element-1 (L1) mRNA expression in rat L6 myotubes. A and B: mRNA expression for L1-Tot (A) and L1–3 (B) following 3-h treatments with a variety of drugs described in Table 2 (n = 4 wells/drug/vehicle treatment). C: phosphorylation of AMP-activated protein kinase (AMPK; Thr172) with increasing doses of 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) along with an accompanying Western blot representative image. D and E: effects of different AICAR doses on L1-Tot (D) and L1–3 (E) mRNA expression. F and G: L1-Tot (F) and L1–3 methylation (G) measured via methylated DNA immunoprecipitation (MeDIP). Data in A–G are presented as individual values, and all treatments were statistically compared with the DMSO control (DMSO presented as solid line at 1.0 ± SD; n = 6 wells/AICAR/vehicle treatment in C–G). 5-AZA, 5-azacytidine; CAF, caffeine; RES, resveratrol; ROT, rotenone; TA, trichostatin A. 1, 2, and 4 mM refer to the increasing concentrations of AICAR.
AICAR, which is a known stimulator of the AMP-dependent protein kinase (AMPK) pathway, was the only compound that appeared to numerically decrease L1-Tot and L1–3 mRNA expression, albeit not significantly. Because of this finding, along with the involvement of increased AMPK activity during endurance exercise, we performed a follow-up experiment using increasing concentrations of AICAR. Notably, all cells in this follow-up experiment were treated simultaneously in the incubator to minimize batch-to-batch treatment effects. Using treatments of 1, 2, and 4 mM concentrations of AICAR in rat L6 myotubes [t(6) = 6.047, P = 0.001 (Welch’s t test), d = 3.49, t(6) = 27.794, P < 0.001 (Welch’s t test), d = 16.05, t(5) = 17.484, P < 0.001, d = 10.09, Fig. 3C], we did not observe a significant decrease in L1-Tot or L1–3 at the 1 mM dose [P = 0.423 (Mann-Whitney U test), d = 0.93 and P = t(10) = 1.501, P = 0.164, d = 0.87, respectively] or the 2 mM dose [t(10) = 0.556, P = 0.590, d = 0.32 and P = 0.873 (Mann-Whitney U test), d = 0.33, respectively]. We did, however, observe significantly lower L1–3 and L1-Tot mRNA expression in cells treated with 4 mM of AICAR [t(10) = 2.423, P = 0.036, d = 1.40 and P = 0.010 (Mann-Whitney U test), d = 2.50, respectively, Fig. 3, D and E].
We then sought to examine whether DNA methylation was different between the AICAR-treated versus vehicle-treated cells. Using the same L1 primers, we did not observe a significant difference between the control condition and 1 mM of AICAR treatment for L1-Tot or L1–3 [t(9) = 0.885, P = 0.399, d = 0.54 and t(9) = 0.970, P = 0.357, d = 0.59, respectively; Fig. 3, F and G]. Paradoxical to what we anticipated, however, L1 methylation for L1-Tot and L1–3 was significantly lower in both the 2 mM [t(5) = 2.940, P = 0.032 (Welch’s t test), d = 1.70 and t(5) = 2.981, P = 0.030 (Welch’s t test), d = 1.72, respectively] and the 4 mM AICAR condition [t(5) = 2.733, P = 0.040 (Welch’s t test), d = 1.58 and t(5) = 2.797, P = 0.036 (Welch’s t test), d = 1.61, respec tively]. Thus, AICAR seemingly reduced L1 mRNA levels in a dose-dependent manner, although this is seemingly independent of L1 DNA methylation.
DISCUSSION
As described before, L1 has the potential to be detrimental to an organism; however, only a handful of papers investigating L1 activity in muscle exist (2, 34, 38, 44, 55). In line with our previous human work, we provide evidence in rats suggesting that long-term voluntary wheel running downregulates L1 mRNA expression. Furthermore, L1–3 mRNA expression was negatively associated with two markers of mitochondrial capacity (CS activity and cytochrome c protein), which strengthens the relationship between decrements in L1 mRNA expression with increased muscle fitness. Our current data suggest voluntary wheel running may decrease L1 mRNA expression by increasing 5′-UTR methylation and reducing chromatin accessibility in regions containing L1 DNA. Furthermore, the in vitro data add to the current body of literature by suggesting that exercise may also decrease L1 expression through increased AMPK signaling. However, these two mechanisms may be independent of each other, and this is discussed in greater detail below.
A burgeoning field of research links cellular metabolism to epigenetic control with direct implications in vivo such as cell fate decisions and various diseased states (6, 14, 37, 52). Histone acetyltransferases/deacetylases and histone/DNA methyltransferases/demethylases are directly influenced by metabolic intermediates and electron carriers, including S-adenosyl methionine (DNMTs and HMTs), acetyl-CoA (HATs), α-ketoglutarate (TETs, JmjCs), FAD (LSD), and NAD+ (SIRTs), among others (3, 16, 52). Furthermore, several studies suggest that α-ketoglutarate and succinate concentrations (activating substrate and inhibitory product of the TETs and JmjCs and DNA and histone demethylases, respectively) are altered in skeletal muscle after an intense bout of endurance exercise (3, 16, 18, 19, 52). Whether these substrate-enzyme interactions are involved during or acutely after exercise to affect L1 mRNA expression remains to be determined. However, it is notable that exercised rats in the current study presented higher levels of DNMT activity and L1 DNA methylation as well as lower levels of L1 DNA in euchromatin regions. Thus, it is quite possible that several of the exercise-induced substrate-enzyme interactions mentioned above may be responsible for creating this molecular milieu.
Given our in vitro findings in L6 myotubes, one exercise-specific pathway that we speculate does downregulate L1 mRNA expression in vivo is increased AMPK activity. Numerous studies have demonstrated that endurance exercise increases skeletal muscle AMPK activation during and acutely following exercise (30). Additionally, and as mentioned before, long-term caloric restriction, which is a well-known activator of skeletal muscle AMPK, has been shown to downregulate L1 mRNA in rodent skeletal muscle (5, 7). Although we did observe lower L1 expression in the 4 mM AICAR conditions, this also resulted in lower L1 DNA methylation, which is a paradoxical finding. There are data suggesting that AMPK interacts with both DNMT and TET enzymes; specifically, DNMT1 activity is inhibited upon phosphorylation by AMPK, while TET2 activity appears to increase after phosphorylation by AMPK (40, 64). These data seemingly support our findings regarding lower L1 DNA methylation in AICAR-treated myotubes. However, it is currently difficult to explain why AICAR and increased AMPK activation decreased L1 mRNA expression as well as L1 DNA methylation, and we speculate that AMPK may either 1) affect L1 mRNA expression through other pathways to reduce L1 DNA chromatin accessibility (e.g., HDAC regulation) or 2) decrease L1 mRNA stability. This hypothetical working model is presented in Fig. 4. The former hypothesis is not far-fetched given that others have demonstrated that exercise-induced increases in AMPK act to alter nuclear HDAC levels and affect GLUT4 mRNA expression patterns (42).
Fig. 4.
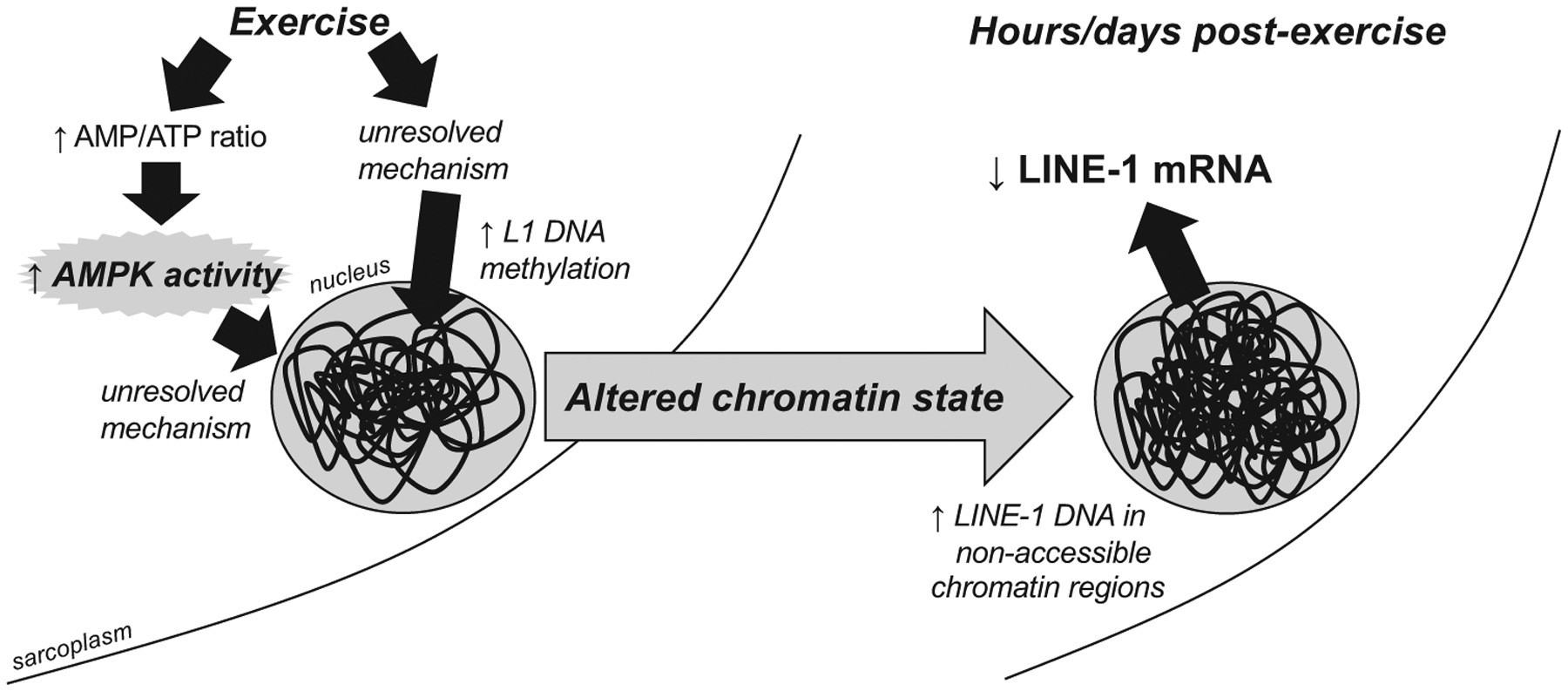
Hypothetical model of how exercise may decrease skeletal muscle long interspersed element-1 (L1) mRNA expression. A series of experiments from our laboratory have determined 1) resistance and endurance exercise and training decrease skeletal muscle L1 mRNA in rats and humans, 2) endurance training in rats reduces L1 chromatin accessibility, and 3) stimulation of AMP-activated protein kinase (AMPK) via 5-aminoimidazole-4-carboxamide ribonucleotide reduces L1 mRNA in vitro, which is independent of L1 DNA methylation. Given these observations, our working model entails exercise upregulating skeletal muscle AMPK activity as well as L1 DNA methylation, which in turn decreases L1 transcription.
Although we did not expect the observed results, our data collectively suggest that L1 DNA methylation, chromatin accessibility, and mRNA expression can be regulated by a variety of mechanisms, and these potential mechanisms are ripe for further interrogation.
We recently facilitated the transfer of tetORFeus mice breeders (48) to John McCarthy’s laboratory, who then crossed multiple tetORFeus mice with mice harboring the human skeletal actin (HSA)-rtTA transgene (28). Although the intent of this cross was to generate offspring capable of experiencing increases in skeletal muscle L1 mRNA (and potentially retrotransposition) via low doses of doxycycline (DOX) administration, a series of pilot DOX administration experiments failed to accomplish these objectives in mice that were positively genotyped for both transgenes (unpublished observations from C. Brooks Mobley, University of Kentucky). Thus, we were unable to examine how the muscle-specific overexpression of L1 affected skeletal muscle physiology, given that DOX did not drive the expression of the transgene. Given the above-mentioned shortcomings, we have not linked exercise-induced alterations in skeletal muscle L1 markers with physiological outcomes. Hence, we can only speculate that this is a favorable outcome given the prior literature, which has demonstrated that markers of increased L1 activity are associated with various genetic maladies and upregulated in skeletal muscle with aging (7, 44). It is notable that recent reports have suggested increased L1 mRNA accumulation in the cytoplasm leads to an increase in L1 cytoplasmic cDNA accumulation and stimulation of the cGAS-STING pathway (59). These events can ultimately result in enhanced proinflammatory cytokine expression. Thus, we speculate that the exercise-induced downregulation in L1 DNA accessibility and L1 mRNA may favorably affect skeletal muscle by either 1) reducing L1 retrotransposition events or 2) reducing the accumulation of cytoplasmic L1 cDNA. Indeed, future innovative research using animal and human models is needed to validate this hypothesis.
Limitations.
There are limitations to the current study. First, only a single muscle was analyzed between groups. This can make interpreting results from a single tissue difficult and does not lend itself well to making whole body inferences. Because of the fact that animals were euthanized the day following the last voluntary wheel running bout, it is also possible that we observed the acute effects of exercise on skeletal muscle L1 markers. However, it is notable that we have previously reported that L1 mRNA is downregulated 48–72 h following a resistance training bout in humans (58). Furthermore, we recently reported that an acute bout of exercise, while modestly downregulating skeletal muscle L1 mRNA in humans, does not affect L1 DNA methylation levels up to 8 h following exercise (55). Thus, we believe that our prior human data support the notion that the observations herein were due to the chronic effects of wheel running rather than transient effects. We also used only female rats for our analysis given that this was the only tissue available through our collaboration. Although there is the possibility that sex differences may be at play, we have reason to believe that L1 responses to exercise training are similar between sexes. First, these female rodent data follow the same trend from our previous studies in which muscle from college-aged male participants was used (58). Likewise, our recent publication in humans also suggests that no sex differences existed between males and females regarding the ability of one exercise bout to transiently downregulate skeletal muscle L1 expression (55). Our in vitro drug screen for exercise-specific pathways was limited due to the fact that dose-response curves and exposure times were not investigated. In this regard, some of the compounds aside from AICAR could possibly have an effect on L1 expression if different doses or exposure times were used. For example, although caffeine showed a surprising result (trending toward increasing L1-Tot and L1–3 mRNA expression), we chose not to pursue caffeine in downstream analyses. Finally, it is possible that the effects of voluntary exercise were not fully manifested because the rats were genetically predisposed for exercise. Even with these limitations, our data suggest that “exercise signals” in skeletal muscle have the ability to alter transcriptional control of L1, and future research is needed to investigate how this is related to skeletal muscle physiology.
In conclusion, this is the first report demonstrating that long-term voluntary wheel running downregulates skeletal muscle L1 gene expression. We speculate that this may occur through changes in L1 5′-UTR methylation and chromatin accessibility as well as increased AMPK activity. However, these two mechanisms may operate independently to downregulate L1 expression.
GRANTS
Reagent costs were funded through discretionary laboratory funds of M. D. Roberts. M. A. Romero was supported by a William Townsend Porter Predoctoral Fellowship from the American Physiological Society.
Footnotes
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
REFERENCES
- 1.Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 15: 405–411, 2012. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res 38: 3909–3922, 2010. doi: 10.1093/nar/gkq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger SL, Sassone-Corsi P. Metabolic signaling to chromatin. Cold Spring Harb Perspect Biol 8: a019463, 2016. doi: 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns KH, Boeke JD. Human transposon tectonics. Cell 149: 740–752, 2012. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantó C, Auwerx J. Calorie restriction: is AMPK a key sensor and effector? Physiology (Bethesda) 26: 214–224, 2011. doi: 10.1152/physiol.00010.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518: 413–416, 2015. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 5: 867–883, 2013. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Yang L. ALUternative regulation for gene expression. Trends Cell Biol 27: 480–490, 2017. doi: 10.1016/j.tcb.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigó R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wier-zbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES; Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562, 2002. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 10.Dai L, LaCava J, Taylor MS, Boeke JD. Expression and detection of LINE-1 ORF-encoded proteins. Mob Genet Elements 4: e29319, 2014. doi: 10.4161/mge.29319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Rica L, Deniz Ö, Cheng KC, Todd CD, Cruz C, Houseley J, Branco MR. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol 17: 234, 2016. doi: 10.1186/s13059-016-1096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deniz Ö, Frost JM, Branco MR. Regulation of transposable elements by DNA modifications. Nat Rev Genet 20: 417–431, 2019. [Erratum in Nat Rev Genet 20: 432, 2019.] doi: 10.1038/s41576-019-0106-6. [DOI] [PubMed] [Google Scholar]
- 13.Dugdale HF, Hughes DC, Allan R, Deane CS, Coxon CR, Morton JP, Stewart CE, Sharples AP. The role of resveratrol on skeletal muscle cell differentiation and myotube hypertrophy during glucose restriction. Mol Cell Biochem 444: 109–123, 2018. doi: 10.1007/s11010-017-3236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim MS, Manda SS, Abril G, Pereira G, Makohon-Moore A, Looijenga LH, Gillis AJ, Hruban RH, Anders RA, Romans KE, Pandey A, Iacobuzio-Donahue CA, Vogelstein B, Kinzler KW, Kazazian HH Jr, Solyom S. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res 25: 1536–1545, 2015. doi: 10.1101/gr.196238.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finley LWS, Vardhana SA, Carey BW, Alonso-Curbelo D, Koche R, Chen Y, Wen D, King B, Radler MR, Rafii S, Lowe SW, Allis CD, Thompson CB. Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat Cell Biol 20: 565–574, 2018. doi: 10.1038/s41556-018-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flasch DA, Macia Á, Sánchez L, Ljungman M, Heras SR, García-Pérez JL, Wilson TE, Moran JV. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell 177: 837–851.e28, 2019. doi: 10.1016/j.cell.2019.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frederick DW, Loro E, Liu L, Davila A Jr, Chellappa K, Silverman IM, Quinn WJ III, Gosai SJ, Tichy ED, Davis JG, Mourkioti F, Gregory BD, Dellinger RW, Redpath P, Migaud ME, Nakamaru-Ogiso E, Rabinowitz JD, Khurana TS, Baur JA. Loss of NAD homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab 24: 269–282, 2016. doi: 10.1016/j.cmet.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Perez JL, Widmann TJ, Adams IR. The impact of transposable elements on mammalian development. Development 143: 4101–4114, 2016. doi: 10.1242/dev.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibala MJ, MacLean DA, Graham TE, Saltin B. Tricarboxylic acid cycle intermediate pool size and estimated cycle flux in human muscle during exercise. Am J Physiol 275: E235–E242, 1998. doi: 10.1152/ajpendo.1998.275.2.E235. [DOI] [PubMed] [Google Scholar]
- 20.Gibala MJ, Tarnopolsky MA, Graham TE. Tricarboxylic acid cycle intermediates in human muscle at rest and during prolonged cycling. Am J Physiol 272: E239–E244, 1997. doi: 10.1152/ajpendo.1997.272.2.E239. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R,Celera, Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F; Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521, 2004. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 22.Grefte S, Wagenaars JA, Jansen R, Willems PH, Koopman WJ. Rotenone inhibits primary murine myotube formation via Raf-1 and ROCK2. Biochim Biophys Acta 1853: 1606–1614, 2015. doi: 10.1016/j.bbamcr.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Hancks DC, Kazazian HH Jr. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 22: 191–203, 2012. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7: 143–148, 1994. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 25.Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 63: 635–646, 2014. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyatt HW, Kephart WC, Holland AM, Mumford P, Mobley CB, Lowery RP, Roberts MD, Wilson JM, Kavazis AN. A ketogenic diet in rodents elicits improved mitochondrial adaptations in response to resistance exercise training compared to an isocaloric Western diet. Front Physiol 7: 533, 2016. doi: 10.3389/fphys.2016.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda K, Ito A, Imada R, Sato M, Kawabe Y, Kamihira M. In vitro drug testing based on contractile activity of C2C12 cells in an epigenetic drug model. Sci Rep 7: 44570, 2017. doi: 10.1038/srep44570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivics Z, Izsvak Z, Minter A, Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci USA 93: 5008–5013, 1996. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwata M, Englund DA, Wen Y, Dungan CM, Murach KA, Vechetti IJ Jr, Mobley CB, Peterson CA, McCarthy JJ. A novel tetracycline-responsive transgenic mouse strain for skeletal muscle-specific gene expression. Skelet Muscle 8: 33, 2018. doi: 10.1186/s13395-018-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, O’Donnell D, Gordon JI, Wang T. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet 51: 611–617, 2019. [Erratum in Nat Genet 51: 920, 2019.] doi: 10.1038/s41588-019-0373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jørgensen SB, Richter EA, Wojtaszewski JF. Role of AMPK in skeletal muscle metabolic regulation and adaptation in relation to exercise. J Physiol 574: 17–31, 2006. doi: 10.1113/jphysiol.2006.109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur K, Yang J, Eisenberg CA, Eisenberg LM. 5-Azacytidine promotes the transdifferentiation of cardiac cells to skeletal myocytes. Cell Reprogram 16: 324–330, 2014. doi: 10.1089/cell.2014.0021. [DOI] [PubMed] [Google Scholar]
- 33.Kazazian HH Jr, Moran JV. Mobile DNA in health and disease. N Engl J Med 377: 361–370, 2017. doi: 10.1056/NEJMra1510092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, Löwer R, Buschmann C, Horn AV, Traub P, Schumann GG. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res 36: 648–665, 2008. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH Jr, Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci USA 103: 8036–8041, 2006. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 13: 655–660, 2006. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 37.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J; International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409: 860–921, 2001. [Errata in Nature 412: 565, 2001; and Nature 411: 720, 2001.] doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 38.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478, 2012. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucchinetti E, Feng J, Silva R, Tolstonog GV, Schaub MC, Schumann GG, Zaugg M. Inhibition of LINE-1 expression in the heart decreases ischemic damage by activation of Akt/PKB signaling. Physiol Genomics 25: 314–324, 2006. doi: 10.1152/physiolgenomics.00251.2005. [DOI] [PubMed] [Google Scholar]
- 40.Malki S, van der Heijden GW, O’Donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell 29: 521–533, 2014. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marin TL, Gongol B, Zhang F, Martin M, Johnson DA, Xiao H, Wang Y, Subramaniam S, Chien S, Shyy JY. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci Signal 10: eaaf7478, 2017. doi: 10.1126/scisignal.aaf7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClintock B The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA 36: 344–355, 1950. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGee SL, van Denderen BJ, Howlett KF, Mollica J, Schertzer JD, Kemp BE, Hargreaves M. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes 57: 860–867, 2008. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 44.Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5: 5011, 2014. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumford PW, Romero MA, Osburn SC, Roberson PA, Vann CG, Mobley CB, Brown MD, Kavazis AN, Young KC, Roberts MD. Skeletal muscle LINE-1 retrotransposon activity is upregulated in older versus younger rats. Am J Physiol Regul Integr Comp Physiol 317: R397–R406, 2019. doi: 10.1152/ajpregu.00110.2019. [DOI] [PubMed] [Google Scholar]
- 46.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468: 443–446, 2010. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 19: 1002–1007, 2009. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Musova Z, Hedvicakova P, Mohrmann M, Tesarova M, Krepelova A, Zeman J, Sedlacek Z. A novel insertion of a rearranged L1 element in exon 44 of the dystrophin gene: further evidence for possible bias in retroposon integration. Biochem Biophys Res Commun 347: 145–149, 2006. doi: 10.1016/j.bbrc.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 49.O’Donnell KA, An W, Schrum CT, Wheelan SJ, Boeke JD. Controlled insertional mutagenesis using a LINE-1 (ORFeus) gene-trap mouse model. Proc Natl Acad Sci USA 110: E2706–E2713, 2013. doi: 10.1073/pnas.1302504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res 28: 1418–1423, 2000. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paixão-Côrtes VR, Salzano FM, Bortolini MC. Origins and evolvability of the PAX family. Semin Cell Dev Biol 44: 64–74, 2015. doi: 10.1016/j.semcdb.2015.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev 20: 149–155, 2010. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid MA, Dai Z, Locasale JW. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 19: 1298–1306, 2017. doi: 10.1038/ncb3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The Influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr 3: MDNA3-0061-2014, 2015. doi: 10.1128/microbiolspec.MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberson PA, Romero MA, Osburn SC, Mumford PW, Vann CG, Fox CD, McCullough DJ, Brown MD, Roberts MD. Skeletal muscle LINE-1 ORF1 mRNA is higher in older humans but decreases with endurance exercise and is negatively associated with higher physical activity. J Appl Physiol (1985) 127: 895–904, 2019. doi: 10.1152/japplphysiol.00352.2019. [DOI] [PubMed] [Google Scholar]
- 56.Roberts MD, Brown JD, Company JM, Oberle LP, Heese AJ, Toedebusch RG, Wells KD, Cruthirds CL, Knouse JA, Ferreira JA, Childs TE, Brown M, Booth FW. Phenotypic and molecular differences between rats selectively bred to voluntarily run high vs. low nightly distances. Am J Physiol Regul Integr Comp Physiol 304: R1024–R1035, 2013. doi: 10.1152/ajpregu.00581.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts MD, Toedebusch RG, Wells KD, Company JM, Brown JD, Cruthirds CL, Heese AJ, Zhu C, Rottinghaus GE, Childs TE, Booth FW. Nucleus accumbens neuronal maturation differences in young rats bred for low versus high voluntary running behaviour. J Physiol 592: 2119–2135, 2014. doi: 10.1113/jphysiol.2013.268805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romero MA, Mobley BC, Mumford PW, Roberson PA, Haun CT, Kephart WC, Healy JC, Beck DT, Young KC, Martin JS, Lockwood CM, Roberts MD. Acute and chronic resistance training downregulates select LINE-1 retrotransposon activity markers in human skeletal muscle. Am J Physiol Cell Physiol 314: C379–C388, 2018. doi: 10.1152/ajpcell.00192.2017. [DOI] [PubMed] [Google Scholar]
- 59.Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, De Cecco M, Leonova KI, Kogan V, Helfand SL, Neretti N, Roichman A, Cohen HY, Meer MV, Gladyshev VN, Antoch MP, Gudkov AV, Sedivy JM, Seluanov A, Gorbunova V. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab 29: 871–885.e5, 2019. doi: 10.1016/j.cmet.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, Zine El Aabidine A, Pioger L, Nigumann P, Saccani S, Andrau J-C, Gilbert N, Cristofari G. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell 74: 555–570.e7, 2019. doi: 10.1016/j.molcel.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 61.Taylor MS, LaCava J, Dai L, Mita P, Burns KH, Rout MP, Boeke JD. Characterization of L1-ribonucleoprotein particles. Methods Mol Biol 1400: 311–338, 2016. doi: 10.1007/978-1-4939-3372-3_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toedebusch RG, Ruegsegger GN, Braselton JF, Heese AJ, Hofheins JC, Childs TE, Thyfault JP, Booth FW. AMPK agonist AICAR delays the initial decline in lifetime-apex Vo2 peak, while voluntary wheel running fails to delay its initial decline in female rats. Physiol Genomics 48: 101–115, 2016. doi: 10.1152/physiolgenomics.00078.2015. [DOI] [PubMed] [Google Scholar]
- 63.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982, 2007. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 64.Wu D, Hu D, Chen H, Shi G, Fetahu IS, Wu F, Rabidou K, Fang R, Tan L, Xu S, Liu H, Argueta C, Zhang L, Mao F, Yan G, Chen J, Dong Z, Lv R, Xu Y, Wang M, Ye Y, Zhang S, Duquette D, Geng S, Yin C, Lian CG, Murphy GF, Adler GK, Garg R, Lynch L, Yang P, Li Y, Lan F, Fan J, Shi Y, Shi YG. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 559: 637–641, 2018. doi: 10.1038/s41586-018-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


