Abstract
The embryonic rat dorsal root ganglion (DRG) neuron-derived 50B11 cell line is a promising sensory neuron model expressing markers characteristic of NGF and GDNF-dependent C-fibre nociceptors. Whether these cells have the capacity to develop into distinct nociceptive subtypes based on NGF- or GDNF-dependence has not been investigated. Here we show that by augmenting forskolin (FSK) and growth factor supplementation with NGF or GDNF, 50B11 cultures can be driven to acquire differential functional responses to common nociceptive agonists capsaicin and ATP respectively. In addition, to previous studies, we also demonstrate that a differentiated neuronal phenotype can be maintained for up to 7 days. Western blot analysis of nociceptive marker proteins further demonstrates that the 50B11 cells partially recapitulate the functional phenotypes of classical NGF-dependent (peptidergic) and GDNF-dependent (non-peptidergic) neuronal subtypes described in DRGs. Further, 50B11 cells differentiated with NGF/FSK, but not GDNF/FSK, show sensitization to acute prostaglandin E2 treatment. Finally, RNA-Seq analysis confirms that differentiation with NGF/FSK or GDNF/FSK produces two 50B11 cell subtypes with distinct transcriptome expression profiles. Gene ontology comparison of the two subtypes of differentiated 50B11 cells to rodent DRG neurons studies shows significant overlap in matching or partially matching categories. This transcriptomic analysis will aid future suitability assessment of the 50B11 cells as a high-throughput nociceptor model for a broad range of experimental applications. In conclusion, this study shows that the 50B11 cell line is capable of partially recapitulating features of two distinct types of embryonic NGF and GDNF-dependent nociceptor-like cells.
Keywords: 50B11, dorsal root ganglia, nociception, nerve growth factor, glial derived neurotrophic factor, ATP, capsaicin, calcium activity, RNA-seq
Introduction
The E14.5 rat dorsal root ganglia (DRG)-derived 50B11 neuron cell line was developed with the aim of investigating neuroprotective strategies for peripheral neuropathies targeting nociceptive cell types.1 The initial characterisation and subsequent reports1–4 have identified the 50B11 cells comparable to a subclass of small, unmyelinated, C-fibre nociceptors, that play a key role in pain signalling via the transduction of high threshold signals from the periphery.5 Other DRG/neuroblastoma-derived cells such as the F-116 and ND7/237 have recently been shown to express markers for both myelinated and unmyelinated neurons and were used as models for sensory neurons and nociceptors. However, they lack key features and nociceptive markers such as the transient cation channel subfamily V member 1 (TrpV1).8,9 In a similar fashion, the E12.5 DRG imposturous-derived MED17.11 cells represent another promising model for the study of nociceptive neurons. The cells express markers of committed sensory neuron progenitors. Upon differentiation MED17.11 cells express multiple markers of maturing DRG neurons including voltage gated sodium channels NaV1.7 and NaV1.9, but not NaV1.8.10
The 50B11 cell line is the only DRG-derived cell line that appears to express markers specific for both nerve growth factor (NGF) and glial-derived neurotrophic factor (GDNF) signaling.1–4,8 Response to these growth factors is an accepted and common way of identifying subtypes of DRG sensory and nociceptive neurons. During the perinatal and postnatal period, prospective nociceptors undergo two distinct differentiation pathways that lead to the formation of NGF-dependent peptidergic and GDNF-dependent non-peptidergic nociceptors. These two major classes of nociceptors express distinct combinations of ion channels and receptors and innervate distinct peripheral and central targets.11–14 Reliance on multiple growth factors is critical for the timely and target-specific development of nociceptors,15,16 and interestingly virtually all DRG neurons normally lost after neonatal axotomy can be saved by either NGF or GDNF.17 All developing nociceptors depend on NGF for survival up until E15.5, when about half of developing nociceptors switch off the expression of NGF-receptor TrkA and begin to express Ret, the transmembrane signaling receptor for GDNF.16 These neurons become GDNF-dependent non-peptidergic nociceptors, most of which bind isolectin B4 (IB4). The remaining nociceptors retain TrkA (a few also co-express Ret) and develop into the peptidergic class of nociceptors that express calcitonin gene-related peptide (CGRP) and substance P (SP) and do not bind IB4.18,19 More importantly, GDNF-dependent neurons differ from NGF-dependent neurons functionally, having longer duration action potentials, higher densities of tetrodotoxin-resistant sodium currents and smaller heat-activated currents.20
Although DRG isolation techniques consistently produce highly enriched primary neuronal cultures, depending on the isolation technique and the species of origin, these cultures can vary in the percentage of neuronal and non-neuronal cells typically they contain.21–24 These heterogeneous populations include several subtypes of nociceptors, and have been of extraordinary value for the study of neuronal anatomy, cell biology, and physiology.25–28 However, the ability to culture and isolate homogeneous primary neurons from the DRG in a subtype-specific manner, or with distinct sensory modalities is fraught with technical difficulties and remains a challenge to investigators.29 Given that the 50B11 cells have been shown to differentiate in response to both NGF and GDNF, our aim was to determine how the timing of exposure to these growth factors is capable of differentiating these cells into a peptidergic-like and non-peptidergic like phenotypes. This capability would provide a readily available and limitless resource of homogeneous nociceptor like cells, currently not feasible with primary culture methods.
Previous characterization of the 50B11 cells1–3 did not investigate their expression profiles of peptidergic and non-peptidergic nociceptive markers. Here we describe a modified protocol for longer-term (7 day) differentiation of 50B11 cells in an NGF- and GDNF-dependent manner into capsaicin- and ATP-responsive cell types, resembling peptidergic and non-peptidergic DRG phenotypes respectively. We also show that 50B11 cells to undergo limited sensitization in response acute prostaglandin E2 (PGE2) but not to interleukin-1 beta (IL1β), which are commonly associated with inflammatory responses in nociceptors. In addition, we present a detailed analysis of nociceptor specific markers in undifferentiated, as well as NGF and GDNF differentiated cells, including RNA-Seq analysis and qRT-PCR validation of key peptidergic and non-peptidergic nociceptive markers. Our results demonstrate the 50B11 cells are a suitable and readily available model for investigations of specific molecular pathways in embryonic nociceptor subtypes. Additionally, we provide for the first time, a comprehensive transcriptomic comparison of between 50B11 cells rodent DRG neurons, highlighting the similarities and differences relating to peptidergic and non-peptidergic nociceptive markers.
Materials and methods
Cell culture, differentiation and growth factor treatments
50B11 cells, a gift from Dr. Ahmet Hoke, were maintained in plated in Neurobasal medium (Life Technologies, Gibco) supplemented with 10% FBS (Life Technologies, Gibco), 1% B27 (Life Technologies), 20 mM D-glucose (Life Technologies, Gibco) and 1% glutaMAX™ media supplement (Life Technologies, Gibco). Cells were not used for experiments after passage 20.
For neurite outgrowth assays cells were plated at different densities including low densities optimal for visualizing individual neurites. 24 h after plating cells were differentiated by adding forskolin (FSK; Sigma–Aldrich, 0.1–100µM) to the medium. Based on observations by Chen et al.1 and our preliminary studies, neuronal phenotype assessed by neurite outgrowth and changes to rounded soma morphology was best established between about 1 and 7 days in vitro (DIV) post-FSK and growth factor treatments (50 ng/ml recombinant mNGF, Peprotech), GDNF (50 ng/ml recombinant hGDNF, Peprotech), and all subsequent protocols were designed to be completed within this time frame. Cells were then used for either immunohistochemistry, lysed for Western blotting or qRT-PCR.
Neurite outgrowth assays
Following culture and treatments described above, 50B11 cells were photographed live on an Olympus (IX71) microscope. Measurements of neurite length (minimum 100 cells) were performed on 16-bit TIFF format images using the NeuronJ plugin written for Fiji (Image J, NIH). Single neurons with minimal or no overlapping of neurite arbours with adjacent cells were analysed using NIH ImageJ software with the NeuronJ Plugin. Distances from soma perimeter to neurite tips were measured by tracing arbours and expressed as the summed length of neurite outgrowth and as length of the longest axon. Phase annuli counts of the cell soma within each image were performed to determine % of differentiated cell in total populations. All data are presented as ± standard error of mean (SEM), and treatment effects were compared by Mann–Whitney rank sum tests.
Western blot
Treated 50B11 cells were lysed using chilled lysis buffer containing 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 1% Triton X-100, 10% glycerol, 1 mM phenylmethanesulfonyl fluoride, 1 mM sodium orthovanadate, 1 µM batimastat (BB-94), and 1% Roche protease inhibitor cocktail (2). For Western blots, lysates were solubilized in an equal volume of 2 × SDS sample buffer containing 4% SDS, 2% glycine, 0.015% bromophenol blue, 20% glycerol, and 10% β-mercaptoethanol in 100 mM Tris-HCl buffer, pH 6.8. Protein quantification for all samples was performed with the BCA Protein Assay Kit (Pierce - Thermo Scientific). Cell lysates were electrophoresed through 4–12% Bis-Tris buffered SDS gels (Life Sciences). Proteins were transferred onto PVDF or nitrocellulose membrane at 100 V for 1 h. The membranes were blocked in 4% skim milk powder for transmembrane protein detection, or 3% bovine serum albumin (BSA; Sigma) for phosphorylated proteins, in 0.1% Tween-20, and 0.02% NaN3 in TBS, pH 8.0, for 1 h at room temperature and incubated overnight with primary antibodies at 4°C. The following antibodies were used for western blotting: rabbit anti-p75NTR #9992 (1:5000, M. Chao), rabbit anti-TrkA (1:500; Biosensis), anti-TrpV1 antibodies (1:500; Biosensis), c-Ret (1:2000; Cell Signalling), anti-P2X3 (1:500; Cell Signalling), GFRα1 (1:200, goat antiserum, R&D Systems) and mouse anti-β-III tubulin (1:2000; Promega). Membranes were then washed three times in TBS-Tween 20 (TBST), pH 8.0, for 10 min and incubated for 1 h with donkey anti-rabbit 680 or donkey anti-mouse 800 secondary (1:50000; Invitrogen) in TBS at room temperature and then washed three times in TBS for 10 min. For stripping and re-probing Western blot membranes were treated with 10 ml of Restore™ PLUS Western blot stripping buffer (Thermo-Fisher) for 30 min at room temperature on an orbital shaker, washed three times with TBST and blocked with either 5% skim milk powder or 3% BSA in PBST for 1 h and then incubated with appropriate primary and secondary antibodies as described above. Western blots for individual markers were performed on separate membranes, which means that the comparison may include differences of antibody binding across the different treatments.
Isolation of total RNA and reverse transcription
Undifferentiated 50B11 cells and those differentiated with NGF (50 ng/ml), GDNF (50 ng/ml) and FSK (10 µM) were harvested and stored in Trizol (Sigma) at −80°C. Samples were mechanically homogenized using a tissue lyser (Qiagen). Total RNA was isolated using a column-based method (Zymo-Spin ICC Columns, Zymogen, Irvine, CA, USA). DNA contamination was removed by on-column DNA digestion. The concentration of total RNA was determined using standard photospectrometry (Nanodrop 2000, Thermo-Scientific, Australia), quality of RNA was determined using a lab-on-chip system (Bioanalyser, Agilent). Only samples with RNA integrity numbers (RIN ≥ 7) were used for subsequent analysis. One microgram of total RNA was reverse transcribed into cDNA according to the manufacturer protocol (SuperscriptII, Bio-Rad, Australia).
q-RT-PCR
The RNA was reverse transcribed into cDNA according to the manufacturers protocol (SuperscriptII, Bio-Rad, Australia). qPCR analysis of the relative mRNA expression levels in undifferentiated and differentiated 50B11 cells was performed using the QuantStudio3 cycler (Applied Biosciences). TaqMan primers (Life Technologies) were used for the detection of molecules characteristic for nociceptors. Hypoxanthine phosphoribosyl transferase 1 (Hprt1) was used as a reference gene.30 The efficiencies of all primer-pairs were determined by 1/5 to 1/625 dilutions in a qPCR and a satisfying efficiency was determined with Q-Gene.31 The primers and efficiencies are listed (Table 1). The final volume for qPCR was 20 µl of which 8 µl were water, 10 µl mastermix (Life Technologies), 1 µl assay-mix (Life Technologies) and 1 µl cDNA. Each qPCR was done in duplicate. The Ct values were determined for each product and normalised as pairwise comparisons against the Ct value of the reference gene and calculated as Mean Normalized Expression (MNE).31
Table 1.
TaqMan primer assay efficiency measurements.
| Primer assay (gene name) | Assay code | Amplicon length (bp) | Efficiency (R2) |
|---|---|---|---|
| Hprt1 | Rn1527840_m1 | 64 | 0.995 |
| P2rx3 | Rn00579301_m1 | 63 | 0.998 |
| Trpv1 | Rn00583117_m1 | 66 | 0.998 |
| Ntrk1 | Rn00572130_m1 | 65 | 0.974 |
| Ngfr | Rn00561634_m1 | 60 | 0.997 |
| Ptger2 | Rn00579419_m1 | 71 | 0.992 |
| Il1rl1 | Rn01640664_m1 | 121 | 0.99 |
| Calca | Rn01511353_g1 | 129 | 0.98 |
| Calcb | Rn00593383_m1 | 63 | 0.986 |
| Ret | Rn01463098_m1 | 97 | 0.991 |
| Gfra3 | Rn01760829_m1 | 71 | 0.989 |
| Scn11a | Rn00570487_m1 | 110 | 0.97 |
| Scn10a | Rn00568393_m1 | 69 | 0.982 |
| Trpa1 | Rn01473803_m1 | 104 | 0.995 |
| Mrgprd | Rn01785783_s1 | 69 | 0.995 |
RNA-Seq
RNA-Seq was employed for whole transcriptome analysis and was conducted in the Flinders Genomics Facility. Libraries were prepared using the TruSeq Stranded TotalRNA Sample preparation kit (Illumina) in combination with the Ribo-Zero Human/Mouse/Rat kit (Illumina) to remove ribosomal RNA. One microgram of total RNA was used to generate each library. Libraries were validated on the LabChip GX Touch 24 (Perkin Elmer) and quantified using Qubit 2.0 (Thermofisher), then diluted and pooled. Sequencing was conducted on the Illumina NextSeq 500 platform providing an average of approximately 60 million reads per library.
The reads were analysed based on the Rattus norvegicus genome (Rnor_6) available at the Ensembl database (https://www.ensembl.org/Rattus_norvegicus/Info/Index). Initially, the reads were subjected to quality control, adapter sequence removal and later mapped onto the rat genome. For the PCA plot, counts were normalized using the RPKM method (reads per kilobase of transcript per million mapped reads). For analysis of differentially expressed genes, the DESeq2 algorithm was used.32 Genes were counted as differentially expressed at an adjusted p-value of <0.05.
The transcripts studied were differentially expressed between undifferentiated and NGF/FSK or GDNF/FSK differentiated cells. Differentially expressed RNAs were functionally annotated using the DAVID bioinformatics resource (david.ncifcrf.gov).33
In addition, genes that were induced from undetectable levels, or completely down regulated to near undetectable levels (0–5 counts), in response to differentiation cues, were also identified. Furthermore, expression patterns of primary microRNA transcripts (pri-miRNA) and long non-coding RNAs (lncRNA) were also compared.
Gene ontology analysis
Gene Ontology (GO) enrichment performed on differential expressed genes to identify the relationship between the genes and GO terms. To compare the GO terms of published data of sensory neuron types34 with the list of differentially expressed genes in this experiment, followed similar workflow focusing only biological process terms. The upregulated genes with 2-fold and p-value of 0.05, of GDNF and NGF (in comparison to UND), samples considered for the GO analysis. Cytoscape 3.5.135 with ClueGo app36 used with the following settings: Markers list of Rattus norvegicus; GO terms levels: 3–8; GO term restriction: 2 genes and 4% evidence and a significance threshold of 0.05. If the GO terms are not found similar to the generalised sensory groups, according to Usoskin et al.,34 the significance threshold is relaxed. Due to the difference in experimental methods, tabulated information corresponds to the GO terms published.
Mapping and differential expression
The reads were mapped against the Rat reference genome (Rnor_6.0) from Ensembl (https://asia.ensembl.org/Rattus_norvegicus/Info/Index) and STAR spliced alignment algorithm37 (version 2.7.0e with default parameters) used for mapping with default parameters. The featureCounts38 program is used to assign the sequence reads to genomic features and counts the reads assigned to each exonic region. For the differential expression, the genes retained which had Counts Per Million (CPM) > 1 in at least half of the libraries. The Differential gene expression analysis carried out using the DESeq2 statistical tool32 in the R statistical program (https://cran.r-project.org/).
Imaging of intracellular calcium concentration
Undifferentiated and differentiated cells were loaded with the cell-permeant Ca2+ indicator, fura-2 AM. 50B11 cells were loaded for 45 min at 37°C. Cells were then washed with HEPES buffer (composition in mM: 138 NaCl, 5 KCl, 1.2 MgCl2, 2 CaCl2, 10 glucose, 10 HEPES, pH 7.4) and incubated at room temperature for a minimum of 30 min prior to starting an experiment to allow intracellular hydrolysis of fura-2 AM.
Coverslips were placed in a small laminar flow perfusion chamber (approximate volume 200 µl; Warner Instrument Corporation) and continuously perfused (flow rate 2 ml/min) with HEPES-buffered saline. Capsaicin (500 nM-5nM), PGE2 (100 nM) and Il-1ß (1 µM) were applied locally (flow rate approximately 3 ml/min) via a fine pipette positioned close to the cells being studied. Cells were sequentially illuminated at 340 nm and 380 nm via two monochromators (PTI, New Jersey) and images of emission fluorescence at 520 nm for each excitation wavelength captured with a cooled CCD camera connected to a PC. Analysis of ratios of emission intensity at 340 nm: 380 nm excitation and subtraction of background fluorescence and baseline for each cell, including ratio values in individual cells determined by defining regions of interest (ROIs) with the Workbench 6.0 analysis software. The interval between live measurements was approximately 2 s.
Ca2+ flow cytometry analysis
Undifferentiated and differentiated 50B11 cells at days 1, 2 and 7 DIV were centrifuged at 500 x g for 4 min, then resuspended in 1 mL of Hank’s Balanced Salt Solution (HBSS, Sigma) supplemented with 1 mM Ca2+ and 20 mM HEPES. Cells were then incubated at 37°C for 35 min in the presence of the Ca2+ indicators, Fluo-3 and Fura Red, washed once with HBSS, centrifuged at 450 x g for 4 min, then resuspended in HBSS at a final volume of 150 µL per FACS tube. Intracellular Ca2+ (Ca2+(i)) flux determined using BD FACSCanto II (BD Biosciences). Intact, single cells were gated based on their forward (FSC)/side scatter (SSC) characteristics and area scaling. Intact, single cells were gated based on their forward (FSC)/side scatter (SSC) characteristics and area scaling. Following a 10 s baseline recording, thapsigargin (1 µM), capsaicin (5–500nM) and ATP (1–50µM) was added, and recording continued for a further 150 s. The Ca2+-ATPase blocker, thapsigargin was used as a positive control to test the ability of cells to respond and for proper Ca2+ indicator loading. To demonstrate TrpV1 activation specificity we used 10 µM the competitive TRPV1 antagonist capsazepine (N-[2–(4-Chlorophenyl)ethyl]-1,3,4,5-tetrahydro-7,8-dihydroxy-2H-2-benzazepine-2-carbothioamide; Merck, Cat# C191), and 1 µM AMG517 (Cayman Chemicals; Cat# 26191).
Data was expressed as the relative change in the ratio of Fluo-3/Fura Red over time using FlowJo V10 (LLC, USA) for analyses. Changes in Fluo-3/Fura Red were compared to the baseline time period for each recording and compared to responses in unstimulated conditions (control). Baseline-subtracted net area under the curve (AUC) was quantified using GraphPad PRISM 5.04 software. Gating strategy from Giles.
Patch-clamp recordings
Rheobase was determined by application of a series of depolarizing pulses from holding potential –70 mV (10 or 25 pA increments (480 ms) followed by hold at –70 mV (100 ms). Borosilicate glass pipettes were fire-polished to 5–10 MΩ. Current-clamp intracellular solution contained (in mM): 135 KCl; 2 MgCl2; 2 MgATP; 5 EGTA-Na; 10 HEPES-Na; adjusted to pH 7.3, 275 mOsm. Extracellular solution contained (in mM): 140 NaCl; 4 KCl; 2 MgCl2; 2 CaCl2; 10 HEPES; 5 glucose, adjusted to pH 7.4, approximately 285 mOsm.
Voltage-clamp recordings: Current-voltage (INa-V) relationship was determined by application of a pre-pulse to –100 mV (100 ms), followed by a series of step pulses from –70 mV to +60 mV (5 mV increments (100 ms)), before returning to hold at –70 mV (repetition interval of 3 sec, P/8 leak subtraction). Pipettes were fire-polished to 1 – 2 MΩ and ≥75% of series resistance was compensated. Voltage-clamp intracellular solution contained (in mM): 60 CsF; 45 CsCl; 2 MgCl2; 5 EGTA-Na; 10 HEPES-Cs; 30 TEA-Cl; 2 MgATP; adjusted to pH 7.2 with CsOH, 280 mOsm. Extracellular solution contained (in mM): 70 NaCl; 50 NMDG; 40 TEA-Cl; 4 CsCl; 2 MgCl2; 2 CaCl2; 10 HEPES; 5 Glucose; adjusted to pH 7.4, approximately 300 mOsm.
Statistical analyses
For in vitro experiments, n represents the number of individual experiments, each with 4 replicates. p < 0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism (GraphPad Software) with individual post-hoc tests outlined in figure legends.
Results
Effects of forskolin and growth factor concentrations on 50B11 morphological differentiation
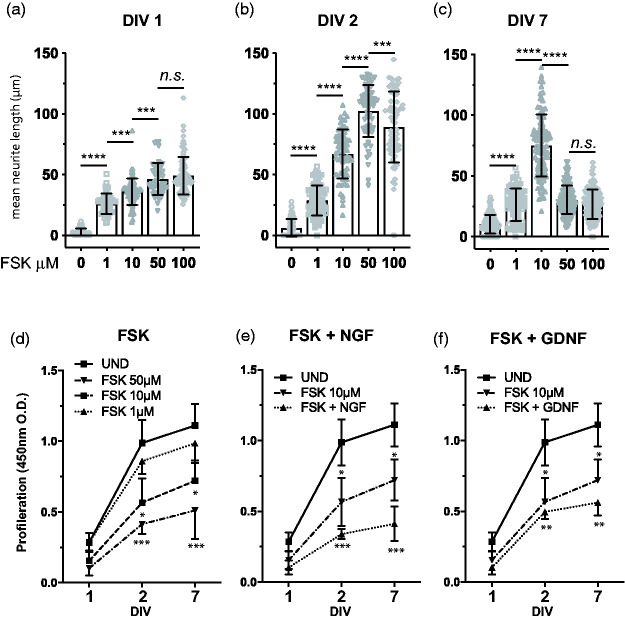
Previous studies characterising the 50B11 cells have reported that both NGF and GDNF had the capacity to synergistically increase the differentiation of 50B11 cell treated with 50 µm forskolin.1,2 However, these reports did not provide detailed comparison of the contribution of FSK or the growth factors in a concentration dependent manner. A fundamental limitation outlined in the initial 50B11 characterisation was the ability to maintain neurites for longer than 3 days.1 To determine whether 50B11 cells could maintain a differentiated phenotype with extended neurites for longer than 3 days we titrated the amount of FSK ranging from 0–100 µM and analysed the neurite outgrowth over 1, 2 and 7 days. Consistent with previous reports, concentrations above 10 µM FSK initiate neuritogenesis within 4 hours (data not shown). Our results show that at 1 day in vitro (1DIV) all concentrations of FSK produce similar neuronal outgrowth (Figure 1(a)). However, this changes significantly at 2DIV where concentrations ranging from 10–100 µM of FSK induce longer neurite processes compared to undifferentiated cells and cells differentiated with 10 µM FSK (Figure 1(b)). As with previous observations, higher concentrations of FSK (50–100µM) caused neurite retraction at 2-3DIV (Figure 1(c)). In contrast, cells cultured in presence of 1–10 µM FSK maintained neurites up to 7DIV (Figure 1(c)), with subsequent de-differentiation in most of the cells by 10DIV (data not shown). One critical observation during the FSK differentiation timeline was the presence of undifferentiated cells across all treatments. To determine whether this was a reflection of cells that failed to differentiate and continued to proliferate over 7DIV, we performed cell proliferation assays in cells treated with FSK only (Figure 1(d)) as well as in combination with NGF (Figure 1(e)) or GDNF (Figure 1(F)). Our data show that increasing concentrations of FSK reduce cell proliferation in a dose-dependent manner, however at 7DIV cells cultured with 1–50 µM FSK are still proliferating, and hence do not completely reach a state of terminal differentiation (Figure 1D). Addition of 50 ng/ml NGF (Figure 1(e)) or GDNF (Figure 1(f)) to cells in presence of 10 µM FSK further reduced proliferation of 50B11 cells in a similar manner to 50–75 µM concentrations of FSK alone (Figure 1(d). Our data indicate that the 50B11 cells can maintain a differentiated morphology for a significantly longer time by decreasing the amount of FSK to 10 µM and addition of nociceptor relevant growth factors.
Figure 1.
Effects of forskolin and growth factor concentrations on 50B11 morphological differentiation. Analysis of mean neurite length (a–c) of 50B11 cells treated with increasing concentrations of forskolin (FSK) ranging from 0–100 µM as indicated on the graph. Measurements represent same cultures measured across DIV 1, 2 and 7 (n = 4 experiments, neurite length analysis expressed as mean ± S.E.M; *, p < 0.01; **, p < 0.05, ***, p < 0.001; one-way ANOVA with Tukey’s multiple comparisons test). 50B11 cell proliferation measured by acid phosphatase to determine the effects of FSK alone (d) and in combination with 50 ng/ml NGF (e) or 10 ng/ml GDNF (n = 4 experiments, neurite length analysis expressed as mean ± S.E.; *, p < 0.01; **, p < 0.05, ***, p < 0.001; one-way ANOVA).
Reduced FSK levels abolish synergistic increases in NGF and GDNF mediated neurite outgrowth
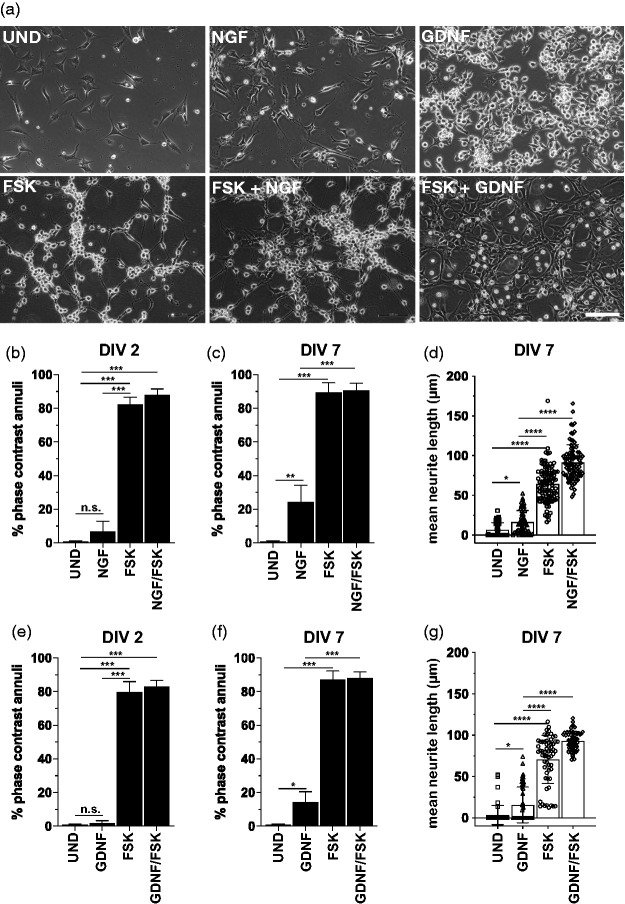
We next wanted to determine whether the reduction of FSK also affected neurite outgrowth. The loss or reduction in the proportion of differentiated cells may affect high through-put assays, and can lead to distorting effects on functional analysis over the period of 2-7DIV. To determine the overall percentage of differentiated cells, we performed phase contrast annuli counts of cell soma and mean neurite length analyses of 50B11 cells treated with FSK only, NGF and GDNF only and FSK in combination with NGF and GDNF. A representative micrograph for each treatment condition is shown in Figure 2(a). Our data shows that FSK alone as well as in combination with NGF or GDNF produce a significant increase in the proportion of differentiated cells (Figure 2(b), (d), (e) and (g) when compared to undifferentiated cells or cells treated with NGF or GDNF only at 2DIV (Figure 1(b)). The addition of NGF or GDNF to FSK treatments does not result in the previously described synergistic effect in relation to cell differentiation when measured cell soma annuli or neurite outgrowth at 2DIV and 7DIV. There are also no differences in total percentage of differentiated cells when comparing FSK only or NGF/FSK and GDNF/FSK treatments over 2 and 7DIV. Interestingly, although neither NGF-only, nor GDNF-only treatments resulted in significant cell differentiation at DIV2 (Figure 2(b) and (e)), both growth factors induced differentiate of about 20–25% of the total cell population at DIV7 (Figure 2(c), (d), (f) and (g)). This data shows that reduction of FSK to 10 µM does not affect the overall percentages of differentiated cells over 7 days, however it does abolish NGF and GDNF-mediated increases in neurite length. These results confirm that cells differentiated under reduced FSK and corresponding growth factors are capable of retaining a similar level of neurite length reported previously.1,2 However, the trade-off in acquiring longer term cultures is the loss of additional growth factor-mediated neurite outgrowth.
Figure 2.
NGF or GDNF do not contribute significantly to the morphological differentiation of 50B11 cells. (a) Hoffman modulation contrast (HMC) micrographs showing 50B11 cells cultured in the absence of any mitogenic agents or growth factors (no treatment; NT) and presence of 10 µM forskolin (FSK), 50 ng/ml NGF, 50 ng/ml, 10 µM FSK and 50 ng NGF or 10 µM FSK and 10 ng GDNF (Scale bar = 100 µm). (b to g) Analysis of neurite length and phase contrast annuli in 50B11 cells showing the differences in morphological characteristics of undifferentiated cells compared to cell differentiated with cells treated with 10 µM FSK only, 50 ng NGF or 10 ng GDNF only, and cell differentiated with FSK and NGF or GDNF. Data showing that both NGF and GDNF have a significant effect at DIV 7 but not DIV 2 (b to c and e to f), however they fail to provide any additional increase in the mean neurite length or the percentage of cells differentiated when compared to FSK treatments only at DIV 2 and 7 (n = 6 experiments, neurite length and phase annuli analysis expressed as mean ± S.E.M; *p < 0.01; **p < 0.05, ***p < 0.001; one way ANOVA with Tukey’s multiple comparisons test).
Differentiation of 50B11 cells with NGF or GDNF results in altered functional responses to capsaicin and ATP
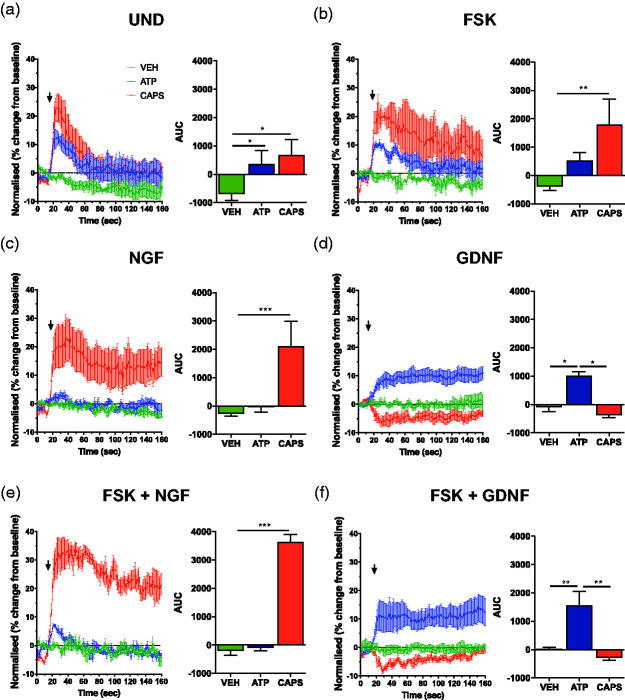
Previous functional characterisation of 50B11 cells,1,2 has been limited to cells cultured over 2–3 days and has not always included analysis of NGF- and GDNF-dependent responses.1,3 To determine whether 50B11 cells responded differently to nociceptive stimuli when differentiated with NGF or GDNF we analysed real-time calcium activity analysis by flow cytometry in responses to 500 nM capsaicin (CAPS) and 50 µM adenosine triphosphate (ATP). Analysis of undifferentiated 50B11 cells (Figure 3(a)) shows that both CAPS and ATP lead to calcium responses (Figure 3(a)). Interestingly, FSK-only differentiated cells show increased response to CAPS, whereas the response to ATP remains similar to undifferentiated cells (Figure 3(b)). Cells differentiated with NGF-only show a significantly increased response to capsaicin when compared to undifferentiated cells (Figure 3(a)) but lost the response to ATP (Figure 3(c)). Similarly, cell differentiated with GDNF-only (Figure 3(d)) maintained a response to ATP but did not elicit a response to CAPS when compared to undifferentiated and FSK-only differentiated cells (Figure 3(a) and (b)). Combined NGF/FSK (Figure 3(e)) and GDNF/FSK (Figure 3(f)) cells retained differential responses to capsaicin and ATP, with significantly increased sensitivity for CAPS in the NGF/FSK differentiated cells when compared to FSK and NGF-only treatment (Figure 3(b) and (c)). Similarly, the reposes to ATP were significantly increased in GDNF/FSK cells when compared to FSK and GDNF-only differentiated cells (Figure 3(b) and (d)). These data shows that although NGF and GDNF alone do not appear to contribute significantly to the neurite outgrowth in 50B11 cells, they have significant effect on their functional phenotype. In this case, NGF appears to dampen the response to ATP and significantly increase sensitivity to CAPS. In contrast, GDNF appears to have the opposite effect and increases responsiveness to ATP but reduces sensitivity to capsaicin. Therefore, NGF and GDNF confer functionally discriminating phenotypes in sensitivity to capsaicin and ATP respectively.
Figure 3.
Differentiation of 50B11 cells with NGF or GDNF results in altered functional responses to capsaicin and ATP. Calcium flow cytometry analysis of 50B11 cells using Fluo-3 signals acquired in FL1 and Fura Red in FL3 showing the differential responses to 500 nM capsaicin and 50 µM adenosine triphosphate (ATP) in undifferentiated 50B11 cells (a), differentiated with FSK (b), 50 ng/ml NGF only (c), 50 ng/ml NGF + 10 µM FSK (d), 10 ng/ml GDNF (e) and 10 ng/ml GDNF + 10 µM FSK (f) (n = 4 experiments, AUC analysis expressed as mean ± S.E.M; *p < 0.01; ***p < 0.001; ANOVA). Treatments represented by colours are Vehicle (green), ATP (blue) and Capsaicin (red).
Differential expression of nociceptive neuron markers in response to NGF and GDNF
To date, protein expression analysis of nociceptive markers in 50B11 cells has received limited attention.1,2,39–41 Differentiation in the presence of NGF or GDNF leads to the establishment of a functionally different phenotypes, which are partially consistent with DRG neuron subtype development.42 To determine whether NGF and GDNF-specific differentiation-dependent responses to CAPS and ATP were accompanied by changes in nociceptive markers, we analysed the relative protein expression of relevant C-fiber nociceptor markers by Western blot at 7DIV. Markers include the NGF cognate receptor TrkA, the common neurotrophin receptors p75NTR, the GDNF receptors GFR alpha 1 and c-Ret, the transient receptor potential cation channel subfamily V member 1 (TrpV1) receptor, as well as the ATP receptor P2X3, in relation to ßIII-tubulin. Representative Western blots show that all of these markers can be detected under undifferentiated, FSK-only, as well as FSK supplemented with NGF or GDNF treatment conditions (Figure 4(a) and (b)). TrkA expression remains relatively constant, as is p75NTR expression in NGF/FSK or in NGF-only or GDNF-only differentiated cells (Figure 4(a) and (b)). Both c-Ret and GFR alpha 1 expression is be lower in NGF treated groups, and expressed at the highest level in GDNF and GDNF/FSK differentiated cells (Figure 4(b)). TrpV1 expression also appears to be differentially regulated by NGF and GDNF, with highest expression seen in NGF and NGF/FSK differentiated cells (Figure 4(a)), and lowest in the GDNF and GDNF/FSK group (Figure 4(b)). The ATP binding purinergic receptor P2X3 shows differential expression between the NGF and GDNF treatments, with its expression increasing in GDNF and GDNF/FSK differentiated cells (Figure 4(b)) and decreasing in the NGF and NGF/FSK group (Figure 4(a)). These results show that both NGF and GDNF induce changes in classical nociceptive markers, and these changes are partly in line with the observed functional responses to capsaicin and ATP (Figure 3(a) to (e)). Specifically, the NGF differentiated and capsaicin responsive cells show an increase in TrpV1, and a decrease in ATP binding P2X3 receptor (Figure 3(c) and (e)), whereas GDNF differentiated and ATP responsive cells (Figure 3(d) and (f)) show a decrease in TrpV1 expression and an increase in P2X3 receptor expression levels.
Figure 4.
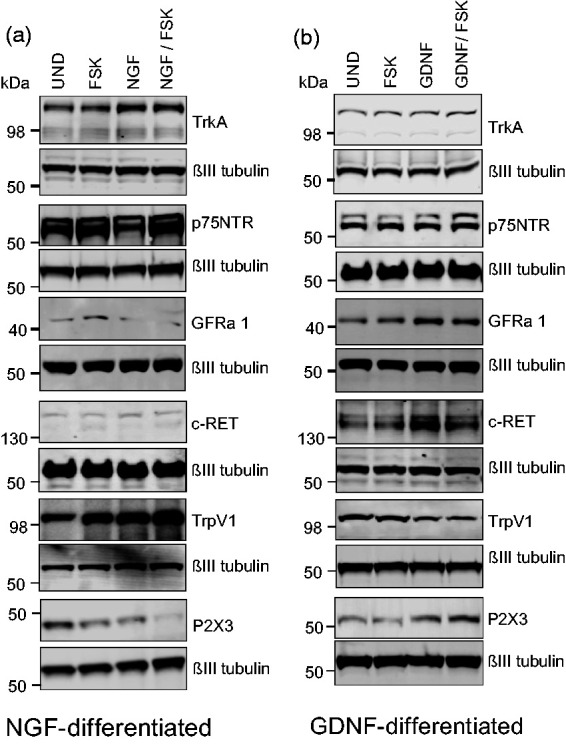
Western blot analysis of nociceptive neuron cell surface markers on 50B11 cells. Western blot images showing the relative protein expression of nociceptive neuron cell surface markers including the NGF cognate receptor TrkA, the common neurotrophin receptors p75NTR, the GDNF receptors GFR alpha 1 and c-Ret, the transient receptor potential cation channel subfamily V member 1 (TrpV1) receptor, as well as the ATP receptor P2X3 in undifferentiated, FSK, NGF, GDNF, NGF/FSK and GDNF/FSK differentiated cells (a and b), in relation to ßIII-tubulin. (n = 3).
Sensitization-like responses of NGF/FSK and GDNF/FSK differentiated 50B11 cells following prostaglandin E2 (PGE2) exposure
We next investigated whether the 50B11 cells were capable of sensitization like responses prototypical of nociceptive C-fibre neurons. Sensitization of nociceptive neurons to stimulation plays a critical role in the enhanced perception of sensation and pain,43–45 and mechanisms of peripheral sensitization may involve increased transduction that is secondary to repeated stimulation or an increase in the excitability of the afferent nerves by molecules that decrease the excitation threshold.14,46,47 Sensitization can also develop in response to inflammation although basic mechanisms of this process are poorly understood. Prostaglandin E2 (PGE2) commonly found at elevated levels in injured tissues have been shown to directly activate as well as sensitize DRG neurons,48 including via the activation TrpV1 receptor by capsaicin.33,48–50 Analysis of calcium increase following acute exposure to capsaicin and ATP in naïve and PGE2 treated (2 min at 1 µM) cells showed that NGF/FSK-differentiated cells show a significantly increased responses to 50 nM capsaicin but not ATP (Figure 5(c) and (d)) when compared to NGF/FSK differentiated controls (Figure 5(a) and (b)). In GDNF/FSK-differentiated cells PGE2 failed to have the same effect on both capsaicin and ATP showing a similar calcium response when compared to naïve cells treated with 50 nM capsaicin (Figure 5(e) to (h)). Additional experiments investigating whether proinflammatory cytokine interleukin-1 (Il-1β), an important mediator in pain sensitization,51,52 was capable of a similar sensitizing effect on both NGF/FSK and GDNF/FSK differentiated cells did not observe any sensitization-like responses (data not shown). These experiments show that 50B11 cells generate acute sensitization-like functional responses to PGE2 stimulation but not Il-1β.
Figure 5.
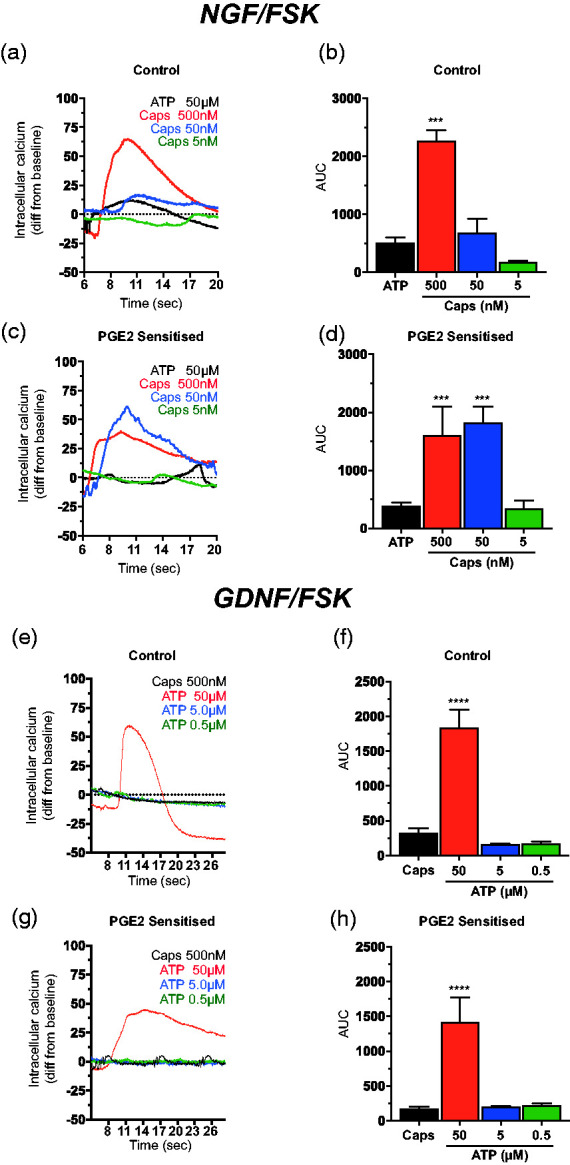
Sensitization-like responses of NGF/FSK but not GDNF/FSK differentiated 50B11 cells following prostaglandin E2 (PGE2) exposure. (a) Representative micrographs showing calcium increase following acute exposure to capsaicin and ATP in naïve (top row) and PGE2 (bottom row) treated (2 min, 1 µM) as indicated (scale bar = 200 µm). (b and c) Representative analysis of control and (d and e) prostaglandin E2 (PGE2) treated 50B11 cells showing an increased Ca2+ response when compared to naïve cells treated with to 5 µM capsaicin. (e and f) Representative calcium influx Analysis of control and (g and h) prostaglandin E2 (PGE2) treated 50B11 cells showing a similar Ca2+ response when compared to naïve cells treated with a range of capsaicin concentrations (500 nM – 5 nM; n = 4 experiments, AUC expressed as mean ± S.E.M; ***p < 0.001; one way ANOVA with Tukey’s comparison).
50b11 cells did not generate action potentials or sodium currents
The initial report by Chen et al. described the ability of the 50B11 cells to generate action potentials following differentiation with 75 µM FSK and 10 ng/ml NGF.1 To determine whether the reduction of FSK concentrations that extended the time 50B11 cells maintain a differentiated phenotype had any impact of the cells generating action potentials and sodium currents, we performed a comprehensive patch clamping screen at 3DIV and 7DIV days following differentiation with 1, 10, 50 or 100 µM FSK alone, or with combination ranging across 10, 50, and 100 ng/ml NGF or GDNF. Effect of positive current injection on membrane potential and voltage-dependence of activation was attempted, however no action potentials or sodium currents were observed in any of the cells, traces of all cells recorded are given in Supplementary Figure 1. These results indicate that the 50B11 cells may have drifted from their original phenotype since being introduced to the field. All cells used for patch clamping electrophysiology experiments were from passage 11 or lower.
PCA analysis and heatmap distribution tables of differentially expressed nociceptive phenotype genes in 50B11 cells
The 50B11 cell line is to our knowledge the only sensory neuron model capable of acquiring differential functional phenotype in a growth factor-depended manner. We performed RNA-Seq screening of undifferentiated and NGF and GDNF differentiated cells to establish a comprehensive gene expression profile of each 50B11 cell subtype. The data allow an understanding how growth factors and FSK modulate the RNA expression profile and connect RNA expression to functional phenotypes. Recent studies by multiple groups have contributed to the ongoing classification of nociceptive subtypes,5,34,53–55 with at least 11 different nociceptor subtypes characterised by large-scale single-cell RNA sequencing.34
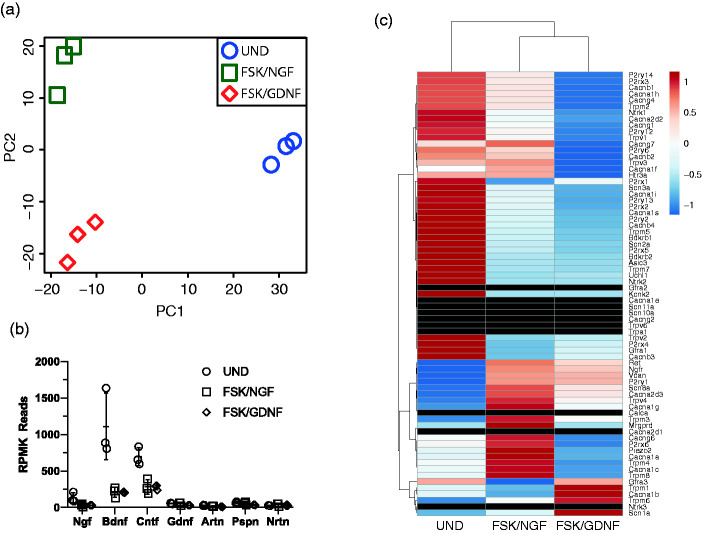
We used a read of about 60 million which allows for a comprehensive analysis of expression patterns. Thus far our data show that the 50B11 cells reflect NGF- and GDNF-mediated differential expression profiles, which partially align with a peptidergic and non-peptidergic DRG expression-type respectively.19,42,55 Principle component analysis (PCA) plot showing RNA expression profile differences between 50B11 cells grown in the presence of NGF/FSK, GDNF/FSK and undifferentiated (UND) cells (Figure 6(a)) demonstrates the distinct expression profiles induced by different growth factors in comparison to undifferentiated cells.
Figure 6.
Analysis of genetic variance including the expression of nociceptor relevant growth factors and common cell surface markers. Principle component analysis (PCA) plot showing RNA expression profile differences between 50B11 cells grown in the presence of NGF/FSK, GDNF/FSK and undifferentiated (UND) cells (a). Graph showing the relative expression of growth factor genes know to be involved in the development and maintenance of nociceptor phenotypes (b; Ngf- nerve growth factor; Bdnf – brain derived neurotrophic factor; Cntf – ciliary neurotrophic factor; Gdnf – glial derived neurotrophic factor; Artn – artemin; Pspn – persepin; Nrtn - neurturin). (c) Heat map of expression of the most significantly enriched genes for each population in all treatment conditions (UND, NGF and GDNF), with neurons grouped according to assigned population (Black bars in the heatmap represent markers that were not detected in RNA-Seq and confirmed to be absent in qRT-PCR).
RNA-Seq screening of undifferentiated and NGF & GDNF differentiated 50B11 cells
On average 70.96% (±0.073, n = 3) from an average of 62 million total reads (62,218,538.22 ± 13,646,681.37, n = 3) were uniquely mapped reads. The number of uniquely mapped reads were similar between samples. The fast majority of genes were protein-coding (93.03% ± 0.01) followed by mitochondrial rRNA (1.66% ± 1.21), ribosomal RNA (1.58% ± 0.51) and long non-coding RNA (0.79% ± 0.23). Principal component analysis (PCA) showed a clear separation of samples generated from undifferentiated 50B11 cells and 50B11 cells differentiated with NGF/FSK and GDNF/FSK for 7 days (Figure 6(c) and (d)).
Analysis of mRNA expression for genes that are relevant for nociceptor function
Major growth factors such as Ngf, Bdnf, Cntf and Gdnf are expressed in undifferentiated and differentiated 50B11 cells after 7d of differentiation. Interestingly in relation to receptors for growth factors, only trkA (Ntrk1) but not trkB (Ntrk2) or C (Ntrk1) were detected. The p75 neurotrophin receptor (Ngfr) was significantly upregulated in response to differentiation (4.06 fold, p adj. 2.07 x 10–12). Persepin (Pspn) and its receptor Gfra4 were expressed under all conditions. Artemin (Artn) and neurturin (Nrtn) were expressed under all conditions. mRNAs for GFRs other than Gfra4 could not be detected.
Peptidergic nociceptors contain characteristic peptides such as substance P or calcitonin-gene-related peptide. None of the genes related to major peptides found in nociceptors such as the preprotachykinin (Tac1) relevant for substance P, calcitonin-gene-related peptide alpha (Calca), somatostatin (Sst) or vasoactive intestinal polypeptide (Vip) were expressed in 50B11 cells.
Transient receptor channels of the vanilloid type (TrpV) are expressed from nociceptors with TrpV1 a characteristic channel expressed from peptidergic nociceptors.54,56,57 TrpV channel expression was restricted to Trpv1-4 (rank order Trpv4 > Trpv2 > Trpv3 = Trpv1) whereas Trpv5 and 6 were not detected independent of differentiation. The TrpA1 receptor is also expressed and characteristic for nociceptors58 but was absent in 50B11 cells.
Mas-related GPR family members (Mrgprs) are almost exclusively expressed in peripheral sensory neurons and are related to itch.59 Only Mrgprd, Mrgpre and Mrgprf were present in 50B11 cells. The mRNA for Mrgprf was induced in response to differentiation. The mRNA is absent in undifferentiated cells but present in NGF and GDNF differentiated cells.
Receptors for ATP are expressed in nociceptor and the P2X3 receptor subtype is characteristic for non-peptidergic neurons.60 Ionotropic receptors for ATP are expressed in 50B11 cells. The P2X3 receptor (P2rx3) was expressed under all conditions, the P2X1 was very low, the P2X7 receptor was not expressed similar to previous reports.59 The rank order of expression was P2X4 > P2X5 > P2X2 > P2X6 > P2X3.
Non-peptidergic receptors bind the IB4 and the nociceptor binding partner for this lectin has been shown to be versican V2 a splice variant of versican (Vcan).61 Vcan expression is significantly higher in GDNF/FSK differentiated cells compared to undifferentiated 50B11 cells.
Voltage gated sodium and calcium channels are key components in action potential generation in nociceptors and differentiated 50B11 cells responded to capsaicin and ATP with an increase in intracellular calcium.62 Classic nociceptor sodium channel alpha subunits of the tetrodotoxin-resistant Nav1.8 (Scn10a) and of Nav1.9 (Scn11a) could not be detected in 50B11 cells whereas voltage gated calcium channels such as Cav2.1 (Cacna1a), Cav2.2 (Cacna1b), Cav1.2 (Cacna1c), including auxiliary β and α2δ2, 4 subunits were expressed.
Analysis of growth factor expression profile in undifferentiated and differentiated cells shows that BDNF and CNTF expression is a characteristic of the 50B11 cell line, however the expression of both growth factors decreases in the presence of FSK/NGF and FSK/GDNF (Figure 6(b)).
To provide a better overview of the nociceptive markers that were differentially expressed or absent Figure 6(c) provides a heat map summary of primary nociceptor genes including neurotrophin receptors, calcium channels, sodium channels, potassium channels and receptors activated as a result heat, cold, mechanical and cellular damage stimulus. Heat map of expression of the most significantly enriched genes for each population in all treatment conditions (UND, NGF and GDNF), with neurons grouped according to assigned population. Comparison to the comprehensive non-biased analysis of mouse DRG neurons by single-cell-RNA-Seq shows that the 50B11 cells share many of the nociceptor specific genes, however classification into a peptidergic and non-petidergic peptide expression profiles was not possible due to their embryonic origin, and the differences in cell numbers analysed (Supplementary Tables 1 and 2). However, the p-values of the GO terms related to the peptidergic and non-petidergic like cells demonstrate high similarities to specific GO terms reflecting sensory properties or functions of nociceptors in Supplementary Tables 1 and 2.
qRT-PCR validation of RNA-Seq data for peptidergic and non-peptidergic nociceptive neuron markers
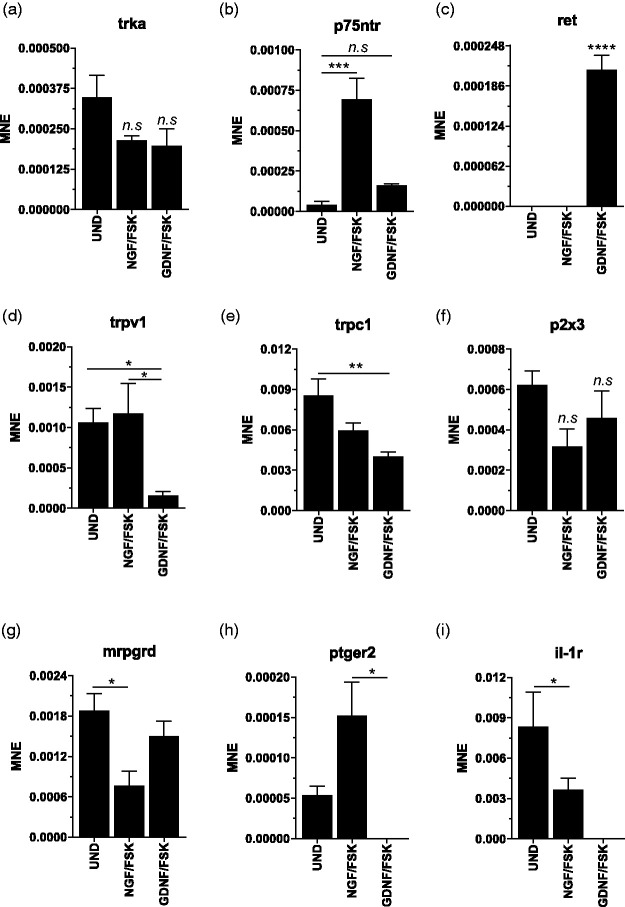
Expression of peptidergic and non-peptidergic phenotype classification markers was validated following RNA-Seq analysis. Relative mRNA-expression levels as mean normalized expression (MNE) for undifferentiated (UND), NGF/FSK differentiated and GDNF/FSK differentiated cells showing in agreement with the Western blot data (Figure 4(a) and (b)) that TrkA (Figure 7(a)) expression levels were not different between treatment groups. The neurotrophin receptor p75 (p75ntr) mRNA expression was significantly upregulated in response to NGF only (Figure 7(b)). Interestingly mRNA for the GDNF receptor GFR alpha-1 and 3 could not be detected (data not shown), whereas c-RET RNA was detectable by qRT-PCR only in GDNF/FSK differentiated cells (Figure 7(c)). Relative mRNA expression levels for the TrpV1 receptor were significantly lower in GDNF/FSK differentiated cells (Figure 7(d)) in agreement with the Western blot data (Figure 4(b)), as was the transient receptor potential channel 1 (trpc1) receptor (Figure 7(e)). Interestingly, no difference in RNA expression was detected in ligand-gated ion P2X3 purinoceptor (Figure 7(f)). Expression levels for the G protein-coupled receptor Mas-related G-protein coupled receptor member D (mrpgrd) which were not detected in the RNA-Seq samples were readily detected in qRT-PCR and were significantly lower in NGF/FSK differentiated cells compared to undifferentiated and GDNF/FSK differentiated cells (Figure 7(g)). Expression levels for PGE2 receptor 2 (ptger2) was significantly upregulated in NGF/FSK cells and undetectable in GDNF/FSK cells (Figure 7(h)). Interleukin-1 receptor (il-1r) RNA expression level was significantly decreased in response to NGF/FSK and also undetectable in GDNF/FSK differentiated cells (Figure 7(i)). (n = 5, expressed as mean ± SEM; one-way ANOVA, *p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001). Similar to the RNA-Seq results precursor mRNAs for CGRP, Calca (and Calcb), could not be detected in 50B11 cells (data not shown). mRNAs for the TRPA1 receptor and for sodium channel subunits Scn10a and Scn11 could not be detected confirming RNA-Seq data. The comparison between PCR and RNA-Seq is summarized in (Table 2).
Figure 7.
qRT-PCR validation of RNA-Seq data for peptidergic and non-peptidergic nociceptive neuron markers. Expression of peptidergic and non-peptidergic phenotype classification markers. Relative mRNA-expression levels as mean normalized expression (MNE) for in undifferentiated (UND) NGF/FSK differentiated and GDNF/FSK differentiated cells showing (a) tropomyosin receptor kinase A (trkA); (b) neurotrophin receptor p75 (p75ntr); (c) c-RET proto-oncogene encodes a receptor tyrosine kinase (ret); (d) the transient receptor potential cation channel subfamily V member 1 (trpV1); (e) transient receptor potential channel 1 (trpc1); (f) ligand-gated ion channel P2X purinoceptor 3 (p2x3); (g) G protein-coupled receptor Mas-related G-protein coupled receptor member D (mrpgrd); (h) Prostaglandin E2 receptor 2 (ptger2); and (i) interleukin-1 receptor (il-1r). (n = 5, expressed as mean ± SEM; one-way ANOVA, Bonferroni’s post hoc test, *p < 0.05; **p < 0.01; ***p < 0.005 and ****p < 0.0001).
Table 2.
Relative mRNA expression data for nociceptor related genes in 50B11 cells and RNAseq in brackets and notable absence of nociceptive markers in grey.
| Gene | Expression (7 d) |
||
|---|---|---|---|
| Undifferentiated | NGF/FSK | GDNF/FSK | |
| Ngfr (p75 neurotrophin receptor) | Low expression (low expression) | Present (present) | Present (present) |
| Ntrk1 (tyrosine kinase receptor A) | Present (low expression) | Present (low expression) | Present (low expression) |
| Ret (Ret proto-oncogene) | Absent (absent) | Absent (low expression) | Present (low expression) |
| Gfra3 (Gdnf family receptor alpha) | Absent | Absent | Absent |
| P2X3 (purinergic receptor P2X3) | Present (low expression) | Present (low expression) | Present (low expression) |
| Trpv1 (transient receptor potential cation channel V member 1) | Present (low expression) | Present (absent) | Low expression (low expression) |
| Trpa1 (transient receptor potential cation channel A member 1) | Absent (absent) | Absent (absent) | Absent (absent) |
| Mrgprd (Mas related GPR family member D) | Present (low expression) | Present (low expression) | Present (low expression) |
| Ptger2 (prostaglandin E receptor 2) | Present (low expression) | Present (low expression) | Absent (low expression) |
| Il1r1 (interleukin 1 receptor type 1 | Present (present) | Present (present) | Absent (present) |
| Calca (calcitonin related peptide alpha) | Absent (absent) | Absent (absent) | Absent (absent) |
| Calcb (calcitonin related peptide alpha) | Absent (absent) | Absent (absent) | Absent (absent) |
| Scn11a (sodium voltage-gated channel alpha subunit 11) | Absent (absent) | Absent (absent) | Absent (absent) |
| Scn10a (sodium voltage-gated channel alpha subunit 10) | Absent (absent) | Absent (absent) | Absent (absent) |
The table summarizes the detection of mRNAs of genes related to nociceptors using qRT-PCR and RNAsq (in brackets) in 50B11 cells. Comparison of expression in undifferentiated cells and cells differentiated for 7 days with NGF and forskolin (NGF/FSK) or GDNF and forskolin (GDNF/FSK)..
Discussion
Anatomical and physiologic changes that occur within nociceptive DRG neurons affirm that these neurons participate in the development and maintenance of different types of pain.11,14,47 The diversity of DRG neuron subtypes,34 displaying unique molecular, morphological, and functional characteristics, is a fundamental feature enabling the discrimination between various types of sensations. Nociceptive neurons are also emerging as relevant therapeutic targets, based on their unique accessibility and increasing clinical evidence through systemic and local neuromodulation approaches.63,64 To address different types of pain, more focussed therapeutic options are needed with selective nociceptor subpopulations as targets. It is well known that nociceptor subpopulations impact differently on the development of pain. For example, TRPV1 which is found primarily on the subpopulation of peptidergic nociceptive neurons65,66 is activated by noxious thermal and chemical (eg, capsaicin) stimuli and is integrally involved in the development of inflammatory hyperalgesia.66 On the other hand, the purinergic receptor P2X3 is found predominantly in GDNF-sensitive non-peptidergic nociceptors, is also important in mechanical and chemical stimuli and mechanical hypersensitivity.67,68 The aim of this study was to determine whether immortalised 50B11 embryonic DRG neurons retained the capacity to differentiate into growth factor-dependent nociceptive subtypes observed in vivo.
Characterisation of the 50B11 cells has been limited to reports describing the generation and initial characterization, including their suitability for the study of excitability, neurodegeneration, neuritogenesis, and transcriptomic comparison to nociceptive cells.1–3,9,69 Most studies relied on differentiating their cultures in the presence of 75 µM of forskolin supplemented with NGF or GDNF, however no titration of forskolin or growth factor was described.1,2 More importantly, the only transcriptomic analysis to date has only been performed on undifferentiated cells.9 Immortalized neuronal cultures of various origins differentiate and maintain neurites under mitogenic agents and/or growth factor stimulation for over a week.70–72 Wanting to determine whether 50B11 cell could maintain a differentiated morphology for a longer period of time than previous reports,1,2 we titrated the amount of FSK and established that reducing the concentration did not significantly reduce the ability of the 50B11 cells to differentiate when assessed by neurite length measurements. Contrary to previous reports1,2 the co-treatment with NGF or GDNF 17 hours post-FSK (1–100 µM; Figures 1 and 2) did not yield synergistic effects in neurite outgrowth. Although the cells maintained a differentiated phenotype up to 7 days in a FSK-dependent manner, the addition of NGF and GDNF (Figure 1(a) to (e), the extension in the time that the cells maintain neurites leads to the loss of additional synergistic effects on neurite elongation in combined FSK and NGF or GDNF treatments, most likely due to the neurites reaching maximum length with additional time in culture.
Our observations of NGF inducing neurite outgrowth in 50B11 cells independent of FSK is in line with experiments showing that factors including NGF released from prostate cancer cells promote differentiation of 50B11 cells.4 Collectively, our results demonstrate that FSK is the primary agent driving rapid neurite differentiation in 50B11 cells, and that reducing its concentration extends the maintenance of a differentiated phenotype to 7 days. NGF and GDNF are both independently capable of driving neurite outgrowth in the absence of FSK over 7 days (Figure 2).
Previous studies have concluded that differentiated 50B11 cells uniformly express both NGF and GDNF-family receptors and could not be subdivided based on unique receptor expression,1,2 therefore we investigated whether functional differences to nociceptive agonists capsaicin and ATP could be observed. 50B11 cells have been shown to respond to capsaicin1 and ATP.3 Capsaicin is a potent activator of the TrpV1 receptor, and ATP activates P2X purinoreceptors, both acting as characteristic receptors in nociceptive neuron subtypes.73 As previous studies characterising 50B11 cells relied primarily on morphological and immunohistochemical analysis we used calcium-imaging to confirm functional presence of nociceptor signalling in differentiated 50B11 cells. Capsaicin responses were observed in acutely dissociated DRG cells from E14.5 DRG.54 We observed that compared to undifferentiated cells the NGF/FSK differentiated cells showed a significantly reduced Ca2+ response to ATP while eliciting an increased response to capsaicin (Figure 3(a) to (d)). In contrast, an inverse relationship was observed in GDNF/FSK differentiated cells (Figure 3).
These data in combination with Western blot and RNA-Seq results show that undifferentiated cells, either do not respond or insufficiently respond to nociceptive stimulation. In contrast, differentiation drives neurons towards a nociceptor-like phenotype. NGF/FSK differentiated cells express TrpV1 receptor mRNA and protein, down-regulate P2X3 receptors and respond to capsaicin (Figure 3(d) to (e)). Simultaneously the response to ATP is reduced (Figure 3(c) and (d)) indicating a functional capsaicin-sensitive/ATP-insensitive phenotype similar to peptidergic nociceptors.67,73,74 Using a different combination of FSK with the growth factor GDNF induces a different phenotype. 50B11 cells lose their ability to respond to capsaicin but increase their sensitivity for ATP (Figure 3(e) and (f)) similar to non-peptidergic nociceptors.66,75
NGF and GDNF have been shown to differentially regulate the expression of TrpV1, where upregulation of TrpV1 expression in DRG neurons is dependent on NGF.76,77 The role of GDNF in the regulation of TrpV1 in 50B11 cell is less clear. Our mRNA and protein studies showed that both c-Ret and GFRα1 are expressed at the highest level in GDNF and GDNF/FSK differentiated cells, In addition, GDNF-only or GDNF/FSK treatment causes a decrease in TrpV1 expression (Figure 4(a) and (b)). These results suggest that 50B11 cells respond to exogenous growth factors similar to developing nociceptive neurons.19 Depending on the growth factor combination with low-concentration FSK cells acquire functional characteristics specific for subtypes of nociceptors. However, nociceptors respond to more than algogenic stimuli. One hallmark of nociceptors is the ability to be sensitized. Given that peripheral sensitization is a key feature of nociceptors and is a key factor in the development of chronic pain, we next investigated whether the 50B11 cells were capable of sensitization-like behavior. Plasticity of DRG nociceptors induced by pro-inflammatory mediators contributes to sensitization. Prostaglandin E2 (PGE2) enriched in injured tissues is known not only directly to sensitize DRG neurons, but also to potentiate sensitizing effects of other pain mediators such as capsaicin via its TrpV1 receptor.50 In addition, cytokines such as interleukin-1β (Il-1β) also contribute to pain hypersensitivity by targeting excitatory and inhibitory synaptic transmission mechanisms in sensory neurons.78,79
Our qRT-PCR results showed presence of the PGE2 receptor (Ptger2) in NGF/FSK-differentiated 50B11 cells but interestingly a significant down-regulation of Il1r in response to differentiation with GDNF. This indicates a growth factor or phenotype-specific receptor expression for inflammatory mediators.
NGF/FSK differentiated cells with increased expression of TrkA and TrpV1 and absence of functional P2X receptors, showed significantly increased responses to 10-fold lower concentration of capsaicin (Figure 5(c) and (d)) but not ATP (data not shown) following acute stimulation with PGE2 (Figure 5(a) and (b)).
Functional data are supported by comprehensive analysis of RNA expression. In contrast to other DRG-derived cell lines, 50B11 cells do not contain chromosomes and genes of other species and are therefore suitable for RNA analysi.8
RNA-Seq in combination with qRT-PCR showed the expression of nociceptor related genes such as TRPV1 and P2X3 in 50B11 cells. Importantly, differentiation with NGF/FSK generated cells with different expression profiles that expressed genes characteristic for non-peptidergic nociceptors whereas differentiation with GDNF/FSK induced expression of mRNAs characteristic for non-peptidergic nociceptors.
The GDNF receptor c-Ret was expressed in GDNF/FSK differentiated cells whereas TRPV1 was highly expressed in NGF- but not GDNF- differentiated cells. In addition, versican was higher expressed in GDNF/FSK differentiated cells. The V2 splice variant of versican is the binding partner for IB4, a marker for non-peptidergic nociceptors.61
Taken together, analysis of mRNA expression suggests that differentiation with NGF or GDNF drives 50B11 cells towards an NGF-dependent or GDNF-dependent DRG phenotype.
However, our data show that 50B11 cells resemble but do not completely reflect gene expression patterns of adult rat nociceptors. The absence of mRNA expression for nociceptor-related neuropeptides such as SP, CGRP or NPY might be related to the embryonic origin of 50B11 cells since these cells were generated from DRG neurons at E14.5. Peptide expression in DRG neurons starts around day E15,42,80,81 therefore absence of peptide expression indeed reflect the embryonic origin of the cells.
In contrast to previous reports,1 we could not detect nociceptor-generated action potentials (Supplementary Figure 1). This finding was supported by absence of expression of voltage-gated sodium channel α-subunits characteristic for Nav1.8 and 1.9 channels in 50B11 cells. Ion channels such as Nav1.8 are expressed by only 10% of trkA positive DRG neurons at E15 and Nav1.9 is absent at this time point and expression starts at E17 (see review42,53,82,83) suggesting a low or absent expression of channels at E14.5. Our patch clamping results aligned with the RNA-Seq data showing the absence of sodium channels, and without having access to alternate stocks of 50B11 cells. One possible reason is that the cells might have drifted from their original phenotype, given that the original report did not provide detailed data on what percentage of cells elicited action potentials.1 However, the absence of sodium channel expression in the 50B11 cells makes them ideal candidates for investigating transfection of either individual or combinations of sodium channels providing a unique toolset to investigate their function in a nociceptive microenvironment. Advances in transfection technology developed for primary neurons,84 could be suitable for transfection of these cells following differentiation, Alternatively, further investigations with long term cultures in presence of growth factor cocktails may yield a more mature phenotype with endogenously induced expression of nociception related channels, as observed for other cell lines.85 To this end, we are applying our phenotypic differentiation strategy to other nociceptor-derived cell lines.
The RNA-Seq data show presence of calcium channels including voltage-gated calcium channels. Therefore Ca2+ responses can be used to demonstrate activation and consequently function of 50B11 cells. The Ca2+ response profile is greatly aligned with protein expression profiles that show an upregulation of TrkA and TrpV1, and downregulation of P2X3 in response to NGF/FSK treatment (Figure 4(a)). Conversely, TrkA and TrpV1 levels decreased and P2X3 expression levels were increased in GFNF/FSK differentiated cells (Figure 4(a) and (b)). Protein expression and Ca2+ responses changed simultaneously in response to individual growth factors further demonstrating a phenotype switch at transcriptional, translational and functional levels.
In conclusion, here we provide detailed characterisation of the DRG-derived 50B11 cells line, showing that these cells recapitulate numerous characteristics of C-fibre nociceptors aligned with their embryonic origin, and are capable of acquiring early phenotypes resembling both peptidergic and non-peptidergic sensory neurons in a growth factor-specific manner. More importantly, we show that this cell line can maintain a sensory phenotype in a FSK-, NGF- and GDNF-dependent manner for up to 7 days by decreasing the amount of FSK in cultures. Interestingly, unlike previous reports, our study indicates that FSK is the primary factor responsible for the neuronal differentiation of these cells, whereas NGF and GDNF are essential for the development of a functionally specific phenotype, sensitive to capsaicin or ATP respectively. Additionally, this functional separation can provide a useful platform for the use of 50B11 cells in investigating NGF/TrpV1/capsaicin and GDNF/P2X/ATP-specific signalling in a homogeneous population of neuronal cells. Interestingly, the 50B11 cells show sensitization to PGE2 only in NGF differentiated cells, suggesting that a more comprehensive test of the subtype specificity of inflammatory mediators may be warranted. More importantly, our RNA-Seq data supports our functional and proteomic analysis of the 50B11 cells, clearly showing that undifferentiated cells, NGF- and GDNF-differentiated cells can be separated by gene expression profile. Subsequent validation of nociceptor-specific markers also confirms growth factor-dependent changes in expression profiles related to both peptidergic and non-peptidergic nociceptors. The study of receptor biology may not have to rely on the complete expression of phenotype-specific nociceptive markers, and should always be validated in naïve DRG neurons. These findings provide detailed validation of undifferentiated and differentiated 50B11 cells and their suitability as a useful tool for investigating mechanisms regulating nociceptor biology.
Accession codes
Gene Expression Omnibus: raw data for individual plates (libraries), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147186. Additional external resources are available as supplementary data and include normalized data for all sequenced wells (expressed as reads per million, RPM), with full sample annotation; the expression profiles for all genes and neuronal populations in line with NGF and GDNF differentiation, allowing visualization of scatter plots of expression (RPM) for any gene; visualizing tool for expression level posteriors; and online search engine for scatter plots of expression for any gene.
Acknowledgements
Biosensis Pty. Ltd. kindly provided the TrkA and TrpV1 antibodies used for western blotting experiments. 50B11 cells were kindly provided by Ahmet Höke at the Department of Neurology, School of Medicine, Johns Hopkins University, Baltimore, MD, USA. We would like to thank Dr Renee Smith and Ms Letitia Pimlott from the Flinders University Genomics Facility for preparing and performing the RNA-Seq experiments, Prof Nicolas Spencer and Mr Lee Travis from the Visceral Neurophysiology Lab for the use of their calcium imaging microscope, Miss Yvette DeGraaf and Patricia Vilmas in the Flinders University Microscopy Facility for technical assistance and maintenance, and Ms Jennifer Washington and Dr Sheree Bailey from the Flinders Medical Centre Flow Cytometry Facility for technical guidance and support. We also thank other members of the Anatomy and Histology department past and present for helpful discussions and Flinders University College of Medicine and Public Health colleagues for critical reading of the manuscript and editorial assistance.
Authors’ Contributions: All authors executed and interpreted experiments and contributed to the drafting of the article. M.D., H.V.R., C.J. O.G.B. and M.Z.M. conceived and designed the experiments. C.J., E.A., M.A., F.K., Y.W. and M.D. contributed to executing all the experiments. M.Z.M., H.V.R., and M.S., provided the RNA-Seq analysis. M.D., H.V.R., M.Z.M., K.J.D., B.C., V.H.N. and B.M.S. provided critical review of the manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Flinders Foundation and the Flinders University Centre for Neuroscience grants.
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs
Matusica Dusan https://orcid.org/0000-0001-6472-7653
Martin M Alyce https://orcid.org/0000-0003-1631-8307
References
- 1.Chen W, Mi R, Haughey N, Oz M, Hoke A. Immortalization and characterization of a nociceptive dorsal root ganglion sensory neuronal line. J Peripher Nerv Syst 2007; 12: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacherjee A, Liao Z, Smith PG. Trophic factor and hormonal regulation of neurite outgrowth in sensory neuron-like 50B11 cells. Neurosci Lett 2014; 558: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vetter I, Lewis RJ. Characterization of endogenous calcium responses in neuronal cell lines. Biochem Pharmacol 2010; 79: 908–920. [DOI] [PubMed] [Google Scholar]
- 4.Pundavela J, Demont Y, Jobling P, Lincz LF, Roselli S, Thorne RF, Bond D, Bradshaw RA, Walker MM, Hondermarck H. ProNGF correlates with Gleason score and is a potential driver of nerve infiltration in prostate cancer. Am J Pathol 2014; 184: 3156–3162. [DOI] [PubMed] [Google Scholar]
- 5.Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci 2007; 8: 114–127. [DOI] [PubMed] [Google Scholar]
- 6.Fan SF, Shen KF, Scheideler MA, Crain SM. F11 neuroblastoma x DRG neuron hybrid cells express inhibitory mu- and delta-opioid receptors which increase voltage-dependent K+ currents upon activation. Brain Res 1992; 590: 329–333. [DOI] [PubMed] [Google Scholar]
- 7.Dunn PM, Coote PR, Wood JN, Burgess GM, Rang HP. Bradykinin evoked depolarization of a novel neuroblastoma x DRG neurone hybrid cell line (ND7/23). Brain Res 1991; 545: 80–86. [DOI] [PubMed] [Google Scholar]
- 8.Haberberger RV, Barry C, Matusica D. Immortalized dorsal root ganglion neuron cell lines. Front Cell Neurosci 2020; 14: 184–Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin K, Baillie GJ, Vetter I. Neuronal cell lines as model dorsal root ganglion neurons: a transcriptomic comparison. Mol Pain .Epub ahead of print 1 May 2016. DOI:10.1177/1744806916646111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doran C, Chetrit J, Holley MC, Grundy D, Nassar MA. Mouse DRG cell line with properties of nociceptors. PLoS One 2015; 10: e0128670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braz JM, Nassar MA, Wood JN, Basbaum AI. Parallel “pain” pathways arise from subpopulations of primary afferent nociceptor. Neuron 2005; 47: 787–793. [DOI] [PubMed] [Google Scholar]
- 12.Snider WD, McMahon SB. Tackling pain at the source: new ideas about nociceptors. Neuron 1998; 20: 629–632. [DOI] [PubMed] [Google Scholar]
- 13.Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to mrgprd. Neuron 2005; 45: 17–25. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Oh U, Wood JN. From transduction to pain sensation: defining genes, cells, and circuits. Pain 2011; 152: S16–S19. [DOI] [PubMed] [Google Scholar]
- 15.Airaksinen MS, Meyer M. Most classes of dorsal root ganglion neurons are severely depleted but not absent in mice lacking neurotrophin-3. Neuroscience 1996; 73: 907–911. [DOI] [PubMed] [Google Scholar]
- 16.Lindsay RM. Role of neurotrophins and trk receptors in the development and maintenance of sensory neurons: an overview. Philos Trans R Soc Lond B Biol Sci 1996; 351: 365–373. [DOI] [PubMed] [Google Scholar]
- 17.Matheson CR, Carnahan J, Urich JL, Bocangel D, Zhang TJ, Yan Q. Glial cell line-derived neurotrophic factor (GDNF) is a neurotrophic factor for sensory neurons: comparison with the effects of the neurotrophins. J Neurobiol 1997; 32: 22–32. [PubMed] [Google Scholar]
- 18.Bennett DL, Averill S, Clary DO, Priestley JV, McMahon SB. Postnatal changes in the expression of the trkA high-affinity NGF receptor in primary sensory neurons. Eur J Neurosci 1996; 8: 2204–2208. [DOI] [PubMed] [Google Scholar]
- 19.Molliver DC, Wright DE, Leitner ML, Parsadanian AS, Doster K, Wen D, Yan Q, Snider WD. IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron 1997; 19: 849–861. [DOI] [PubMed] [Google Scholar]
- 20.Stucky CL, Lewin GR. Isolectin B(4)-positive and -negative nociceptors are functionally distinct. J Neurosci 1999; 19: 6497–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen PL, Doucette JR, Nazarali AJ. A novel method of eliminating non-neuronal proliferating cells from cultures of mouse dorsal root ganglia. Cell Mol Neurobiol 2003; 23: 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delree P, Leprince P, Schoenen J, Moonen G. Purification and culture of adult rat dorsal root ganglia neurons. J Neurosci Res 1989; 23: 198–206. [DOI] [PubMed] [Google Scholar]
- 23.Zuchero JB. Purification of dorsal root ganglion neurons from rat by immunopanning. Cold Spring Harb Protoc 2014; 2014: 826–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R, Lin G, Xu H. An efficient method for dorsal root ganglia neurons purification with a one-time anti-mitotic reagent treatment. PLoS One 2013; 8: e60558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barde YA, Edgar D, Thoenen H. Sensory neurons in culture: changing requirements for survival factors during embryonic development. Proc Natl Acad Sci USA 1980; 77: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burkey TH, Hingtgen CM, Vasko MR. Isolation and culture of sensory neurons from the dorsal-root ganglia of embryonic or adult rats. Methods Mol Med 2004; 99: 189–202. [DOI] [PubMed] [Google Scholar]
- 27.Owen DE, Egerton J. Culture of dissociated sensory neurons from dorsal root ganglia of postnatal and adult rats. Methods Mol Biol 2012; 846: 179–187. [DOI] [PubMed] [Google Scholar]
- 28.Katzenell S, Cabrera JR, North BJ, Leib DA. Isolation, purification, and culture of primary murine sensory neurons. Methods Mol Biol 2017; 1656: 229–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckert SP, Taddese A, McCleskey EW. Isolation and culture of rat sensory neurons having distinct sensory modalities. J Neurosci Methods 1997; 77: 183–190. [DOI] [PubMed] [Google Scholar]
- 30.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3: RESEARCH0034–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 2003; 19: 1439–1440. [DOI] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 34.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015; 18: 145–153. [DOI] [PubMed] [Google Scholar]
- 35.Shannon PT, Grimes M, Kutlu B, Bot JJ, Galas DJ. RCytoscape: tools for exploratory network analysis. BMC Bioinf 2013; 14: 217–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009; 25: 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 39.Van Opdenbosch N, Van den Broeke C, De Regge N, Tabarés E, Favoreel HW. The IE180 protein of pseudorabies virus suppresses phosphorylation of translation initiation factor eIF2alpha. J Virol 2012; 86: 7235–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Carozzi VA, Reed N, Mi R, Marmiroli P, Cavaletti G, Hoke A. Ethoxyquin provides neuroprotection against cisplatin-induced neurotoxicity. Sci Rep 2016; 6: 28861–28806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bestall SM, Hulse RP, Blackley Z, Swift M, Ved N, Paton K, Beazley-Long N, Bates DO, Donaldson LF. Sensory neuronal sensitisation occurs through HMGB-1-RAGE and TRPV1 in high-glucose conditions. J Cell Sci 2018; 131: jcs215939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ernsberger U. Role of neurotrophin signalling in the differentiation of neurons from dorsal root ganglia and sympathetic ganglia. Cell Tissue Res 2009; 336: 349–384. [DOI] [PubMed] [Google Scholar]
- 43.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell 2009; 139: 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baron R, Wasner G, Binder A. Chronic pain: genes, plasticity, and phenotypes. Lancet Neurol 2012; 11: 19–21. [DOI] [PubMed] [Google Scholar]
- 45.Sapunar D, Kostic S, Banozic A, Puljak L. Dorsal root ganglion – a potential new therapeutic target for neuropathic pain. J Pain Res 2012; 5: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horton A, Laramee G, Wyatt S, Shih A, Winslow J, Davies AM. NGF binding to p75 enhances the sensitivity of sensory and sympathetic neurons to NGF at different stages of development. Mol Cell Neurosci 1997; 10: 162–172. [DOI] [PubMed] [Google Scholar]
- 47.Hill RG. Molecular basis for the perception of pain. Neuroscientist 2001; 7: 282–292. [DOI] [PubMed] [Google Scholar]
- 48.Baba H, Kohno T, Moore KA, Woolf CJ. Direct activation of rat spinal dorsal horn neurons by prostaglandin E2. J Neurosci 2001; 21: 1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St-Jacques B, Ma W. Peripheral prostaglandin E2 prolongs the sensitization of nociceptive dorsal root ganglion neurons possibly by facilitating the synthesis and anterograde axonal trafficking of EP4 receptors. Exp Neurol 2014; 261: 354–366. [DOI] [PubMed] [Google Scholar]
- 50.Ma W, St-Jacques B, Rudakou U, Kim YN. Stimulating TRPV1 externalization and synthesis in dorsal root ganglion neurons contributes to PGE2 potentiation of TRPV1 activity and nociceptor sensitization. Eur J Pain 2017; 21: 575–593. [DOI] [PubMed] [Google Scholar]
- 51.Woolf CJ, Allchorne A, Safieh-Garabedian B, Poole S. Cytokines, nerve growth factor and inflammatory hyperalgesia: the contribution of tumour necrosis factor alpha. Br J Pharmacol 1997; 121: 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sommer C, Kress M. Recent findings on how proinflammatory cytokines cause pain: peripheral mechanisms in inflammatory and neuropathic hyperalgesia. Neurosci Lett 2004; 361: 184–187. [DOI] [PubMed] [Google Scholar]
- 53.Felts PA, Yokoyama S, Dib-Hajj S, Black JA, Waxman SG. Sodium channel alpha-subunit mRNAs I, II, III, NaG, Na6 and hNE (PN1): different expression patterns in developing rat nervous system. Brain Res Mol Brain Res 1997; 45: 71–82. [DOI] [PubMed] [Google Scholar]
- 54.Hjerling-Leffler J, Alqatari M, Ernfors P, Koltzenburg M. Emergence of functional sensory subtypes as defined by transient receptor potential channel expression. J Neurosci 2007; 27: 2435–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibanez CF, Ernfors P. Hierarchical control of sensory neuron development by neurotrophic factors. Neuron 2007; 54: 673–675. [DOI] [PubMed] [Google Scholar]
- 56.Hwang SJ, Oh JM, Valtschanoff JG. Expression of the vanilloid receptor TRPV1 in rat dorsal root ganglion neurons supports different roles of the receptor in visceral and cutaneous afferents. Brain Res 2005; 1047: 261–266. [DOI] [PubMed] [Google Scholar]
- 57.Cavanaugh DJ, Chesler AT, Jackson AC, Sigal YM, Yamanaka H, Grant R, O'Donnell D, Nicoll RA, Shah NM, Julius D, Basbaum AI. Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells. J Neurosci 2011; 31: 5067–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi K, Fukuoka T, Obata K, Yamanaka H, Dai Y, Tokunaga A, Noguchi K. Distinct expression of TRPM8, TRPA1, and TRPV1 mRNAs in rat primary afferent neurons with adelta/c-fibers and colocalization with trk receptors. J Comp Neurol 2005; 493: 596–606. [DOI] [PubMed] [Google Scholar]
- 59.Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng HJ, Geng Y, Undem BJ, Kollarik M, Chen ZF, Anderson DJ, Dong X. Sensory neuron-specific GPCR mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 2009; 139: 1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Liu YW, Yue K, Ru Q, Xiong Q, Ma BM, Tian X, Li CY. Differential expression of ATP-gated P2X receptors in DRG between chronic neuropathic pain and visceralgia rat models. Purinerg Signal 2016; 12: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogen O, Bender O, Lowe J, Blenau W, Thevis B, Schroder W, Margolis RU, Levine JD, Hucho F. Neuronally produced versican V2 renders C-fiber nociceptors IB4 -positive. J Neurochem 2015; 134: 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park J, Luo ZD. Calcium channel functions in pain processing. Channels (Austin) 2010; 4: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berta T, Qadri Y, Tan PH, Ji RR. Targeting dorsal root ganglia and primary sensory neurons for the treatment of chronic pain. Expert Opin Ther Targets 2017; 21: 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krames ES. The dorsal root ganglion in chronic pain and as a target for neuromodulation: a review. Neuromodulation 2015; 18: 24–32; discussion 32. [DOI] [PubMed] [Google Scholar]
- 65.Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, Julius D. The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 1998; 21: 531–543. [DOI] [PubMed] [Google Scholar]
- 66.Price TJ, Flores CM. Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J Pain 2007; 8: 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ernsberger U. The role of GDNF family ligand signalling in the differentiation of sympathetic and dorsal root ganglion neurons. Cell Tissue Res 2008; 333: 353–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fabbretti E. ATP P2X3 receptors and neuronal sensitization. Front Cell Neurosci 2013; 7: 236–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Melli G, Hoke A. Dorsal root ganglia sensory neuronal cultures: a tool for drug discovery for peripheral neuropathies. Expert Opin Drug Discov 2009; 4: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA 1976; 73: 2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 1992; 194: 209–221. [DOI] [PubMed] [Google Scholar]
- 72.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem 2000; 75: 991–1003. [DOI] [PubMed] [Google Scholar]
- 73.Dubin AE, Patapoutian A. Nociceptors: the sensors of the pain pathway. J Clin Invest 2010; 120: 3760–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meacham K, Shepherd A, Mohapatra DP, Haroutounian S. Neuropathic pain: central vs. peripheral mechanisms. Curr Pain Headache Rep 2017; 21: 28–04. [DOI] [PubMed] [Google Scholar]
- 75.Donnelly-Roberts D, McGaraughty S, Shieh CC, Honore P, Jarvis MF. Painful purinergic receptors. J Pharmacol Exp Ther 2008; 324: 409–415. [DOI] [PubMed] [Google Scholar]
- 76.Zhang X, Huang J, McNaughton PA. NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. Embo J 2005; 24: 4211–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amaya F, Shimosato G, Nagano M, Ueda M, Hashimoto S, Tanaka Y, Suzuki H, Tanaka M. NGF and GDNF differentially regulate TRPV1 expression that contributes to development of inflammatory thermal hyperalgesia. Eur J Neurosci 2004; 20: 2303–2310. [DOI] [PubMed] [Google Scholar]
- 78.Copray JC, Mantingh I, Brouwer N, Biber K, Kust BM, Liem RS, Huitinga I, Tilders FJ, Van Dam AM, Boddeke HW. Expression of interleukin-1 beta in rat dorsal root ganglia. J Neuroimmunol 2001; 118: 203–211. [DOI] [PubMed] [Google Scholar]
- 79.Kawasaki Y, Zhang L, Cheng JK, Ji RR. Cytokine mechanisms of central sensitization: distinct and overlapping role of interleukin-1beta, interleukin-6, and tumor necrosis factor-alpha in regulating synaptic and neuronal activity in the superficial spinal cord. J Neurosci 2008; 28: 5189–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ai X, MacPhedran SE, Hall AK. Depolarization stimulates initial calcitonin gene-related peptide expression by embryonic sensory neurons in vitro. J Neurosci 1998; 18: 9294–9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fitzgerald M. The development of nociceptive circuits. Nat Rev Neurosci 2005; 6: 507–520. [DOI] [PubMed] [Google Scholar]
- 82.Benn SC, Costigan M, Tate S, Fitzgerald M, Woolf CJ. Developmental expression of the TTX-resistant voltage-gated sodium channels Nav1.8 (SNS) and Nav1.9 (SNS2) in primary sensory neurons. J Neurosci 2001; 21: 6077–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waxman SG, Kocsis JD, Black JA. Type III sodium channel mRNA is expressed in embryonic but not adult spinal sensory neurons, and is reexpressed following axotomy. J Neurophysiol 1994; 72: 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dib-Hajj SD, Choi JS, Macala LJ, Tyrrell L, Black JA, Cummins TR, Waxman SG. Transfection of rat or mouse neurons by biolistics or electroporation. Nat Protoc 2009; 4: 1118–1126. [DOI] [PubMed] [Google Scholar]
- 85.Eggett CJ, Crosier S, Manning P, Cookson MR, Menzies FM, McNeil CJ, Shaw PJ. Development and characterisation of a glutamate-sensitive motor neurone cell line. J Neurochem 2008; 74: 1895–1902. [DOI] [PubMed] [Google Scholar]